Introduction
Prostate cancer (PCa) is common in American males
and is the second leading cause of cancer-related mortality
(1,2).
Prostate specific antigen (PSA) has been widely accepted for
diagnosis in patients with PCa in the early stages of disease.
However, the use of the PSA test is controversial and the subject
of much debate due to its high false positive rate. For example, a
consensus has not been reached on whether PSA serves as an
effective diagnostic marker, and, more importantly, whether PSA
level is associated with the risk of the disease (3). Overall, it is imperative to evaluate the
biological role behind PCa tumorigenesis and progression in order
to identify more effective prognostic biomarkers that can predict
which patients only require active surveillance, and which patients
require more aggressive treatment.
Distinct miRNA profiles have been observed in
different human tumor types (4–7). The
aberrations of certain miRNA signatures have been reported to be
correlated with cancer progression and can predict treatment
response and prognosis (8,9). Our previous studies and others have
demonstrated that the dysregulation of miRNAs are associated with
PCa tumorigenesis and progression (10–13).
However, the precise mechanism governing how PCa-associated miRNAs
affect disease progression, has yet to be completely
elucidated.
In recent years, and increasing amount of research
has demonstrated that miR-505 may serve as the tumor-suppressive
gene in several tumor types. For example, miR-505 may inhibit cell
proliferation and induce cell apoptosis in breast cancer (14). Whereas, another study indicated that
miR-505 depletion enhanced tumorigenesis and epithelial-mesenchymal
transition (EMT) in hepatoma cells (15). A study by Lu et al reported
that decreased levels of miR-505 were observed in endometrial
carcinoma and its restoration inhibited the activities of cell
proliferation, invasion and migration, yet increased apoptosis
(16). However, the tumor-suppressing
function of miR-505 and its mediatory mechanism in PCa require
further investigation.
Neuronal cell adhesion molecule is a member of the
immunoglobulin superfamily (17).
Neuron-glial-related cell adhesion molecule (NRCAM) was initially
identified and has been extensively explored in the peripheral and
central nervous system (18).
Furthermore, the overexpression of NRCAM has been frequently
identified in several malignancies including melanoma, papillary
thyroid cancer, colorectal cancer and PCa (19–22). In
melanoma cell lines, increased NRCAM expression was associated with
increased tumorigenicity (19). In
addition, the aberration of NRCAM significantly promoted cell
viability and invasiveness in thyroid cancer (20). Nevertheless, the oncogenic role of
NRCAM in PCa has not been adequately studied. In summary, the aim
of the present study was to demonstrate the tumor-suppressing
function of miR-505 and highlight its effects on NRCAM in PCa via
in vitro studies.
Materials and methods
Ethical approval
The approval of this research by the Ethical
Committee of Guangzhou First People's Hospital (Guangzhou Medical
University, China) was provided prior to the commencement of the
project. All patients recruited in the present study provided
written informed consent.
Patients and tissues samples
The present study collected 20 pairs of PCa tissues
and adjacent normal tissues following radical prostatectomy at
Guangzhou First People's Hospital. The tumor tissue sections were
frozen in liquid nitrogen. The tissue microarrays (TMAs; PR807c;
Alenabio) contained 50 primary PCa tissues, 10 normal prostate
tissues and 20 benign prostatic hyperplasia (BPH) tissues along
with detailed follow-up data for further immunohistochemical
staining. No previous treatment had been performed on the patients
with PCa. Additional investigations were conducted using The Cancer
Genome Atlas (TCGA) database, which contains 499 human primary PCa
tumors with clinicopathological information, to complement our TMA
results. The TCGA dataset was downloaded from the cBioPortal for
Cancer Genomics. Biochemical recurrence (BCR) survival and
disease-free survival was calculated (11–13). BCR
was defined as postoperative serum prostate-specific antigen (PSA)
>0.2 ng/ml. Disease-free survival was defined as the time to the
first evidence of loco-regional or distant clinical recurrence
after the initial surgery.
Cell culture, cell lines construction
and transfection
PC-3, DU145 and LNCaP cell lines (PCa) and RWPE-1
(normal prostate) were purchased from American Type Culture
Collection (ATCC). Cells were cultured in a humidified incubator at
37°C with 5% CO2 according to the protocol outlined in a
previous study (11–13). Human premicroRNA Expression Construct
Lenti-miR-505 [PMIRH505PA-1; System Biosciences (SBI), LLC] was
used as the pMIRNA1 lentivectors to express miR-505 precursor
(pre-505). Scramble control hairpin in pCDH-CMV-MCS-EF1α-copGFP
(CD511B-1) was designed as a negative control (pre-NC; cat. no.
PMIRH000PA-1; SBI). With pPACKH1 Packaging Plasmid Mix (cat. no.
LV500A-1; SBI), pre-505/pre-NC were transfected into 293TN cells
(SBI) in order to package the construct. By using the
LentiConcentin Virus Precipitation Solution (cat. no. LV810A-1;
SBI), the virus particles were collected according to the
manufacturer's protocol. Following transfection, PC-3 and LNCaP
cells were isolated and then seeded.
NRCAM expression plasmid (pCMV-NRCAM) was obtained
from Biogot Technology Co., Ltd. The cells transfected with
pCMV-NRCAM or pCMV (empty vector) were used as a corresponding
control. The cells were isolated for functional analyses at 48 h
following transfection.
Gene expression profiling
Gene expression profiles of miR-505-overexpressing
LNCaP cells and its corresponding control cells were conducted and
normalized. The experiments were peformed according to the
manufacturer's protocol as outlined in a previous study (12). Briefly, total RNA from the transfected
cells was amplified, labeled and purified. Then array
hybridization, wash and scan were determined. Gene Spring Software
(Agilent Technologies, Inc.) was used to analyze the data, and
genes with a fold change greater than two folds (P<0.05) were
selected.
RT-qPCR
As previously described (11–13), total
miRNA of PCa cells and tissues was isolated with the miRNA
Isolation Kit (BioTek China), and total RNA was extracted with the
RNeasy mini kit (Qiagen GmbH) when detecting the miRNA and mRNA
expression, respectively. The cDNA was synthesized using an RT-qPCR
Detection kit (GeneCopoeia). The primer sequences were obtained
from Thermo Fisher Scientific, Inc. and are presented in Table I. RT-qPCR analysis was performed and
the relative changes of miR-505 and the predictive targets were
normalized to the levels of RNU6B RNA or 18S rRNA. The
2−ΔΔCq values were calculated to the relative expression
(23).
 | Table I.Oligonucleotide sequence for all the
primers used in the present study. |
Table I.
Oligonucleotide sequence for all the
primers used in the present study.
| Name | Oligonucleotide
sequence (5′-3′) |
|---|
| miR-505-F |
ATGGGATGAAGTGATGATGCAAA |
| miR-505-R |
ACGCAAATATTGTGAAACACTGGTA |
| SOX6-F |
GCAGCAACAGATCCAGGTTCA |
| SOX6-R |
CAGAGTCCGCTGGTCATGTG |
| IRF6-F |
GGACGTCATGGACAGAGGAC |
| IRF6-R |
GGTGGGCAATGAGATCGCTA |
| NRCAM-F |
GAGCGAAGGGAAAGCTGAGA |
| NRCAM-R |
ACAATGGTGATCTGGATGGGC |
| NOV-F |
AGCAGCCAACAGATAAGAAAGGA |
| NOV-R |
TATTGTGGGGAGTGCAGCAG |
| LPL-F |
CACCTCATTCCCGGAGTAGC |
| LPL-R |
TCCTGTTACCGTCCAGCCAT |
| GREM1-qF |
GCTTGTGCGTAGTTCGTGTG |
| GREM1-qR |
CCCGCCCCTTTAGATGTGAG |
| AMOT-F |
GCAATCCAGACAAAACAGATGGG |
| AMOT-R |
TCTGCAGCTCTTGATTTGGC |
| CACNA2D3-F |
CAGTTGGTGGCACTCCGATA |
| CACNA2D3-R |
GCTGGATGACAAAGGACTTGGA |
| 18S rRNA-F |
GTAACCCGTTGAACCCCATT |
| 18S rRNA-R |
CCATCCAATCGGTAGTAGCG |
| RNU6B-F |
CTCGCTTCGGCAGCACA |
| RNU6B-R |
AACGCTTCACGAATTTGCGT |
Immunohistochemical staining
Immunohistochemistry was performed on the tissue
sections of TMAs of the patients with PCa to gauge the expression
levels of NRCAM. Following deparaffinization and dehydration,
sections were processed to reveal antigens using a microwave oven.
Briefly, the sections were blocked and then incubated with primary
antibody against NRCAM (1:100, cat. no. ab87427; Abcam). The slides
were subsequently incubated with anti-rabbit secondary antibody and
visualized by DakoCytomation Liquid DAB plus Substrate Chromogen
System (DakoCytomation).
Two pathologists, who were blind to patient data
independently evaluated the TMA samples. The immunohistochemistry
score was calculated by the sum of the staining intensity and the
fraction of positive tumor cells as previously described (11–13).
Target gene prediction programs
The potential targets of miR-505 were determined by
three different predicting programs, incuding TargetScan (24), miRanda (25), and miRWalk (26). The miRanda-miRSVR scores, TargetScan
context score or miRWalk scores were defined to aggregate per gene
and miR-505. The intersection of miRanda (score <-0.5) and
TargetScan (contextscore <-0.2) and miRWalk (score >0.80)
were used to predict the targets of miR-505.
In vitro luciferase assay
As previously described (11–13),
luciferase vectors were designed to express the wild-type (WT) or
mutant (MUT) 3′-UTR NRCAM sequences. The PCa cells were
co-transfected with WT luciferase vector and miR-505 mimic and the
corresponding controls. At 24 h post-transfection, the fluorescence
reader (Promega Corporation) was employed to calculate the
luciferase signals.
Cell proliferation assay
As previously described (11–13), a
CCK-8 assay was performed to monitor the proliferative ability of
PCa cells following transfection. Briefly, cells were incubated
with CCK-8 (Beyotime Institute of Biotechnology) at 37°C containing
5% CO2 for 4 h. The proliferative ability of PC-3/LNCaP
cells was determined at 4, 24, 48 and 72 h.
Wound healing analysis
This assay was performed according to the protocol
outlined in previous studies (11–13). When
LNCaP and PC-3 cells reached confluence, a linear scratch wound was
induced in monolayers with a sterile pipette tip. After washing,
the migrated cells through the scratch wound were observed and
calculated at 0 and 48 h.
Cell invasion analysis
As previously described (11–13), the
CytoSelect Cell Invasion Kit supplied by Cell Biolabs was used to
detect invasive abilities of PCa cells according to the
manufacturer's protocol. Briefly, PC-3/LNCaP cells were collected,
and suspended in serum-free DMEM. These cells were located on the
Transwell inserts coated with Matrigel (BD Biosciences), while the
normal medium was used as an attractant. Following a 48-h
incubation, 4% paraformaldehyde was used to fix the membranes at
room temperature for 15 min, which were then stained with 0.1%
crystal violet at room temperature for 10 min. Finally, the number
of cells that had invaded through the membranes were counted in 16
randomly-selected fields of view under an optical microscope
(Olympus Corporation) using ×100 and ×400 magnifications.
Cell cycle analysis
As previously described (11–13), the
present study trypsinized PC-3/LNCaP cells following transfection
according to the manufacturer's protocol. Briefly, the cells were
washed with phosphate-buffered saline (PBS), stained with propidium
iodide (Sigma-Aldrich; Merck KGaA) and incubated for 30 min at room
temperature. Finally, flow cytometry (BD Biosciences) was conducted
to obtain propidium iodide signals.
Apoptotic assay
The experiment was performed according to the
manufacturer's protocol (11–13). Briefly, cells were collected, washed,
stained with Annexin V and propidium iodide, and then subjected to
the FACScan flow cytometer (BD Biosciences). BD FACSuite™ software
(BD Biosciences) was conducted for further calculation.
Statistical analysis
The results are presented as the mean ± SD and were
analyzed using SPSS version 17.0 for Windows (SPSS, Inc.). The
unpaired Student's t-test or Mann-Whitney U test were conducted to
compare the two groups. One-way analysis of variance (ANOVA) or
two-way ANOVA with Bonferroni's post hoc test were also used when
comparing more than two groups. To determine the associations
between NRCAM and clinicopathological features, Fisher's exact test
or Pearson χ2 test were performed. Kaplan-Meier method
and Cox proportional hazards regression model were conducted for
survival estimation. P<0.05 was considered to indicate a
statistically significant difference.
Results
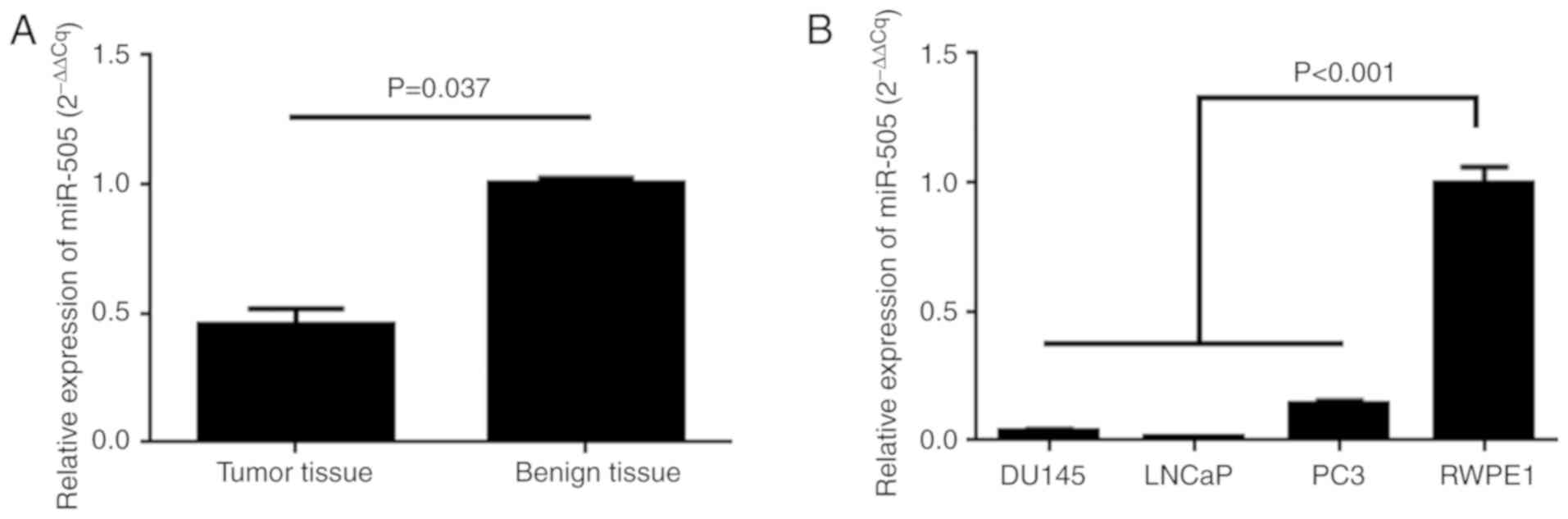
Decreased expression of miR-505 is
observed in PCa
The present study, aimed to validate the expression
of miR-505 with RT-qPCR in PCa cells and tissues, and the RT-qPCR
results demonstrated that miR-505 expression was significantly
decreased in PCa tissues when compared with normal tissues
(P=0.037; Fig. 1A). The data also
revealed that the expression levels of miR-505 were also
significantly reduced in PCa cells (P<0.001; Fig. 1B).
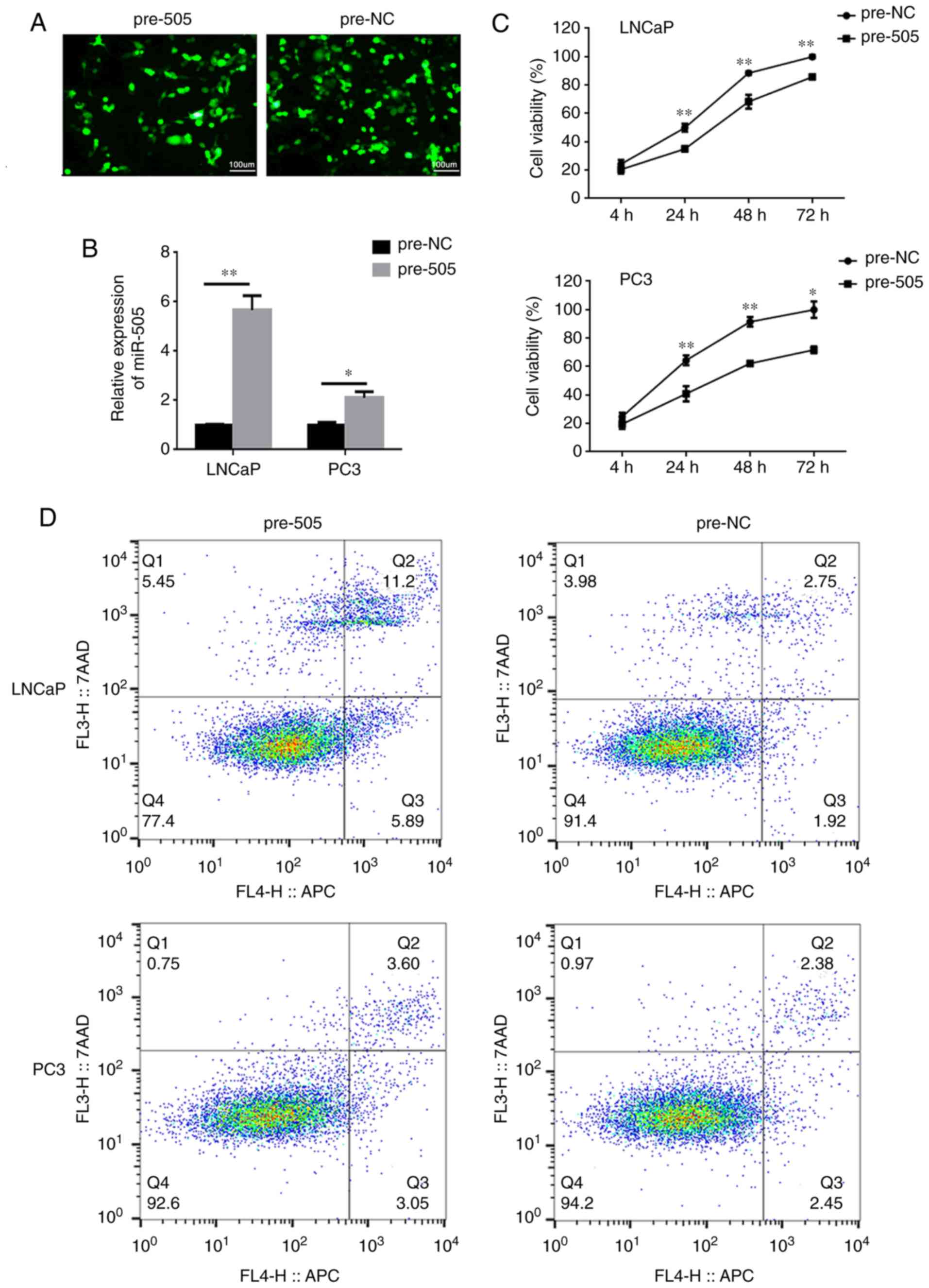
miR-505 suppresses tumorigenic
potential of PCa cells in vitro
The present study then constructed stable LNCaP and
PC-3 cell lines expressing miR-505 following lentiviral
transduction, and the success of this was confirmed via RT-qPCR
(P<0.05; Fig. 2A and B). CCK-8
assays confirmed that the proliferative activities of
miR-505-overexpressing LNCaP and PC-3 cells was significantly
inhibited when compared with control cells at 24, 48 and 72 h
following transfection (all groups P<0.05; Fig. 2C). In addition, flow cytometric
analysis indicated that miR-505 could induce cell apoptosis
(P<0.01; Fig. 2D and E), which
resulted in significant decrease in the number of PCa cells in the
G2+S phase (P< 0.05; Fig. 2F and
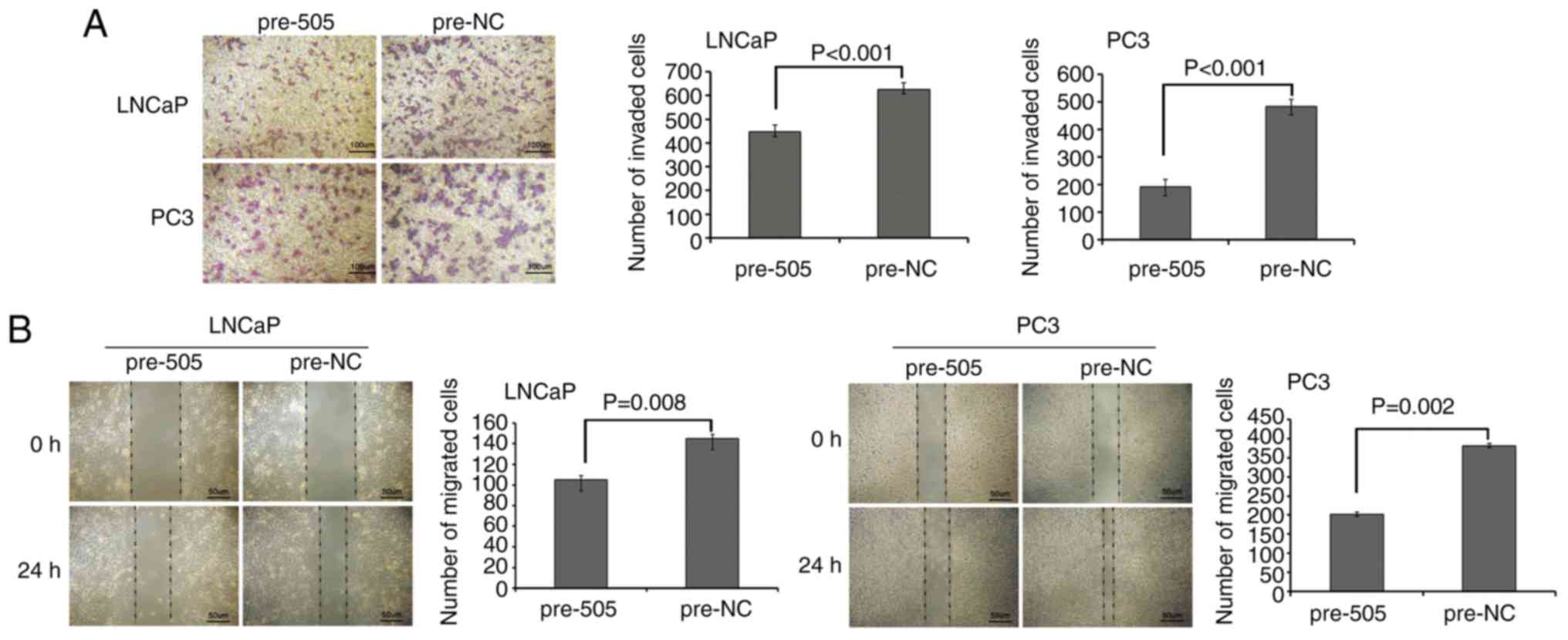
G). Transwell assays revealed that miR-505 suppressed the
invasion of PCa cells (P<0.001; Fig.
3A), and wound healing assays revealed that miR-505 also
markedly inhibited the migration of PCa cells (P<0.01; Fig. 3B).
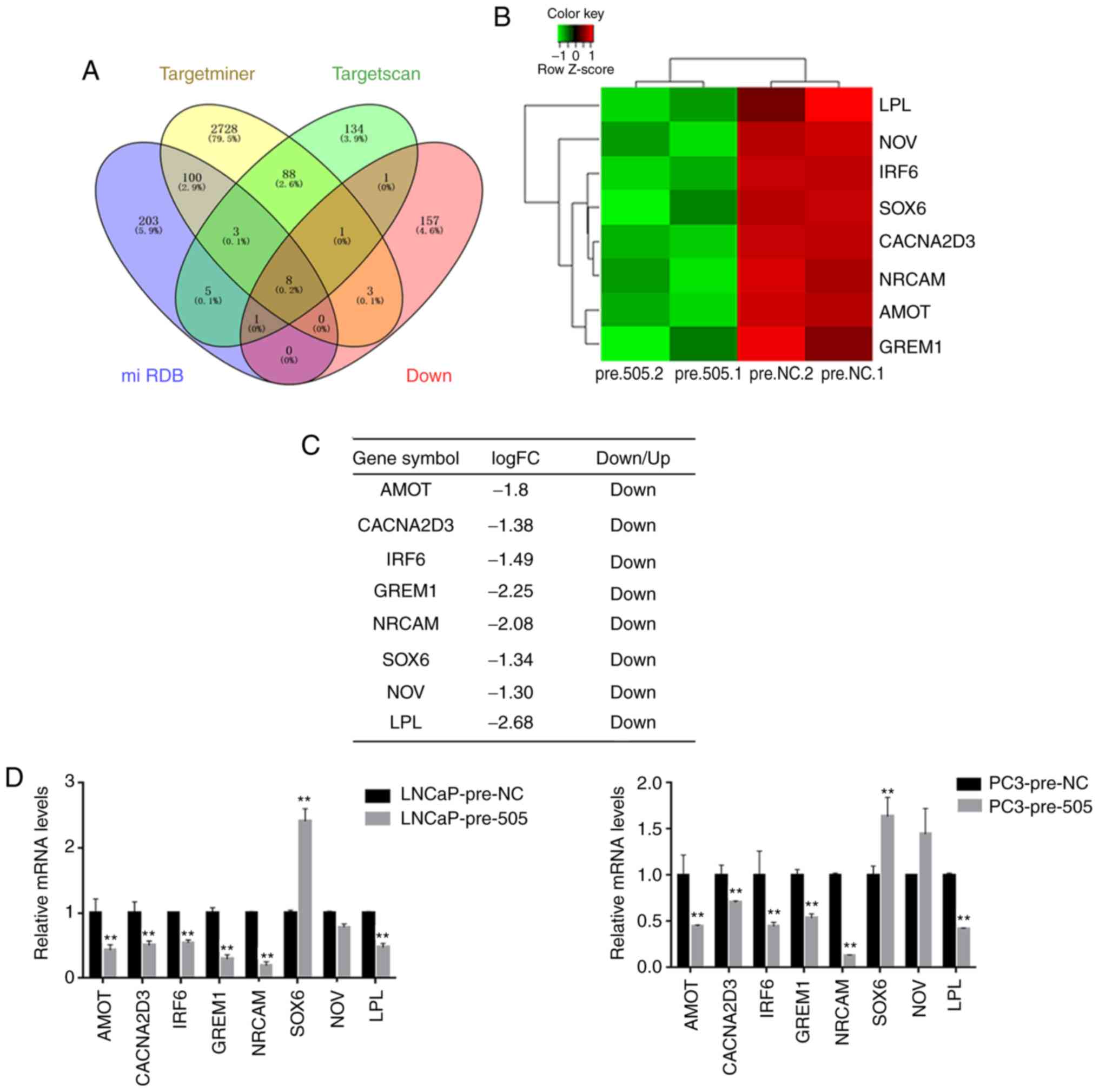
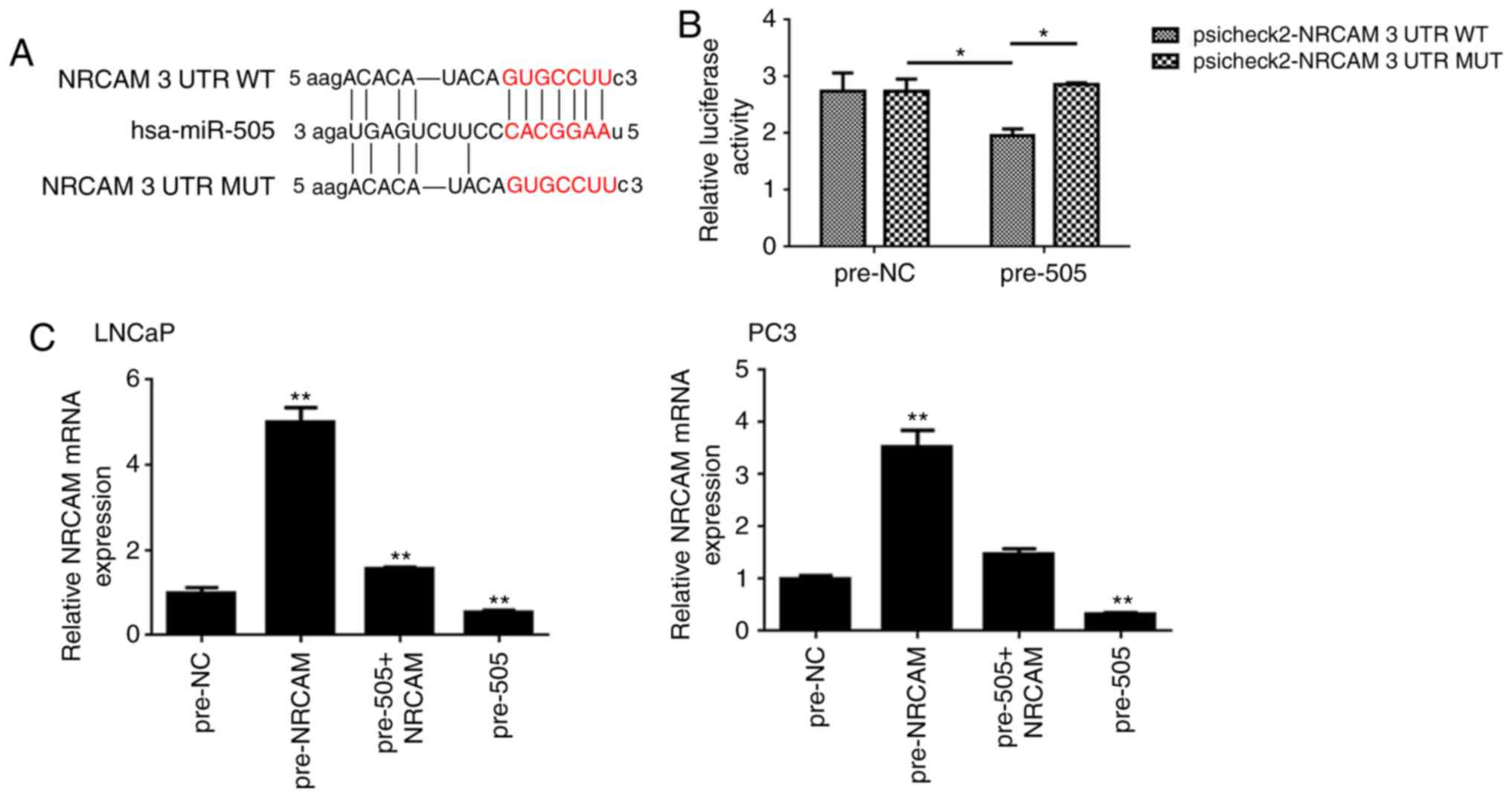
miR-505 directly targets NRCAM in PCa
cells
Gene expression profiles of miR-505-overexpressing
LNCaP cells and its corresponding control cells were compared. From
the microarray data, 406 differentially expressed genes (235 genes
upregulated and 171 genes downregulated) were identified. It is
well established that miRNAs negatively modulate mRNA stability and
translation by directly binding to the 3′UTR of specific target
genes (4). Therefore, the 171
downregulated genes detected were further analyzed using
bioinformatics tools.
The potential targets of miR-505 were determined by
three different types of predicting software. In combination with
our microarray data of miR-505-overexpressing PCa cells, eight
candidate targets were selected, including LPL, NOV, IRF6, SOX6,
CACNA2D3, NRCAM, AMOT, and GREM1, which were downregulated in LNCaP
cells of miR-505 overexpression (Fig.
4A-C, P<0.05 vs. pre-NC; P<0.01 vs. pre-NC). RT-qPCR
confirmed that AMOT, CACNA2D3, NRCAM, IRF6, GREM1 and LPL were
significantly decreased in PCa cells (LNCaP and PC-3) with miR-505
overexpression (Fig. 4D, P<0.01
vs. pre-NC). As the oncogenic role of NRCAM had been observed in
several malignancies (18–21), NRCAM was selected for subsequent
investigation.
In addition, the luciferase reporter assay
demonstrated that miR-505 interacts directly with the 3′UTR of
NRCAM (Fig. 5A and B, P<0.05).
Furthermore, RT-qPCR indicated that restoration of miR-505
attenuated the NRCAM protein expression in PC-3 and LNCaP cells
(Fig. 5C, P<0.01 vs. pre-NC).
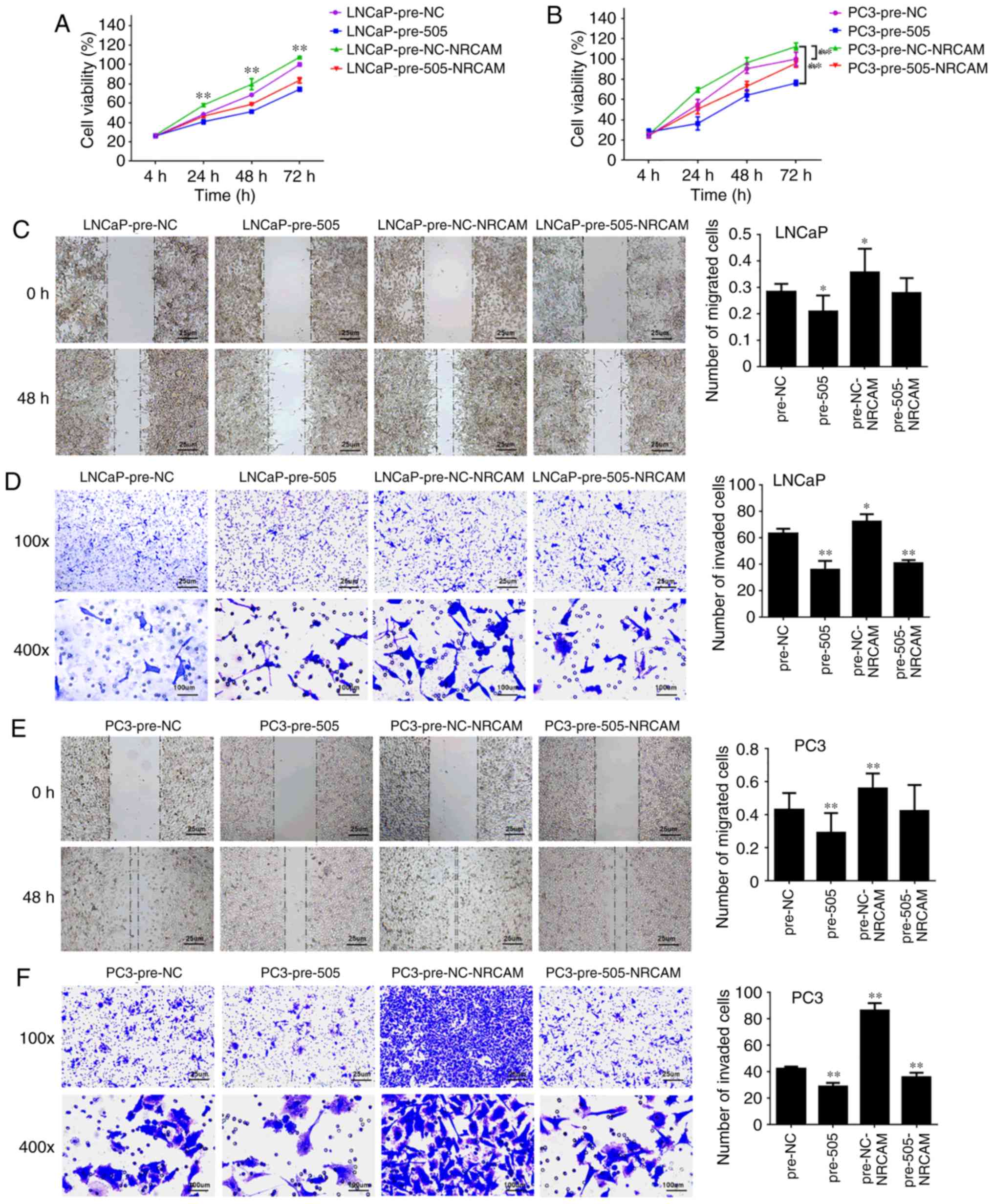
miR-505 suppresses tumorigenicity by
targeting NRCAM in PCa cells
To reveal whether the inhibitory effect of miR-505
in PCa functioned by mediating NRCAM, pCDNA3.1(+)-Vectors
expressing NRCAM were constructed. RT-qPCR revealed that the
tumor-suppressive role of miR-505 on NRCAM protein levels in PCa
cells could be rescued by NRCAM overexpression (Fig. 5C, P<0.01 vs. pre-NC). In addition,
NRCAM stimulation antagonized the inhibitory role of proliferation
(Fig. 6A and B), migration (Fig. 6C and E), and invasion (Fig. 6D and F) of LNCaP and PC-3 cells, which
were induced by miR-505 upregulation.
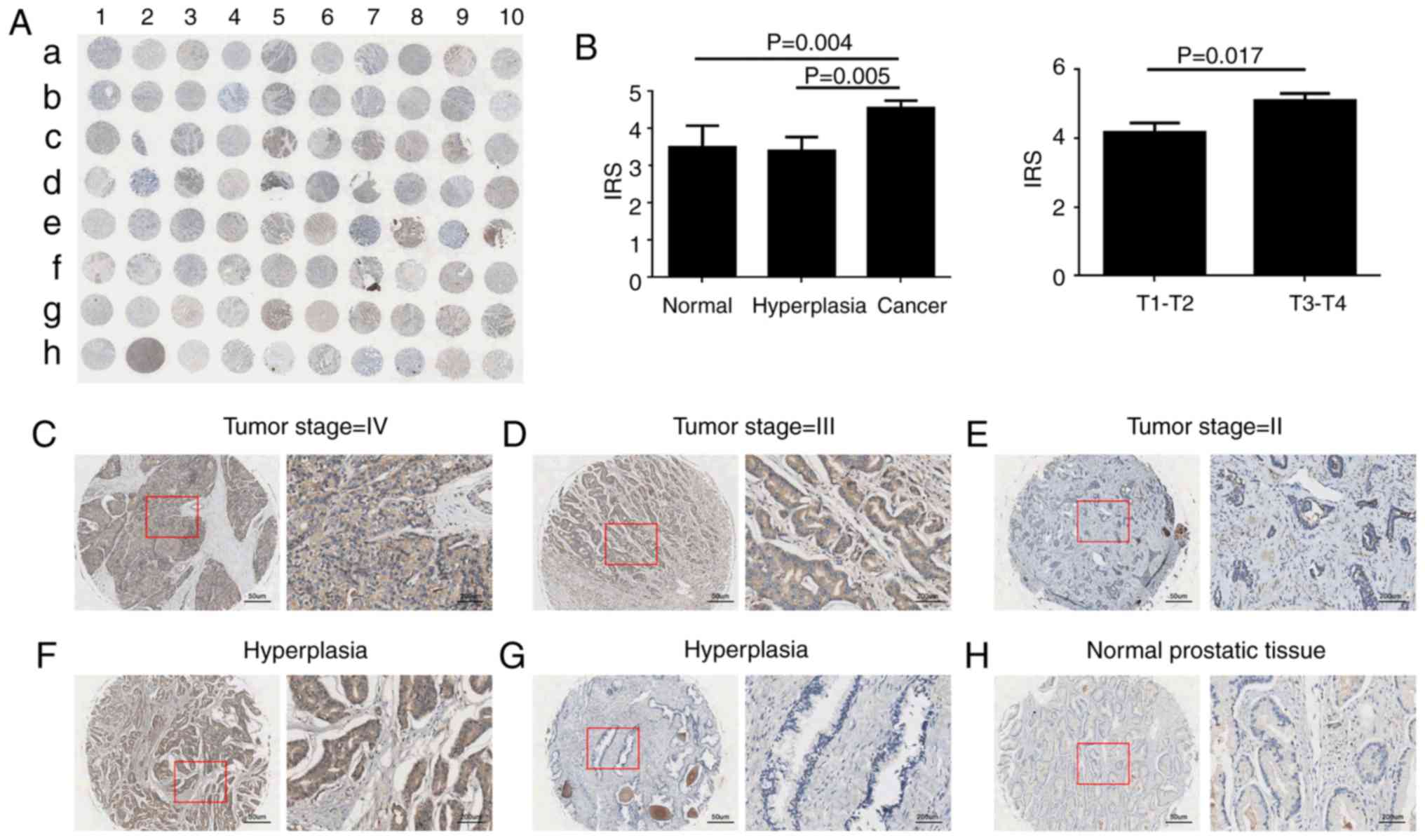
NRCAM is upregulated in PCa
tissues
To analyze the protein expression of NRCMA in PCa, a
TMA was used consisting of 50 PCa tissues, 20 benign hyperplasia
tissues, and 10 normal tissues. The immunohistochemical staining
revealed that NRCAM was primarily expressed in the cytoplasm of the
PCa cells (Fig. 7A, C-F). However,
faint NRCAM staining was also identified in the majority of BPH and
normal tissues (Fig. 7A, G and H).
The present study demonstrated that NRCAM expression was
significantly upregulated in PCa cells when compared with benign
prostate epithelium (Fig.7B,
P<0.05). In addition, the PCa patients with advanced stage
exhibited an increased NRCAM expression when compared with lower
stage tumors (Fig. 7B, P=0.017).
Combined expression of NRCAM and
miR-505 may predict PCa progression
The present study also performed investigations
using The Cancer Genome Atlas (TCGA), which contains data from 499
primary human tumors with PCa-specific mortality and other
clinicopathological information. As presented in Table II, the TMA datasets indicated that
high NRCAM expression was correlated with a higher Gleason score
(P=0.032) and advanced pathological stage (P=0.010), data from TCGA
also revealed NRCAM was strongly associated with lymph node
metastasis (P<0.001) and Gleason score (P=0.013).
 | Table II.Correlation of NRCAM expression with
clinicopathologic characteristics in patients with prostate
cancer. |
Table II.
Correlation of NRCAM expression with
clinicopathologic characteristics in patients with prostate
cancer.
|
|
| TMA |
|
| TCGA |
|
|---|
|
|
|
|
|
|
|
|
|---|
|
|
| NRCAM
expression |
|
| NRCAM
expression |
|
|---|
|
|
|
|
|
|
|
|
|---|
| Clinical
features | Case | Low, n (%) | High, n (%) | P-value | Case | Low, n (%) | High, n (%) | P-value |
|---|
| Tissue |
|
|
|
|
|
|
|
|
|
Cancer | 50 | 17 (34.0) | 33 (66.0) | 0.432 | 499 | 250 (50.1) | 249 (49.9) | – |
|
Non-cancer | 23 | 9 (39.1) | 14 (60.9) |
|
|
|
|
|
| Age |
|
|
|
|
|
|
|
|
|
≤60 | 12 | 3 (25.0) | 9 (75.0) | 0.331 | 222 | 117 (52.7) | 105 (47.3) | 0.132 |
|
>60 | 68 | 25 (36.8) | 43 (63.2) |
| 273 | 129 (47.3) | 144 (52.7) |
|
| Sex |
|
|
|
|
|
|
|
|
|
Male | 80 | 28 (35.0) | 52 (65.0) | – | 499 | 250 (50.1) | 249 (49.9) | – |
| Serum PSA levels
(ng/ml) |
|
|
|
|
|
|
|
|
|
<4 |
|
|
|
| 411 | 198 (48.2) | 213 (51.8) | 0.293 |
| ≥4 |
|
|
|
| 27 | 15 (55.6) | 12 (44.4) |
|
| Gleason score |
|
|
|
|
|
|
|
|
| ≤7 | 31 | 14 (45.2) | 17 (54.8) | 0.032a | 291 | 159 (54.6) | 132 (45.4) | 0.013a |
|
>7 | 19 | 3 (15.8) | 16 (84.2) |
| 204 | 90 (44.1) | 114 (55.9) |
|
| Pathological
grade |
|
|
|
|
|
|
|
|
| ≤2 | 4 | 4 (100.0) | 0 (0.0) | 0.010a |
|
|
|
|
|
>2 | 46 | 13 (28.3) | 33 (71.7) |
|
|
|
|
|
| Tumor stage |
|
|
|
|
|
|
|
|
| T1 | 29 | 11 (37.9) | 18 (62.1) | 0.351 | 176 | 88 (50.0) | 88 (50.0) | 0.470 |
|
T2-T4 | 21 | 6 (28.6) | 15 (71.4) |
| 228 | 116 (50.9) | 112 (49.1) |
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
|
| N0 | 43 | 15 (34.9) | 28 (65.1) | 0.554 | 342 | 190 (55.6) | 152 (44.4) |
<0.001b |
| N1 | 7 | 2 (28.6) | 5 (71.4) |
| 80 | 25 (31.2) | 55 (68.8) |
|
| Distant
metastasis |
|
|
|
|
|
|
|
|
| M0 | 44 | 13 (29.5) | 31 (70.5) | 0.093 | 453 | 223 (49.2) | 230 (50.8) | 0.513 |
| M1 | 6 | 4 (66.7) | 2 (33.3) |
| 3 | 1 (33.3) | 2 (66.7) |
|
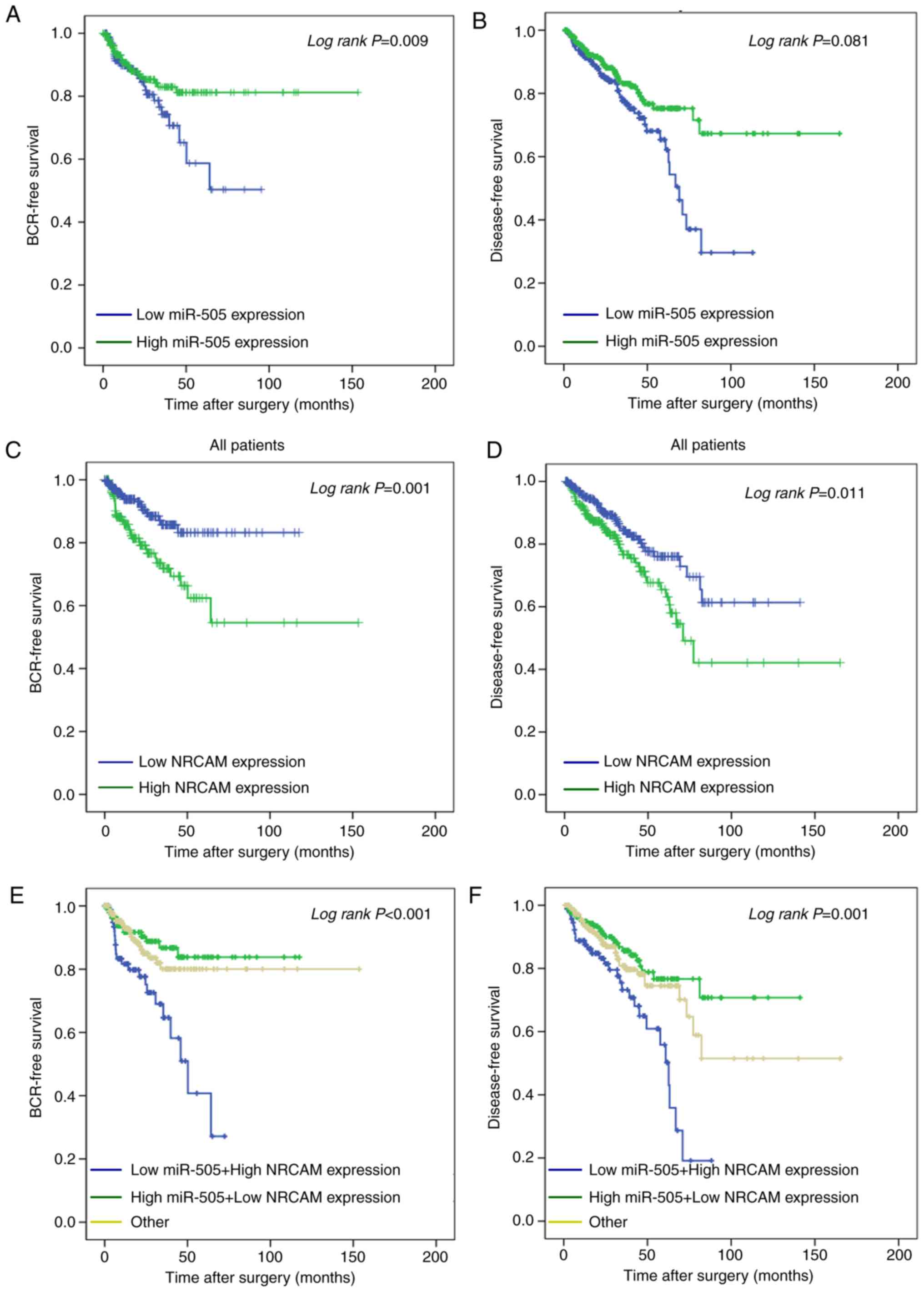
Patients with a low miR-505 expression had a
significantly worse BCR after radical prostatectomy as analyzed by
the Kaplan-Meier method (P=0.009; Fig.
8A); however, a significant trend for disease-free survival was
not observed (P=0.081; Fig. 8B). In
addition, high levels of NRCAM were significantly correlated with
unfavorable BCR-free survival (P=0.001; Fig. 8C) and disease-free survival (P=0.011;
Fig. 8D), respectively. The combined
use of these two biomarkers in prognosis is worthy of further
investigation. Collectively, these results indicated that patients
with PCa with a low expression of miR-505, and a high expression of
NRCAM exhibited poor BCR-free survival (P<0.001; Fig. 8E) and disease-free survival
(P<0.01; Fig. 8F) when compared
with contrasting groups of patients. The multivariate model
revealed that a high expression of NRCAM (Hazard Ratio 3.63;
P=0.016) may serve as an independent prognostic factor for
unfavorable disease-free survival (Table III).
 | Table III.Prognostic value of NRCAM expression
for disease-free survival by Cox proportional hazards model. |
Table III.
Prognostic value of NRCAM expression
for disease-free survival by Cox proportional hazards model.
|
| Disease-free
survival |
|---|
|
|
|
|---|
| Variables | HR (95% CI) | P-value |
|---|
| Univariate
analysis |
|
|
| Gleason
score (≤7 vs. >7) | 3.930
(2.187–7.063) |
<0.001b |
| Tumor
stage (T1 vs. T2-T4) | 3.416
(1.714–6.810) |
<0.001b |
| Distant
metastasis (M0 vs. M1) | 3.536
(0.488–25.641) |
0.212 |
| PSA
(<4 vs. ≥4) | 10.426
(5.309–20.474) |
<0.001b |
| NRCAM
expression (low vs. high) | 0.517
(0.303–0.880) |
0.015a |
| Multivariate
analysis |
| Tumor
stage (T1 vs. T2-T4) | 2.443
(1.037–5.754) |
0.041a |
| NRCAM
expression (low vs. high) | 3.74
(1.68–8.30) |
0.016a |
Discussion
The present study provided novel evidence revealing
decreased levels of miR-505 in PCa. Additionally, miR-505 may exert
a tumor-suppressive role by inhibiting PCa cell viability, invasion
and migration, and inducing cell cycle arrest and increasing
apoptotic activity. The results also indicated that miR-505 could
directly bind to the 3′UTR of NRCAM in PCa cells, and the
inhibitory effect of miR-505 potentially functions by mediating
NRCAM. Based on the TMA and TCGA data, the low expression of
miR-505 and the high expression of NRCAM were associated with the
progression and poor prognosis of PCa following radical
prostatectomy.
Other than in the regulation of physiological
processes in normal tissues, miR-505 has also been revealed to be
implicated in the development of malignant tissues (14–16,27–32).
For example, Verduci et al observed the suppressive role of
miR-505 in a mouse embryonic fibroblast (27). Previous studies indicated that miR-505
was underexpressed and it served as a candidate tumor suppressor in
different malignancies, such as cervical (28), colorectal (29,30),
breast (14) and liver cancer
(15) as well as endometrial
carcinoma (16), which were
corroborated by the results of the present study. However, other
studies have reported contradictory findings in the serum of
certain tumors in which the level of circulating miR-505 was higher
than that in control patients (31,32).
Therefore, additional experiments should be performed in order to
investigate the role of miR-505 in PCa serum.
miRNAs can target 20–30% of mRNA transcripts
(33). Additionally, recent studies
have identified several known miR-505 targets in various
malignancies. For example, enforced expression of miR-505 decreased
tumorigenic activities by targeting transforming growth factor-α
(TGF-α) from in vitro and in vivo assays in
endometrial carcinoma (15). In
addition, cell proliferation and invasion were inversely modulated
by miR-505 by targeting HMGB1 in hepatoma cells (16). Previous studies have also provided
evidence that miR-505 could directly target FZD4 and S100A4 in
cervical and colorectal cancer, respectively (28,30).
The present study combined microarray data of
miR-505-overexpressing cells and three miRNA target prediction
algorithms for a bioinformatics analysis, yielding eight potential
target genes including AMOT, CACNA2D3, NRCAM, IRF6, GREM1 and LPL.
Since the oncogenic role of NRCAM had been observed in several
malignancies (19–22), NRCAM was selected for additional
investigation. NRCAM is a member of the immunoglobulin superfamily,
and is an adhesion molecule which is associated with axonal
guidance and growth (17,18). Notably, NRCAM has been frequently
reported to have an increased expression in human cancers, and can
stimulate cell motility, and promote cell transformation in thyroid
and melanoma cancer by activating the PI3K/AKT and ERK/MAPK
signaling via interactions with α4β1 integrins and EGFR (19,20).
Although a previous study revealed that the increased expression of
NRCAM was evident in PCa (22), its
oncogenic role has yet to be completely elucidated. Cai et
al reported that NRCAM may be targeted by miR-203 in esophageal
cancer as indicated by the results of their bioinformatics analysis
(34). However, to the best of our
knowledge, no studies have stated that NRCAM expression is mediated
by miR-505 in the investigated malignancies. In the present study,
NRCAM was significantly upregulated in PCa tissues as demonstrated
by a TMA assay, and the results verified that NRCAM was targeted by
miR-505 through dual luciferase reporter assays. In addition,
miR-505 restoration reduced NRCAM protein expression from our in
vitro study. To the best of our knowledge, the present study is
the first to establish the association between miR-505 and NRCAM in
PCa.
The roles of the other potential targets (AMOT,
GREM1, CACNA2D3, IRF6, and LPL) with PCa progression were elusive.
AMOT serves a role in PCa proliferation via the Hippo/YAP pathway
(35). miR-205 regulated the
proliferation and the invasion of breast cancer cells by
suppressing the expression of AMOT (36). In addition, another study indicated
that miR-497 could directly target the AMOT gene in human
osteosarcoma cells to inhibit cell proliferation and invasion
(37). GREM1 can serve as the bone
morphogenetic protein antagonist during human cancer progression
(38). miR-128-3p suppressed the
proliferation and metastatic potential of glioma cells by targeting
GREM1 (39). CACNA2D3 may serve as a
pivotal gene in ERG-positive prostate cancer (40). The developmental transcription factor
IRF6 may be associated with cell proliferation, cancer stem cell
properties and chemotherapeutic sensitivity in nasopharyngeal
carcinoma (41). The LPL gene is
commonly methylated in PCa and may be involved in tumor progression
(42). However, the role between
these potential gene targets and miR-505 should be further
revealed.
The biological significance of miR-505 and NRCAM in
the carcinogenesis of PCa is substantiated by our TMA and TCGA
data, in which the expression of miR-505 and NRCAM were closely
associated with PCa recurrence and disease progression. This is
consistent with the finding in other human tumor types. For
example, patients with a low expression of miR-505 had advanced
pathological stage or a poor predicted survival in colon
adenocarcinoma, endometrial cancer and cervical cancer (16,28,29).
Previous studies have suggested that unfavorable tumor phenotype
and disease prognosis are associated with NRCAM overexpression in
several other tumor types, such as colorectal cancer and PCa
(21,22). However, this controversial data has
been also reported by Tsourlakis et al for NRCAM in PCa
tissues, in which the high expression of NRCAM was associated with
a favorable clinical disease course (22). We can infer that this conflicting role
may be due to the tissue heterogeneity and complex interactions
with other molecules.
In conclusion, the present study demonstrated the
inhibitory effects of miR-505 on PCa tumorigenesis, potentially by
targeting NRCAM. The combined analysis of NRCAM and miR-505 may be
associated with an unfavorable progression and prognosis in
PCa.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National
Key Basic Research Program of China (2015CB553706), the Guangzhou
Municipal Science and Technology Project (grant nos. 201803040001
and 201707010291), the Projects of Guangdong Key Laboratory of
Clinical Molecular Medicine and Diagnostics, the Natural Science
Foundation of Hunan Province (grant no. 2016JJ2117), the Science
and Technology Project of Huizhou (grant no. 190409094571998) and
the National Natural Science Foundation of China (grant no.
81571427).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
WDZ, XHL, HF, ZYC participated in the study design
and coordination, analysis and interpretation of data, material
support for obtained funding, and supervised the study. XHL
performed most of the experiments and statistical analysis and
drafted the manuscript. JML, YJZ, JHC and ZJ carried out the
experiments and sample collection. All authors read and approved
the final manuscript and agree to be accountable for all aspects of
the work in ensuring that questions related to the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
The approval of this research by the Ethics
Committee of Guangzhou First People's Hospital (Guangzhou Medical
University, China) was provided prior to the commencement of the
project. All patients recruited in the present study provided
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bostwick DG, Burke HB, Djakiew D, Euling
S, Ho SM, Landolph J, Morrison H, Sonawane B, Shifflett T, Waters
DJ and Timms B: Human prostate cancer risk factors. Cancer. 101 (10
Suppl):S2371–S2490. 2004. View Article : Google Scholar
|
|
3
|
Schröder FH, Hugosson J, Roobol MJ,
Tammela TL, Zappa M, Nelen V, Kwiatkowski M, Lujan M, Määttänen L,
Lilja H, et al: Screening and prostate cancer mortality: Results of
the European randomised study of screening for prostate cancer
(ERSPC) at 13 years of follow-up. Lancet. 384:2027–2035. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He HC, Han ZD, Dai QS, Ling XH, Fu X, Lin
ZY, Deng YH, Qin GQ, Cai C, Chen JH, et al: Global analysis of the
differentially expressed miRNAs of prostate cancer in Chinese
patients. BMC Genomics. 14:7572013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Porkka KP, Pfeiffer MJ, Waltering KK,
Vessella RL, Tammela TL and Visakorpi T: MicroRNA expression
profiling in prostate cancer. Cancer Res. 67:6130–6135. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ozen M, Creighton CJ, Ozdemir M and
Ittmann M: Widespread deregulation of microRNA expression in human
prostate cancer. Oncogene. 27:1788–1793. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bell EH, Kirste S, Fleming JL, Stegmaier
P, Drendel V, Mo X, Ling S, Fabian D, Manring I, Jilg CA, et al: A
novel miRNA-based predictive model for biochemical failure
following post-prostatectomy salvage radiation therapy. PLoS One.
10:e01187452015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Martens-Uzunova ES, Jalava SE, Dits NF,
van Leenders GJ, Møller S, Trapman J, Bangma CH, Litman T,
Visakorpi T and Jenster G: Diagnostic and prognostic signatures
from the small non-coding RNA transcriptome in prostate cancer.
Oncogene. 31:978–991. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Coarfa C, Fiskus W, Eedunuri VK,
Rajapakshe K, Foley C, Chew SA, Shah SS, Geng C, Shou J, Mohamed
JS, et al: Comprehensive proteomic profiling identifies the
androgen receptor axis and other signaling pathways as targets of
microRNAs suppressed in metastatic prostate cancer. Oncogene.
35:2345–2356. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang YQ, Ling XH, Yuan RQ, Chen ZY, Yang
SB, Huang HX, Zhong WD and Qiu SP: miR-30c suppresses prostate
cancer survival by targeting the ASF/SF2 splicing factor
oncoprotein. Mol Med Rep. 16:2431–2438. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin ZY, Chen G, Zhang YQ, He HC, Liang YX,
Ye JH, Liang YK, Mo RJ, Lu JM, Zhuo YJ, et al: MicroRNA-30d
promotes angiogenesis and tumor growth via MYPT1/c-JUN/VEGFA
pathway and predicts aggressive outcome in prostate cancer. Mol
Cancer. 16:482017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cai C, Chen QB, Han ZD, Zhang YQ, He HC,
Chen JH, Chen YR, Yang SB, Wu YD, Zeng YR, et al: miR-195 inhibits
tumor progression by targeting RPS6KB1 in human prostate cancer.
Clin Cancer Res. 21:4922–4934. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamamoto Y, Yoshioka Y, Minoura K,
Takahashi RU, Takeshita F, Taya T, Horii R, Fukuoka Y, Kato T,
Kosaka N and Ochiya T: An integrative genomic analysis revealed the
relevance of microRNA and gene expression for drug-resistance in
human breast cancer cells. Mol Cancer. 10:1352011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen S, Sun KX, Liu BL, Zong ZH and Zhao
Y: MicroRNA-505 functions as a tumor suppressor in endometrial
cancer by targeting TGF-α. Mol Cancer. 15:112016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu L, Qiu C, Li D, Bai G, Liang J and Yang
Q: MicroRNA-505 suppresses proliferation and invasion in hepatoma
cells by directly targeting high-mobility group box 1. Life Sci.
157:12–18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grumet M: Nr-CAM: A cell adhesion molecule
with ligand and receptor functions. Cell Tissue Res. 290:423–428.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Grumet M, Mauro V, Burgoon MP, Edelman GM
and Cunningham BA: Structure of a new nervous system glycoprotein,
Nr-CAM, and its relationship to subgroups of neural cell adhesion
molecules. J Cell Biol. 113:1399–1412. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Conacci-Sorrell M, Kaplan A, Raveh S,
Gavert N, Sakurai T and Ben-Ze'ev A: The shed ectodomain of Nr-CAM
stimulates cell proliferation and motility, and confers cell
transformation. Cancer Res. 65:11605–11612. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Sui F, Ma J, Ren X, Guan H, Yang
Q, Shi J, Ji M, Shi B, Sun Y and Hou P: Positive feedback loops
between NrCAM and major signaling pathways contribute to thyroid
tumorigenesis. J Clin Endocrinol Metab. 102:613–624.
2017.PubMed/NCBI
|
|
21
|
Chan JY, Ong CW and Salto-Tellez M:
Overexpression of neurone glial-related cell adhesion molecule is
an independent predictor of poor prognosis in advanced colorectal
cancer. Cancer Sci. 102:1855–1861. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsourlakis MC, Walter E, Quaas A, Graefen
M, Huland H, Simon R, Sauter G, Steurer S, Schlomm T and Minner S:
High Nr-CAM expression is associated with favorable phenotype and
late PSA recurrence in prostate cancer treated by prostatectomy.
Prostate Cancer Prostatic Dis. 16:159–164. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maragkakis M, Alexiou P, Papadopoulos GL,
Reczko M, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis
K, Simossis VA, et al: Accurate microRNA target prediction
correlates with protein repression levels. BMC Bioinformatics.
10:2952009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Verduci L, Simili M, Rizzo M, Mercatanti
A, Evangelista M, Mariani L, Rainaldi G and Pitto L: MicroRNA
(miRNA)-mediated interaction between leukemia/lymphoma-related
factor (LRF) and alternative splicing factor/splicing factor 2
(ASF/SF2) affects mouse embryonic fibroblast senescence and
apoptosis. J Biol Chem. 285:39551–39556. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma C, Xu B, Husaiyin S, Wang L,
Wusainahong K, Ma J, Zhu K and Niyazi M: MicroRNA-505 predicts
prognosis and acts as tumor inhibitor in cervical carcinoma with
inverse association with FZD4. Biomed Pharmacother. 92:586–594.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu M, Kuang Y, Wang M, Han X and Yang Q: A
microRNA expression signature as a predictor of survival for colon
adenocarcinoma. Neoplasma. 64:56–64. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mudduluru G, Ilm K, Fuchs S and Stein U:
Epigenetic silencing of miR-520c leads to induced S100A4 expression
and its mediated colorectal cancer progression. Oncotarget.
8:21081–21094. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Keller A, Backes C, Leidinger P, Kefer N,
Boisguerin V, Barbacioru C, Vogel B, Matzas M, Huwer H, Katus HA,
et al: Next-generation sequencing identifies novel microRNAs in
peripheral blood of lung cancer patients. Mol Biosyst. 7:3187–3199.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Du M, Shi D, Yuan L, Li P, Chu H, Qin C,
Yin C, Zhang Z and Wang M: Circulating miR-497 and miR-663b in
plasma are potential novel biomarkers for bladder cancer. Sci Rep.
5:104372015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cai X, Yang X, Jin C, Li L, Cui Q, Guo Y,
Dong Y, Yang X, Guo L and Zhang M: Identification and verification
of differentially expressed microRNAs and their target genes for
the diagnosis of esophageal cancer. Oncol Lett. 16:3642–3650.
2018.PubMed/NCBI
|
|
35
|
Zeng H, Ortiz A, Shen PF, Cheng CJ, Lee
YC, Yu G, Lin SC, Creighton CJ, Yu-Lee LY and Lin SH: Angiomotin
regulates prostate cancer cell proliferation by signaling through
the Hippo-YAP pathway. Oncotarget. 8:10145–10160. 2017.PubMed/NCBI
|
|
36
|
Zhang H and Fan Q: MicroRNA-205 inhibits
the proliferation and invasion of breast cancer by regulating AMOT
expression. Oncol Rep. 34:2163–2170. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ruan WD, Wang P, Feng S, Xue Y and Zhang
B: MicroRNA-497 inhibits cell proliferation, migration, and
invasion by targeting AMOT in human osteosarcoma cells. Onco
Targets Ther. 9:303–313. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu Y, Li Y, Hou R and Shu Z: Knockdown
GREM1 suppresses cell growth, angiogenesis, and
epithelial-mesenchymal transition in colon cancer. J Cell Biochem.
120:5583–5596. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fu C, Li D, Zhang X, Liu N, Chi G and Jin
X: LncRNA PVT1 facilitates tumorigenesis and progression of glioma
via regulation of MiR-128-3p/GREM1 axis and BMP signaling pathway.
Neurotherapeutics. 15:1139–1157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee SR, Choi YD and Cho NH: Association
between pathologic factors and ERG expression in prostate cancer:
Finding pivotal networking. J Cancer Res Clin Oncol. 144:1665–1683.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu L, Huang TJ, Hu H, Wang MY, Shi SM,
Yang Q, Lin F, Qiang YY, Mei Y, Lang YH, et al: The developmental
transcription factor IRF6 attenuates ABCG2 gene expression and
distinctively reverses stemness phenotype in nasopharyngeal
carcinoma. Cancer Lett. 431:230–243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim JW, Cheng Y, Liu W, Li T,
Yegnasubramanian S, Zheng SL, Xu J, Isaacs WB and Chang BL: Genetic
and epigenetic inactivation of LPL gene in human prostate cancer.
Int J Cancer. 124:734–738. 2009. View Article : Google Scholar : PubMed/NCBI
|