Introduction
Mucin1 (MUC1), a transmembrane glycoprotein of the
mucin family, is expressed on the apical surface of most glandular
epithelia in normal tissue and is overexpressed in most
adenocarcinomas with aberrant glycosylation and loss of apical
expression (1–5). MUC1 consists of a large extracellular
N-terminal subunit containing a variable number of tandem repeats
(VNTRs) region and a C-terminal subunit that resides on the cell
surface as a heterodimeric complex via a strong noncovalent
interaction. The C-terminal subunit is composed of a 58-amino acid
extracellular domain, a 28-amino acid transmembrane domain (TM) and
a 72-amino acid cytoplasmic tail (CT) (6–8).
MUC1-CT is highly conserved between different species and contains
7 tyrosine residues that can be phosphorylated by multiple kinases
(9,10).
Previous studies have reported that aberrantly
overexpressed MUC1 influences cell adhesion (11), motility, migration, metastasis
(12–14) and cell-cell aggregation (15). Studies have shown that MUC1-CT is
involved in many signaling pathways, including Wnt/β-catenin
(16), c-Src (17), Grb2/Sos (18), p53 (19,20),
GSK3β (16), EGFR (21,22)
and NF-κB (23,24), that regulate the processes of cell
survival, proliferation and apoptosis. β-catenin is a major
effector of the Wnt signaling pathway, and it interacts with
MUC1-CT at an SXXXXXSSLS site. This interaction blocks the
GSK3β-induced degradation of β-catenin and promotes the
translocation of β-catenin to the nucleus. In the nucleus,
β-catenin forms a complex with the transcription factors lymphoid
enhancer factor/T cell factor (LEF/TCF) and activates the
transcription of Wnt-responsive genes such as cyclin D1 and c-Myc
to regulate cell proliferation (25,26).
Further studies have demonstrated that MUC1 plays a key role as an
oncogene in the formation and progression of lung (27), breast (28), ovarian (29) pancreatic and gastric carcinomas
(30,31).
Several reports, including our studies, have shown
that MUC1 is also expressed in hepatocellular carcinoma (HCC) cell
lines, such as SNU-475, SNU-449, Mahlavu (32) and SMMC-7721, and in liver carcinoma,
cirrhotic liver, and normal liver tissues (33). However, the role of MUC1 in HCC
progression remains unclear. In this study, we analyzed whether
MUC1 plays a crucial role in HCC progression.
Materials and methods
Cell culture
The HCC cell line SMMC-7721 was purchased from the
Cell Bank of the Shanghai Institute of Cell Biology, Chinese
Academy of Sciences. Cells were cultured in Iscove’s modified
Dulbecco’s medium (IMDM) supplemented with 100 U/ml penicillin, 100
μg/ml streptomycin, and 10% fetal bovine serum (FBS; Gibco-BRL,
Carlsbad, CA, USA) in an incubator at 37°C and 5% CO2.
The stable MUC1-knockdown cells and negative control cells were
maintained with 600 μg/ml G418 (Sigma).
Generation of stable MUC1 knockdown in
SMMC-7721 cells
Small interfering RNA (siRNA) oligonucleotides were
synthesized to target sequences of the MUC1 gene:
GACTGATGCCAGTAGCACT, GenBank accession No. J05582. The siRNA was
inserted into the expression vector pGCsilencer™ U6.Neo.GFP
(Genechem Co., Ltd., Shanghai, China). A nonspecific siRNA was used
as a negative control. The siRNA expression plasmids were
transfected into SMMC-7721 cells using Lipofectamine 2000
(Invitrogen). Cells were screened with 1,200 μg/ml G418 for 3
weeks. Three independent MUC1-knockdown clones (MR1-C6, MR1-D4 and
MR1-D9) and a negative control clone (NC) were isolated and
identified by reverse transcription-polymerase chain reaction
(RT-PCR) and western blotting.
RT-PCR and quantitative real-time PCR
(qRT-PCR)
Cells were harvested, RNA was isolated using TRIzol
(Invitrogen Life Technologies). Total RNA was converted to cDNA
using M-MLV reverse transcriptase and oligo (dT) primers (Promega
Corporation, Madison, WI, USA) according to the manufacturer’s
protocol. Reverse transcribed products were used to amplify MUC1 by
RT-PCR using Ex Taq DNA polymerase (Takara Bio, Inc., Shiga,
Japan). β-actin was used as an internal control gene. Primers
sequences used for RT-PCR were: 5′-GGTCTTGCTGGTCTTAGGAGAGAC-3′
(forward) and 5′-CTGAAGTCCAGCTGACCCTGTAGCTTCACG-3 (reverse) for
MUC1, and 5′-GTTGCTATCCAGGCTGTGC-3′ (forward) and
5′-AGCACTGTGTTGGCGTAAG-3′ (reverse) for β-actin. The length of
RT-PCR was 30 cycles. Amplification products were separated by 1.5%
agarose gel, and DNA was visualized by a Gel Image System (Tanon).
qRT-PCR was performed using a FastStart Universal SYBR-Green Master
(Rox; Roche) on an ABI7300 instrument. The primer sequences used
for qRT-PCR were: 5′-GCGTTTCCCAGAGTCATC-3′ (forward) and
5′-CCTCCCTTCAACACTTCCT-3′ (reverse) for cyclin D1, and
5′-TACATCCTGTCCGTCCAA-3′ (forward) and 5′-TTTCCTTACGCACAAGAGTT-3′
(reverse) for c-Myc, and 5′-AGTTGCGTTACACCCTTTC-3′ (forward) and
5′-CCTTCACCGTTCCAGTTT-3′ (reverse) for β-actin. The expression of
each investigated gene was normalized to the housekeeping gene
β-actin. Data were calculated using the 2−ΔΔCT method
(34) and are presented as the fold
change in gene expression relative to the negative control sample.
For the negative control sample, 2−ΔΔCT=1.
Cell viability assay
Cell viability was determined at different time
points using a WST-1 cell viability assay according to the
manufacturer’s protocol (Roche Diagnostics, Mannheim, Germany).
Triplicate wells containing 5×103 cells were evaluated
for viability. The absorbance was measured using a microplate
reader at a wavelength of 450 nm (BioTek Instruments, Inc.,
Winooski, VT, USA). The relative cell viability was calculated as
the A450 nm (MUC1-silenced clones at Tn) / A450 nm (NC at Tn) ×
100%.
Colony formation assay
Cells (2×103) were seeded into 6
well-plates for 3 weeks. Cell clones were stained with crystal
violet staining solution (Sigma) and the colony size and number
were observed. Colony numbers that contained >10 cells were
counted and analyzed, the colony formation ratio was calculated as
(colony number / seeded cell number) × 100%. Data are expressed as
a ratio of the results obtained with each MUC1-knockdown clone and
the negative control.
Cell cycle analysis
Cells (1×106) were harvested, washed with
PBS, fixed with 70% ice-cold ethanol for 30 min and washed with
PBS. Subsequently, cells were incubated in staining buffer with
PI/RNase (BD Biosciences) for 30 min at 4°C in the dark. The cell
cycle was analyzed by flow cytometry (FACSCalibur; BD
Biosciences).
Assays for cell migration and
invasion
For migration assay, the scratch test was performed
(35). Matrigel invasion by cells
was studied using transwell chambers with 8-μm pore size filters
(Corning Incorporated, Corning, NY, USA) coated with Matrigel
matrix (BD Biosciences) in a 24-well plate (36).
Cell-cell aggregation assay
Constant stirring method was performed to evaluate
the cell-cell aggregation (37).
Briefly, cells (2.5×105) were seeded into 24-well
plates. Plates were incubated at 37°C with constant stirring (150
rpm) for 2 h. Cells were then fixed with glutaraldehyde at time
zero and at the end of the incubation. Aggregates were photographed
under a microscope, and isolated cells were counted. The cell-cell
aggregation index = 1 - (number of isolated cells at 2 h / number
of isolated cells at 0 h) and was normalized to the data obtained
with the negative control.
Annexin V-phycoerythrin staining
analysis
Cells (1×105) were grown on slides in
triplicate in 24-well plates and cultured for 48 h. Then, cells
were washed with PBS and incubated in 500 μl binding buffer with 1
μl Annexin V-PE (BD Biosciences) for 15 min in the dark at room
temperature. The slides were coverslipped with glycerol and then
examined and imaged on an inverted fluorescence microscope (IX71;
Olympus). Annexin V-positive cell rate was calculated as (Annexin
V-positive cell number / total cell number) × 100%.
Western blot analysis
Western blotting was performed as previously
described (36). Briefly, cells
were lysed with RIPA lysis buffer, and the protein concentrations
in cell lysates were measured using a BCA protein assay Kit
(Beyotime Biotechnology, Jiangsu, China). Nuclear and cytoplasmic
protein extracts were isolated using a cytoplasmic and nuclear
protein extraction kit (Thermo Scientific) according to the
manufacturer’s protocol. Equal amounts of cell lysate protein were
separated by 10% SDS-PAGE and transferred to PVDF membranes
(Millipore, Billerica, MA, USA). The primary antibodies used were
antibodies against MUC1 (GP1.4) (1:2,000; NeoMarkers), c-Myc
(1:1,000), cyclin D1 (1:1,000), β-actin (1:2,000), IκBα (1:2,000)
and Lamin B1 (1:2,000; all from Epitomics, Burlingame, CA, USA),
β-catenin (1:1,000; BD Biosciences), E-cadherin (Proteintech),
caspase-3 (Santa Cruz Biotechnology).
Co-immunoprecipitation analysis
Cell lysates were first precleared with protein G
agarose beads (Promega) for 3 h at 4°C and, subsequently, equal
amounts of sample lysate were incubated with either 1.0 μg of mouse
IgG or anti-MUC1-CT antibody (Ab-5; Neomarker) for 16 h at 4°C,
followed by precipitation with protein G agarose beads.
Immunoprecipitated proteins from cell lysates and total cell
lysates were subjected to immunoblot analysis with
anti-β-catenin.
Luciferase reporter assay
Cells were transfected using Lipofectamine 2000
(Invitrogen) according to the manufacturer’s protocol with 1.0
μg/well of TOPflash and FOPflash plasmids (Upstate Biotechnology,
Inc., Lake Placid, NY, USA) plus 0.05 μg/well of phRL-TK (Promega)
to normalize transfection efficiency (36).
In vivo tumor growth assays and
immunohistochemical staining
BALB/c nude mice (4–6 weeks old) were purchased from
Beijing HFK Bioscience Co., Ltd., China. Animals were maintained in
specific pathogen-free conditions and environment under controlled
conditions of light and humidity. Animal experiments were carried
out in accordance with the National Institutes of Health Guide for
the Care and Use of Laboratory Animals. Mice were randomly divided
into 4 groups (5 animals/group), designated the SMMC-7721 group,
the NC group, the MR1-C6 group and the MR1-D4 group. Cells
(2×106) were subcutaneously injected into the right
flank of each mouse. On day 35 post-injection, the tumors were
dissected, fixed in 10% neutral-buffered formalin and embedded in
paraffin for immunohistochemical staining using primary antibody
mouse anti-MUC1 monoclonal antibody (GP1.4) and an UltraSensitiveTM
SP (Mouse/Rabbit) IHC Kit (MaiXin.BIO., Fuzhou, China).
Global gene expression analysis by
microarray
NC, MR1-C6 and MR1-D4 cells were harvested, and
total RNA was isolated using TRIzol (Invitrogen Life Technologies)
as described above. Global gene expression analysis was performed
using Roche NimbleGen microarrays (Kang Chen Bio-tech, Shanghai,
China).
Statistical analysis
All statistical analyses were performed using
unpaired Student’s t-tests, and P<0.05 was considered to
indicate a statistically significant result.
Results
Establishment of stable MUC1 knockdown in
SMMC-7721 cells
To determine the effect of MUC1 expression in HCC,
we silenced MUC1 in SMMC-7721 cells using RNAi. SMMC-7721 cells
were transfected with MUC1-targeted or nonspecific siRNA in the
expression vector pGCsilencer™U6.Neo.GFP. We identified three
independent SMMC-7721-MUC1-knockdown stable cell clones (termed
MR1-C6, MR1-D4 and MR1-D9) and a negative control clone termed NC
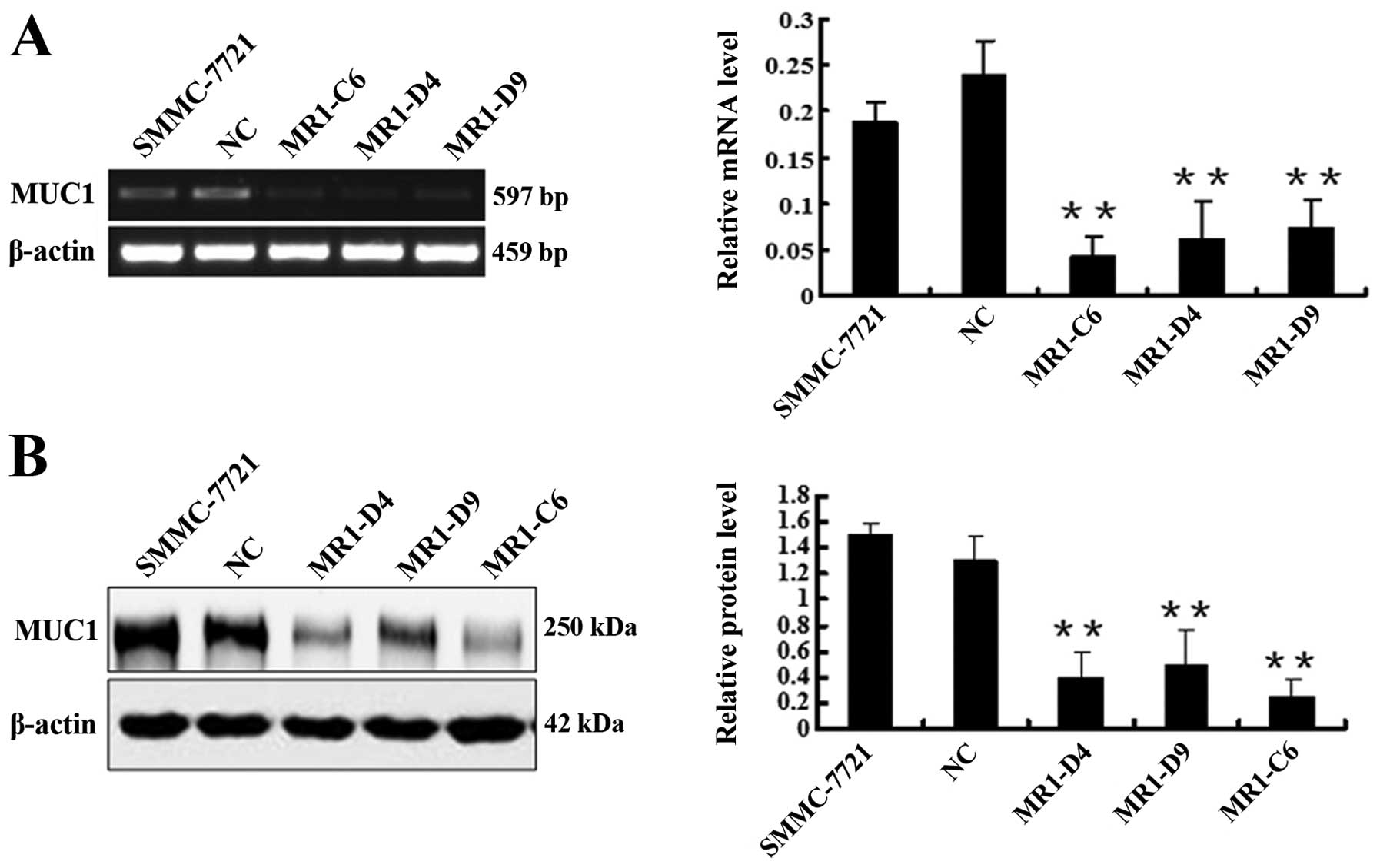
by RT-PCR (Fig. 1A) and western
blotting (Fig. 1B). These results
showed that MUC1 expression in the MR1-C6, MR1-D4 and MR1-D9 cell
clones was significantly decreased when compared to NC or SMMC-7721
cells (P<0.01). The silencing efficiency in clones MR1-C6 and
MR1-D4 reached 82.34 and 74.53%, respectively. Therefore, we
selected the MR1-C6 and MR1-D4 clones for subsequent studies.
Knockdown of MUC1 expression suppresses
SMMC-7721 cell proliferation and arrests cells in S-phase
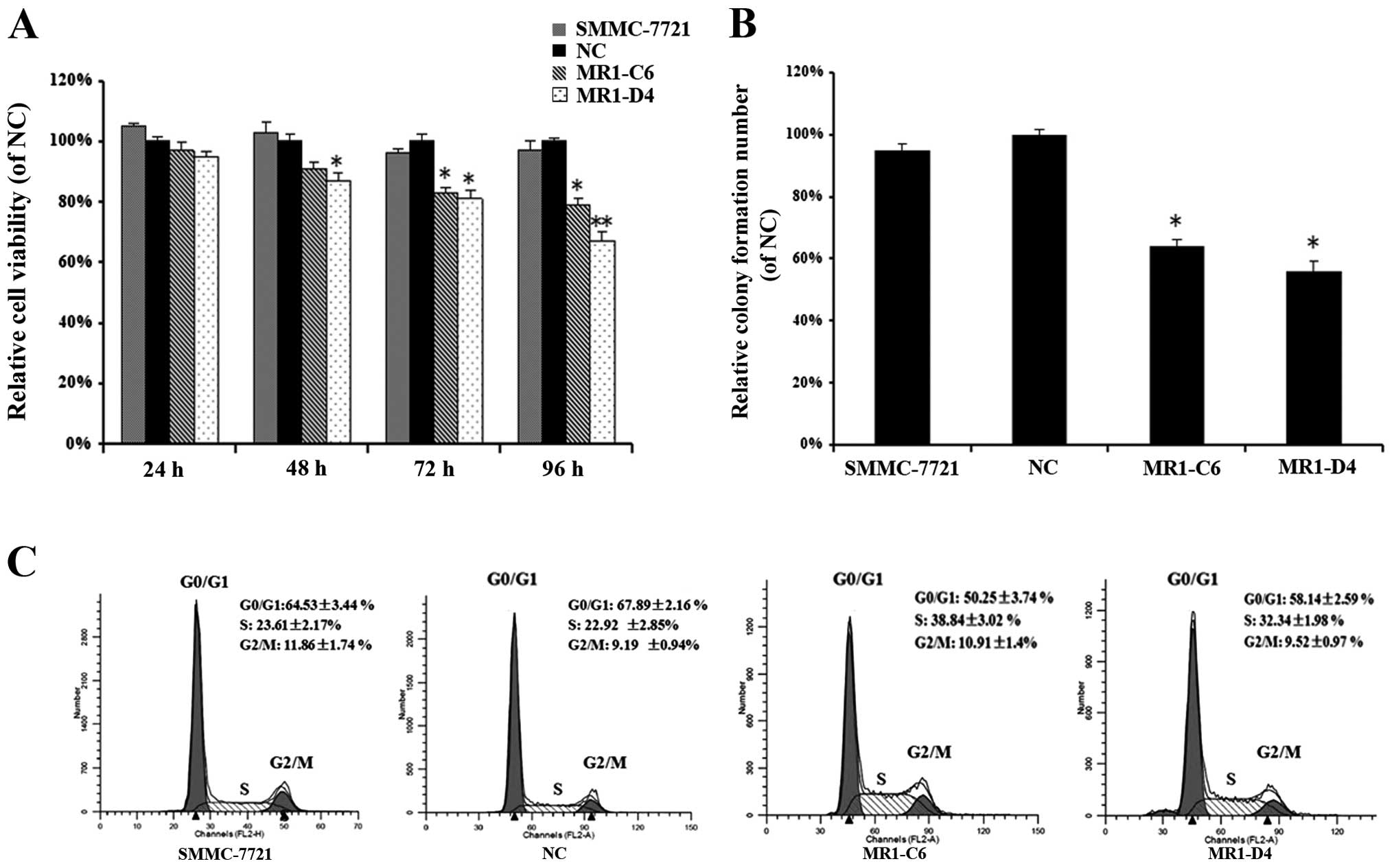
To investigate the influence of MUC1 knockdown on
SMMC-7721 cell proliferation, we performed WST-1 cell viability and
colony formation assays. WST-1 assays showed that the viability of
MR1-C6 and MR1-D4 clones was significantly reduced in a
time-dependent manner when compared to NC or SMMC-7721 cells
(Fig. 2A) (P<0.05). Colony
numbers were decreased in MR1-C6 and MR1-D4 cells when compared to
NC or SMMC-7721 cells (Fig. 2B)
(P<0.05). We also analyzed the cell cycle in MR1-C6, MR1-D4,
SMMC-7721 and NC cells using flow cytometry. These results showed
that MR1-C6 and MR1-D4 cells had a higher percentage of cells in
the S-phase and fewer in the G0/G1 phase when compared to NC or
SMMC-7721 cells (Fig. 2C),
demonstrating that knockdown of MUC1 in SMMC-7721 cells inhibits
cell proliferation and induces cell cycle arrest in the
S-phase.
Knockdown of MUC1 expression alters the
β-catenin signaling pathway by blocking β-catenin translocation to
the nucleus
Numerous reports have shown that MUC1 is involved in
the β-catenin signaling pathway by binding to β-catenin. Therefore,
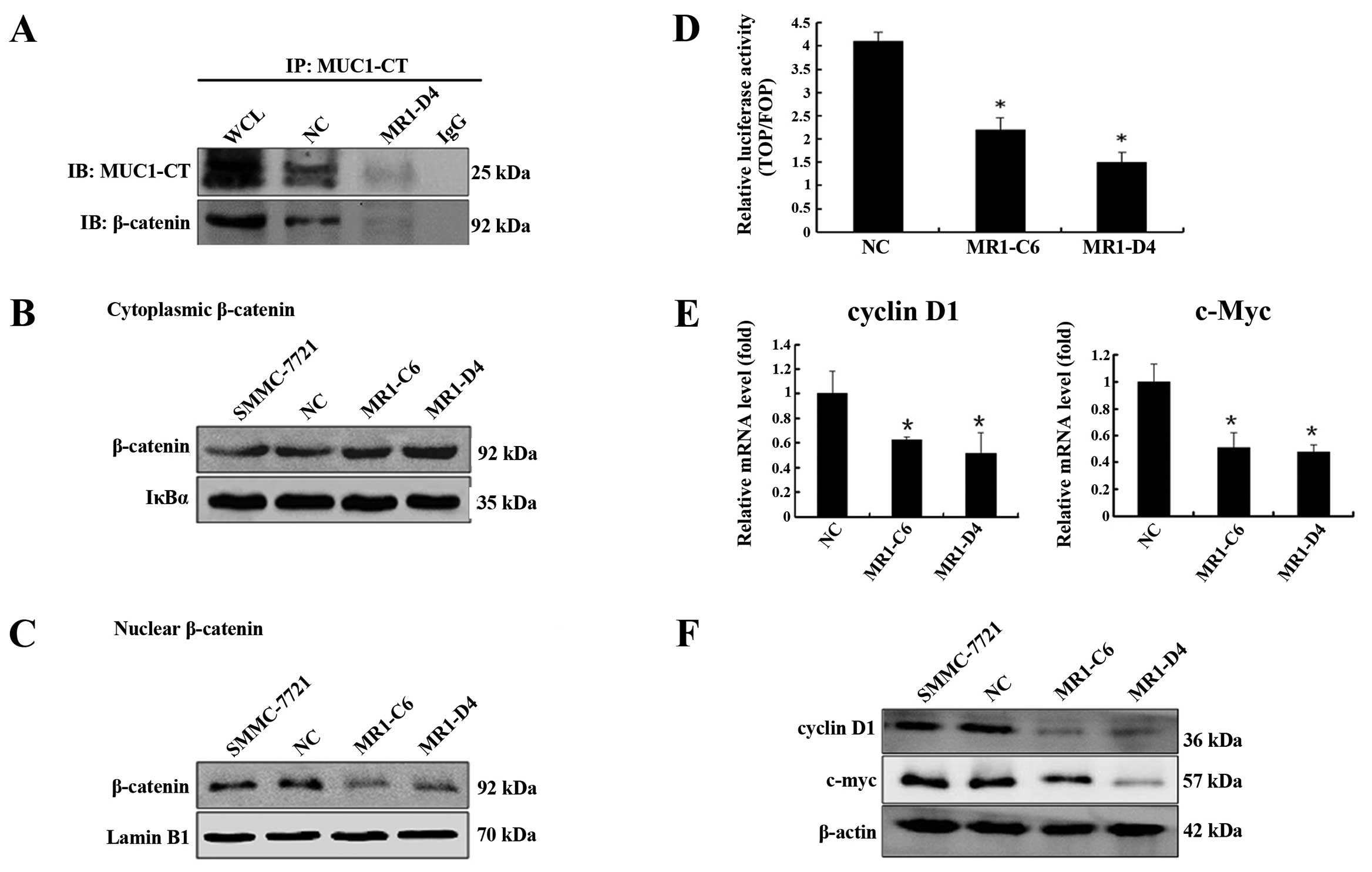
we investigated the interaction between MUC1 and β-catenin in
SMMC-7721 cells by co-immunoprecipitation. As shown in Fig. 3A, β-catenin was detected and was
directly bound to MUC1-CT in SMMC-7721 cells. Furthermore,
knockdown of MUC1 expression decreased the efficiency of the
interaction between MUC1-CT and β-catenin. To evaluate the effect
of MUC1 gene silencing on β-catenin subcellular localization, we
used western blotting and showed that cytoplasmic β-catenin levels
were significantly increased (Fig.
3B) (P<0.05), while nuclear β-catenin levels were
significantly decreased (Fig. 3C)
(P<0.05) in MR1-C6 and MR1-D4 cells when compared to NC or
SMMC-7721 cells. These data indicate that knockdown of MUC1 blocks
the translocation of β-catenin from the cytoplasm to the
nucleus.
As the Wnt pathway is known to be involved in tumor
cell proliferation, we performed a luciferase reporter assay to
determine the effect of the MUC1-CT/β-catenin interaction on the
activation of Wnt signaling in SMMC-7721 cells. The results showed
that TOPflash/FOPflash reporter activity in MR1-C6 and MR1-D4 cells
was significantly lower than the activity in NC cells (Fig. 3D) (P<0.05). We used qRT-PCR and
western blotting to evaluate the expression of cyclin D1 and c-Myc,
which have been confirmed as target genes of the β-catenin
signaling pathway that stimulate cell proliferation. As shown in
Fig. 3E and F, knockdown of MUC1
significantly reduced both the mRNA and protein levels of cyclin D1
and c-Myc (P<0.05). The above data suggest that MUC1 gene
silencing inhibits SMMC-7721 proliferation by ablating the
β-catenin signaling.
Knockdown of MUC1 expression enhances
cell-cell aggregation
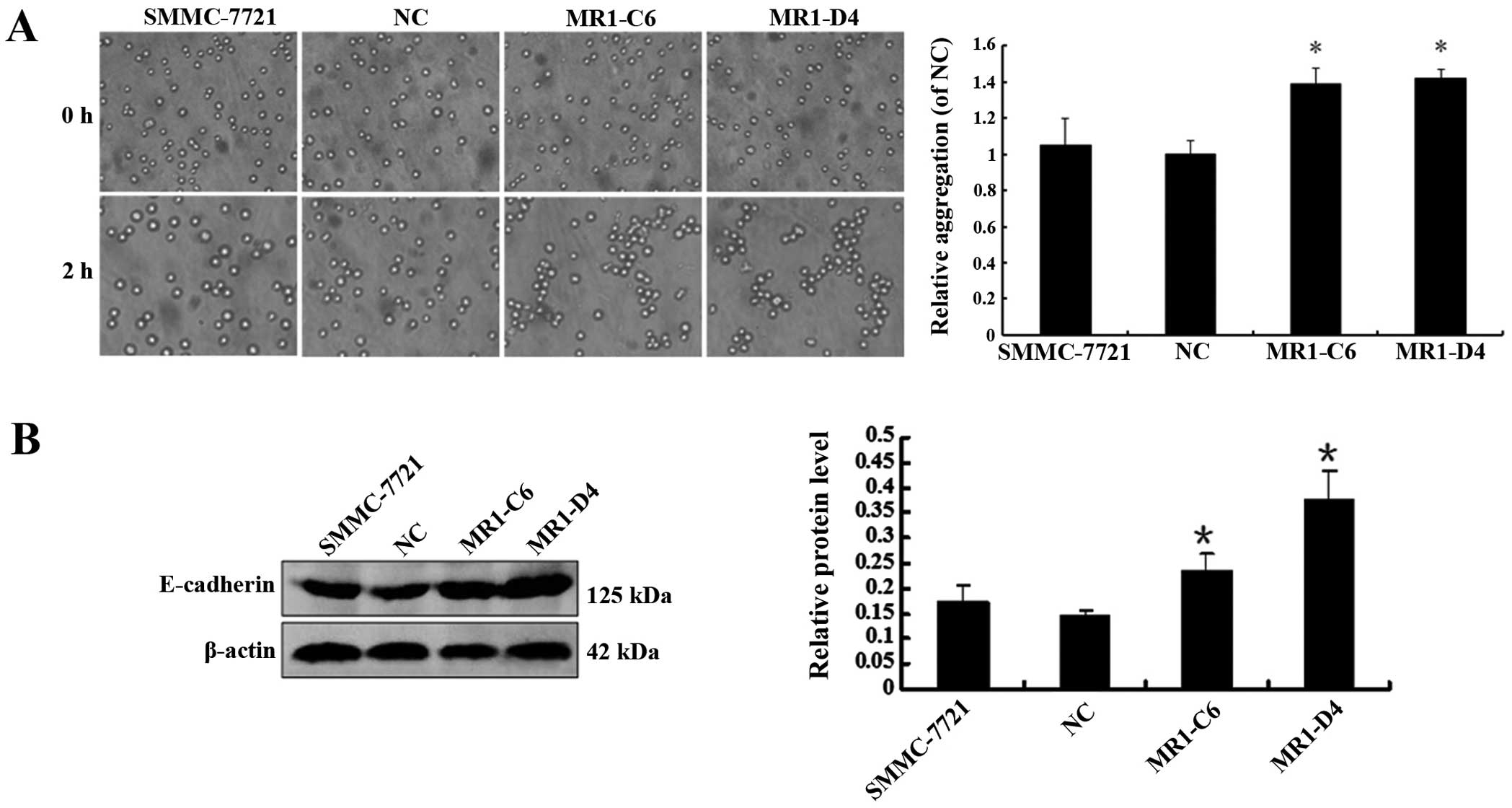
We examined the effect of MUC1 knockdown on the
cell-cell aggregation, migration and invasion of SMMC-7721 cells.
We developed a cell aggregation assay and examined the expression
of E-cadherin, a molecular chaperone of β-catenin that plays an
important role in cell aggregation. The results showed that the
level of cell-cell aggregation and E-cadherin expression in the
MR1-C6 and MR1-D4 cells were significantly enhanced when compared
to NC or SMMC-7721 cells (Fig. 4A and
B) (P<0.05), indicating that knockdown of MUC1 may enhance
cell-cell aggregation by promoting E-cadherin expression. In
addition, we performed scratch test migration and Matrigel invasion
assays and found no significant differences in migration or
invasion between MR1-C6 or MR1-D4 cells and NC or SMMC-7721 cells
(data not shown).
Knockdown of MUC1 expression induces
apoptosis in SMMC-7721 cells
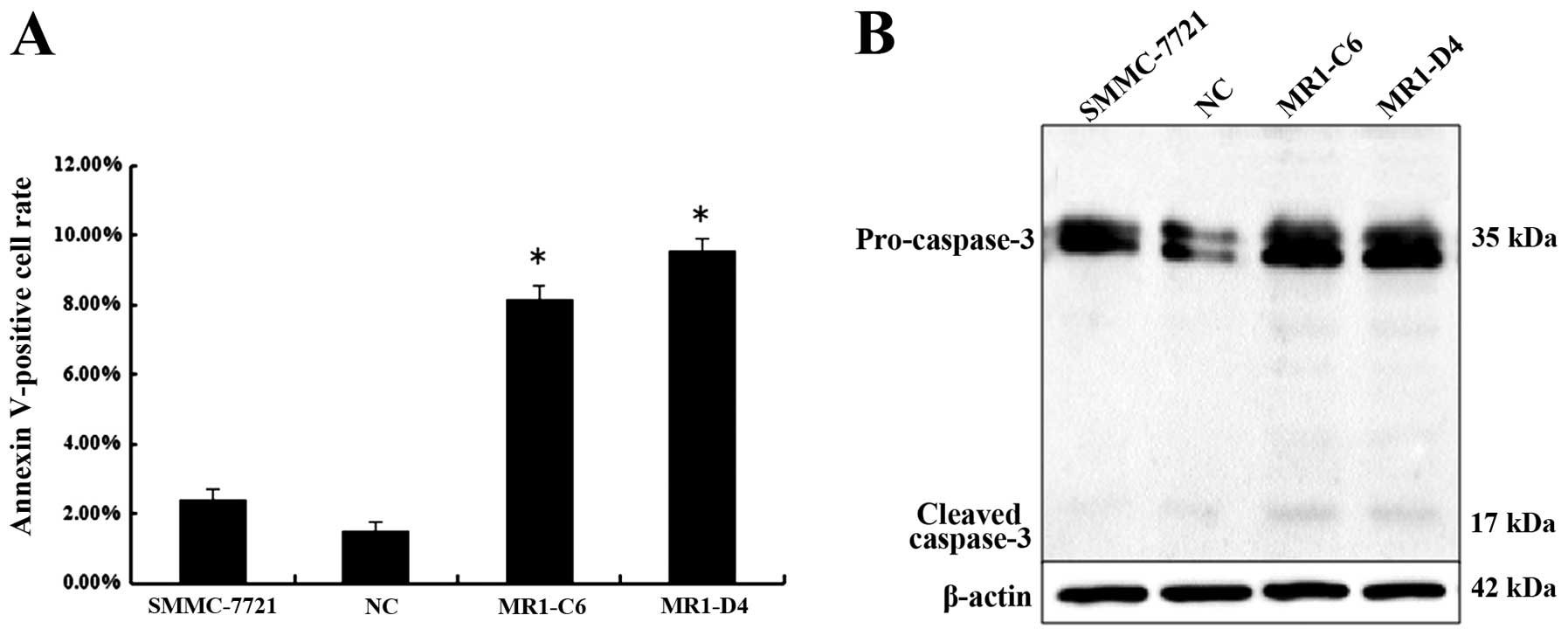
Next, we investigated the effect of MUC1 knockdown
on cell apoptosis by Annexin V-phycoerythrin staining analysis. The
results showed that more MR1-C6 and MR1-D4 cells were stained when
compared with NC or SMMC-7721 cells. The percentage of Annexin
V-positive cell population is plotted in Fig. 5A. Western blotting showed that
caspase-3 cleavage was observed in MR1-C6 and MR1-D4 cells, while
cleavage was not present in NC or SMMC-7721 cells (Fig. 5B). Taken together, these results
indicate that knockdown of MUC1 induces apoptosis in SMMC-7721
cells.
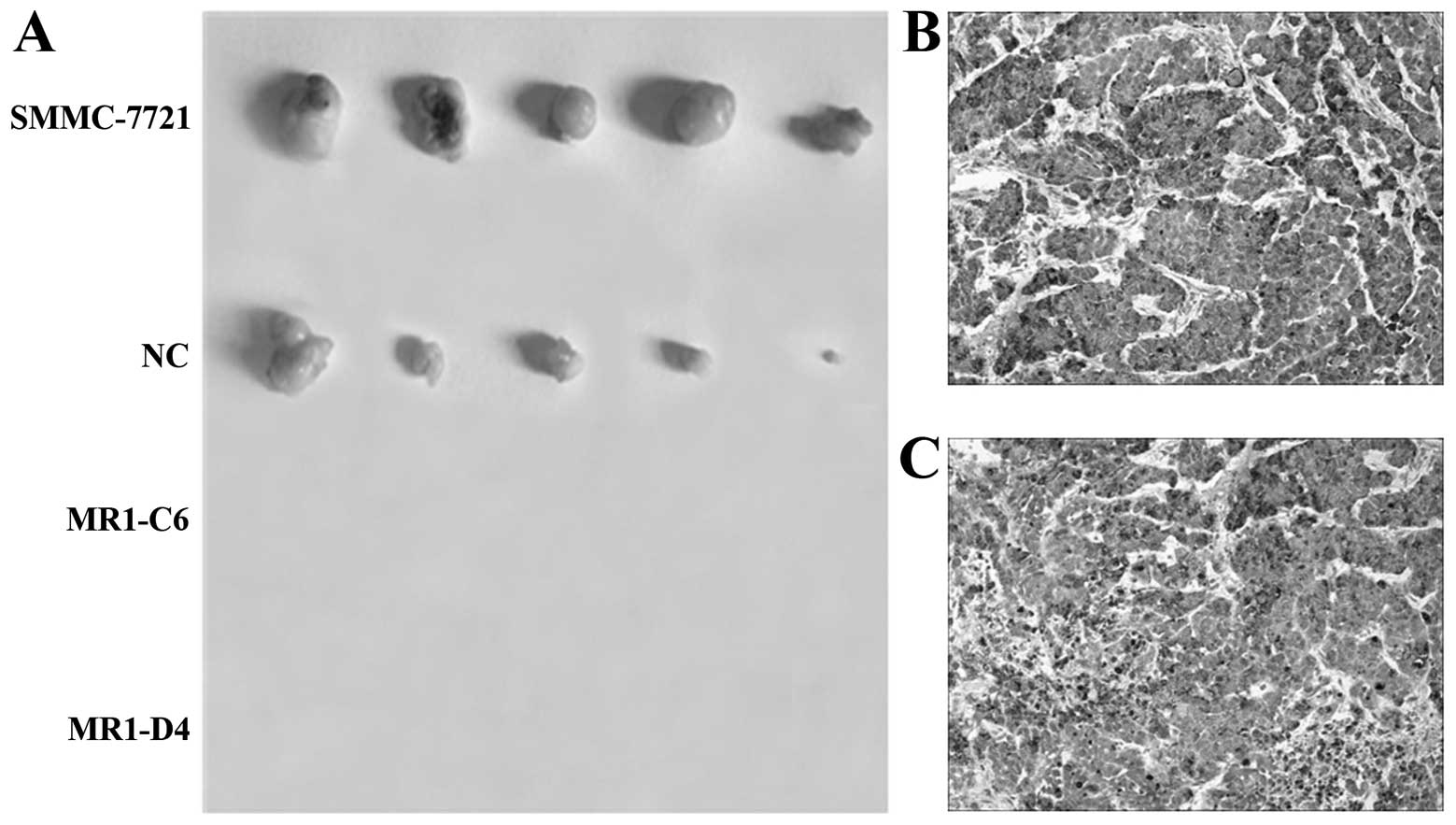
Knockdown of MUC1 expression suppresses
tumor growth in vivo
To evaluate the effects of MUC1 expression on
tumorigenesis in vivo, SMMC-7721, NC, MR1-C6 and MR1-D4
cells were inoculated subcutaneously into BALB/c nude mice to
establish a subcutaneous transplant tumor model. Thirty-five days
after tumor implantation, mice were sacrificed, and the tumors were
dissected. As shown in Fig. 6A,
tumor growth was observed in both the NC and SMMC-7721 groups but
not in the MR1-C6 and MR1-D4 groups. Immunohistochemical staining
showed that MUC1 was significantly expressed by tumors from mice
inoculated with SMMC-7721 and NC cells (Fig. 6B and C). These results indicate that
knockdown of MUC1 significantly suppresses SMMC-7721 tumor growth
in vivo.
Global gene expression analysis in
SMMC-7721 cells
To further explore the mechanisms of MUC1 gene
silencing that lead to inhibited cell proliferation and enhanced
aggregation of SMMC-7721 cells, we utilized gene expression
microarrays to analyze differential gene expression in clones NC,
MR1-C6 and MR1-D4. As shown in Table
I, some genes related to proliferation and aggregation were
differentially expressed between clones MR1-C6 or MR1-D4 and NC. A
series of genes encoding molecules that related to proliferation in
the Wnt/β-catenin, NF-κB, MAPK, insulin, TGF-β and VEGF signaling
pathways were expressed at significantly lower levels in the
MUC1-knockdown clones MR1-C6 and MR1-D4. However,
aggregation-related genes such as ICAM and FAT were expressed at
significantly higher levels. These results indicate that knockdown
of MUC1 influenced a number of signaling pathways related to cell
proliferation and cell-cell aggregation in SMMC-7721 cells,
affecting the occurrence and development of HCC.
 | Table IGene expression microarray results by
comparison between MR1-C6 or MR1-D4 and NC clone, by order of
magnitude. |
Table I
Gene expression microarray results by
comparison between MR1-C6 or MR1-D4 and NC clone, by order of
magnitude.
| Genes downregulated
>2-fold in MUC1-knockdown clones |
|---|
| Wnt/β-catenin
signal |
| Cyclin D1 |
| C-myc |
| Transcription
factor 3 (TCF3) |
| TEA domain family
member1 (TEF1/TCF13) |
| Transcription
factor 7-like 1 (TCF7L1) |
| NF-κB signal |
| Nuclear factor of
κ light polypeptide gene enhancer in B-cells 1 (p105) (NF-κB1) |
| Nuclear factor of
κ light polypeptide gene enhancer in B-cells 2 (p49/p100)
(NF-κB2) |
| Inhibitor of κ
light polypeptide gene enhancer in B-cells, kinase beta
(IκBKB) |
|
Dystrophin-associated glycoprotein
(DAG1) |
| Insulin signal |
| Insulin-like
growth factor 2 receptor (IGF2R) |
| Insulin receptor
substrate 2 (IRS2) |
| Insulin-like
growth factor binding protein 7 (IGFBP7) |
| Eukaryotic
translation initiation factor 4E binding protein 1 (4EBP1) |
| Inositol
polyphosphate phosphatase-like 1 (SHIP2) |
| Ras homolog
enriched in brain (Rheb) |
| TGF-β signal |
| SMAD, mothers
against DPP homolog 2 (Smad2) |
| SMAD, mothers
against DPP homolog 3 (Smad3) |
| Cell division
cycle 42 (GTP binding protein, 25 kDa) (CDC42) |
| Ras homolog gene
family, member A (RhoA) |
| Basic
helix-loop-helix domain containing, class B, 2 (bHLHB2) |
| MAPK signal |
| Mitogen-activated
protein kinase kinase 5 (MAP2K5) |
| Mitogen-activated
protein kinase kinase 6 (MAP2K6) |
| Growth factor
receptor-bound protein 10 (GRB10) |
| RAB31, member RAS
oncogene family (RAB31) |
| VEGF signal |
| Vascular
endothelial growth factor B (VEGFB) |
| Cell adhesion and
aggregation associated |
| Laminin, γ 1
(formerly LAMB2) (LAMC1) |
|
| Genes upregulated
>2-fold in MUC1-knockdown clones |
|
| Cell proliferation
associated |
| Glycoprotein
hormones, α polypeptide (CGA) |
| Growth arrest and
DNA-damage-inducible, |
| γ interacting
protein 1 (GADD45GIP1) |
| Growth
differentiation factor 2 (GDF2 ) |
| DnaJ (Hsp40)
homolog, subfamily B, member 2 (DNAJB2) |
| SRY (sex
determining region Y)-box 17 (SOX17) |
| Netrin 1
(NTN1) |
| Metallothionein 1E
(MT1E) |
| Nuclear factor of
κ light polypeptide gene enhancer in B-cells inhibitor, β
(NFκBIB) |
| Activin A receptor
type II-like 1 (ACVRL1) |
| ADAM
metallopeptidase with thrombospondin type I motif, 15
(ADAMTS15) |
| Cell adhesion and
aggregation associated |
| Intercellular
adhesion molecule 5 (ICAM5) |
| Tumor necrosis
factor receptor superfamily, member 12A (CD266) (FN14) |
| CD36 molecule
(thrombospondin receptor) (CD36) (FAT) |
Discussion
In the present study, we investigated the effects of
MUC1 knockdown on the HCC cell line SMMC-7721.
SMMC-7721-MUC1-knockdown stable clones were generated by RNA
interference. We identified three independent MUC1-knockdown
clones, MR1-C6, MR1-D4 and MR1-D9, and one negative control clone,
NC, via immunofluorescence staining, PCR and western blotting. We
selected the clones MR1-C6 and MR1-D4 due to their higher silencing
efficiency and the NC clone to study in vitro cell
proliferation, colony formation, cell cycle, migration, invasion,
aggregation and cell apoptosis as well as to evaluate in
vivo tumorigenesis.
We found that the MUC1-silenced clones MR1-C6 and
MR1-D4 had significantly inhibited cell proliferation, similar to
results described by Tsutsumida et al (38), who showed that knockdown of MUC1 in
pancreatic carcinoma cells (S2-013) significantly decreased cell
proliferation. Raina et al (39) showed that 3Y1 cells overexpressing
MUC1 could induce malignant transformation and enhance colony
formation. Thus, we also utilized the colony formation assay to
detect the effect of MUC1 gene silencing in MR1-C6 and MR1-D4
cells, showing a significant reduction in colony formation, further
confirming that knockdown of MUC1 negatively regulated HCC cell
proliferation. Subsequently, we performed cell cycle analysis and
found that knockdown of MUC1 induced cell cycle arrest in the
S-phase. In addition, we observed that knockdown of MUC1 induced
apoptosis in SMMC-7721 cells, which is consistent with previous
studies (40–42). Moreover, tumor growth was not
observed in mice injected with MR1-C6 and MR1-D4 cells. These
results suggest that MUC1 plays an important role in HCC
tumorigenesis.
Several published studies (43–45)
have shown that MUC1-CT interacts with β-catenin to form a complex
and regulate the cellular localization of β-catenin. Nuclear
translocation of β-catenin can activate cyclin D1 and c-Myc
expression and stimulate cell proliferation, thus contributing to
tumorigenesis and tumor progression. Our current study showed that
MUC1-CT could interact with β-catenin and that knockdown of MUC1 in
SMMC-7721 cells increased cytoplasmic levels of β-catenin but
decreased the nuclear translocation of β-catenin, reduced the
activity of TCF, downregulated expression of cyclin D1 and c-Myc,
and arrested the cell cycle in S-phase. These results provide a
potential mechanistic explanation of how MUC1 knockdown decreased
the proliferation of SMMC-7721 cells in vitro.
To determine the effect of MUC1 on SMMC-7721 cell
migration, invasion and aggregation, we conducted the scratch test,
Matrigel invasion and aggregation assays, respectively. The results
showed that knockdown of MUC1 promoted cell-cell aggregation when
compared to controls. Cell-cell aggregation is mainly mediated by
the cadherin complex, which can maintain epidermal morphogenesis of
epidermal cells and is involved in cell-cell interactions. Yuan
et al (46) showed that
MUC1-silenced cells could increase the cell-cell aggregation of
breast cancer cells and lead to higher expression of E-cadherin.
Our result showed that E-cadherin expression was significantly
increased in MR1-C6 and MR1-D4 cells, indicating that knockdown of
MUC1 may enhance cell-cell aggregation by promoting E-cadherin
expression. In addition, we found no significant differences in
cell migration or invasion activity between MUC1-silenced clones
and controls, which is consistent with Costa et al (37). The reasons why MUC1 gene silencing
did not affect HCC cell migration and invasion require further
study.
To explore the mechanisms of MUC1 gene silencing
that lead to inhibited cell proliferation and enhanced aggregation
of SMMC-7721 cells, we further utilized microarrays to measure
global gene expression and analyze differential gene expression
between MUC1-silenced MR1-C6 or MR1-D4 cells and the NC cells. We
showed that a series of genes encoding molecules in the
Wnt/β-catenin, NF-κB, MAPK, insulin, VEGF and TGF-β signaling
pathways were influenced by knockdown of MUC1, and these may
contribute to the phenotypic alterations observed. However, the
mechanisms underlying how MUC1 regulates TGF-β signaling pathway
remain unclear and related experiments are being carried out.
In conclusion, our study shows for the first time
that MUC1 expression influences the proliferation, apoptosis and
cell-cell aggregation of HCC cell line SMMC-7721. These results
indicate that MUC1 plays an important role in HCC tumorigenesis,
and further suggest that MUC1 may be a potential target for HCC
treatment.
Acknowledgements
This study was supported by grants from the China
National Natural Science Foundation (No. 31170875) and the Major
Development Programs for New Drugs of the Chinese Academy of
Sciences during the 12th Five-Year Plan Period (No. 2011ZX09102-
001-36).
References
|
1
|
Kufe DW: Mucins in cancer: function,
prognosis and therapy. Nat Rev Cancer. 9:874–885. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Duffy MJ: CA 15-3 and related mucins as
circulating markers in breast cancer. Ann Clin Biochem. 36(Pt 5):
579–586. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Apostolopoulos V, Pietersz GA, Tsibanis A,
et al: Pilot phase III immunotherapy study in early-stage breast
cancer patients using oxidized mannan-MUC1 [ISRCTN71711835]. Breast
Cancer Res. 8:R272006.
|
|
4
|
Sugiura D, Aida S, Denda-Nagai K, et al:
Differential effector mechanisms induced by vaccination with MUC1
DNA in the rejection of colon carcinoma growth at orthotopic sites
and metastases. Cancer Sci. 99:2477–2484. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang S, Zhang HS, Cordon-Cardo C,
Ragupathi G and Livingston PO: Selection of tumor antigens as
targets for immune attack using immunohistochemistry: protein
antigens. Clin Cancer Res. 4:2669–2676. 1998.PubMed/NCBI
|
|
6
|
Gendler S, Taylor-Papadimitriou J, Duhig
T, Rothbard J and Burchell J: A highly immunogenic region of a
human polymorphic epithelial mucin expressed by carcinomas is made
up of tandem repeats. J Biol Chem. 263:12820–12823. 1988.PubMed/NCBI
|
|
7
|
Ligtenberg MJ, Kruijshaar L, Buijs F, van
Meijer M, Litvinov SV and Hilkens J: Cell-associated episialin is a
complex containing two proteins derived from a common precursor. J
Biol Chem. 267:6171–6177. 1992.PubMed/NCBI
|
|
8
|
Kufe DW: Targeting the human MUC1
oncoprotein: a tale of two proteins. Cancer Biol Ther. 7:81–84.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh PK and Hollingsworth MA: Cell
surface-associated mucins in signal transduction. Trends Cell Biol.
16:467–476. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Carraway KL III, Funes M, Workman HC and
Sweeney C: Contribution of membrane mucins to tumor progression
through modulation of cellular growth signaling pathways. Curr Top
Dev Biol. 78:1–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu LG, Andrews N, Zhao Q, et al:
Galectin-3 interaction with Thomsen-Friedenreich disaccharide on
cancer-associated MUC1 causes increased cancer cell endothelial
adhesion. J Biol Chem. 282:773–781. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rahn JJ, Shen Q, Mah BK and Hugh JC: MUC1
initiates a calcium signal after ligation by intercellular adhesion
molecule-1. J Biol Chem. 279:29386–29390. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rahn JJ, Chow JW, Horne GJ, et al: MUC1
mediates transendothelial migration in vitro by ligating
endothelial cell ICAM-1. Clin Exp Metastasis. 22:475–483. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shen Q, Rahn JJ, Zhang J, et al: MUC1
initiates Src-CrkL- Rac1/Cdc42-mediated actin cytoskeletal
protrusive motility after ligating intercellular adhesion
molecule-1. Mol Cancer Res. 6:555–567. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao Q, Barclay M, Hilkens J, et al:
Interaction between circulating galectin-3 and cancer-associated
MUC1 enhances tumour cell homotypic aggregation and prevents
anoikis. Mol Cancer. 9:1542010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang L, Chen D, Liu D, Yin L, Kharbanda S
and Kufe D: MUC1 oncoprotein blocks glycogen synthase kinase
3beta-mediated phosphorylation and degradation of beta-catenin.
Cancer Res. 65:10413–10422. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Kuwahara H, Ren J, Wen G and Kufe D:
The c-Src tyrosine kinase regulates signaling of the human DF3/MUC1
carcinoma-associated antigen with GSK3 beta and beta-catenin. J
Biol Chem. 276:6061–6064. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pandey P, Kharbanda S and Kufe D:
Association of the DF3/MUC1 breast cancer antigen with Grb2 and the
Sos/Ras exchange protein. Cancer Res. 55:4000–4003. 1995.PubMed/NCBI
|
|
19
|
Wei X, Xu H and Kufe D: Human mucin 1
oncoprotein represses transcription of the p53 tumor suppressor
gene. Cancer Res. 67:1853–1858. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Singh PK, Behrens ME, Eggers JP, et al:
Phosphorylation of MUC1 by Met modulates interaction with p53 and
MMP1 expression. J Biol Chem. 283:26985–26995. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schroeder JA, Thompson MC, Gardner MM and
Gendler SJ: Transgenic MUC1 interacts with epidermal growth factor
receptor and correlates with mitogen-activated protein kinase
activation in the mouse mammary gland. J Biol Chem.
276:13057–13064. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lau SK, Shields DJ, Murphy EA, et al:
EGFR-mediated carcinoma cell metastasis mediated by integrin
alphavbeta5 depends on activation of c-Src and cleavage of MUC1.
PLoS One. 7:e367532012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ahmad R, Raina D, Trivedi V, et al: MUC1
oncoprotein activates the IkappaB kinase beta complex and
constitutive NF-kappaB signalling. Nat Cell Biol. 9:1419–1427.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ahmad R, Raina D, Joshi MD, et al: MUC1-C
oncoprotein functions as a direct activator of the nuclear
factor-kappaB p65 transcription factor. Cancer Res. 69:7013–7021.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu XF, Yang E, Li J and Xing PX: MUC1
cytoplasmic tail: a potential therapeutic target for ovarian
carcinoma. Expert Rev Anticancer Ther. 6:1261–1271. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kufe DW: Functional targeting of the MUC1
oncogene in human cancers. Cancer Biol Ther. 8:1197–1203. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Klinge CM, Radde BN, Imbert-Fernandez Y,
et al: Targeting the intracellular MUC1 C-terminal domain inhibits
proliferation and estrogen receptor transcriptional activity in
lung adenocarcinoma cells. Mol Cancer Ther. 10:2062–2071. 2011.
View Article : Google Scholar
|
|
28
|
Hattrup CL, Bradley JM, Kotlarczyk KL, et
al: The MUC1 cytoplasmic tail and tandem repeat domains contribute
to mammary oncogenesis in FVB mice. Breast Cancer (Auckl). 1:57–63.
2008.PubMed/NCBI
|
|
29
|
Wang L, Ma J, Liu F, et al: Expression of
MUC1 in primary and metastatic human epithelial ovarian cancer and
its therapeutic significance. Gynecol Oncol. 105:695–702. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kohlgraf KG, Gawron AJ, Higashi M, et al:
Contribution of the MUC1 tandem repeat and cytoplasmic tail to
invasive and metastatic properties of a pancreatic cancer cell
line. Cancer Res. 63:5011–5020. 2003.PubMed/NCBI
|
|
31
|
Inagaki Y, Tang W, Xu H, et al: Sustained
aberrant localization of KL-6 mucin and beta-catenin at the
invasion front of human gastric cancer cells. Anticancer Res.
31:535–542. 2011.PubMed/NCBI
|
|
32
|
Bozkaya G, Korhan P, Cokakli M, et al:
Cooperative interaction of MUC1 with the HGF/c-Met pathway during
hepatocarcinogenesis. Mol Cancer. 11:642012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yuan SF, Li KZ, Wang L, et al: Expression
of MUC1 and its significance in hepatocellular and
cholangiocarcinoma tissue. World J Gastroenterol. 11:4661–4666.
2005.PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
|
|
35
|
Veeravalli KK, Chetty C, Ponnala S, et al:
MMP-9, uPAR and cathepsin B silencing downregulate integrins in
human glioma xenograft cells in vitro and in vivo in nude mice.
PLoS One. 5:e115832010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang F, Li Q, Ni W, et al: Expression of
human full-length MUC1 inhibits the proliferation and migration of
a B16 mouse melanoma cell line. Oncol Rep. 30:260–268.
2013.PubMed/NCBI
|
|
37
|
Costa NR, Paulo P, Caffrey T,
Hollingsworth MA and Santos-Silva F: Impact of MUC1 mucin
downregulation in the phenotypic characteristics of MKN45 gastric
carcinoma cell line. PLoS One. 6:e269702011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tsutsumida H, Swanson BJ, Singh PK, et al:
RNA interference suppression of MUC1 reduces the growth rate and
metastatic phenotype of human pancreatic cancer cells. Clin Cancer
Res. 12:2976–2987. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Raina D, Kharbanda S and Kufe D: The MUC1
oncoprotein activates the anti-apoptotic phosphoinositide
3-kinase/Akt and Bcl-xL pathways in rat 3Y1 fibroblasts. J Biol
Chem. 279:20607–20612. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Raina D, Ahmad R, Chen D, Kumar S,
Kharbanda S and Kufe D: MUC1 oncoprotein suppresses activation of
the ARF-MDM2-p53 pathway. Cancer Biol Ther. 7:1959–1967. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Agata N, Ahmad R, Kawano T, Raina D,
Kharbanda S and Kufe D: MUC1 oncoprotein blocks death
receptor-mediated apoptosis by inhibiting recruitment of caspase-8.
Cancer Res. 68:6136–6144. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ahmad R, Alam M, Rajabi H and Kufe D: The
MUC1-C oncoprotein binds to the BH3 domain of the pro-apoptotic BAX
protein and blocks BAX function. J Biol Chem. 287:20866–20875.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li Y, Liu D, Chen D, Kharbanda S and Kufe
D: Human DF3/MUC1 carcinoma-associated protein functions as an
oncogene. Oncogene. 22:6107–6110. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wen Y, Caffrey TC, Wheelock MJ, Johnson KR
and Hollingsworth MA: Nuclear association of the cytoplasmic tail
of MUC1 and beta-catenin. J Biol Chem. 278:38029–38039. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li Y, Yi H, Yao Y, et al: The cytoplasmic
domain of MUC1 induces hyperplasia in the mammary gland and
correlates with nuclear accumulation of beta-catenin. PLoS One.
6:e191022011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yuan Z, Wong S, Borrelli A and Chung MA:
Downregulation of MUC1 in cancer cells inhibits cell migration by
promoting E-cadherin/catenin complex formation. Biochem Biophys Res
Commun. 362:740–746. 2007. View Article : Google Scholar : PubMed/NCBI
|