Introduction
Long intergenic noncoding RNAs (lincRNAs) are
non-protein coding transcripts >200 nucleotides in length. They
have been identified for decades, but they failed to draw attention
until now, mainly due to the limitation of technology and
traditional conceptual understanding of molecular biology. LincRNAs
are located and transcribed within the intergenic stretches with
the exception that a minority of them are within protein coding
genes; their length varies from several hundred to tens of
thousands of bases (1,2). Large scale transcriptomic sequencing
by next generation sequencing indicated that lincRNA number in the
order of tens of thousands in mammals. However, despite
accumulating evidence suggesting that the majority of these are
likely to be functional, only a relatively small proportion has
been functionally annotated (3).
From collected data, lincRNAs regulate target gene expression at
both the transcriptional and post-transcriptional level. LincRNAs
regulate gene transcription through either targeting specific
transcriptional factors or targeting the general transcriptional
machinery (4). LincRNAs also
regulate the transcription of large numbers of genes by chromatin
modifications (5). In addition to
regulating transcription, lincRNAs also control various aspects of
post-transcriptional mRNA processing, including splicing,
transport, translation and degradation of target mRNAs (6,7).
LincRNAs not only play critical roles in physical function
regulation, such as imprinting (8),
but they are also involved in the progress of various diseases
including cancer (9–11). At present, lincRNAs are emerging as
either oncogenes or tumor suppressor genes (11–15).
Previous evidence demonstrated the aberrant expression of lincRNAs
in digestive system carcinomas (16–21).
However, the biological function of lincRNAs in the vast majority
of digestive system carcinomas remains unclear. LincRNA-p21 has
been found to be a downstream target of p53 and to modulate the
expression of numerous genes at the transcriptional level (22). The expression of lincRNA-p21 is
found to be downregulated in several types of tumor (22,23),
suggesting that lincRNA-p21 may function as a tumor suppressor.
Recently, lincRNA-p21 was found to inhibit the expression of
β-catenin at the post-transcriptional level (24). It has been reported that the
elevation of the Wnt/β-catenin signaling pathway is one of the
common features of colorectal cancer (CRC) (25). Recent studies also demonstrated that
Wnt/β-catenin signaling is one of the effective targets for
chemotherapy and chemoprevention of CRC (26–28). A
recent study showed that expression of lincRNA-p21 was lower in
human CRC tissue compared to paired normal tissue (23). However, the pathological role of
decreased lincRNA-p21 in CRC was not detected. Since lincRNA-p21
has been reported to inhibit the translation of β-catenin in HeLa
cells (24), and β-catenin aberrant
activation affected CRC treatment, we hypothesized that lincRNA-p21
may affect the treatment of CRC through regulating β-catenin
activity. To test this hypothesis, we detected the expressional
change of lincRNA-p21 and evaluated the role of lincRNA-p21 in CRC
radiotherapy; the regulation role of lincRNA-p21 in the
Wnt/β-catenin signaling pathway in CRC cells as a candidate for the
molecular mechanism of lincRNA-p21 regulating the radiosensitivity
of CRC radiotherapy was also detected.
Materials and methods
Patient samples
The present study consisted of 30 CRC tissues and
their adjacent normal mucosa, which were resected at the Department
of Gastrointestinal Surgical Oncology (The Affiliated Tumor
Hospital of Harbin Medical University) between 2011 and 2012. None
of the patients received preoperative treatment such as irradiation
or chemotherapy. All research protocols in the present study were
approved by the Ethics Committee of the Cancer Research Institute
at Harbin Medical University. Staging of the tumors was performed
according to NCCN Guidelines.
Cell lines and cell culture
The normal colorectal cell line, FHC, and the CRC
cell lines SW1116, SW620, LS 174T, HT29 and LOVO were provided by
the Shanghai Institute of Cell Biology (Shanghai, China). The cell
lines were maintained in Dulbecco’s modified Eagle’s medium or
RPMI-1640, respectively, containing 10% FBS with 100 U/ml
penicillin and 100 μg/ml streptomycin, and were cultured in a
humidified 5% CO2 incubator at 37°C. The medium was
changed every 2 days (29).
Exponentially growing cells were used for experiments.
Irradiation treatment of cells
The 3×105 cells were plated in triplicate
in 25 cm2 tissue culture dishes. After transfecting for
24 h, cells were irradiated with various single radiation doses (0,
2 and 4 Gy) X-ray. Then, cells were harvested at 0, 0.5, 1, 2, 6,
12, 24 and 48 h after irradiation for further analyses.
Total RNA preparation and reverse
transcription
Total RNA from colorectal and CRC epithelial cells
and tissues was extracted using TRIzol as previously described
(30). Agarose gel electrophoresis
identified integrity of total RNA. The ratio of A260:A280 was used
to indicate the purity of total RNA. cDNA was generated using the
First Strand cDNA Synthesis kit (Roche), according to the
manufacturer’s instructions.
Quantitative real-time polymerase chain
reaction (qRT-PCR) assay
To detect and compare gene expression, qRT-PCR was
performed with a Power SYBR-Green PCR Master Mix (Roche,
Switzerland) and an Applied Biosystems 7500 Fast System (ABI;
Foster City, CA, USA) as described by Wang et al (31). Relative levels of gene expression
were determined with glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) as the control. The cycle number at which the reaction
crossed an arbitrarily placed threshold (Ct) was determined for
each gene and the ΔΔCt method (2−ΔCt) was used to
determine the target gene levels relative to those of GAPDH. The
primers used in this study are summarized in Table I.
 | Table IThe primer sequences used in the
present study. |
Table I
The primer sequences used in the
present study.
| Name | Sequence |
|---|
| LincRNA-p21 | F:
GGGTGGCTCACTCTTCTGGC |
| LincRNA-p21 | R:
TGGCCTTGCCCGGGCTTGTC |
| β-Catenin | F:
ATTGTCCACGCTGGATTTTC |
| β-Catenin | R:
TCGAGGACGGTCGGACT |
| GAPDH | F:
AGCCACATCGCTCAGACAC |
| GAPDH | R:
GCCCAATACGACCAAATCC |
| Noxa | F:
ATGAATGCACCTTCACATTCCTCT |
| Noxa | R:
TCCAGCAGAGCTGGAAGTCGAGTGT |
| c-Myc | F:
CGCTTCTCTGAAAGGCTCTCCTTG |
| c-Myc | R:
GAGTCGTAGTCGAGGTCATAGTTC |
| Cyclin | D1 F:
AGGAGAACAAACAGATCA |
| Cyclin | D1 R:
TAGGACAGGAAGTTGTTG |
| TCF4 | F:
GAGAAT-TCATGCCGCAGCT |
| TCF4 | R:
CAGATATCTTTTTAAACGCTACA |
Western blot assay
Total protein was extracted using SDS protein lysis
buffer. Protein (80 μg) for each sample was resolved by 8% SDS-PAGE
and then transferred to PVDF membranes. The membranes were blocked
by TBST buffer (TBS plus 0.1% Tween-20) containing 5% w/v skimmed
milk and hybridized with primary antibody including rabbit
anti-β-catenin (1:500), rabbit anti-c-myc (1:750; both from Abcom,
Hong Kong), mouse anti-β-actin (1:5,000; Santa Cruz), rabbit
anti-cyclin D1 (1:50; Abcom), followed by incubation with specific
HRP-conjugated secondary antibody (ZSGB-BIO, Beijing). Protein
bands were visualized by the ECL detecting system (Applygen,
Beijing).
RNA interference and overexpression
RNA interference for lincRNA-p21 was performed as
previously described (22). Several
siRNA oligos targeting lincRNA-p21 (#1 UGA AAAGAGCCGUGAGCUA, #2
AAAUAAAGAUGGUGGA AUG and #3 AGUCAAAGGCAAUGAGCAU) and Lipofectamine
2000 were purchased from Invitrogen. The siRNA transfections were
performed with 50 nM of siRNA and Lipofectamine 2000 in serum-free
culture media following the manufacturer’s instructions as
previously described (32,33). A plasmid expressing lincRNA-p21 was
used to construct plasmid plincRNA-p21pcDNA3.1 (+). The plasmid was
previously reported (24) and
kindly provided by Prof. Myriam Gorospe.
Analysis of cell apoptosis
Cells undergoing early and late apoptosis were
quantified using flow cytometry analysis (Cytomics FC500; Beckman
Coulter, Miami, FL, USA), following the staining with Annexin
V-FITC and propidium iodide (PI; BD Biosciences, San Jose, CA,
USA). All experiments were performed in triplicate.
Statistical analysis
Statistical analysis was performed with SPSS13.0
software. Results are expressed as the means ± standard deviation
from at least 3 separate experiments. Standard statistical tests
including Student’s t-test and one-way ANOVA were used to evaluate
the significance. P<0.05 was considered to indicate a
statistically significant difference.
Results
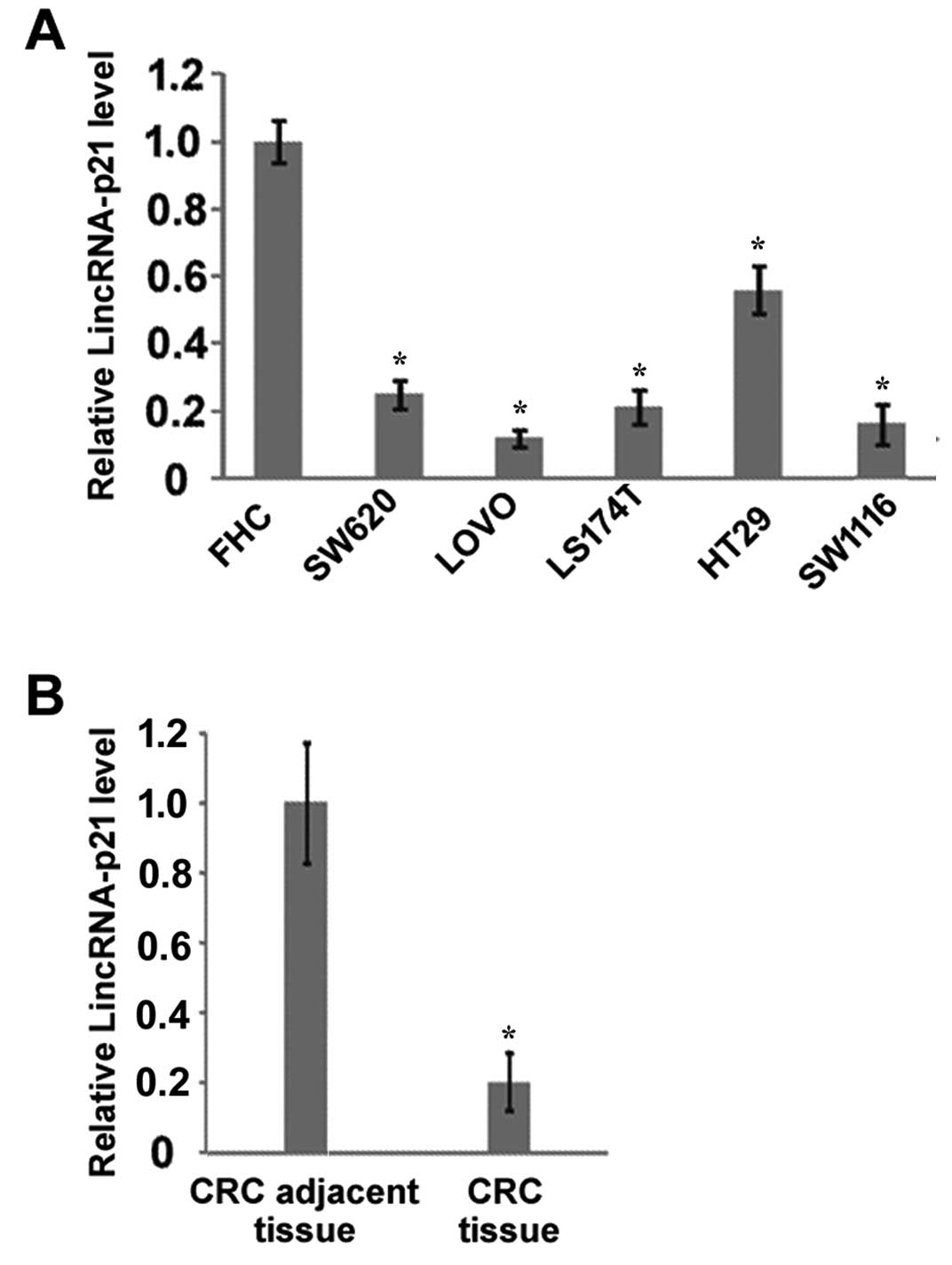
LincRNA-p21 is downregulated in CRC cell
lines and CRC tumor tissues
To examine the potential roles of lincRNA-p21 in
CRC, we first compared the expression of lincRNA-p21 between normal
colorectal epithelial cells and CRC cell lines. Our results showed
that the expression of lincRNA-p21 in all the tested CRC cell lines
(SW620, LOVO, LS174T, HT29 and SW116) was lower than in the normal
colorectal cell line FHC (Fig. 1A).
Furthermore, we compared the expression of lincRNA-p21 in 30 CRC
tissues and their paired adjacent tissues. The results demonstrated
that the expression of lincRNA-p21 in CRC tissues was significantly
decreased compared with their adjacent tissues (Fig. 1B). These results suggest that
lincRNA-p21 is downregulated in CRC, which may contribute to the
development of CRC and/or affect the treatment of CRC.
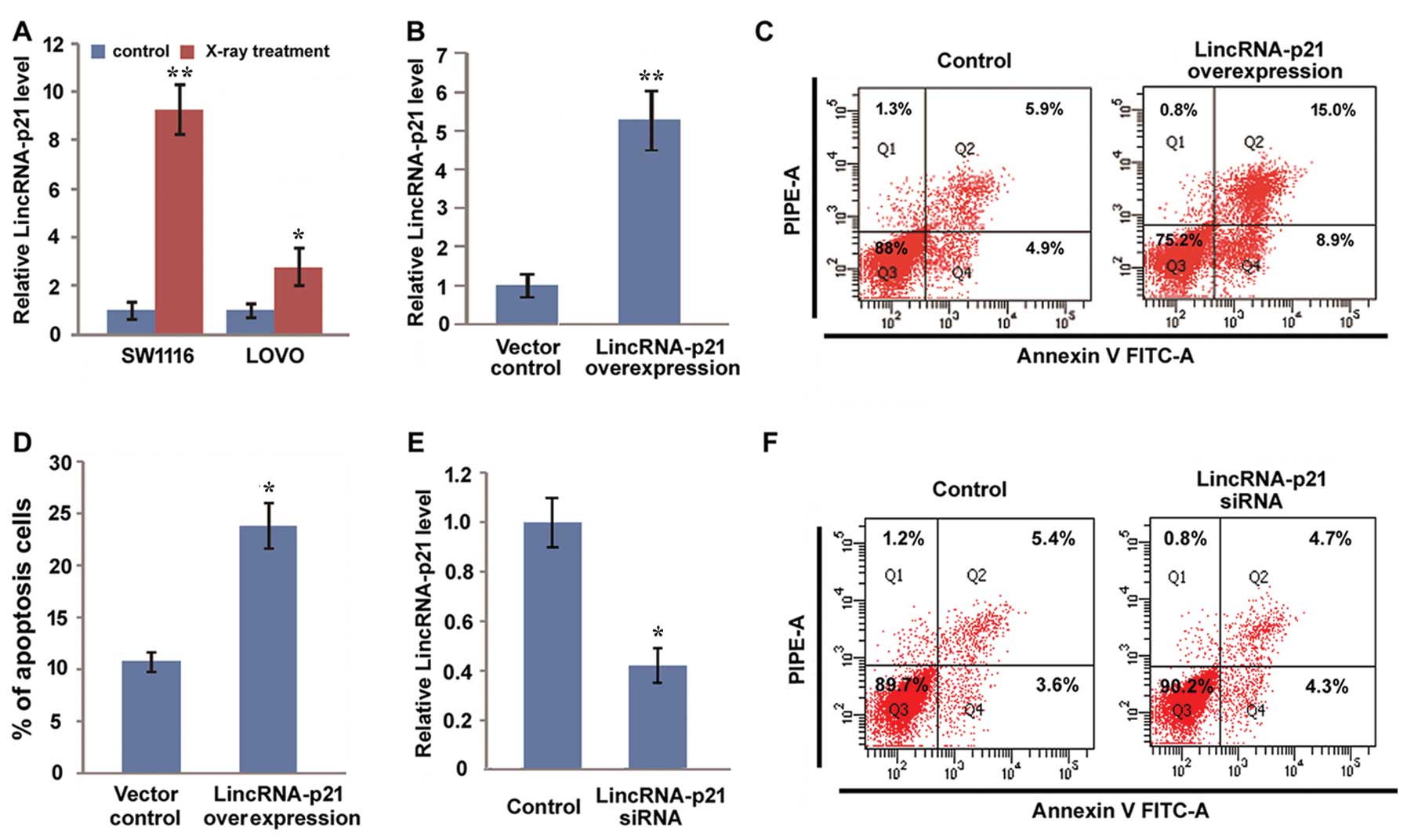
LincRNA-p21 enhances radiosensitivity by
promoting apoptosis
Radiotherapy is considered as a standard
preoperative treatment approach to reduce local recurrence for
locally advanced rectal cancers (34–37).
To detect the potential role of lincRNA-p21 in CRC radiotherapy, we
first detected the effect of radiotherapy on the expression of
lincRNA-p21. We showed that X-ray treatment elevated the expression
of lincRNA-p21 in both SW1116 and LOVO cells (Fig. 2A). Then, we overexpressed
lincRNA-p21 in SW1116 cells (Fig.
2B) and treated lincRNA-p21 overexpressed cells and control
cells with X-ray. Subsequently, the apoptosis of both group cells
was detected with Annexin V and PI staining by fluorescence
activated cell sorting (FACS). Our results showed that the
apoptosis rate of lincRNA-p21 overexpressed cells was ~25%, while
it was only 10% in control cells (Fig.
2C and D), which suggested that overexpression of lincRNA-p21
increases the sensitivity of CRC radiotherapy by promoting the
apoptosis of CRC cells. Furthermore, we inhibited endogenous
lincRNA-p21 expression by using siRNA. To avoid the off-target
effect of siRNA, we transfected SW1116 cells with a mixture of
three synthesized lincRNA-p21 siRNAs; the results showed endogenous
lincRNA-p21 decreased 60% after transfection with lincRNA-p21
siRNAs, which suggests that lincRNA-p21 was successfully inhibited
by siRNA in SW1116 cells (Fig. 2E).
Then, we evaluated the effect of lincRNA-p21 knockdown on the
apoptosis of SW1116 cells induced by X-ray irradiation. We did not
observe a significant difference in the apoptosis rate between
lincRNA-p21 knockdown cells and control cells. Our results indicate
that the expression level of lincRNA-p21 in CRC may affect the
sensitivity of CRC treatment.
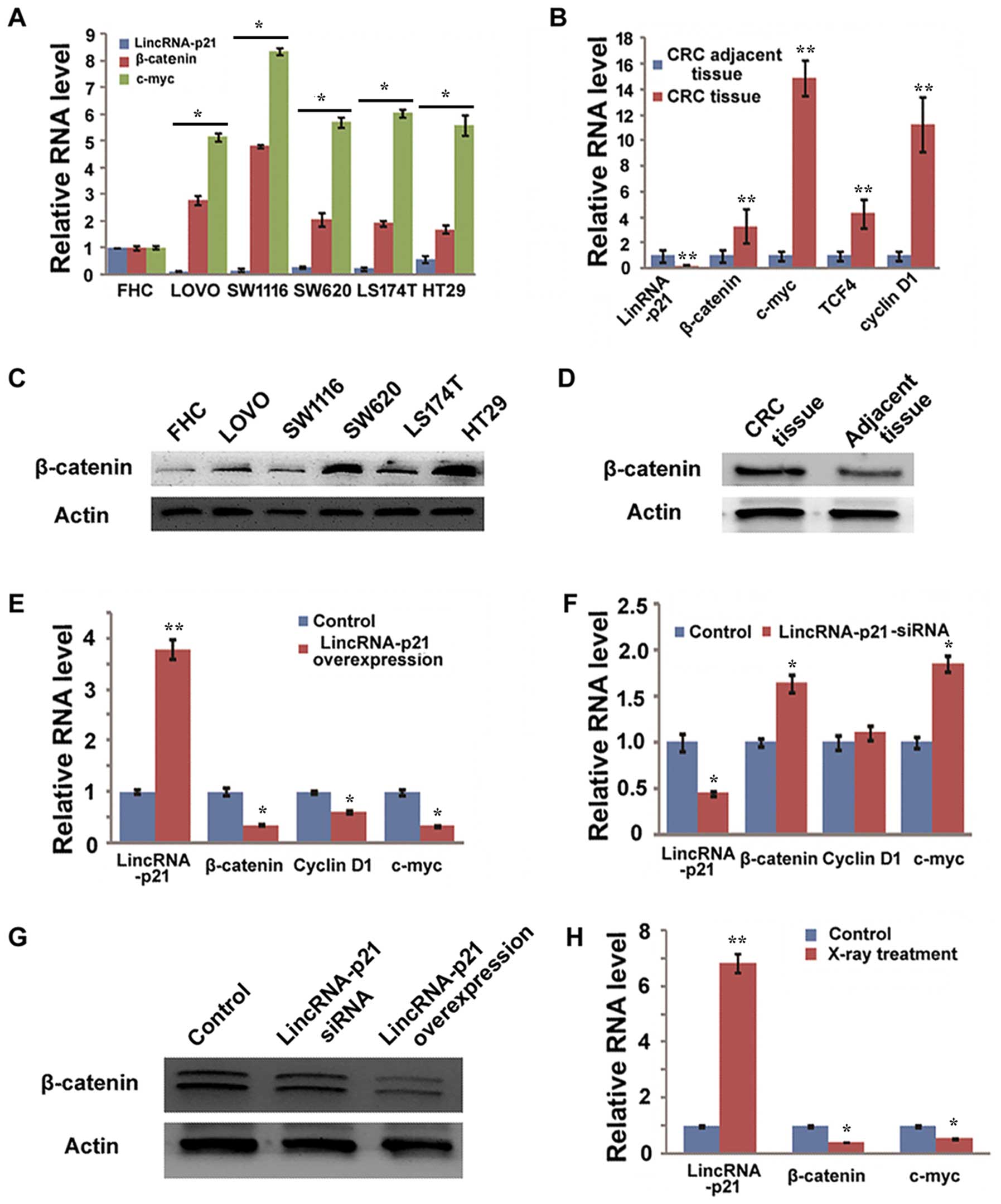
LincRNA-p21 inhibits the Wnt/β-catenin
signaling pathway
LincRNA-p21 has been reported to inhibit the
translation of β-catenin in HeLa cells (24); however, whether lincRNA-p21 also
inhibits the translation of β-catenin in CRC remains unknown. We
first detected the relationship between lincRNA-p21 and the
Wnt/β-catenin signaling pathway in normal colorectal epithelial
cells and CRC cell lines. Our results showed that β-catenin was
highly expressed in all the tested CRC cell lines, at both the mRNA
and the protein level, compared with FHC cells (Fig. 3A and C). LincRNA-p21 was lower in
these CRC cell lines than in the FHC cells (Fig. 3A). Furthermore, the activity of
Wnt/β-catenin signaling pathway was investigated in these cell
lines by detecting expression of c-myc, one of the target genes of
β-catenin, and the results showed that c-myc level was higher in
all the CRC cell lines than in the FHC cells (Fig. 3A). The relationship between
lincRNA-p21 and β-catenin was also detected in CRC tissues and CRC
adjacent tissues. The total RNA of 30 CRC tissues and 30 adjacent
tissues were mixed together, respectively. Then, the expression of
lincRNA-p21, β-catenin and Wnt/β-catenin target genes including
c-myc, cyclin D1 and TCF4 was detected using qRT-PCR. The results
showed that β-catenin and its target genes were significantly
increased in CRC tissues when compared with CRC adjacent tissues
(Fig. 3B). Consistently, the
protein level of β-catenin was also confirmed to be elevated in CRC
tissues (Fig. 3D). These results
suggest that there is an inverse correlation between the expression
of lincRNA-p21 and the expression and activity of β-catenin in the
CRC cell lines and tissues, and it suggests that lincRNA-p21 may
regulate the expression and activity of β-catenin in CRC.
We further detected the direct impact of lincRNA-p21
on the expression of β-catenin by gain-of-function and
loss-of-function strategy, respectively. We demonstrated that
lincRNA-p21 overexpression in SW1116 cells significantly inhibited
the expression of β-catenin at both the mRNA and the protein level
(Fig. 3E and G). Furthermore, the
overexpression of lincRNA-p21 inhibited Wnt/β-catenin signaling
activity as evidenced by the decrease of β-catenin target genes,
such as cyclin D1 and c-myc (Fig.
3E). However, the knockdown of lincRNA-p21 failed to promote
the expression of β-catenin at both the mRNA and the protein level
(Fig. 3F and G). The obvious
activation of Wnt/β-catenin signaling pathway caused by the
knockdown of lincRNA-p21 was also not observed (Fig. 3F).
Finally, to investigate whether the inhibition of
Wnt/β-catenin is involved in the mechanism of lincRNA-p21 enhanced
sensitivity of CRC radiotherapy, we detected the expression and
activity of β-catenin in SW1116 cells after X-ray treatment. We
showed that the irradiation with X-ray decreased the expression and
activation of β-catenin (Fig. 3H).
This result suggests that the inhibition of β-catenin is involved
in the lincRNA-p21 regulating sensitivity of CRC radiotherapy.
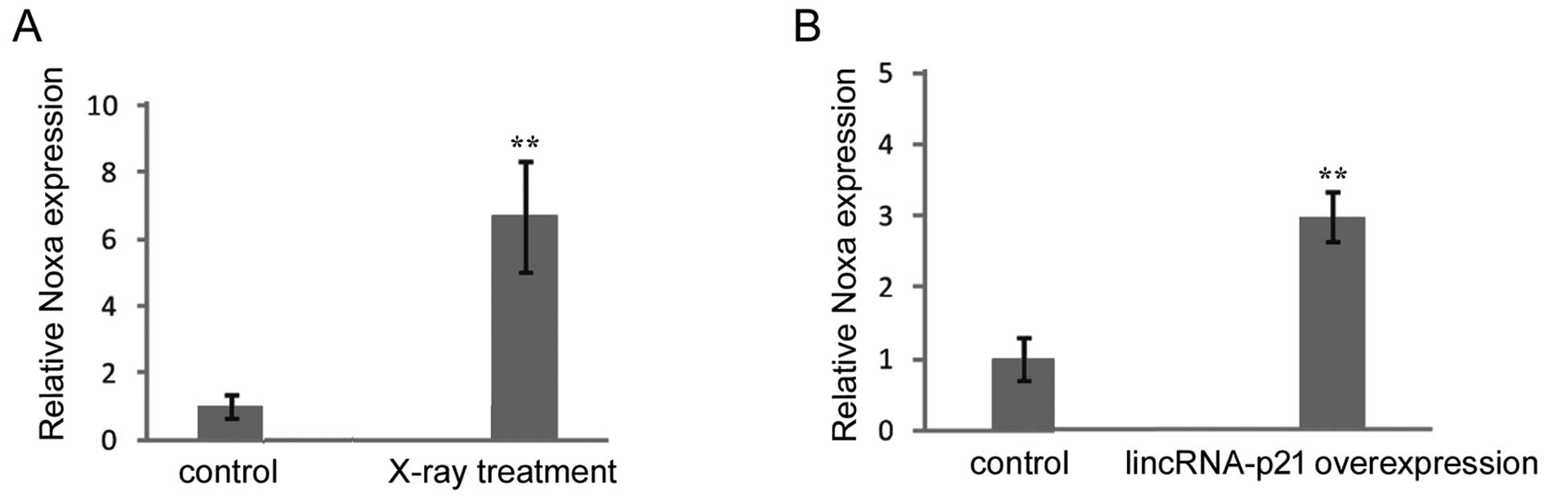
LincRNA-p21 promotes pro-apoptosis gene
Noxa expression
Noxa, a pro-apoptosis gene, is positively regulated
by lincRNA-p21 and contributes to cell apoptosis after p53
activation (22). We detected the
effect of X-ray irradiation and lincRNA-p21 on the expression of
Noxa. We showed that X-ray treatment resulted in the upregulation
of Noxa expression in SW1116 cells (Fig. 4A). The overexpression of lincRNA-p21
also led to increase of Noxa expression (Fig. 4B). These results suggested that the
upregulated pro-apoptosis gene Noxa resulting from lincRNA-p21
elevation after X-ray irradiation may also be involved in the
molecular mechanism of lincRNA-p21 enhancing sensitivity of CRC
radiotherapy.
Discussion
Recent studies have shown that lincRNA-p21 levels
may significantly influence the biological properties of cells,
including tumor cell proliferation and apoptosis (22,38).
CRC is the third most common form of cancer and the fourth most
frequent cause of cancer-related mortality, with >1 million new
cases each year worldwide (39).
However, current knowledge about lincRNA-p21 function in human CRC
is still preliminary. Therefore, determining whether lincRNA-p21 is
essential for the progression of CRC may provide a promising
therapeutic target. Our findings showed that the expression level
of lincRNA-p21 was significantly lower in CRC tissue compared with
the adjacent normal tissue from the same patient. It is well known
that ~90% of CRC cases originate from the constitutive activation
of Wnt/β-catenin signaling pathway. Previous reports demonstrated
that the Wnt/β-catenin signaling pathway is a crucial pathway in
the initiation and progression of CRC (37,40–42).
Disruption of the Wnt/β-catenin signaling pathway caused by either
mutant Wnt or stabilization of the β-catenin protein resulted in
activation of the TCF-4 transcription factor and increased
expression of target genes including c-myc and cyclin D1 (43). The c-myc and cyclin D1 are
identified as target genes in the Wnt/β-catenin signaling pathway
and overexpress in CRC (44,45).
In the present study, we showed that lincRNA-p21 was inversely
correlated with Wnt/β-catenin signaling pathway target genes in
both clinical tissue specimens and human CRC cell lines, and the
overexpression of lincRNA-p21 significantly inhibited Wnt/β-catenin
signaling activity through directly inhibiting β-catenin stability
and/or translation. Our results are consistent with recent findings
that β-catenin translation is suppressed by lincRNA-p21 in human
HeLa cell line (24). However,
obvious activation of the Wnt/β-catenin signaling pathway caused by
the knockdown of lincRNA-p21 was not observed in this study. Our
results showed that lincRNA-p21 level was very low in CRC cell
lines, thus we speculate that these models are not the most
suitable for knockdown models in this setting. This could not
exclude that lincRNA-p21 may be a necessary but not sufficient
factor for β-catenin inhibition.
Radiotherapy is considered as a standard
preoperative treatment approach to reduce local recurrence for
locally advanced rectal cancers (34,35).
However, considerable rectal cancers are resistant to preoperative
radiotherapy (46) and radiotherapy
may result in strong side-effects in some patients. Therefore,
increasing radiosensitivity may be very useful for enhancing the
effect of therapy while reducing the side-effects of X-ray
irradiation. A previous study showed that anti-vascular endothelial
growth factor therapy may enhance radiosensitization in patients
with locally advanced inoperable CRC (36). Radiosensitization may occur through
a direct effect on CRC cells followed by suppression of some
transcription factors or genes (37). Whether the transcription factor
β-catenin is one of the targets for CRC radiotherapy and if
lincRNA-p21 affects radiosensitivity is unknown. Our results showed
that β-catenin is decreased while lincRNA-p21 is increased after
X-ray irradiation, and the enforced expression of lincRNA-p21
decreases the surviving fraction of tumor cells in response to
X-ray irradiation and enhances radiosensitivity. We further showed
that lincRNA-p21 is likely to enhance radiosensitivity by inducing
tumor cell apoptosis. However, there is no significant change of
cell apoptosis after X-ray irradiation in cells treated with
lincRNA-p21 siRNA compared with those treated with negative siRNA
control. This result is consistent with the expression change of
β-catenin following lincRNA-p21 knockdown.
Pro-apoptotic protein Noxa initiates apoptosis by
binding to regulatory sites on anti-apoptotic Bcl-2 protein and
directly neutralizing its function. A previous study showed that a
chemotherapy drug may induce the pro-apoptotic protein Noxa in CRC
cells (47). Other studies
indicated that the loss of Noxa may enhance resistance to X-ray
irradiation-induced apoptosis (48,49).
Consistent with these previous studies, our results showed that
X-ray treatment may activate lincRNA-p21 and Noxa expression and
then induce CRC cell apoptosis. Moreover, we found that lincRNA-p21
can promote cell apoptosis after X-ray irradiation by increasing
Noxa expression. Our data provide an interesting link between
lincRNA-p21 and Noxa, and suggest that Noxa acts as a downstream
target of lincRNA-p21. This result is consistent with a previous
report in which the expression of Noxa was found to change
according to lincRNA-p21 status identified by gene expression
microarray (22).
In conclusion, we first found that lincRNA-p21
increases the sensitivity of radiotherapy for CRC by targeting the
β-catenin signaling pathway. The present study not only deepens our
understanding of the mechanism of radiotherapy for CRC, but it also
provides a potential target for CRC radiotherapy. Our results also
suggest that lincRNA-p21 itself and compounds that can induce the
expression of lincRNA-p21 may be useful for enhancing the
sensitivity of CRC radiotherapy.
Acknowledgements
The authors thank Professor Myriam Gorospe of the
National Institutes of Health Laboratory of Molecular Biology and
Immunology for kindly providing lincRNA-p21 expression plasmid. The
authors also acknowledge the technical assistance of Yuyan Ma and
Yanmei Yang of the Cancer Research Institute of Harbin Medical
University. This study was supported by the National Natural
Science Foundation of China (grant no. 81172265).
References
|
1
|
Guttman M, Amit I, Garber M, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Katayama S, Tomaru Y, Kasukawa T, et al:
Antisense transcription in the mammalian transcriptome. Science.
309:1564–1566. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Amaral PP, Clark MB, Gascoigne DK, Dinger
ME and Mattick JS: lncRNAdb: a reference database for long
noncoding RNAs. Nucleic Acids Res. 39:D146–D151. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Goodrich JA and Kugel JF: Non-coding-RNA
regulators of RNA polymerase II transcription. Nat Rev Mol Cell
Biol. 7:612–616. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen X, Xu H, Yuan P, et al: Integration
of external signaling pathways with the core transcriptional
network in embryonic stem cells. Cell. 133:1106–1117. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Beltran M, Puig I, Peña C, et al: A
natural antisense transcript regulates Zeb2/Sip1 gene expression
during Snail1-induced epithelial-mesenchymal transition. Genes Dev.
22:756–769. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Centonze D, Rossi S, Napoli I, et al: The
brain cytoplasmic RNA BC1 regulates dopamine D2
receptor-mediated transmission in the striatum. J Neurosci.
27:8885–8892. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Braidotti G, Baubec T, Pauler F, et al:
The Air noncoding RNA: an imprinted cis-silencing
transcript. Cold Spring Harb Symp Quant Biol. 69:55–66. 2004.
|
|
9
|
Fu X, Ravindranath L, Tran N, Petrovics G
and Srivastava S: Regulation of apoptosis by a prostate-specific
and prostate cancer-associated noncoding gene, PCGEM1. DNA
Cell Biol. 25:135–141. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin R, Maeda S, Liu C, Karin M and
Edgington TS: A large noncoding RNA is a marker for murine
hepatocellular carcinomas and a spectrum of human carcinomas.
Oncogene. 26:851–858. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calin GA, Liu CG, Ferracin M, et al:
Ultraconserved regions encoding ncRNAs are altered in human
leukemias and carcinomas. Cancer Cell. 12:215–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Braconi C, Valeri N, Kogure T, et al:
Expression and functional role of a transcribed noncoding RNA with
an ultraconserved element in hepatocellular carcinoma. Proc Natl
Acad Sci USA. 108:786–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas.
Molecular Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qi P and Du X: The long non-coding RNAs, a
new cancer diagnostic and therapeutic gold mine. Mod Pathol.
26:155–165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Weakley SM, Wang H, Yao Q and Chen C:
Expression and function of a large non-coding RNA gene XIST in
human cancer. World J Surg. 35:1751–1756. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang J, Liu X, Wu H, et al: CREB
up-regulates long non-coding RNA, HULC expression through
interaction with microRNA-372 in liver cancer. Nucleic Acids Res.
38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yuan S-X, Yang F, Yang Y, et al: Long
noncoding RNA associated with microvascular invasion in
hepatocellular carcinoma promotes angiogenesis and serves as a
predictor for hepatocellular carcinoma patients’ poor
recurrence-free survival after hepatectomy. Hepatology.
56:2231–2241. 2012.PubMed/NCBI
|
|
18
|
Yang F, Xue X, Bi J, et al: Long noncoding
RNA CCAT1, which could be activated by c-Myc, promotes the
progression of gastric carcinoma. J Cancer Res Clin Oncol.
139:437–445. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu W, Bhagat TD, Yang X, et al:
Hypomethylation of noncoding DNA regions and overexpression of the
long noncoding RNA, AFAP1-AS1, in Barrett’s esophagus and
esophageal adenocarcinoma. Gastroenterology. 144:956–966. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tahira AC, Kubrusly MS, Faria MF, et al:
Long noncoding intronic RNAs are differentially expressed in
primary and metastatic pancreatic cancer. Mol Cancer. 10:1412011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Merchant JL: Inflammation, atrophy,
gastric cancer: connecting the molecular dots. Gastroenterology.
129:1079–1082. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huarte M, Guttman M, Feldser D, et al: A
large intergenic noncoding RNA induced by p53 mediates global gene
repression in the p53 response. Cell. 142:409–419. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhai H, Fesler A, Schee K, Fodstad Ø,
Flatmark K and Ju J: Clinical significance of long intergenic
noncoding RNA-p21 in colorectal cancer. Clin Colorectal Cancer.
12:261–266. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoon JH, Abdelmohsen K, Srikantan S, et
al: LincRNA-p21 suppresses target mRNA translation. Mol Cell.
47:648–655. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Samuels Y and Velculescu VE: Oncogenic
mutations of PIK3CA in human cancers. Cell Cycle. 3:1221–1224.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu K, Yang Q, Mu Y, Zhou L, Liu Y, Zhou Q
and He B: Berberine inhibits the proliferation of colon cancer
cells by inactivating Wnt/β-catenin signaling. Int J Oncol.
41:292–298. 2012.PubMed/NCBI
|
|
27
|
Dihlmann S, Klein S and Doeberitz Mv:
Reduction of β-catenin/T-cell transcription factor signaling by
aspirin and indomethacin is caused by an increased stabilization of
phosphorylated β-catenin. Mol Cancer Ther. 2:509–516. 2003.
|
|
28
|
Boon EM, Keller JJ, Wormhoudt TA, et al:
Sulindac targets nuclear β-catenin accumulation and Wnt signalling
in adenomas of patients with familial adenomatous polyposis and in
human colorectal cancer cell lines. Br J Cancer. 90:224–229.
2004.
|
|
29
|
Ji BC, Hsu WH, Yang JS, et al: Gallic acid
induces apoptosis via caspase-3 and mitochondrion-dependent
pathways in vitro and suppresses lung xenograft tumor growth in
vivo. J Agric Food Chem. 57:7596–7604. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xi Y, Nakajima G, Gavin E, et al:
Systematic analysis of microRNA expression of RNA extracted from
fresh frozen and formalin-fixed paraffin-embedded samples. RNA.
13:1668–1674. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang K, Zhang R, He F, et al:
Electroacupuncture frequency-related transcriptional response in
rat arcuate nucleus revealed region-distinctive changes in response
to low- and high-frequency electroacupuncture. J Neurosci Res.
90:1464–1473. 2012. View Article : Google Scholar
|
|
32
|
Kalin TV, Wang IC, Ackerson TJ, et al:
Increased levels of the FoxM1 transcription factor accelerate
development and progression of prostate carcinomas in both TRAMP
and LADY transgenic mice. Cancer Res. 66:1712–1720. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim IM, Ackerson T, Ramakrishna S, et al:
The Forkhead Box m1 transcription factor stimulates the
proliferation of tumor cells during development of lung cancer.
Cancer Res. 66:2153–2161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Glimelius B and Oliveira J; ESMO
Guidelines Working Group Rectal cancer. ESMO clinical
recommendations for diagnosis, treatment and follow-up. Ann Oncol.
20(Suppl 4): S54–S56. 2009. View Article : Google Scholar
|
|
35
|
Sauer R, Becker H, Hohenberger W, et al:
Preoperative versus postoperative chemoradiotherapy for rectal
cancer. N Engl J Med. 351:1731–1740. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Koukourakis MI, Giatromanolaki A, Sheldon
H, et al: Phase I/II trial of bevacizumab and radiotherapy for
locally advanced inoperable colorectal cancer:
vasculature-independent radiosensitizing effect of bevacizumab.
Clin Cancer Res. 15:7069–7076. 2009. View Article : Google Scholar
|
|
37
|
Kendziorra E, Ahlborn K, Spitzner M, et
al: Silencing of the Wnt transcription factor TCF4 sensitizes
colorectal cancer cells to (chemo-) radiotherapy. Carcinogenesis.
32:1824–1831. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Özgür E, Mert U, Isin M, Okutan M, Dalay N
and Gezer U: Differential expression of long non-coding RNAs during
genotoxic stress-induced apoptosis in HeLa and MCF-7 cells. Clin
Exp Med. 13:119–126. 2013.PubMed/NCBI
|
|
39
|
Jemal A, Center MM, Ward E and Thun MJ:
Cancer occurrence. Methods Mol Biol. 471:3–29. 2009. View Article : Google Scholar
|
|
40
|
Bordonaro M: Crosstalk between Wnt
signaling and RNA processing in colorectal cancer. J Cancer.
4:96–103. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li X, Pu J, Jiang S, et al: Henryin, an
ent-kaurane diterpenoid, inhibits Wnt signaling through
interference with β-catenin/TCF4 interaction in colorectal cancer
cells. PLoS One. 8:e685252013.PubMed/NCBI
|
|
42
|
Waaler J, Machon O, von Kries JP, et al:
Novel synthetic antagonists of canonical Wnt signaling inhibit
colorectal cancer cell growth. Cancer Res. 71:197–205. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bright-Thomas RM and Hargest R: APC,
β-Catenin and hTCF-4; an unholy trinity in the genesis of
colorectal cancer. Eur J Surg Oncol. 29:107–117. 2003.
|
|
44
|
He TC, Sparks AB, Rago C, et al:
Identification of c-MYC as a target of the APC pathway.
Science. 281:1509–1512. 1998.PubMed/NCBI
|
|
45
|
Khor TO, Gul YA, Ithnin H and Seow HF: A
comparative study of the expression of Wnt-1, WISP-1, survivin and
cyclin-D1 in colorectal carcinoma. Int J Colorectal Dis.
21:291–300. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cunningham D, Atkin W, Lenz HJ, et al:
Colorectal cancer. Lancet. 375:1030–1047. 2010. View Article : Google Scholar
|
|
47
|
Raats DA, de Bruijn MT, Steller EJ, Emmink
BL, Borel-Rinkes IH and Kranenburg O: Synergistic killing of
colorectal cancer cells by oxaliplatin and ABT-737. Cell Oncol
(Dordr). 34:307–313. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shibue T, Takeda K, Oda E, et al: Integral
role of Noxa in p53-mediated apoptotic response. Genes Dev.
17:2233–2238. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Miyoshi-Imamura T, Kakinuma S, Kaminishi
M, et al: Unique characteristics of radiation-induced apoptosis in
the postnatally developing small intestine and colon of mice.
Radiat Res. 173:310–318. 2010. View Article : Google Scholar : PubMed/NCBI
|