Introduction
Global cancer statistics report that lung cancer has
been the primary cause of cancer-associated mortality in males and
females in the past two decades (1,2). In China,
lung cancer mortality remains high and is likely to continue to
rise (3). Currently, genetic and
genomic methods are important tools, and have been utilized for
investigating non-small cell lung cancer (NSCLC) pathogenesis,
diagnosis and treatment (4–6).
Long non-coding RNAs (lncRNAs) are RNA molecules of
typically >200 nucleotides, but with no protein-coding
capability (7). It has been reported
that some lncRNAs can act as carcinogenic or cancer-suppressive
factors associated with tumour cell invasion and metastasis in
NSCLC, including HOX transcript antisense RNA (8), prostate cancer associated transcript 1
(9), PCGEM1 prostate-specific
transcript (10) and transcribed
ultra-conserved region 338 (11). On
the contrary, various other lncRNAs, such as growth arrest specific
5 (12), tumour protein p53 pathway
corepressor 1 (13), maternally
expressed 3 (14) and phosphatase and
tensin homolog pseudogene 1 (15),
can reduce the metastasis and invasion of tumour cells through
suppressive actions. Therefore, the role of lncRNAs in tumours is
complex and diverse. Additionally, the signalling pathways that
lncRNAs act on are also varied, including the phosphoinositide
3-kinase/protein kinase B and transforming growth factor-β
(TGF-β)/SMAD3 pathways (16,17). Certain types of lncRNA can be used as
biomarkers for clinical diagnosis and prognosis assessment,
including metastasis associated lung adenocarcinoma transcript 1
(18). Competing endogenous RNAs
(ceRNAs) are subset of lncRNAs that have an important role in cell
cycle progression; however, there are only a few reports available
in the literature regarding their role in tumourigenesis (19).
In this study, a lncRNA array was used to screen a
group of lncRNAs associated with lung cancer metastasis. The role
LINC00887 and its associated ceRNA in NSCLC invasion and metastasis
were systematically investigated. Lung cancer cell lines, 95-C and
95-D were cultured and analysed with respect to LINC00887 and its
associated microRNAs (miRNAs/miRs). Subsequently, the
proliferation, apoptosis and migration of lung cancer cells, and
role of LINC00887 were examined.
Materials and methods
Cell culture and RNA extraction
The 95-C cell line is a low metastatic ability human
lung cancer cell, and 95-D is a high metastatic cell line, both of
which were purchased from the Cell Bank of the Chinese Academy of
Sciences (Shanghai, China). RPMI-1640 (Thermo Fisher Scientific,
Inc., Waltham, MA, USA) medium containing 10% foetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% dual antibiotic
(chloramphenicol and streptomycin) was used for cell culture at
37°C and 5% CO2 in a saturated humidity incubator. The
cells were cultured at a 10×109/ml density, and then
harvested for RNA extraction.
Total RNA was extracted from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), following the
manufacturer's instructions. The homogenate samples were incubated
at 15–30°C for 5 min to completely dissociate the nucleoprotein
complex. For every 1 ml TRIzol reagent, 0.2 ml chloroform was
added, and the samples were shaken vigorously for 15 sec, then
incubated at 20°C for 3 min. The samples at were centrifuged at 4°C
and 12,000 × g for 15 min. RNA was in the upper aqueous phase, and
the volume of the aqueous phase was ~60% of the total volume of
TRIzol reagent used. After incubation at 20°C for 10 min, the
samples underwent centrifugation at 4°C and 12,000 × g for 10 min.
The RNA solution obtained was stored at −70°C. Each sample was
quantified, and the purity was determined by the absorbance ratio
at 260 and 280 nm (A260/A280).
LncRNA array analysis
Total RNA from 95-C and 95-D cells were analysed
using the GeneSpring GX v12.0 software (Agilent Technologies, Inc.,
Santa Clara, CA, USA) and lncRNA microarray (Human LncRNA Array
v3.0; Arraystar, Inc., Rockville, MD, USA), and scanned with the
Agilent G2505C scanner (Agilent Technologies, Inc.). Differentially
expressed lncRNAs were selected by fold change filtering (fold
change ≥2.0), and then, for those that were statistically
significant, volcano plot filtering (fold change ≥2.0, P≤0.05).
Subsequently, LINC00887 was selected with the highest statistical
score and validated as the research object of the study. The
datasets generated and/or analysed during the current study are
available in the Gene Expression Omnibus repository (ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124619).
Bioinformatic analysis of LINC00887
and reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) validation
Bioinformatic analysis of LINC00887 was performed
using the Ensembl (asia.ensembl.org) and eBioPortal (cbioportal.org/) databases. PrimeScript®
First Strand cDNA Synthesis Kit to perform RT. TaqMan®
MicroRNA Reverse Transcription Kit was used for microRNA RT, with a
incubation at 30°C for 10 min and 42°C and 30 min. PCR was
conducted using SYBR Select Master Mix (Thermo Fisher Scientific,
Inc.) and a 7500 Fast Real-Time PCR system at 50°C for 2 min and
95°C for 10 min, followed by cycles of 95°C for 15 sec and 60°C for
1 min. LINC00887 primers were as follows: Forward,
5′-TGGCCAGTGTTTCACCTGTT-3′ and reverse, 3′-TGATTTCCTCCAACGTGCCA-5′.
β-actin primers were as follows: Forward,
5′-ATCCAGGCTGTGCTATCCCT-3′ and reverse, 3′-GGGCATACCCCTCGTAGATG-5′
(2−ΔΔCq) (20).
RNA interference and cell
proliferation
The types of LINC00887 small interfering RNA
sequences (siRNA; 5′-GGCCTTTGCAGTTATTAGGAA-3′ and control RNA
sequence 5′-GGCCTTTGCGTCACGCCTTAG-3′; Guangzhou RiboBio Co., Ltd.,
Guangzhou, China) were synthesized and the suppressive effect in
95-D cells was analysed by RT-qPCR. siRNA2 was transfected into
95-D cell lines. Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.)
was added when the cell confluence reached 70–80%. 95-C-pGL3
vector, 95-C-LINC00887, 95-D-control and 95-D-siRNA were
transfected (2 µg plasmid or 100 pmol siRNA per well) using
Lipofectamine® 3000 for 8 h, and then continuously
cultured for 24 h. The cell number in each group was controlled at
~3×104/ml. Each well was topped up with 50 µl MTT
solution and maintained for 4 h. After 4 h, the culture solution in
the 96-well plate was aspirated with a pipette. Dimethyl sulfoxide
(150 µl) was added to each well, and the culture plate was placed
on a microplate reader. The setup procedure involved shaking for 5
min and optical density (OD) was recorded at a wavelength of 490
nm. The tumour cell growth inhibition rate (IR) was calculated
using the formula: Cell growth IR (%)=[1-(OD value of experimental
group-OD value of blank groups)/(OD value of control group-OD value
of blank groups) ×100.
Detection of cell invasion and
migration ability
Cells were cultured and transfected for 48 h, then a
cell suspension (1×106 cells/ml) was produced in 1X
binding buffer. The cell cycle was analysed in flow cytometer with
an Annexin V-fluorescein isothiocyanate (FITC) and Annexin
V-propidium iodide (PI) reagent kit (BestBio Science).
Subsequently, the 95-C cell concentration was reduced to
3×104 cell/ml and a Transwell assay was performed using
Millipore chambers (8-µm diameter). DMEM (500 µl) without FBS was
used to dilute the Matrigel (BD Biosciences; Becton-Dickinson and
Company, Franklin Lakes, NJ, USA; 1:3) and spread on the upper
chambers (10% FBS in DMEM; 50 µl/well). DMEM solution containing
10% FBS (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) was
used to dilute the fibronectin (1:2,000) and 50 µl/well was added
to lower chambers. The above reagents were also applied to the 95-D
cell chambers. Cells in both chambers were cultured for a further
24 h. Finally, the remaining cells were counted under a microscope
after fixing in 75% methanol and staining (3 min, 25°C).
In vitro tumour sphere formation
assays
95-D cells (1×106/ml) were cultured in
6-well dishes. Tumour sphere nutrient solution was prepared using a
formula consisting of 500 ml DMEM/F12, 20 ng/ml epidermal growth
factor, 10 ng/ml basic fibroblast growth factor and 5 µg insulin,
and diluted with B27 supplement at 1:50. Samples (20 µl) of each
group were collected (3×104 cells/ml) and stained with
trypan blue (0.4%, 5 min). Cellular morphology was observed using a
bright field microscope, at a diluted concentration of one cell per
µl.
Construction of the overexpression
plasmid and LINC00887 promoter reporter
The LINC00887 gene was inserted into pcDNA3.1+
vector and LINC00887 enhancer into the pGL3 basic vector. Takara
reverse transcription kit (Takara Biotechnology Co., Ltd., Dalian,
China) was used to prepare the LINC DNA from lung adenocarcinoma
tissue specimens. The LINC00887 gene was amplified by PCR, 2X Taq
MasterMix (Tiangen Biotech Co., Ltd., Beijing, China; Tm at
66–65°C). The restriction endonuclease was used to hydrolyze the
product and vector of PCR [pcDNA3.1 (+)], and 95-D cell genome was
extracted using the QIAamp DNA Mini Kit (Qiagen, Inc., Valencia,
CA, USA). The LINC00887 gene does not have specific promoter, but
an enhancer was found in the Ensembl database (http://useast.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000214145;r=3:194296465-194312803).
Online analysis in the Ensembl database indicated that the enhancer
sequence contains a TPA-responsive element binding sequence
[TGA(C/G) TCA] and SMAD binding element binding motifs (GTCT/AGAC).
The enhancer sequence was amplified by Q5 PCR polymerase chain
reaction. The recombinants and PRL-TK plasmids were transformed
into DH5α competent cells. Plasmid Mini Kit (Omega Bio-Tek, Inc.,
Norcross, GA, USA) was used to extract plasmids and they were
identified by electrophoresis and sequencing.
LINC00887 intracellular location and
TGF-β signal pathway regulation
95-C and 95-D cells (1×106 cells/ml) were
cultured on glass slides, and an 18S probe (Guangzhou RiboBio Co.,
Ltd., Guangzhou, China) was prepared for fluorescence in
situ hybridization. A laser scanning confocal microscope (LSCM;
OLYMPUS Fluoview FV1000; Olympus Corporation, Tokyo, Japan) was
used to observe and locate LINC00887 in 95-C and 95-D cells. The
cells were initially cultured in Opti-MEM solution with
Lipofectamine® 3000 transfection reagent for 48 h, then
continuously cultured for a further 48 h with 10 µg/ml TGF-β and 5
µm ITD-1. When analysing the TGF-β pathway, pcDNA3.1-LINC00887
plasmid was also transfected for 48 h in the indicated groups (2
µg/well in 6-well plates). Luciferase Assay System (Promega
Corporation, Madison, WI, USA) and luminometer (Hitachi, Ltd.,
Tokyo, Japan) was used to examine the association between TGF-β and
LINC00887 expression. Cells were transfected with pGL3 basic vector
(Firefly luciferase) and pRL-TK plasmid (Renilla
luciferase). LINC promoter was synthesized and insert into pGL3
basic and the empty vector was used as control.
Prediction of LINC00887-targeting
miRNAs and validation
LNCipedia (lncipedia.org/db/search) database was used to predict
and screen the miRNAs with mutual downstream genes. miRNA
precursors (CCTCGAGCTATTCTCATTCATTATG XhoI) and pGL3 empty
vector or pGL3-LINC00887 were transfected into 293T cells (Cell
Bank of the Chinese Academy of Sciences, Shanghai, China) for 24 h,
and then tested for luciferase activity.
miRNA expression and regulation by
LINC00887
Three LINC00887 siRNAs were synthesized (Guangzhou
RiboBio Co., Ltd.) and the suppression effect of Linc00887 on 95-D
cells was analysed by RT-qPCR. LINC00887 was overexpressed in the
95-C cell line using the pcDNA3.1-LINC0088 vector. siRNA2 was
transfected into 95-D cell lines, and the expression levels of
miRNAs were detected using a TaqMan probe. The argonaute-2 (Ago2)
antibody was sequentially processed for immunoprecipitation, nuclei
acid purification and quantification. The details of the experiment
are as follows: 95-D cells were digested and collected in Eppendorf
tubes, washed in PBS, and the Magna RIP RNA-Protein
Immunoprecipitation Kit (EMD Millipore) was used for
immunoprecipitation of RNA-binding proteins. Cell lysate was
prepared by 200 µl RIP lysis Buffer, 1 µl Protease Inhibitor
Cocktail and 0.5 µl RNase inhibitor. Magnetic bead tubes were
vortexed for 30 sec for full suspension; 50 µl magnetic bead
suspension, 100 µl RIP Wash Buffer heavy suspension beads, 5 µg
Ago2 antibody (cat. no. 2897; Cell Signaling Technology, Inc.,
Danvers, MA, USA) was added to clean the beads twice.
Immunoprecipitation of RNA-binding Protein-RNA complexes was
performed. The magnetic beads and cell lysate were centrifuged at
4°C, 21,480 × g for 10 min. The beads were added to the magnetic
beads tube and incubated overnight on the suspension apparatus at
4°C. RNA was purified and precipitated using 80% ethanol. The
supernatant was discarded by centrifugation at 21,480 × g for 15
min and then dried. RNA was dissolved in 10–20 µl RNase-free water,
placed on ice and the RT-PCR was performed using the
TaqMan® MicroRNA Reverse Transcription Kit, with all
operations performed according to the manufacturer's instructions.
The contents of LINC00887 and levels of miR-613 (forward,
5′-CCGCTCGAGTCTACTAGGTGTGGGCTTTA-3′; reverse,
3′-GGCAAGCTTCTGTGGCCTTCCTTACTCTT-5′); miR-206 (forward,
5′-TGGAATGTAAGGAAGTGTGTGG-3′; reverse,
3′-ACCTGACCGGCCGTAACAACCC-5′) and miR-1-2 (forward,
5′-UGUAUGAAGAAAUGUAGGUAU-3′; reverse, 3′-ACAUACUUCUUUACACCCAUA-5′)
were detected by conventional qPCR using TaqMan probes.
TaqMan® 2X Universal PCR Master Mix was used under the
following conditions: Initial denaturation at 95°C, for 10 min
followed by 40 cycles of 95°C for 15 sec, 60°C for 1 min and 72°C
for 45 sec. The level of LINC00887 was examined by using qPCR with
X inactive specific transcript (XIST) as the negative control
(forward, 5′-CTTGGATGGTTGGTTGCCAGCTA-3′; reverse,
3′-TCATGCCATCCACCTA-5′); U6 RNA (forward, 5′-TGCTCGCTTCGGCAGC-3′;
reverse, 3′-ACTACATGTGCATGCT-5′) was used as the internal reference
for miRNA. TaqMan probe was utilized to detect the level of
miR-613, miR-206 and miR-1-2, with the U6b promoters the negative
control.
Prediction and expression regulation
of miRNA downstream targeting genes
The downstream interacting genes of miR-613, miR-206
and miR-1-2 were predicted using microRNA.org
(microrna.org/microrna/getMirnaForm.do) and screening
the mutually interacting genes fibronectin 1 (FN1), MET
proto-oncogene, receptor tyrosine kinase (MET) and SMAD4 (according
to the mirSVR score <-1.0). Then, LINC00887 was overexpressed in
95-C cell lines, and knocked down in 95-D cell lines using siRNA2.
Changes of gene expression products of three genes were analysed by
qPCR. TB Green Advantage qPCR premixes (Takara Biotechnology Co.,
Ltd.) was used in qPCR for these genes. qPCR was performed using
the 7500 fast system with as previously described above. The
primers of these were as follows: MET, forward,
5′-TCCTCTGGGAGCTGATGACA-3′ and reverse, 3′-CTGGGCAGTATTCGGGTTGT-5′;
FN1, forward, 5′-AGCCTGGGAGCTCTATTCCA-3′ and reverse,
3′-CTTGGTCGTACACCCAGCTT-5′; SMAD4, forward,
5′-GTCAGCTGCTGCTGGAATTG-3′ and reverse 3′-GTCTTGGGTAATCCGGTCCC-5′.
β-actin was used as the internal reference. Accordingly, changes in
the genomic products of FN1, MET and SMAD4 were observed.
Statistical analysis
SPSS statistical software (version 18.0; SPSS, Inc.,
Chicago, IL, USA) used to perform statistical analysis. Data are
expressed as the mean ± standard deviation from at least three
experiments. Statistical comparisons were based on the Student's
t-test or one-way analysis of variance (ANOVA). The Student's
t-test was used to compare the differences between two groups of
mean values. The ANOVA test followed by a post-hoc test (least
significance difference test, Student-Newman-Keuls) was used for
evaluating the difference among multiple groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Association between LINC00887 and
clinical overall survival time
RT-qPCR validation results suggested that LINC00887
is highly expressed in 95-D cells. In a sample containing of 522
cases of adenocarcinoma of the lung in the eBioPortal database
(cbioportal.org), LINC00887 was overexpressed in
11% (Z score ±1) and the survival time of the patient in these
cases was significantly lower than in patients without LINC
overexpression (P=0.0141). In another dataset containing of 504
cases of lung carcinoma, LINC00887 gene expression was found in 38%
of the cases. Although only 9% had an overexpressed LINC00887 level
(Z score ±1), the survival time of patient was also significantly
reduced in these patients, which implies the potential importance
this LINC00887 in the development of lung cancer.
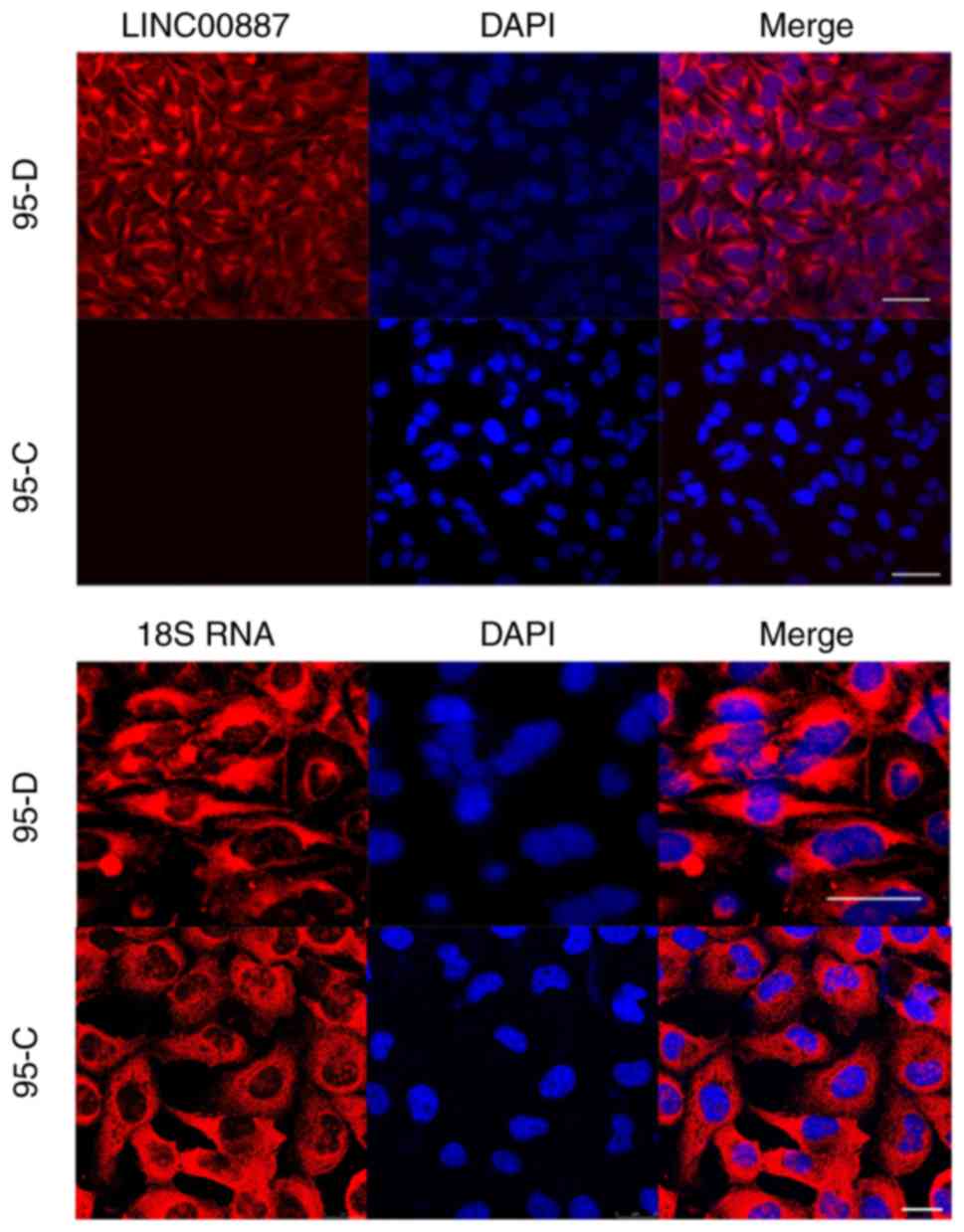
In-cell localization of LINC00887
The results of LSCM observation revealed that 18S
RNA was present in the cytoplasm of 95-D and 95-C cells (Fig. 1). Therefore, LINC00887 was also
located in the cytoplasm, with almost no expression found in the
cell nuclei of 95-D cells. The LINC00887 expression level in 95-D
cells was far higher than in 95-C cells.
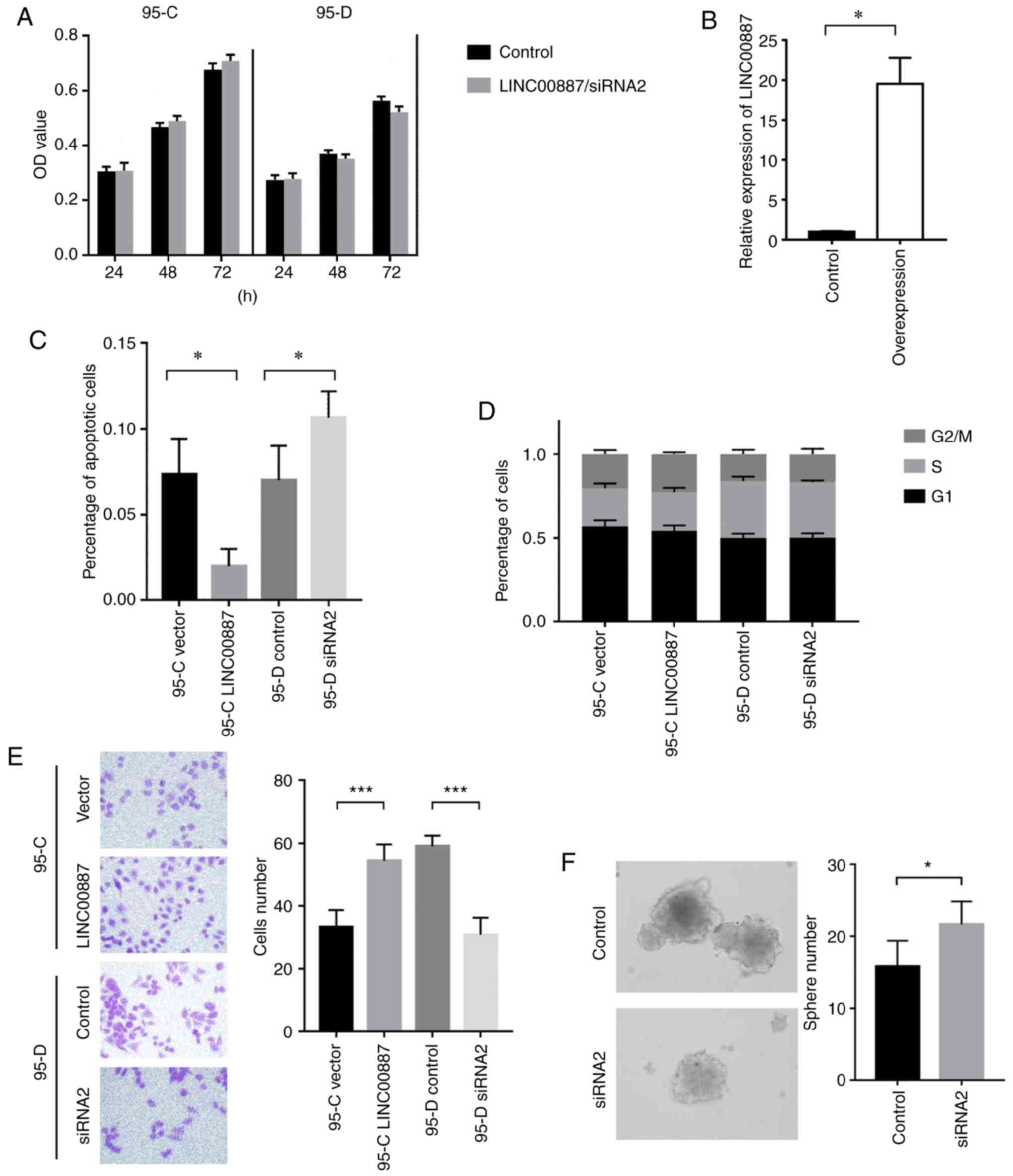
LINC00887 influences the cell cycle
and apoptosis of lung cancer cell lines
LINC00887 was transfected and overexpressed in 95-C
cells and reduced by siRNA 95-D cells, and MTT assay results showed
only a small difference, but no statistical significance in these
two groups when the cells were cultured for 24 and 72 h. It is
suggested that LINC00887 has no significant effect on the
proliferation of 95-C and 95-D cells (Fig. 2A). Meanwhile, in the overexpressed
group of LINC00887 in 95-C cells, the LINC00887 RNA level was
significantly higher than that of the control group, indicating
that the upregulation was successful and effective (Fig. 2B). The 95-C/95-D cells were prepared
using Annexin-FITC and Annexin-PI reagent kits. The distribution
and proportion of cells in stage of the cell cycle was detected via
flow cytometry, and the number of apoptotic cells was calculated.
Apoptosis analysis showed that increase in LINC00887 expression
reduced apoptosis of 95-C cells, while removing LINC00887 knockdown
induced a higher apoptosis rate in 95-D cells (Fig. 2C). No difference detected between the
G2/M, S and G1 phases of the cycle after flow cytometry examination
(Fig. 2D). Furthermore, Transwell
assays revealed that LINC00887 overexpression enhances the invasion
of lung cancer cells (Fig. 2E). When
LINC00887 was silenced from 95-D cells, the spheroidisation ability
of 95-D cells was significantly reduced (Fig. 2F).
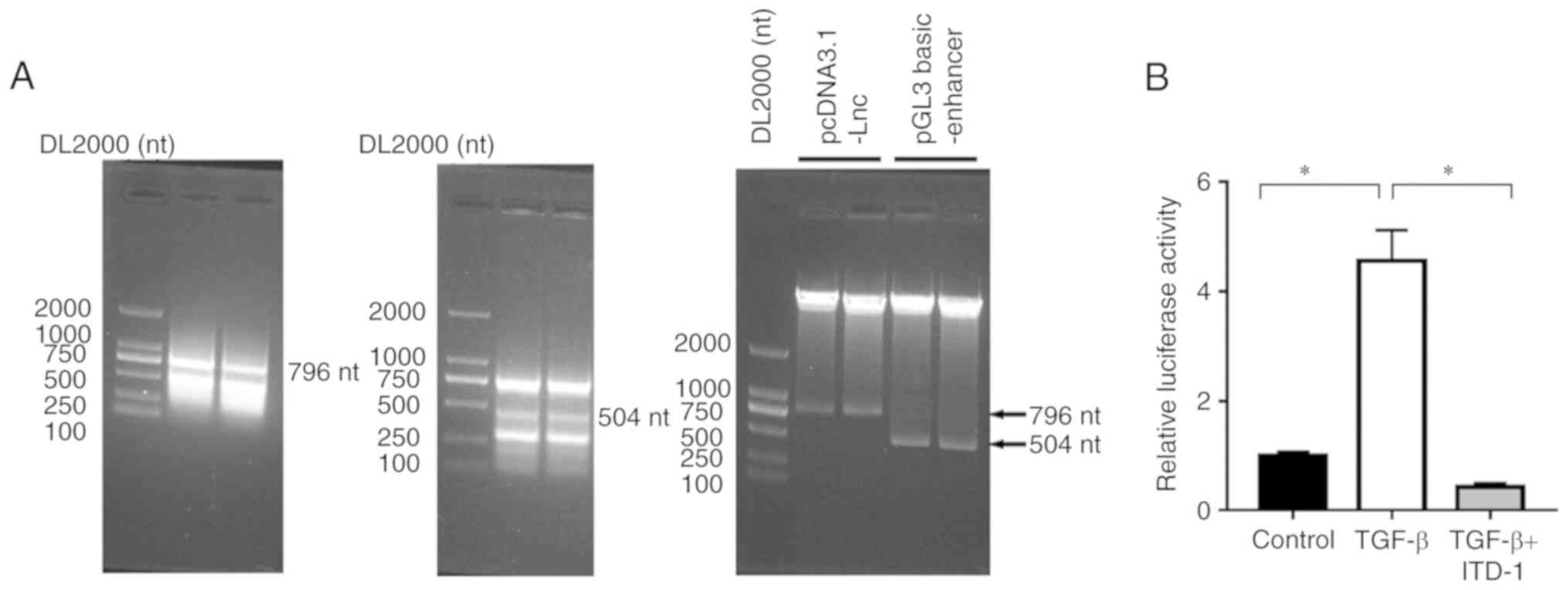
Interaction between LINC00887and genes
in the TGF-β signal transduction pathway
On the basis of bioinformatics analysis of the
Ensembl database, enhancers are present in the binding sequence
(TGAC/GTCA) of a TPA-responsive element and in the binding motif
sequence (GTCT/AGAC) of a SMAD binding element. Two plasmids of
pcDNA3.1-LINC00887 and pGL3 basic-enhancer, which was contained the
LINC00887 sequence and the luciferase, were generated for use in
reporter system. Restriction enzyme analysis was performed, and the
plasmids were expressed on 796 nt and 504 nt bands on the PCR
adhesive strip and the results as shown in (Fig. 3A). The transcriptional activity of
LINC00887 in 95-D cells with added TGF-β was significantly
increased, and the relative luciferase activity was significantly
increased compared with the control 95-D cell group (P=0.0004;
Fig. 3B). When ITD-1 was added to the
cells, the TGF-β pathway was suppressed and the relative luciferase
activity associated with LINC00887 expression was significantly
reduced, (TGF + ITD-1 vs. TGF-β; P=0.0002; Fig. 3B).
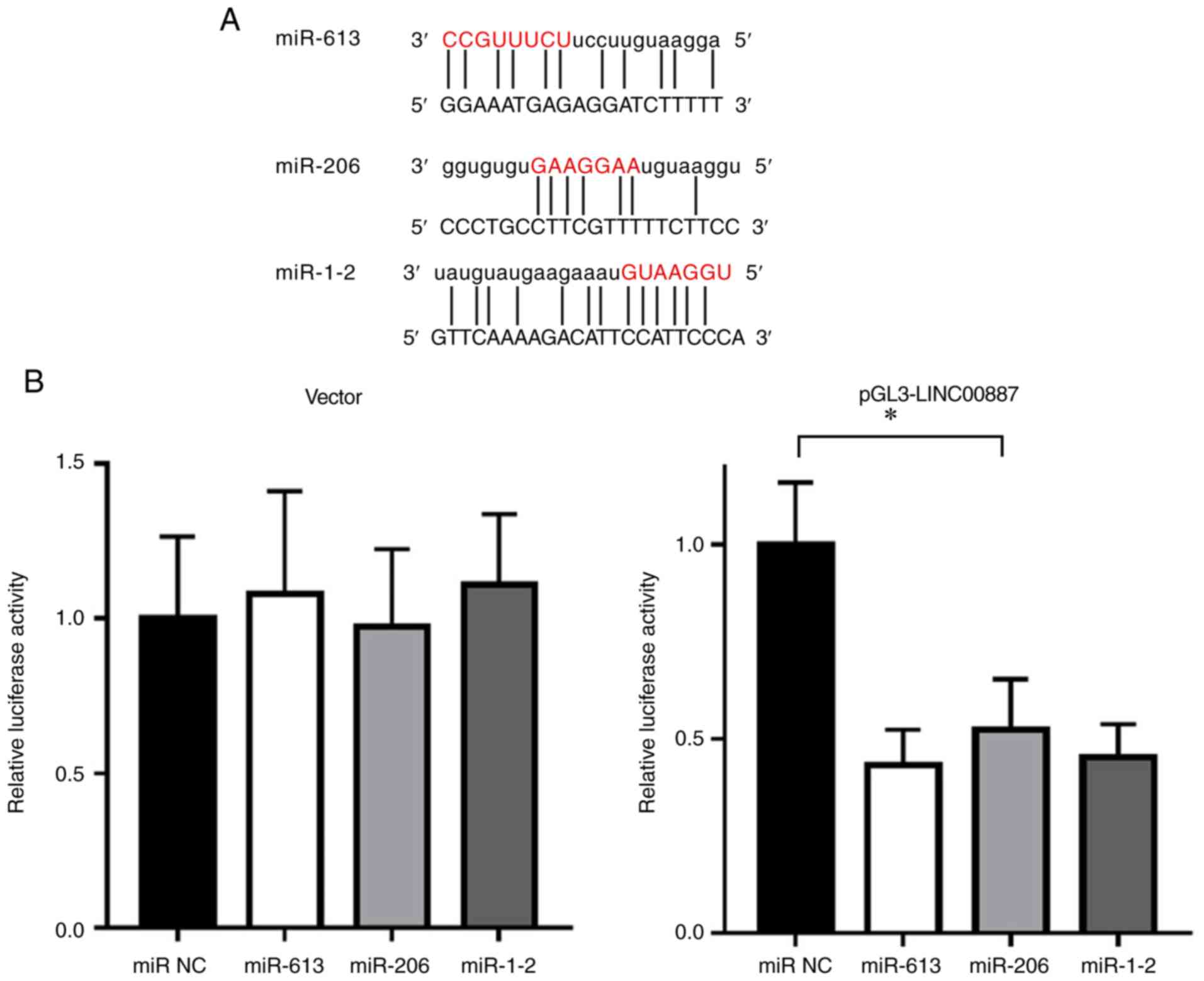
miR-613, miR-206 and miR-1-2 are
ceRNAs associated with LINC00887
The online searching results for LINC00887
(http://www.lncipedia.org/db/search)
indicated that the five top rated miRNAs were miR-1227, miR-613,
miR-206, miR-5581-3p and miR-1-2. The base pairing ability of each
miRNA to LINC00887 was evaluated as 77.67, 73.48, 73.29, 67.50 and
65.30, respectively. It was noted that miR-613, miR-206 and miR-1-2
have similar downstream target genes. Simulated hybridization
suggested that the seed sequences in these targeting miRNAs were
fully integrated with LINC00887, as shown in Fig. 4A. When pGL3-LINC00887 or empty vector
were co-transfected into 293T cells with miRNA precursors for 24 h
and luciferase activity was tested, the miRNA precursors had no
effect on the luciferase activity in the empty vector group.
However, these miRNAs suppressed the luciferase activity of
pGL3-LINC00887. Therefore, it can be concluded that miR-613,
miR-206 and miR-1-2 bind with LINC00887 act as ceRNAs (Fig. 4B).
LINC00887 can combine with miRNAs in
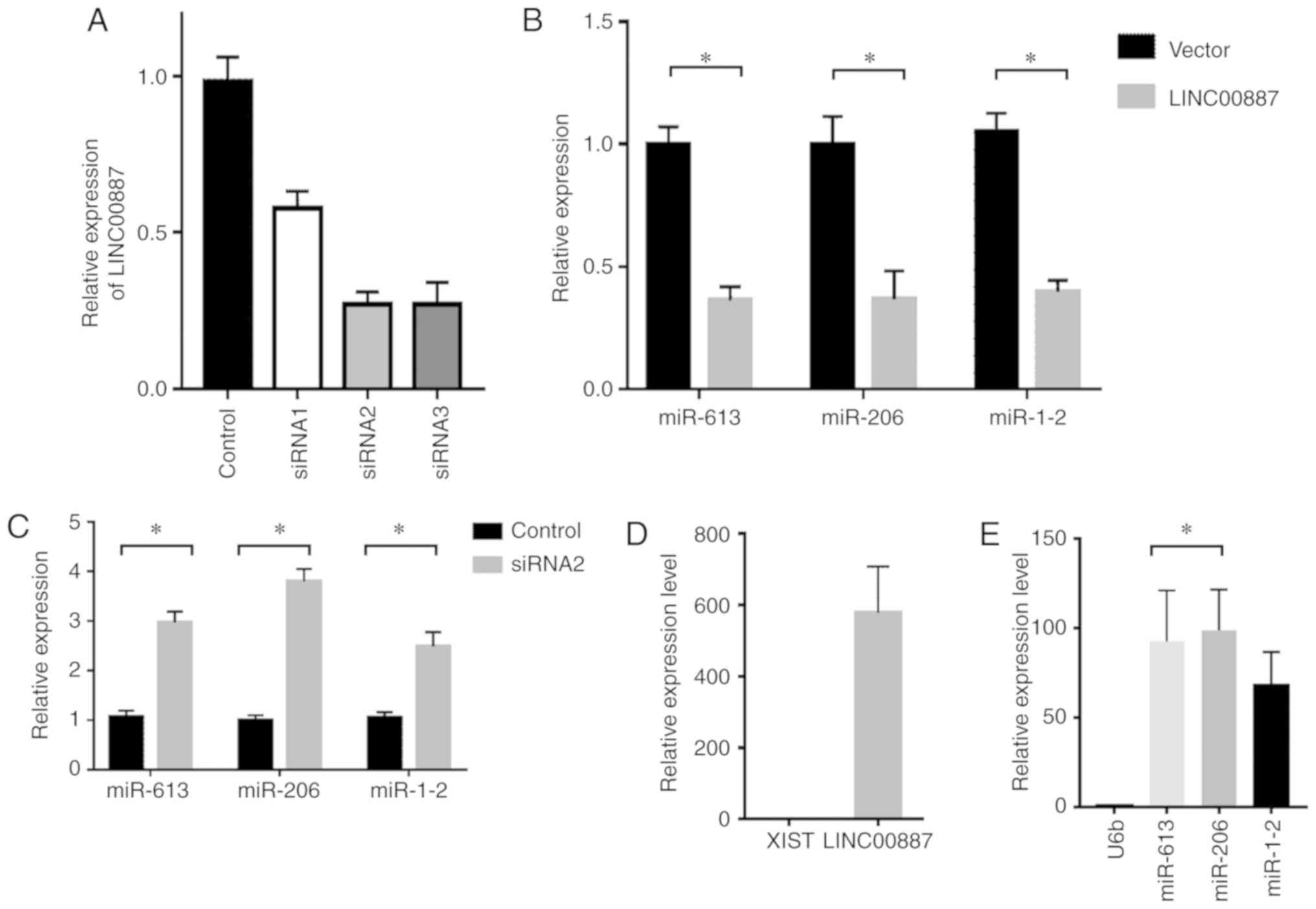
95-C/95-D cells resulting in RNA degradation
By synthesizing three interference sections of
LINC00887 (Guangzhou RiboBio Co., Ltd.), i.e. antagomir 1, 2 and 3,
it was found that siRNA2 had the best interference results in 95-D
cells (Fig. 5A). Empty vector and
LINC00887 overexpression vector were transfected into 95-C cells,
and the expression levels of miRNAs were analysed using a TaqMan
probe. The results indicated LINC00887 overexpression reduced the
levels of miR-613, miR-206 and miR-1-2 (Fig. 5B). This suggested a linear association
between the miRNAs and LINC00887.
Similarly, these miRNAs were increased in 95-D cell
lines when the expression of LINC00887 was silenced using by siRNA
(Fig. 5c). Immunoprecipitation was
performed out using an Ago2 antibody; the nucleic acid content of
the products bound to Ago2 was subsequently purified and
quantified. The levels of XIST (negative control) and LINC00887
associated with Ago2 were analysed by RT-qPCR. The internal
reference gene U6b (negative control) and the levels of miR-613,
miR-206 and miR-1-2 were also analysed using TaqMan probes. The
results suggested that LINC00887 and its associated miRNAs
interacted with Ago2, indicating that degradation was triggered by
the binding with miRNAs (Fig. 5d and
e).
LINC00887 and miRNA hybrids degrade
and inactivate the downstream targeting genes FN1, MET and
SMAD4
Using a mirSVR score of <-1.0 as the criterion,
FN1, MET and SMAD4 were ranked as the top three downstream genes
targeted by miR-613, miR-206 and miR-1-2 in the microRNA.org database (Fig.
6A). When LINC00887 was overexpressed in 95-C cell lines the
mRNA levels FN1, MET and SMAD4 significantly increased compared
with the control group (empty vector). This indicated that
LINC00887 effectively combines with and degrade miR-613, miR-206
and miR-1-2s, and therefore suppresses their actions on the
downstream target genes (Fig. 6B).
Similarly, in 95-D cell lines with initial high expression of
LINC00887, FN1, MET and SMAD4 were reduced by LINC00887 siRNA
(P<0.0001; Fig. 6C).
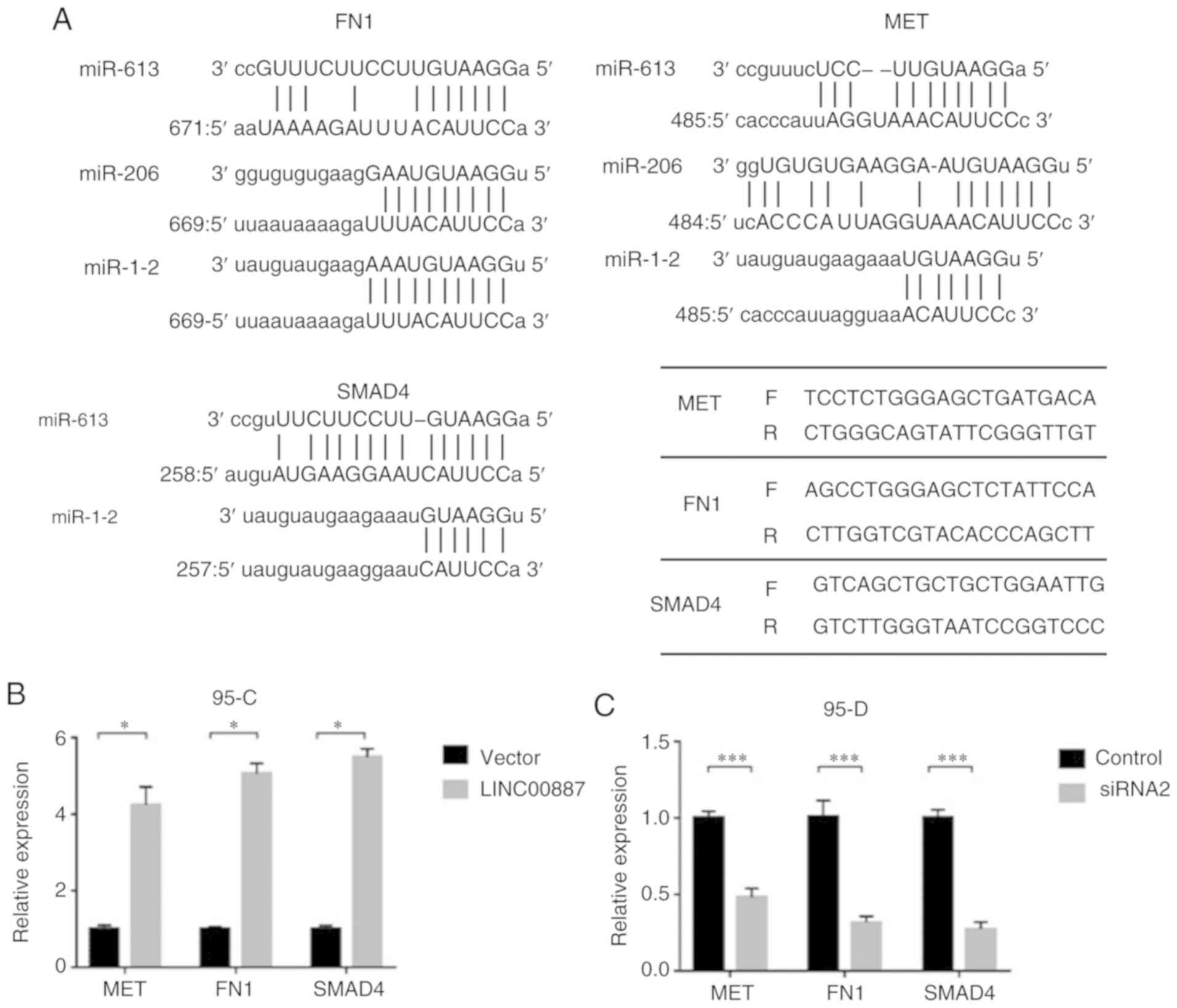 | Figure 6.LINC00887 can regulate downstream
genes MET, FN1 and SMAD4. (a) LINC00887 and the three miRNAs
interact with each other. In 95-C cell lines, LINC00887 can
effectively combine and degrade these miRNAs, and therefore
suppress and regulate the downstream genes (b) Overexpression of
LINC00887 increased MET, FN1 and SMAD4 levels (*P<0.05). (c)
LINC00887 siRNA reduced the expression of MET, FN1 and SMAD4
(t-test, ***P<0.0001). miR, microRNA; MET, MET proto-oncogene,
receptor tyrosine kinase; FN1, fibronectin 1; mothers against
decapentaplegic homolog 4; LINC00887, long intergenic non-protein
coding RNA 887. |
Discussion
Although the function of most lncRNAs remains
unknown thus far, it is of great interest to find out whether they
have could be used for diagnosing and treating clinic diseases. In
the current study, 95-D and 95-C cells, with different expression
levels of LINC00887, were used to investigate the role of
LINC00887. According to the data from eBioPortal, patients with a
high level of LINC00887 generally have shorter overall survival
time, indicating LINC00887 has specific functions and clinical
value for patients with NSCLC. High levels of LINC00887 may
potentially accelerate the malignant evolution of NSCLC cells, and
therefore, induce the metastasis and spread of tumor cells to the
rest of the body. In addition, Transwell experimental results
suggested that LINC00887 significantly enhanced the invasion of
lung cancer cells. By silencing LINC00887 in 95-D cells, their
spheroidisation ability was significantly reduced, which indicates
a reduced potential for spreading and survival at other sites.
These results suggest that LINC00887 is a persistent and functional
lncRNA.
From the results of the current study, it can be
concluded that LINC00887 exerts its function by interacting with
miRNAs, and these interact as ceRNAs (21). The ceRNA theory suggests that
downstream targeting gene functions are affected by changes in
miRNA quantity induced by bas pairing between lncRNAs and miRNAs.
ceRNAs generally include miRNAs, pseudogene transcripts and
circular RNAs (22). The most common
way for lncRNAs to interact with miRNAs is through a so-called RNA
sponge effect, which is determined by stable expression of lncRNAs
and miRNAs and their integration with miRNA response elements (MRE)
(23). In a study of gallbladder
carcinoma (24), lncRNA MATLAT1 acted
as a ceRNA regulating miR-206 through a molecular sponge mechanism
and promoted the development of cancer. In this study, LINC00887
was identified in the cytoplasm only and expressed highly in
nuclei.
Hybridization simulation suggested many miRNAs can
interact with LINC00887, among which miR-613, miR-206 and miR-1-2
were ranked as the top three candidates. When LINC00887 was
silenced, the levels of miR-613, miR-206 and miR-1-2 increased
significantly; however, when LINC00887 was overexpressed in 95-C
cells, the levels of miR-613, miR-206 and miR-1-2 were
significantly reduced, indicating an inversely proportional
relationship between LINC00887 and certain miRNAs. It remains
unclear if the association between LINC00887 and miR-613, miR-206
and miR-1-2 temporarily suppresses their activity or induced RNA
degradation. By examining the RNA pulldown products isolated by
Ago2 immunoprecipitation, it was found that the concentrations of
LINC00887, miR-613, miR-206 and miR-1-2 were all higher than the
negative control RNAs (XIST and U6b), indicating that interaction
between the LINC00887 can induced degradation of miRNAs. These
experiments suggested that there is a linear relationship between
LINC00887 and miR-613, miR-206 and miR-1-2, and that binding with
LINC00887 can induce the degradation of miRNAs by Ago2. Therefore,
miR-613, miR-206 and miR-1-2 can act as ceRNAs of LINC00887.
The metastasis of lung cancer is a multifaceted
process (25). Metastasis involves
the migration of lung cancer cells towards areas of higher oxygen
tension and changing of cellular phenotype through EMT, changes in
the expression of vascular endothelial growth factor (VEGF) and its
receptors (26), stroma interaction
(27) and interfering with immune
system communication (28). Recent
studies have shown that lncRNAs can control protein transcription,
translation and function. It is now believed that dozens of lncRNAs
are involved in lung cancer cell invasion and metastasis (29,30).
In this study, it was demonstrated that
LINC00887-miRNA interaction regulates downstream genes including
FN1, MET and SMAD4, which are associated with the spread of lung
cancer. The results suggested that overexpression of LINC00887 in
95-C cell lines can reduce the expression of miR-613, miR-206 and
miR-1-2. In contrast, FN1, MET and SMAD4 mRNA expression was
increased by overexpression of LINC00887, which indicated an
interaction between LINC00887 and three associated miRNAs and that
this induced their degradation. The regulatory effects on
downstream genes are therefore released, which potentially
increases the motility and invasion of 95-C cells.
FN1 is a component of the extracellular matrix (ECM)
and is widely distributed in smooth muscle cell layers (31). Abnormal expression of FN1 has been
identified in several human diseases, including cancers, and can
act as an important marker of EMT (32). miR-206 is involved in the formation of
smooth muscle in the airway and is reported to be associated with
FN1, interact, with ECM proteins such as VEGF and TGF-β1, and
contribute to the development of diseases including
bronchopulmonary dysplasia (33).
Relevant studies on miR-1 and MET genes revealed that miR-1
inhibited cell proliferation, and reduced migration and motility of
A549 cells. The downregulation of MET could be a potential
mechanism by which miR-1 regulates the growth and metastatic
potential of these cells (34). SMAD4
is a key regulator of TGF-β signalling. SMAD4 not activates the
expression and increases the activity of EMT transcription factors
(35–37).
Therefore, interactions between LINC00887-miRNAs and
regulatory target genes may have an important roles in the invasion
and metastasis of lung cancer cells; LINC00887-miRNA associations
can affect NSCLC cell EMT by altering the TGF-β signalling pathway.
In this study, the transcription of LINC00887 was significantly
increased by adding TGF-β to 95-D cells, this effect was suppressed
by the specific TGF-β pathway inhibitor, ITD-1. The cell cycle and
proliferation of 95-C cells was unaffected by overexpression and
silencing of LINC00887; however, cell apoptosis was altered by
changes in LINC00887 expression. Of course, in vivo
experiments are required to further validate the role of LINC00887.
We plan to use a lentivirus packaging vector to knock out the
expression of LINC00887 in 95-D cells, and inoculate the cell line
into nude mice to observe the growth of tumours, and detect the
changes in associated metastasis indicators in metastatic
tumours.
In summary, although the mechanism of NSCLC
metastasis is complex, the present study established an interactive
association between LINC00887 and miR-613, miR-206 and miR-1-2,
providing a new path for studying metastasis mechanisms. LINC00887,
and miR-613, miR-206 and miR-1-2 act as ceRNAs, and potentially
regulate the EMT transition of NSCLC cells through the TGF-β
pathway, therefore promoting cell migration and the acquisition of
stem cell features. It should be noted that this study is a
preliminary investigation into the function of LINC00887,
particularly with respect to signal transduction pathways. EMT
mechanisms via TGF-β signalling are complex, and further study is
required.
Acknowledgements
Not applicable.
Funding
This study was funded by The Program for Research
Projects of the Shaanxi Provincial Health Department (grant no.
2016D036), Shaanxi Provincial Traditional Chinese Medicine
Administration Chinese Medicine Research Project (grant no.
JCPT039), Research for Projects of the Xi'an technology office
program (grant no. 2016047SF/YX03(4), Key Projects of Social
Development Research Programs of Shaanxi Province (grant no.
2017SF-199), Natural Science Basic Research Program of Shaanxi
Province (grant no. 2017JM8195, 2018JM7044) and National Natural
Science Foundation (grant no. 81672300).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YT and SY made substantial contributions to
conception and design. YT, MY, LS and SH performed acquisition of
data. SS, WS and JW analysed and interpreted data. LL, QH, YD and
JZ have been involved in drafting the manuscript or revising it
critically for important intellectual content. SS and XR
contributed to the checking of article data and the calculation of
statistical methods. JS provided experimental technical support,
and checked the integrity of the data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hashim D, Boffetta P, La Vecchia C, Rota
M, Bertuccio P, Malvezzi M and Negri E: The global decrease in
cancer mortality: Trends and disparities. Ann Oncol. 27:926–933.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang L, Yu C, Liu Y, Wang J, Li C, Wang Q,
Wang P, Wu S and Zhang ZJ: Lung cancer mortality trends in China
from 1988 to 2013: New challenges and opportunities for the
government. Int J Environ Res Public Health. 13:E10522016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stratton MR, Campbell PJ and Futreal PA:
The cancer genome. Nature. 458:719–724. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ansari J, Yun JW, Kompelli AR, Moufarrej
YE, Alexander JS, Herrera GA, Herrera GA and Shackelford RE: The
liquid biopsy in lung cancer. Genes Cancer. 7:355–367.
2016.PubMed/NCBI
|
|
6
|
MacConaill LE and Garraway LA: Clinical
implications of the cancer genome. J Clin Oncol. 28:5219–5228.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Prensner JR, Iyer MK, Balbin OA,
Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso
CS, Kominsky HD, et al: Transcriptome sequencing across a prostate
cancer cohort identifies PCAT-1, an unannotated lincRNA implicated
in disease progression. Nat Biotechnol. 29:742–749. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Braconi C, Valeri N, Kogure T, Gasparini
P, Huang N, Nuovo GJ, Terracciano L, Croce CM and Patel T:
Expression and functional role of a transcribed noncoding RNA with
an ultraconserved element in hepatocellular carcinoma. Proc Natl
Acad Sci USA. 108:786–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang X, Gejman R, Mahta A, Zhong Y, Rice
KA, Zhou Y, Cheunsuchon P, Louis DN and Klibanski A: Maternally
expressed gene 3, an imprinted noncoding RNA gene, is associated
with meningioma pathogenesis and progression. Cancer Res.
70:2350–2358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Poliseno L, Salmena L, Zhang J, Carver B,
Haveman WJ and Pandolfi PP: A coding-independent function of gene
and pseudogene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hao Y, Yang X, Zhang D, Luo J and Chen R:
Long noncoding RNA LINC01186, regulated by TGF-β/SMAD3, inhibits
migration and invasion through Epithelial-Mesenchymal-Transition in
lung cancer. Gene. 608:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang C, Li X, Wang Y, Zhao L and Chen W:
Long non-coding RNA UCA1 regulated cell cycle distribution via CREB
through PI3-K dependent pathway in bladder carcinoma cells. Gene.
496:8–16. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang JZ, Xiang JJ, Wu LG, Bai YS, Chen ZW,
Yin XQ, Wang Q, Guo WH, Peng Y, Guo H and Xu P: A genetic variant
in long non-coding RNA MALAT1 associated with survival outcome
among patients with advanced lung adenocarcinoma: A survival cohort
analysis. BMC Cancer. 17:1672017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA Hypothesis: The Rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Venables JP, Klinck R, Koh C, Gervais-Bird
J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe
E, et al: Cancer-associated regulation of alternative splicing. Nat
Struct Mol Biol. 16:670–676. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang SH, Zhang WJ, Wu XC, Zhang MD, Weng
MZ, Zhou D, Wang JD and Quan ZW: Long non-coding RNA Malat1
promotes gallbladder cancer development by acting as a molecular
sponge to regulate miR-206. Oncotarget. 7:37857–37867.
2016.PubMed/NCBI
|
|
24
|
Popper HH: Progression and metastasis of
lung cancer. Cancer Metastasis Rev. 35:75–91. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Decaussin M, Sartelet H, Robert C, Moro D,
Claraz C, Brambilla C and Brambilla E: Expression of vascular
endothelial growth factor (VEGF) and its two receptors
(VEGF-R1-Flt1 and VEGF-R2-Flk1/KDR) in non-small cell lung
carcinomas (NSCLCs): Correlation with angiogenesis and survival. J
Pathol. 188:369–377. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Suzuki K, Sun R, Origuchi M, Kanehira M,
Takahata T, Itoh J, Umezawa A, Kijima H, Fukuda S and Saijo Y:
Mesenchymal stromal cells promote tumor growth through the
enhancement of neovascularization. Mol Med. 17:579–587. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiang R, Luo Y, Niethammer AG and Reisfeld
RA: Oral DNA vaccines target the tumor vasculature and
microenvironment and suppress tumor growth and metastasis. Immunol
Rev. 222:117–128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang C, Li X, Zhao H and Liu H: Long
non-coding RNAs: Potential new biomarkers for predicting tumor
invasion and metastasis. Mol Cancer. 15:622016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rokavec M, Horst D and Hermeking H:
Cellular model of colon cancer progression reveals signatures of
mRNAs, miRNA, lncRNAs, and epigenetic modifications associated with
metastasis. Cancer Res. 77:1854–1867. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang X, Xu J, Wang J, Gortner L, Zhang S,
Wei X, Song J, Zhang Y, Li Q and Feng Z: Reduction of MicroRNA-206
contributes to the development of bronchopulmonary dysplasia
through up-regulation of fibronectin 1. PLoS One. 8:e747502013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yen CY, Huang CY, Hou MF, Yang YH, Chang
CH, Huang HW, Chen CH and Chang HW: Evaluating the performance of
fibronectin 1 (FN1), integrin α4β1 (ITGA4), syndecan-2 (SDC2), and
glycoprotein CD44 as the potential biomarkers of oral squamous cell
carcinoma (OSCC). Biomarkers. 18:63–72. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Duan J, Zhang X, Zhang S, Hua S and Feng
Z: miR-206 inhibits FN1 expression and proliferation and promotes
apoptosis of rat type II alveolar epithelial cells. Exp Ther Med.
13:3203–3208. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nasser MW, Datta J, Nuovo G, Kutay H,
Motiwala T, Majumder S, Wang B, Suster S, Jacob ST and Ghoshal K:
Downregulation of Micro-RNA-1 (miR-1) in lung cancer. Suppression
of tumorigenic property of lung cancer cells and their
sensitization to doxorubicin-induced apoptosis by miR-1. J Biol
Chem. 283:33394–33405. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Grelet S, McShane A, Geslain R and Howe
PH: Pleiotropic roles of non-coding RNAs in TGF-β-mediated
epithelial-mesenchymal transition and their functions in tumor
progression. Cancers (Basel). 9(pii): E752017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Moustakas A and Heldin CH: Mechanisms of
TGF-β-induced epithelial-mesenchymal transition. J Clin Med.
5:E632016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee JK, Joo KM, Lee J, Yoon Y and Nam DH:
Targeting the epithelial to mesenchymal transition in glioblastoma:
The emerging role of fMET signaling. Onco Targets Ther.
7:1933–1944. 2014.PubMed/NCBI
|