Introduction
Colon cancer is one of the most common malignancies
in the world and results from an accumulation of genetic and
epigenetic aberrations (1). It is
the second leading cause of cancer mortality in the United States
with ~52,000 deaths and 143,000 cases expected in 2012 (2) and ranks the third leading cause of
cancer mortality worldwide (3). In
addition, changes in lifestyle and nutrition have led to the rise
in colon cancer incidence in China and other economically
transitioning countries over the past few decades (4). In spite of advances in screening and
prevention, early detection, adjuvant therapy and treatment of
metastatic disease, colon cancer remains the major cause of cancer
morbidity and mortality. Evidence has been provided by molecular
investigations that multiple alterations in genes of the striking
signaling pathway, including adenomatous polyposis coli protein
(APC), β-catenin and c-myc, are involved in colonic carcinogenesis
(5,6). Despite existing intensive study, the
molecular mechanisms underlying the development and progression of
colon cancer remain poorly understood. It is of great clinical
importance to further investigate the molecular mechanisms of this
cancer and to find valuable early diagnostic markers with high
specificity and sensitivity as well as novel therapeutic
targets.
The ubiquitin proteasome system (UPS) regulates the
ubiquitination, and thus the degradation and turnover, of many
proteins crucial to cellular regulation and function (7). Aberrancies within the UPS pathway can
result in a malignant cellular phenotype which can lead to several
types of human malignant cancer (8,9).
Fundamental to the specificity of this system are ubiquitin-protein
ligases (E3s) (10). Ubiquitin-like
with PHD and ring finger domains 2 (UHRF2), a member of the UHRF
[ubiquitin PHD really interesting new gene (RING) finger] family,
is a nuclear E3 ubiquitin ligase mapped to 9p23–24.1 (11,12).
UHRF2 comprises diverse domains including the
ubiquitin-like (UBL) domain, tandem Tudor domain (TTD), plant homeo
domain (PHD) finger domain, SET and RING associated (SRA) domain
and RING finger domain (11–13).
Due to its multiple domains, UHRF2 has such a complex function that
it is involved in cell cycle network, the epigenetic system and the
UPS. On account of its three different network modules, UHRF2 plays
a contradictory role in tumorigenesis. Using a cancer outlier
profile analysis identified that DNA copy number loss of the UHRF2
gene in a variety of malignancies related to the brain (14). On the contrary, UHRF2 has been
proven to play a role as an oncogene in breast cancer cells
(15). However, the
clinicopathological significance and mechanism of UHRF2 involvement
in the aggression of colon cancer is not completely understood.
The present study sought to assess the hypothesis
that UHRF2 expression is dysregulated in colon cancer at both the
mRNA and protein levels and its predictive value in colon cancer.
Immunohistochemistry was used to explore the expression of UHRF2
both in primary colon cancer specimens and paired adjacent normal
mucosa tissue and investigated whether UHRF2 may be used as an
independent biomarker to predict metastasis and prognosis in
patients with colon cancer.
Materials and methods
Patients and specimens
The Ethics Committee of Shanghai Jiaotong University
Affiliated First People’s Hospital approved this study. A total of
203 colon cancer patients were enrolled; all provided informed
consent according to a protocol approved by the Institutional
Review Board of the Shanghai First People’s Hospital. The patients
permitted surgical resection of tumors by the same surgical team at
the Shanghai Jiaotong University affiliated Shanghai First People’s
Hospital Gastrointestinal Cancer Center between January 2001 and
December 2003. No patients had received therapy prior to surgery.
There were 86 men and 117 women with a mean age of 65±15 years
(range, 22–95 years). Formalin-fixed, paraffin-embedded samples for
immunohistochemistry were obtained from the 203 colon carcinoma
tissues and paired normal mucosal tissue taken from a segment of
the resected specimens that was the farthest from the tumor (>10
cm). All tissues were histologically confirmed to be adenocarcinoma
of the colon. Patient follow-up was carried out according to the
National Comprehensive Cancer Network Practice Guidelines in colon
cancer (Engstrom PF.2005; 3:468–91). Disease-free survival (DFS)
and overall survival (OS) rates were defined as the interval from
the initial surgery to clinically or radiologically proven
recurrence/metastasis and death, respectively. The final follow-up
was on June 29, 2008 with a median patient follow-up time for
survivors of 61 months (range, 9–89 months). Detailed patient
demographic information is shown in Table I. Vascular invasion was defined as
vessel wall occlusion or destruction, accompanied by a surrounding
fibroinflammatory reaction (16).
 | Table IExpression of UHRF2 in normal and
colon cancer tissues. |
Table I
Expression of UHRF2 in normal and
colon cancer tissues.
| Expression of
UHRF2 | Normal tissue
(%) | Tumor tissue (%) | P-value |
|---|
| All subjects |
| No. of subjects | 203 | 203 | <0.001a |
| Negative | 132 (65.0) | 70 (34.5) | |
| Weak | 38 (18.7) | 57 (28.1) | |
| Positive | 33 (16.3) | 76 (37.4) | |
| Subjects without
LNM |
| No. of subjects | 108 | 108 | 0.003a |
| Negative | 77 (71.3) | 53 (49.1) | |
| Weak | 16 (14.8) | 32 (29.6) | |
| Positive | 15 (13.9) | 23 (21.3) | |
| Subjects with
LNM |
| No. of subjects | 95 | 95 | <0.001a |
| Negative | 55 (57.9) | 18 (18.9) | |
| Weak | 18 (23.2) | 25 (26.3) | |
| Positive | 22 (18.9) | 52 (54.7) | |
| P-value | 0.131 | <0.001a | |
RNA extraction, reverse transcription PCR
and quantitative real-time PCR
Total RNA in 40 paired, frozen primary colon cancer
tissues, and adjacent normal mucosa were extracted according to the
manufacturer’s instructions (Qiagen, Hilden, Germany). One
microgram of total RNA from each sample was subjected to
first-strand complementary DNA synthesis using an A3500 RT-PCR
System® according to the recommendations of the
manufacturer (Promega Corporation, Madison, WI, USA). To confirm
UHRF2 gene expression in colon tumors, relative UHRF2 mRNA levels
were assessed by quantitative real-time PCR (qPCR) using
Mastercycler ep realplex® (Eppendorf, Hamburg, Germany)
with an IQTM SYBR-Green Supermix kit (Bio-Rad, Berkeley, CA, USA)
according to the manufacturer’s instructions and using the
following thermal cycling conditions: initial denaturation (10 min
at 95°C) followed by 40 cycles of denaturation (10 sec at 95°C),
annealing (15 sec at 58°C) and elongation (1 min at 72°C).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an
internal control. The primers for qPCR were: UHRF2 sense,
5′-TTCTTGCTCCTGTCGTGTATGT-3′ and antisense,
5′-CTTGAGTCTTTCACCAGCCTTT-3′; GAPDH sense,
5′-GTCCACCACCCTGTTGCTGTA-3′ and antisense,
5′-CTTCAACAGCGACACCCACTC-3′. Each reaction was repeated at least
three times, and then the mean UHRF2 mRNA level for each tumor was
compared with the level of its matched non-tumorous tissue. The
fold-change (2−ΔΔCt) (17) in UHRF2 expression in each paired
sample was calculated using the formulas: UHRF2ΔCt = (Avg.UHRF2_Ct
− Avg.GAPDH_Ct), UHRF2ΔΔCt = (UHRF2ΔCt_tumor −
UHRF2ΔCt_non-tumor).
Western blot analysis
Total proteins were extracted from 8 paired frozen
colon tumor and adjacent normal tissues using
radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris pH
7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate and 0.1% SDS)
with a protease inhibitor cocktail (0.89 μg/μl; Sigma-Aldrich) and
phenylmethanesulfonyl fluoride (17.4 μg/μl) and quantified using a
BCA protein assay kit according to the manufacturer’s instructions
(Beyotime Biotechnology Co., Jiangsu, China). Equivalent amounts of
protein were separated by electrophoresis on a 9.0% sodium dodecyl
sulphatepolyacrylamide gels and transferred onto polyvinylidene
difluoride (PVDF) membranes, which were blocked with 5% non-fat dry
milk in 0.05% PBS-T for 1 h at room temperature, followed by
incubation with the rabbit anti-UHRF2 polyclonal antibody (1:500)
or anti-β-actin monoclonal antibody (1:2,000) (both from Abcam,
Cambridge, UK) overnight at 4°C. After washing with TBST buffer,
the membranes were incubated with a goat-anti-rabbit IgG
HRP-conjugated secondary antibody (1:5,000; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) for 2 h. The membranes
were washed, and bound antibodies were detected by enhanced
chemiluminescence (Millipore) according to the manufacturer’s
instructions and were exposed to X-ray film. The abundance of each
protein was determined and normalized against β-actin
expression.
Tissue microarray (TMA) construction and
immunohistochemistry
TMA construction was undertaken as previously
reported (18). Briefly,
paraffin-embedded TMA sections were dewaxed and rehydrated before
antigen retrieval was carried out. Immunohistochemistry was
performed to further evaluate the histological expression of UHRF2.
UHRF2 and Ki-67 expression were detected on the TMAs following
preheated (95–98°C) citrate buffer (pH 6.0) for 10 min aimed at
antigen retrieval. Immunolabeling was carried out using a primary
antibody against UHRF2 (1:50; Abcam) and the proliferation index
Ki-67 (1:50; Dako Cytomation, Copenhagen, Denmark). Sections were
incubated overnight at 4°C and then incubated with the secondary
antibody (EnVision System, Dako) for 1 h at room temperature. After
rinsing three times in PBS for 10 min each, the sections were
incubated with 3,3-diaminobenzidine tetrahydrochloride (DAB) liquid
for 1 min, counterstained with Mayer hematoxylin, dehydrated and
then mounted. The negative control was prepared with normal tissue
and without anti-UHRF2 antibody incubation.
Evaluation of immunohistochemical
staining
Immunoreactivity was evaluated independently by
three researchers who were blinded to patient outcomes
(double-blinded) on the basis of the intensity and extent of
staining (19). The staining
intensity for UHRF2 was graded as 0 (no staining), 1 (mild
staining), 2 (moderate staining) and 3 (intense staining). The
staining extent was scored using the scale as follows: 0 (no
staining of cells), 1 (<10% of tissue stained positive), 2
(10–50% stained positive), 3 (>50% stained positive). The final
staining score was defined as the sum of the intensity and extent
scores. The specimens were divided into three groups according to
their overall scores as follows: 0–2, expression; 3–4, weak
expression; 5–6, strong expression. The Ki-67 proliferation index
was on the basis of the percentage of cells with positive nuclear
staining and was divided into two groups selecting 10% positively
staining nuclei as the cutoff point: negative (≤10% of cells with
positive nuclei) and positive (>10% of cells with positive
nuclei) (20). In cases of
discrepant assessments, the sections were re-investigated by both
pathologists under a multi-head microscope until consensus was
achieved.
Statistical analyses
All statistical analyses were performed using the
SPSS 16.0 statistics software package (SPSS Inc., Chicago, IL,
USA). Paired t-tests were carried out to indicate the expression
(ΔCT) of UHRF2 in normal and adjacent cancer tissues. The
χ2 test or Fisher’s exact test, where appropriate, for
proportionality was used to analyze the relationship between the
expression of UHRF2, the Ki-67 proliferation index and the
clinicopathological variables. The Kaplan-Meier method was applied
to calculate the survival rates and the differences between the
survival curves were examined by the log-rank test. Univariate Cox
proportional hazards regressions were used to estimate the
individual hazard ratio (HR) for the DFS and OS. The significant
variables in the univariate analyses (P<0.05) were then put into
the multivariate analysis. A P-value of <0.05 was considered to
indicate a statistically significant difference.
Results
Upregulation of UHRF2 expression in
primary colon cancer as compared with adjacent normal mucosa
UHRF2 gene expression was confirmed by real-time PCR
analysis of primary colon cancer and adjacent normal mucosa from 40
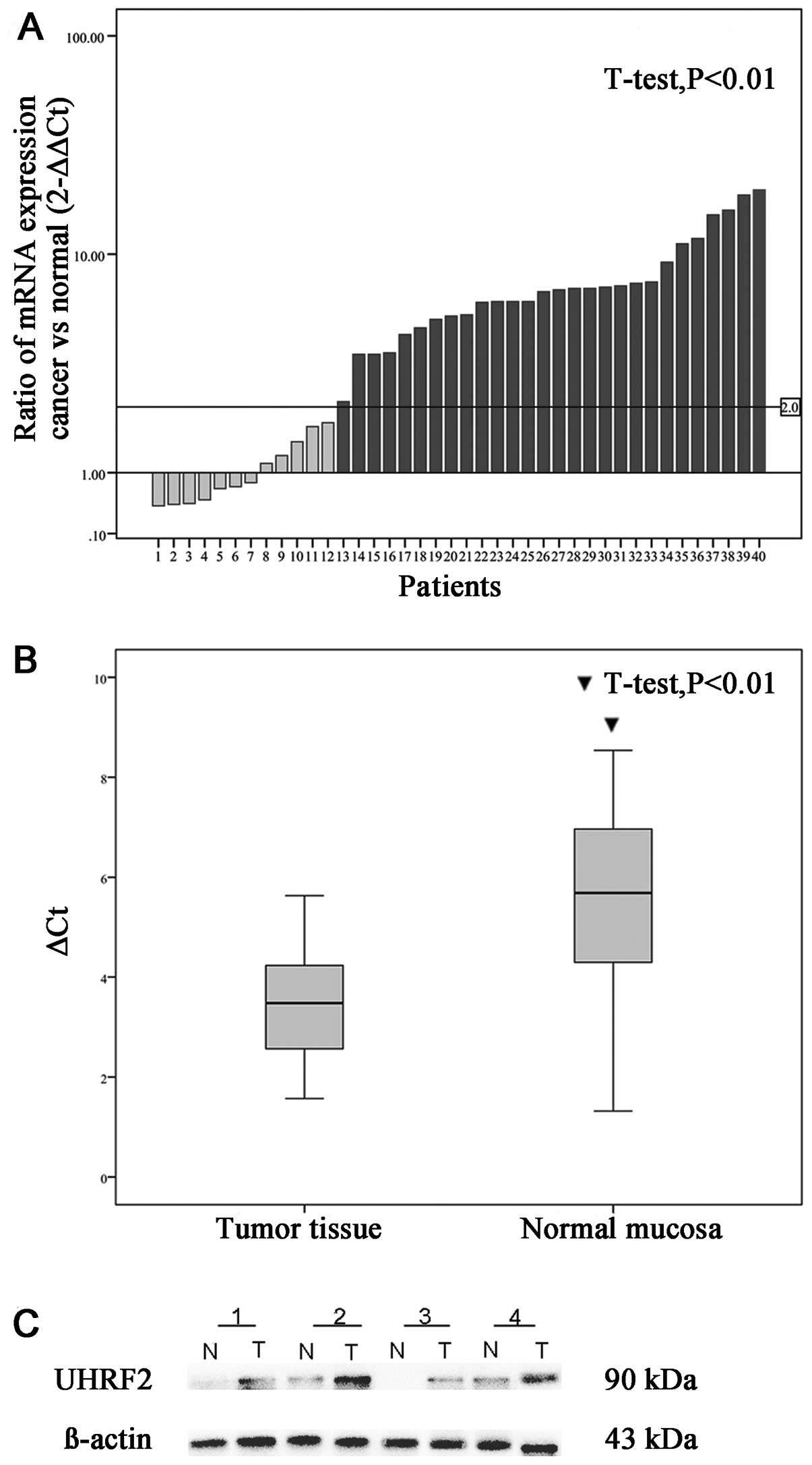
patients with colon cancer with GAPDH as the internal reference. Of
the 40 paired cases, 28 (68%) colon cancer tissues showed at least
a two-fold increase in UHRF2 mRNA level compared with that of the
adjacent non-cancerous mucosa (Fig.
1A). In patients with colon cancer, the expression (ΔCt) of
UHRF2 was 3.51±1.072 in tumor tissue and 5.4530±1.772 in normal
tissue (P<0.001; Fig. 1B). The
2−ΔΔCt was 5.78±4.93 (range 0.58–19.70). This difference
in UHRF2 mRNA expression was significant (P<0.001; Fig. 1). This suggests that UHRF2 mRNA
level was upregulated in cancerous tissues as compared with
adjacent normal mucosa. Subsequent western blotting confirmed that
UHRF2 protein levels were significantly upregulated in cancerous
tissues as compared with adjacent normal mucosa (Fig. 1C).
Association of UHRF2 TMA
immunohistochemical staining with patient clinicopathological
parameters
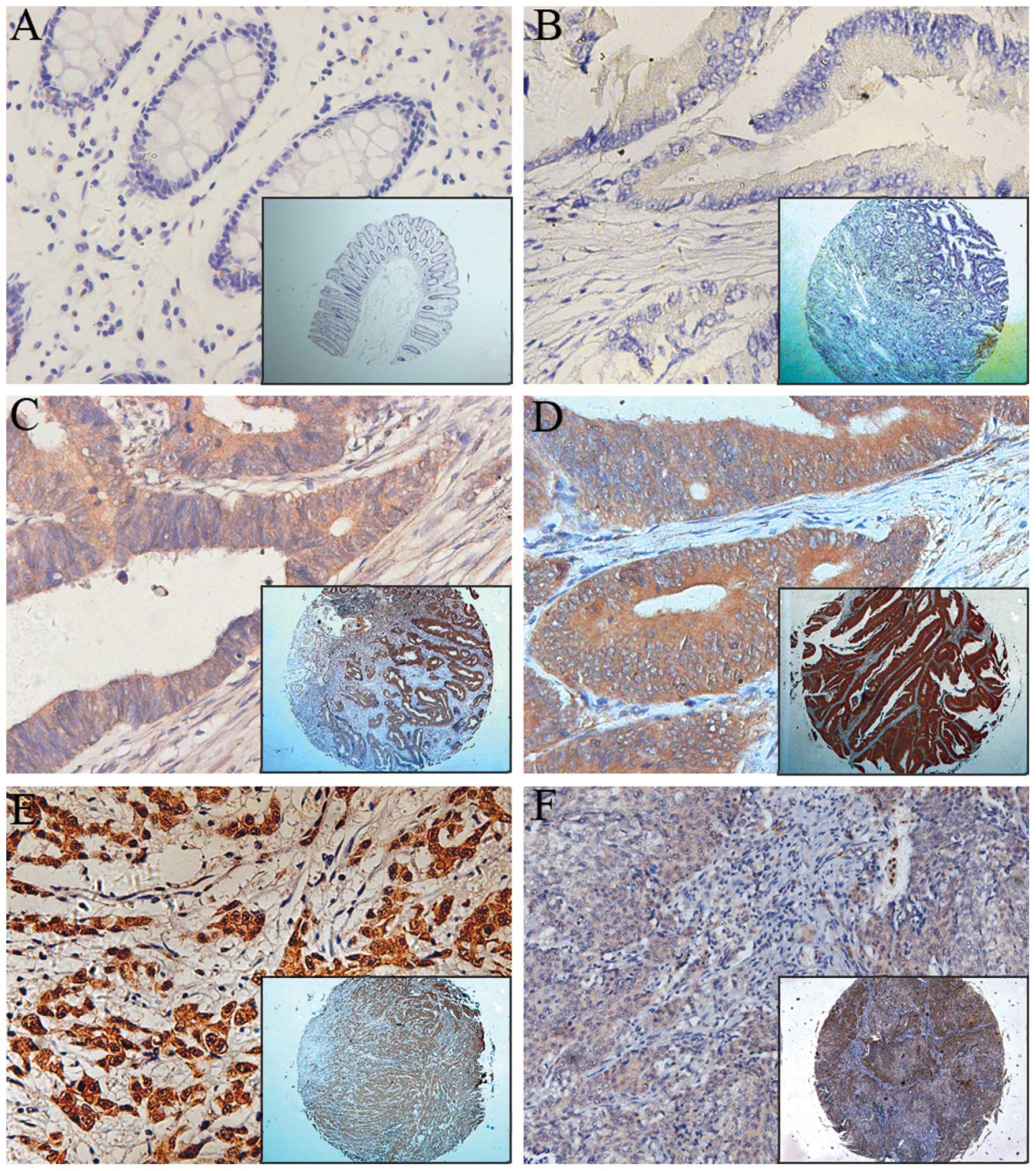
UHRF2 was observed mainly in the cytoplasm of
colonic epithelial tumor cells, with nuclear staining only rarely
observed by immunohistochemistry (Fig.
2). As shown in Table I, the
distribution of UHRF2 expression was significantly different
between normal and tumor tissues (P<0.001). Among the 203 normal
mucosa specimens on the paired TMA, 132 (65.0%) showed negative
UHRF2 expression with weak staining in 38 (18.7%) cases and strong
staining in 33 (16.3%) additional cases. However, UHRF2 expression
was obvious in the majority of colon tumor specimens, with weak
staining in 57 (28.1%) cases, strong staining in 75 (36.9%) cases
and negative staining in 71 (35%) cases. This was also observed in
patients with LNM or without LNM (Table
I). The relationship between the expression of UHRF2
(immunohistochemical staining) and clinicopathological features is
summarized in Table II. Expression
(staining) of UHRF2 was highly correlated with the American Joint
Committee on Cancer (AJCC) stage (P<0.001), T classification
(P<0.001), nodal involvement (P<0.001), extent of tumor
differentiation, recurrence and metastasis and the presence of
distant metastasis (P<0.001). Factors not significantly
associated with staining included age, gender, location of the
tumor and vascular invasion.
 | Table IIAssociation between
clinicopathological features and UHRF2 or Ki-67 protein
expression. |
Table II
Association between
clinicopathological features and UHRF2 or Ki-67 protein
expression.
| | UHRF2 expression | |
|---|
| |
| |
|---|
| n | Negative (n=71)
(%) | Weak (n=57) (%) | Strong (n=75)
(%) | P-value |
|---|
| Age, years (n,
%) | | | | | 0.919 |
| <65 | 81 | 29 (40.8) | 23 (40.4) | 29 (38.7) | |
| ≥65 | 122 | 42 (59.2) | 34 (59.6) | 46 (61.3) | |
| Gender (n,%) | | | | | 0.547 |
| Male | 86 | 33 (46.5) | 21 (36.8) | 32 (42.7) | |
| Female | 117 | 38 (53.5) | 36 (63.2) | 43 (57.3) | |
| Location (n,%) | | | | | 0.439 |
| Right | 84 | 30 (42.3) | 21 (36.8) | 33 (44.0) | |
| Transverse | 19 | 9 (12.7) | 4 (7.0) | 6 (8.0) | |
| Left | 20 | 7 (9.9) | 9 (15.8) | 4 (5.3) | |
| Sigmoid colon | 80 | 25 (35.2) | 23 (40.4) | 32 (42.7) | |
| T stage
(n,%)b | | | | | <0.001a |
| T1 | 8 | 3 (4.2) | 2 (3.5) | 3 (4.0) | |
| T2 | 23 | 9 (12.7) | 10 (17.5) | 4 (5.3) | |
| T3 | 76 | 38 (53.5) | 20 (35.1) | 18 (24.0) | |
| T4 | 96 | 21 (29.6) | 25 (43.9) | 50 (66.7) | |
| N stage (n,%) | | | | | <0.001a |
| N0 | 108 | 53 (76.4) | 32 (56.1) | 23 (30.7) | |
| N1 | 61 | 15 (21.1) | 14 (24.6) | 32 (42.7) | |
| N2 | 34 | 3 (4.2) | 11 (19.3) | 20 (26.7) | |
| M stage
(n,%)b | | | | | 0.018a |
| M0 | 185 | 69 (97.2) | 53 (93.0) | 63 (84.0) | |
| M1 | 18 | 2 (2.8) | 4 (7.0) | 12 (16.0) | |
| AJCC stage
(n,%) | | | | | <0.001a |
| I | 24 | 10 (14.1) | 10 (17.5) | 4 (5.3) | |
| II | 81 | 42 (59.2) | 21 (36.8) | 18 (24.0) | |
| III | 80 | 17 (23.9) | 22 (38.6) | 41 (54.7) | |
| IV | 18 | 2 (2.8) | 4 (7.0) | 12 (16.0) | |
| Differentiation
(n,%) | | | | | <0.001a |
| High | 99 | 46 (64.8) | 29 (50.9) | 24 (32.0) | |
| Moderate | 74 | 22 (31.0) | 20 (35.1) | 32 (42.7) | |
| Low | 30 | 3 (4.2) | 8 (14.0) | 19 (25.3) | |
| Vascular invasion
(n,%)b | | | | | 0.05 |
| Yes | 189 | 71 (98.6) | 55 (96.5) | 63 (85.1) | |
| No | 14 | 1 (1.4) | 2 (3.5) | 11 (14.9) | |
| Ki-67 index
(n,%) | | | | | 0.165 |
| Negative | 43 | 16 (22.5) | 16 (28.1) | 11 (14.7) | |
| Positive | 160 | 55 (77.5) | 41 (71.9) | 64 (85.3) | |
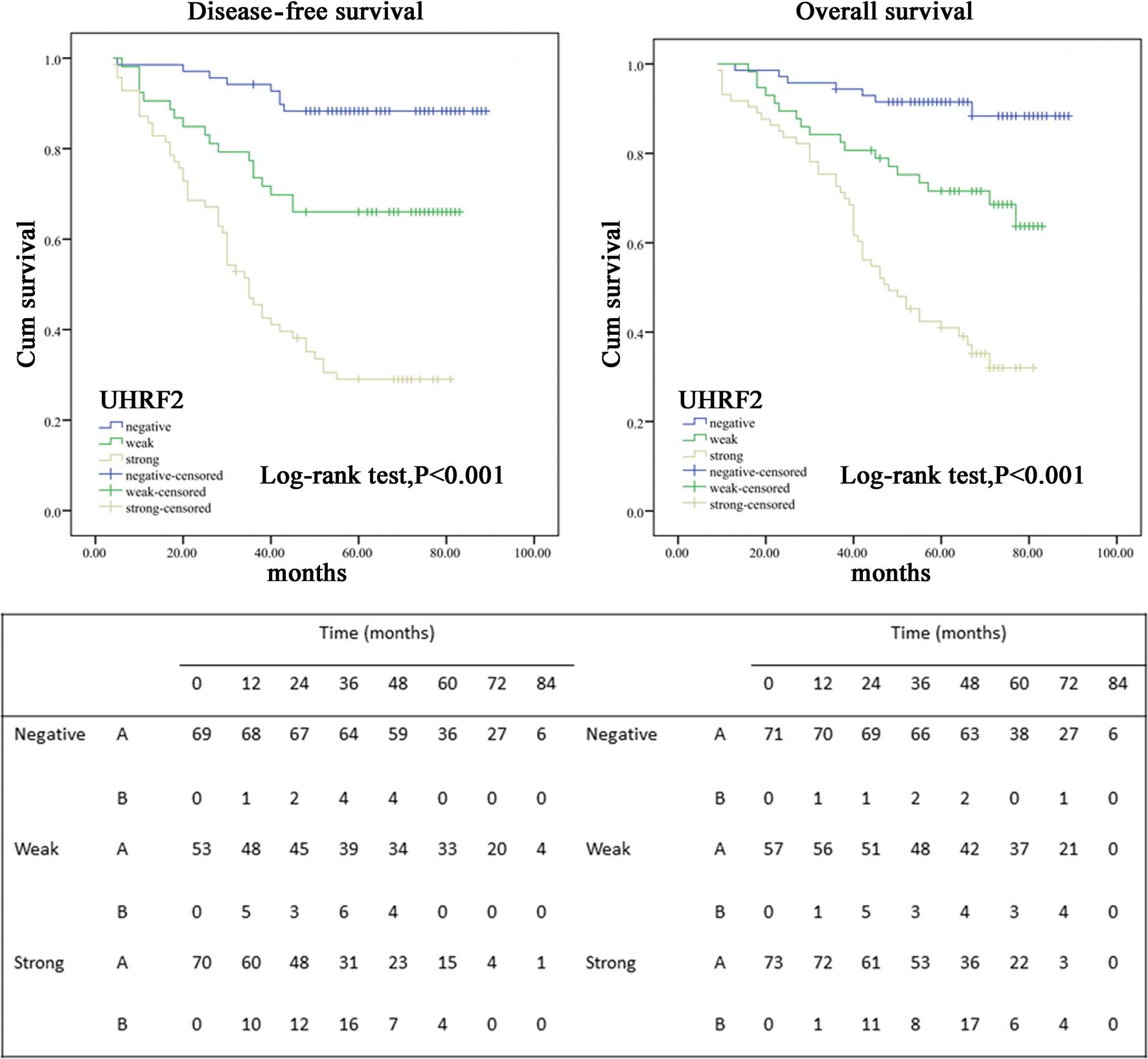
Survival analysis and prognostic
significance of UHRF2 expression
To assess the possible association between tumor
UHRF2 expression and patient survival, Kaplan-Meier curves with a
log-rank test for OS and DFS were undertaken (Fig. 3). The 5-year OS rate of the 203
patients who underwent curative surgery with primary colon cancer
was 68%, with 65 deaths occurring during the follow-up. However,
the 5-year DFS rate was 62%, with 74 events occurring. Fifty-four
patients (27%) developed distant metastases and 20 patients (9%)
were diagnosed with local tumor recurrence. There was a
considerable difference in the proportion of samples with
metastasis or local recurrence from primary colon cancer between
the UHRF2-positive and UHRF2-negative groups. More patients with
UHRF2-positive tumors subsequently developed metastases or local
recurrence than those with UHRF2-weak or UHRF2-negative tumors
(P<0.001) [UHRF2-positive, 49 (65.3%) of 70 patients;
UHRF2-weak, 35 (29.9%) of 53; UHRF2-negative, 8 (10.7%) of 69].
Patients with negative tumor UHRF2 expression had a better 5-year
DFS and OS rate than did the group with positive-UHRF2 expression
(DFS, 88.4% negative vs. 30% positive; OS, 91.4 vs. 29.5%;
P<0.001, respectively). Kaplan-Meier curves showed that the rate
of recurrence was significantly elevated with positive-UHRF2
expression (Fig. 3). The estimated
mean OS was significantly different between patients with
UHRF2-positive and UHRF2-negative tumors (71.5±2.2 and 58.6±5.0
months, respectively; P<0.001). The estimated mean DFS time was
66.2±2.4 and 54.2±5.9 months for subjects with UHRF2-positive and
UHRF2-negative tumors (P<0.001). The DFS and OS rates were
significantly decreased with increasing UHRF2 expression (Fig. 3).
In univariate analysis, patients whose localized
colon tumors were UHRF2-positive had a significantly lower 5-year
DFS than those with UHRF2-negative tumors [HR 9.511 (95% CI,
4.496–20.118)] (Fig. 3; Table III). The 5-year OS was also
significantly lower in patients with UHRF2-positive tumors than in
those with UHRF2-negative tumors [HR 9.820 (95% CI, 4.405–21.891)]
(Fig. 3; Table IV). In addition, pT stage
(P<0.001), pN stage (P<0.001), AJCC stage (P<0.001),
vascular invasion, Ki-67 expression and level of tumor
differentiation (P<0.001) were associated with OS and DFS. To
further define increased UHRF2 expression as an independent factor
influencing tumor recurrence, multivariate analysis was performed
using the Cox proportional hazards model for all of the significant
variables in the univariate analysis (Tables III and IV); this demonstrated that positive tumor
UHRF2 expression remained a significant independent prognostic
factor for increased disease recurrence and decreased survival.
 | Table IIICox proportional hazards model
univariate and multivariate analyses of individual parameters for
correlations with disease-free survival (DFS). |
Table III
Cox proportional hazards model
univariate and multivariate analyses of individual parameters for
correlations with disease-free survival (DFS).
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| HR | CI | P-value | HR | CI | P-value |
|---|
| Age, years |
| <65 |
| ≥65 | 1.063 | 0.647–1.749 | 0.808 | | | |
| Gender |
| Male |
| Female | 1.221 | 0.745–2.002 | 0.427 | | | |
| Tumor location |
| Right |
| Transverse | 0.859 | 0.328–2.252 | 0.758 | | | |
| Left | 1.135 | 0.489–2.634 | 0.768 | | | |
| Sigmoid colon | 1.251 | 0.731–2.140 | 0.413 | | | |
| T stage |
| T1 | 0.383 | 0.093–1.580 | 0.184 | 0.418 | 0.096–1.827 | 0.247 |
| T2 | 0.125 | 0.030–0.516 | 0.004a | 0.313 | 0.072–1.372 | 0.123 |
| T3 | 0.357 | 0.204–0.625 | <0.001a | 0.439 | 0.243–0.793 | 0.006a |
| T4 |
| N stage |
| N0 |
| N1 | 3.433 | 1.809–6.515 | <0.001a | 0.129 | 0.064–0.263 | <0.001a |
| N2 | 14.180 | 7.477–26.892 | <0.001a | 0.246 | 0.133–0.453 | <0.001a |
| M stage |
| M0 |
| M1 | 9.028 | 4.322–18.855 | <0.001a | 0.295 | 0.135–0.649 | 0.002a |
| AJCC stage |
| I+II |
| III+IV | 5.830 | 3.264–10.415 | <0.001a | | | |
|
Differentiation | | | | | | |
| High |
| Moderate | 2.340 | 1.306–4.193 | 0.004a | | | |
| Low |
| Vascular
invasion | 6.363 | 3.350–12.087 | <0.001a | | | |
| No |
| Yes | 5.162 | 2.735–9.742 | <0.001a | | | |
| UHRF2 |
| Negative |
| Weak | 4.352 | 1.660–12.7366 | 0.003a | 0.145 | 0.056–0.378 | <0.001a |
| Strong | 12.991 | 5.144–32.813 | <0.001a | 0.483 | 0.268–0.872 | 0.016a |
| Ki-67 |
| Negative |
| Weak | 2.090 | 0.914–4.778 | 0.081 | | | |
| Positive | 2.102 | 1.023–4.321 | 0.043a | | | |
 | Table IVCox proportional hazards model
univariate and multivariate analyses of individual parameters for
correlations with overall survival (OS). |
Table IV
Cox proportional hazards model
univariate and multivariate analyses of individual parameters for
correlations with overall survival (OS).
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| HR | CI | P-value | HR | CI | P-value |
|---|
| Age |
| <65 |
| ≥65 | 1.038 | 0.653–1.651 | 0.875 | | | |
| Gender |
| Male |
| Female | 0.747 | 0.463–1.197 | 0.223 | | | |
| Tumor location |
| Right |
| Transverse | 0.940 | 0.568–1.554 | 0.808 | | | |
| Left | 0.751 | 0.314–1.797 | 0.520 | | | |
| Sigmoid colon | 0.901 | 0.397–2.401 | 0.802 | | | |
| T stage |
| T1 | 0.356 | 0.087–1.461 | 0.152 | 0.211 | 0.047–0.950 | 0.043a |
| T2 | 0.108 | 0.026–0.443 | 0.002a | 0.291 | 0.067–1.266 | 0.100 |
| T3 | 0.337 | 0.197–0.578 | <0.001a | 0.410 | 0.230–0.732 | 0.003a |
| T4 |
| N stage |
| N0 |
| N1 | 0.071 | 0.038–0.133 | <0.001a | 2.351 | 1.211–4.562 | 0.012a |
| N2 | 0286 | 0.167–0.488 | <0.001a | 7.142 | 3.640–14.743 | <0.001a |
| M stage |
| M0 |
| M1 | 0.068 | 0.037–0.123 | <0.001a | 7.528 | 3.823–14.820 | <0.001a |
| AJCC stage |
| I+II |
| III+IV | 6.663 | 3.755–11.821 | 0.000a | | | |
|
Differentiation |
| High |
| Moderate | 2.368 | 1.342–4.178 | 0.003a | | | |
| Low | 7.499 | 4.112–13.678 | <0.001a | | | |
| No |
| Yes | 4.677 | 2.545–8.595 | <0.001a | | | |
| UHRF2 |
| Negative |
| Weak | 3.651 | 1.518–8.783 | 0.004a | 2.361 | 0.965–5.778 | 0.060 |
| Strong | 9.820 | 4.405–21.891 | <0.001a | 4.205 | 1.805–9.793 | <0.001a |
| Ki-67 |
| Negative |
| Weak | 2.302 | 0.917–5.775 | 0.076a | | | |
| Positive | 3.096 | 1.406–6.817 | <0.005a | | | |
Discussion
The present study shows for the first time that
overexpression of UHRF2 was significantly associated with cancer
progression and metastasis independent of pathological
Tumor-Node-Metastasis staging. The data support UHRF2 as a novel
prognostic indicator of colon cancer outcomes; individuals with
UHRF2-strong tumors have poorer OS and DFS as compared to those
with UHRF2-negative tumors. Correlations of UHRF2 expression with
advancing tumor stages suggest that UHRF2 may contribute to the
progression of colon carcinogenesis.
In the fresh colon tissues examined in the present
study, real-time PCR, immunostaining and western blotting showed
that the elevated expression of UHRF2 occurred both at the
transcriptional and post-transcriptional levels. In addition, we
found that elevated expression of UHRF2 correlated with several
clinicopathological factors including AJCC stage (P<0.001), T
classification (P<0.001), nodal involvement (P<0.001), extent
of differentiation (P<0.001) and the presence of distant
metastasis (P=0.018).
The UHRF family, including UHRF2 and UHRF1, is
considered to be involved in carcinogenesis (11). UHRF2 is a multidomain protein with
802 amino acid residues that shows high structural similarity to
its close homolog UHRF1, a putative oncogenic factor, suggested to
be an important biomarker to discriminant several types of cancer
(21–23). However, in sharp contrast to UHRF1,
the UHRF2 gene is frequently discrepant in tumorigenesis. In the
present study, UHRF2 acted as an oncogene in the development and
progression of colon cancer, which is in accordance with the role
UHRF2 plays in breast cancer. However, other reports have
elucidated its role of tumor suppression in malignant glioma
(24). This apparent contradiction
arises since UHRF2 is a multiple functional protein that is
involved in the coordination of three different network modules as
previously demonstrated. It is common knowledge that cancer results
from an accumulation of genetic and epigenetic aberrations
(1). Thus, we speculate that UHRF2
may have an oncogenic role in colon cancer by mediating tumor
suppressor gene inactivation via both DNA methylation and histone
modification pathway development and may promote tumor progression
through ubiquitin-mediated degradation of the suppressor such as
P53 in colon cancer. It is known that inhibiting the proteasome
pathways is a notable strategy for anticancer drug development
(25) and UHRF2 is a ubiquitin E3
ligase. Targeting UHRF2 would selectively stabilize a specific
cellular protein regulated by it, thus avoiding unwanted effects on
other cellular proteins. In brief, UHRF2 is a potential novel
therapeutic target in colon cancer.
In recent years, our understanding of the metastatic
process has significantly evolved. However, the mechanisms involved
in colon cancer metastasis are not fully understood as metastasis
is a multistep process and requires altered expression of a
spectrum of genes (26). In the
present study, we examined the expression of UHRF2 in 95 samples of
primary colon cancer with metastasis and 108 samples of primary
colon cancer without metastasis. There is a significant difference
between them (P<0.001). The expression of UHRF2 was upregulated
in the specimens with metastasis compared with those without
metastasis (54.7 vs. 21.3%). Finally, our results provide the first
evidence that the expression of UHRF2 may be a potential molecular
marker. UHRF2 expression was associated with an increased risk of
metastasis/local recurrence and was strongly linked to poor
survival outcomes, with hazard ratios of 9.511 for DFS and 9.820
for OS in the univariate analysis. In multivariate analysis, UHRF2
expression appeared to be an independent prognostic factor for OS
and DFS in colon cancer.
The present study found that increased UHRF2
expression was observed in colon cancer tissue and was associated
with multiple clinicopathological factors as well as patient OS and
DFS. Further studies are necessary to evaluate the mechanism by
which UHRF2 is upregulated in colon cancer, the role of UHRF2 in
colon cancer progression as well as its value in the prognosis of
this disease.
Acknowledgements
This study was supported by the Key Basic Research
Project of the Science and Technology Commission of Shanghai
(11JC1410200), the National Natural Science Foundation of China
(81072008, 81172328), the Medical Guidance Project of Shanghai
Science and Technology Commission (114119a4600, 124119a1700).
References
|
1
|
Vogelstein B and Kinzler KW: Cancer genes
and the pathways they control. Nat Med. 10:789–799. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
3
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar
|
|
4
|
Zhang S, Cui Y, Weng Z, Gong X, Chen M and
Zhong B: Changes on the disease pattern of primary colorectal
cancers in Southern China: a retrospective study of 20 years. Int J
Colorectal Dis. 24:943–949. 2009.PubMed/NCBI
|
|
5
|
Petrova TV, Nykänen A, Norrmén C, et al:
Transcription factor PROX1 induces colon cancer progression by
promoting the transition from benign to highly dysplastic
phenotype. Cancer Cell. 13:407–419. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sansom OJ, Meniel VS, Muncan V, et al:
Myc deletion rescues Apc deficiency in the small intestine.
Nature. 446:676–679. 2007. View Article : Google Scholar
|
|
7
|
Micel LN, Tentler JJ, Smith PG and
Eckhardt GS: Role of ubiquitin ligases and the proteasome in
oncogenesis: novel targets for anticancer therapies. J Clin Oncol.
31:1231–1238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bedford L, Lowe J, Dick LR, Mayer RJ and
Brownell JE: Ubiquitin-like protein conjugation and the
ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov.
10:29–46. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reinstein E and Ciechanover A: Narrative
review: protein degradation and human diseases: the ubiquitin
connection. Ann Intern Med. 145:676–684. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Naujokat C and Sarić T: Concise review:
role and function of the ubiquitin-proteasome system in mammalian
stem and progenitor cells. Stem Cells. 25:2408–2418. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bronner C, Achour M, Arima Y, Chataigneau
T, Saya H and Schini-Kerth VB: The UHRF family: oncogenes that are
drugable targets for cancer therapy in the near future? Pharmacol
Ther. 115:419–434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hopfner R, Mousli M, Jeltsch JM, et al:
ICBP90, a novel human CCAAT binding protein, involved in the
regulation of topoisomerase IIα expression. Cancer Res. 60:121–128.
2000.
|
|
13
|
Mori T, Li Y, Hata H, Ono K and Kochi H:
NIRF, a novel RING finger protein, is involved in cell-cycle
regulation. Biochem Biophys Res Commun. 296:530–536. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kotliarov Y, Steed ME, Christopher N, et
al: High-resolution global genomic survey of 178 gliomas reveals
novel regions of copy number alteration and allelic imbalances.
Cancer Res. 66:9428–9436. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu J, Liu S, Liu G, et al: Identification
and functional analysis of 9p24 amplified genes in human breast
cancer. Oncogene. 31:333–341. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li DW, Tang HM, Fan JW, et al: Expression
level of Bmi-1 oncoprotein is associated with progression and
prognosis in colon cancer. J Cancer Res Clin Oncol. 136:997–1006.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan DW, Fan JW, Yu ZH, et al:
Downregulation of metallothionein 1F, a putative oncosuppressor, by
loss of heterozygosity in colon cancer tissue. Biochim Biophys
Acta. 1822:918–926. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Troup S, Njue C, Kliewer EV, et al:
Reduced expression of the small leucine-rich proteoglycans,
lumican, and decorin is associated with poor outcome in
node-negative invasive breast cancer. Clin Cancer Res. 9:207–214.
2003.PubMed/NCBI
|
|
19
|
Bachmann IM, Puntervoll HE, Otte AP and
Akslen LA: Loss of BMI-1 expression is associated with clinical
progress of malignant melanoma. Mod Pathol. 21:583–590. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hashimoto Y, Skacel M, Lavery IC,
Mukherjee AL, Casey G and Adams JC: Prognostic significance of
fascin expression in advanced colorectal cancer: an
immunohistochemical study of colorectal adenomas and
adenocarcinomas. BMC Cancer. 6:2412006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lorenzato M, Caudroy S, Bronner C, et al:
Cell cycle and/or proliferation markers: what is the best method to
discriminate cervical high-grade lesions? Hum Patho. 36:1101–1107.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Crnogorac-Jurcevic T, Gangeswaran R,
Bhakta V, et al: Proteomic analysis of chronic pancreatitis and
pancreatic adenocarcinoma. Gastroenterology. 129:1454–1463. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Unoki M, Kelly JD, Neal DE, Ponder BA,
Nakamura Y and Hamamoto R: UHRF1 is a novel molecular marker for
diagnosis and the prognosis of bladder cancer. Br J Cancer.
101:98–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu TF, Zhang W, Su ZP, et al: UHRF2 mRNA
expression is low in malignant glioma but silencing inhibits the
growth of U251 glioma cells in vitro. Asian Pac J Cancer Prev.
13:5137–5142. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun Y: E3 ubiquitin ligases as cancer
targets and biomarkers. Neoplasia. 8:645–654. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma C, Rong Y, Radiloff DR, et al:
Extracellular matrix protein βig-h3/TGFBI promotes metastasis of
colon cancer by enhancing cell extravasation. Genes Dev.
22:308–321. 2008.
|