Introduction
Non-small cell lung cancer (NSCLC) represents
approximately 85% of all lung cancer cases and remains the leading
cancer killer with globally increasing incidence (1). Despite public and medical endeavors to
vanquish this disease, NSCLC has risen during the last decade, and
>40% of the patients diagnosed with NSCLC have distant
metastases, which are associated with unsatisfactory prognosis and
high mortality (2). NSCLC is also
considered as a highly heterogeneous disease that differs in
epidemiological, histological, molecular and phenotypic
characteristics (3), rendering it
an urgent challenge for early detection and effective treatment of
disease. Nowadays, it has become clear that epigenetic and genetic
modifications occurring in a number of genes play an important role
for the maintenance of normal cellular homeostasis and in the
occurrence of numerous pathologies; on the other hand, the
intervention on such modifications may improve the treatment and
prevention of numerous diseases, including NSCLC.
DNA methylation is a fundamental process for
eukaryotic development and leads to long term-repression of genome
expression (4). DNA methylation
changes are common and relatively stable in various types of
cancers, providing useful tools for diagnostic, prognostic and even
therapeutic intervention (5). It
has been reported that a wide spectrum of aberrantly methylated
genes in NSCLC regulate proliferation and maintenance of genome
stability (6,7). Notably, tobacco smoking, a major
etiological factor for NSCLC, may predispose to aberrant
methylation of key regulatory genes (8).
FBJ murine osteosarcoma viral oncogene homolog B
(FOSB), a member of the FOS family, heterodimerizes with JUN family
proteins to form activator protein-1 (AP-1) transcription factor
complexes, which play a central role in the transcriptional
regulation of numerous genes that are associated with cell
proliferation, differentiation, migration, metastasis, and
apoptosis (9). Recent data
indicated that FOSB can act as a common target for anticancer drugs
(10,11). In contrast to the amount of data on
the function of c-FOS, far less is known about the role of FOSB
(12). In addition, despite the
numerous studies that have revealed its oncogenic function in tumor
formation (13–17), FOSB has been revealed to be
downregulated in breast, gastric, and pancreatic cancer (18,19,20).
Furthermore, there are controversial data reporting the expression
and functional role of FOSB in lung cancer (11,21).
Therefore, to understand the biological role of FOSB and its
clinical significance in NSCLC progression, the methylation status
of the promoter and exon 4 of the FOSB gene was evaluated in
176 specimens from patients with NSCLC using bisulfite sequencing
and its association with patient outcomes was investigated.
Materials and methods
NSCLC tissue samples
Fresh tissue samples were obtained from 176 patients
(aged 35 to 83 years) with primary NSCLC and corresponding normal
tissues. The present study was approved by the Institutional Review
Board (IRB) of Kyungpook National University Hospital (KNUH; Daegu,
Korea) (No. 2014-04-210) and all patients provided signed informed
consent. The clinical characteristics of the patients are
summarized in Table I. NSCLC and
normal tissue samples were obtained at the time of surgery, and
were immediately frozen in liquid nitrogen and stored at −80°C
until DNA extraction. Only tumors with >80% of tumorous
components were used for methylation analysis and the histological
adequacy of tissue specimens was verified by hematoxylin-eosin
(H&E) staining. Tumors were staged according to the American
Joint Cancer Committee (AJCC) criteria.
 | Table I.Comparison of methylation status of
the FOSB exon 4 according to characteristics of NSCLC
patients. |
Table I.
Comparison of methylation status of
the FOSB exon 4 according to characteristics of NSCLC
patients.
| Variables | Unmethylation, n
(%) | Methylation, n
(%) |
P-valuea |
|---|
| All subjects | 18 (10.2) | 158 (89.8) |
|
| Age (years) |
|
|
|
|
≤64 | 7
(8.9) | 72
(91.1) | 0.589 |
|
>64 | 11 (11.3) | 86
(88.7) |
|
| Sex |
|
|
|
|
Men | 15 (12.2) | 108 (87.8) | 0.189 |
|
Women | 3
(5.7) | 50
(94.3) |
|
| Smoking status |
|
|
|
|
Ever | 15 (12.6) | 104 (87.4) | 0.132 |
|
Never | 3
(5.3) | 54
(94.7) |
|
| Histological
types |
|
|
|
|
SQC | 11 (13.1) | 73
(86.9) | 0.230 |
|
ADC | 7
(7.6) | 85
(92.4) |
|
| Lymph node
metastasis |
|
|
|
| N0 | 10 (7.7) | 120 (92.3) | 0.062 |
| N1 and
N2 | 8
(17.4) | 38
(82.6) |
|
| Pathologic
stage |
|
|
|
| Stage
I | 6
(6.6) | 85
(93.4) | 0.100 |
| Stage
II–IIIA | 12 (14.1) | 73
(85.9) |
|
Cell culture, total RNA isolation, and
reverse transcription PCR (RT-PCR)
The NCI-H157 and NCI-H187 cell lines were obtained
from the American Type Culture Collection and cultured in
Dulbecco's modified Eagle's medium/F12 or RPMI-1640 medium (Thermo
Fisher Scientific, Inc.) with 10% fetal bovine serum and 1%
penicillin-streptomycin. Genetic characteristics of the NCI-H157
cell line were determined by PCR-single-locus-technology at
Eurofins Genomics. Cells were maintained in an incubator at 37°C in
5% CO2. 5-Aza-2′-deoxycytidine (5-AzadC) was added daily
to the culturing medium at the indicated concentration for 72 h.
Total RNA was isolated from clinical tissue samples and cell lines
using TRIzol (Thermo Fisher Scientific, Inc.) and then first-strand
cDNA was synthesized using the SuperScript First-Strand Synthesis
Kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The relative expression of FOSB
mRNA was measured with semi-quantitative RT-PCR using GAPDH
as an internal control for normalization. All primer sequences are
listed in Table SI. The PCR
thermal cycling began with initial denaturation for 2 min at 95°C,
followed by 30 amplification cycles (1 min at 95°C, 1 min at 54°C,
1 min at 72°C) and a final 5 min incubation at 72°C. PCR products
were electrophoresed on 2% agarose gel, stained with ethidium
bromide (0.5 µg/ml) for 15 min at room temperature and visualized
using the Syngene DigiGenius Gel Documentation system
(Syngene).
DNA extraction, bisulfite treatment,
and methylation analysis
DNA samples were extracted using the QIAamp DNA Mini
Kit (Qiagen, Inc.) and bisulfited using the EZ DNA Methylation-Gold
Kit (Zymo Research) following the manufacturer's instructions. The
converted DNA was pyrosequenced using a PyroMark Q96MD system
(Qiagen, Inc.) as previously described (22). PCR primer sequences and
pyrosequencing primers are presented in Table SI. FOSB methylation for each
sample was calculated from the average value of examined CpGs
[mC/total C ×100 (%)] and represented as a mean methylation index
(MI).
Statistical analysis
Differences in the methylation level of FOSB
promoter and exon 4 between tumor and matched normal tissues were
analyzed using paired t-tests. A comparison of unmethylation
proportion of FOSB exon 4 according to the clinical
characteristics was evaluated using chi-square test. Overall
survival (OS) was estimated with Kaplan-Meier method (log-rank
test) and multivariate Cox regression analysis. A P-value <0.05
was considered to indicate a statistically significant difference.
All analyses were performed using SAS ver. 9.4 (SAS, Inc.).
Results
Methylation status and expression of
the FOSB gene in NSCLC samples
Bisulfite pyrosequencing was performed to analyze
the methylation status of the human FOSB gene in malignant
and adjacent non-malignant lung tissue of 176 NSCLC patients. Due
to DNA methylation changes in the promoter and gene body regions of
FOSB (23,24), pyrosequencing primers encompassing
nine and seven CpGs within the promoter and exon 4 regions of the
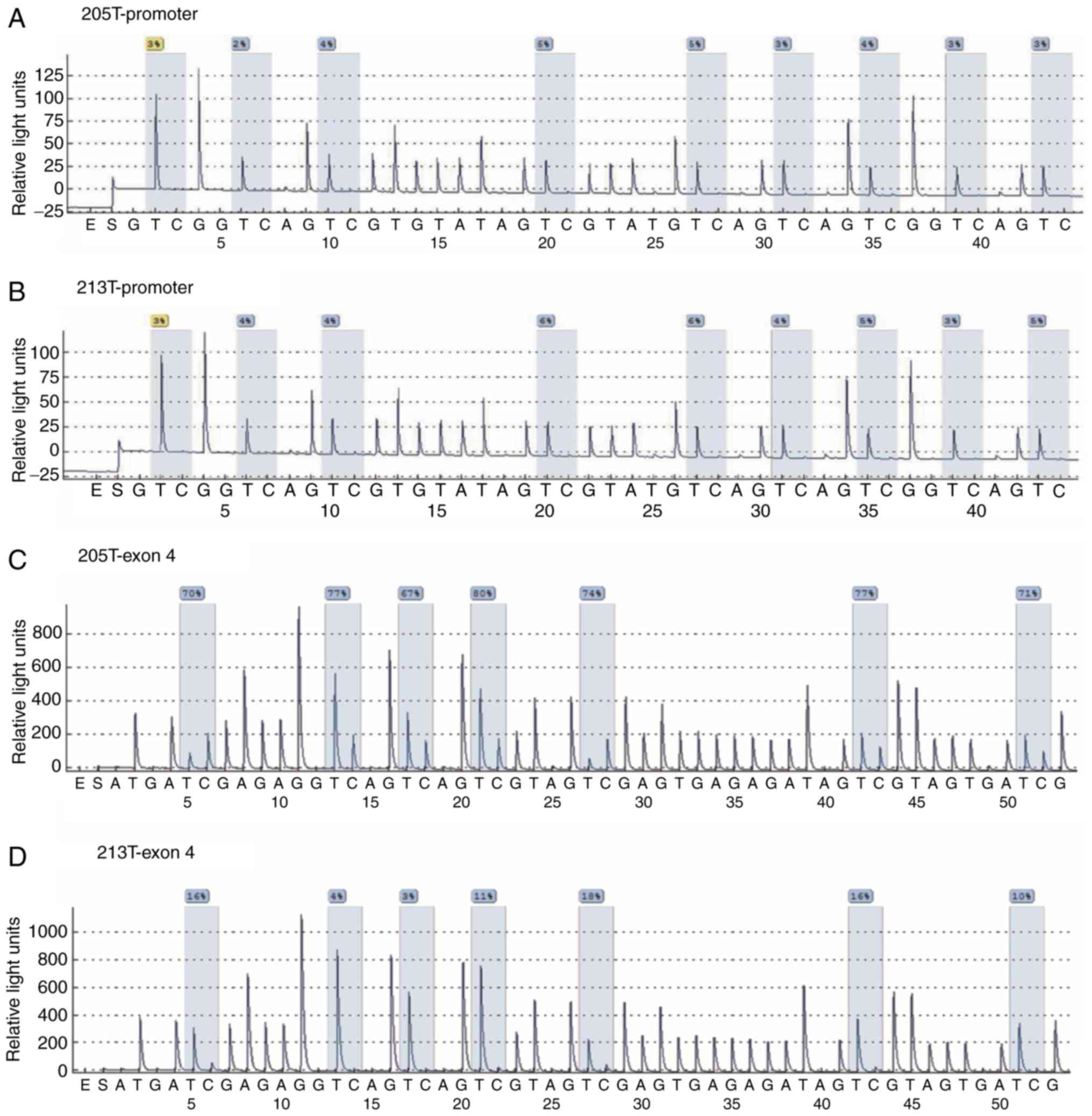
FOSB gene were designed, respectively. Representative
examples of pyrosequencing are presented in Fig. 1 and revealed that non-CpG cytosine
residues were converted to thymine, indicating the completeness of
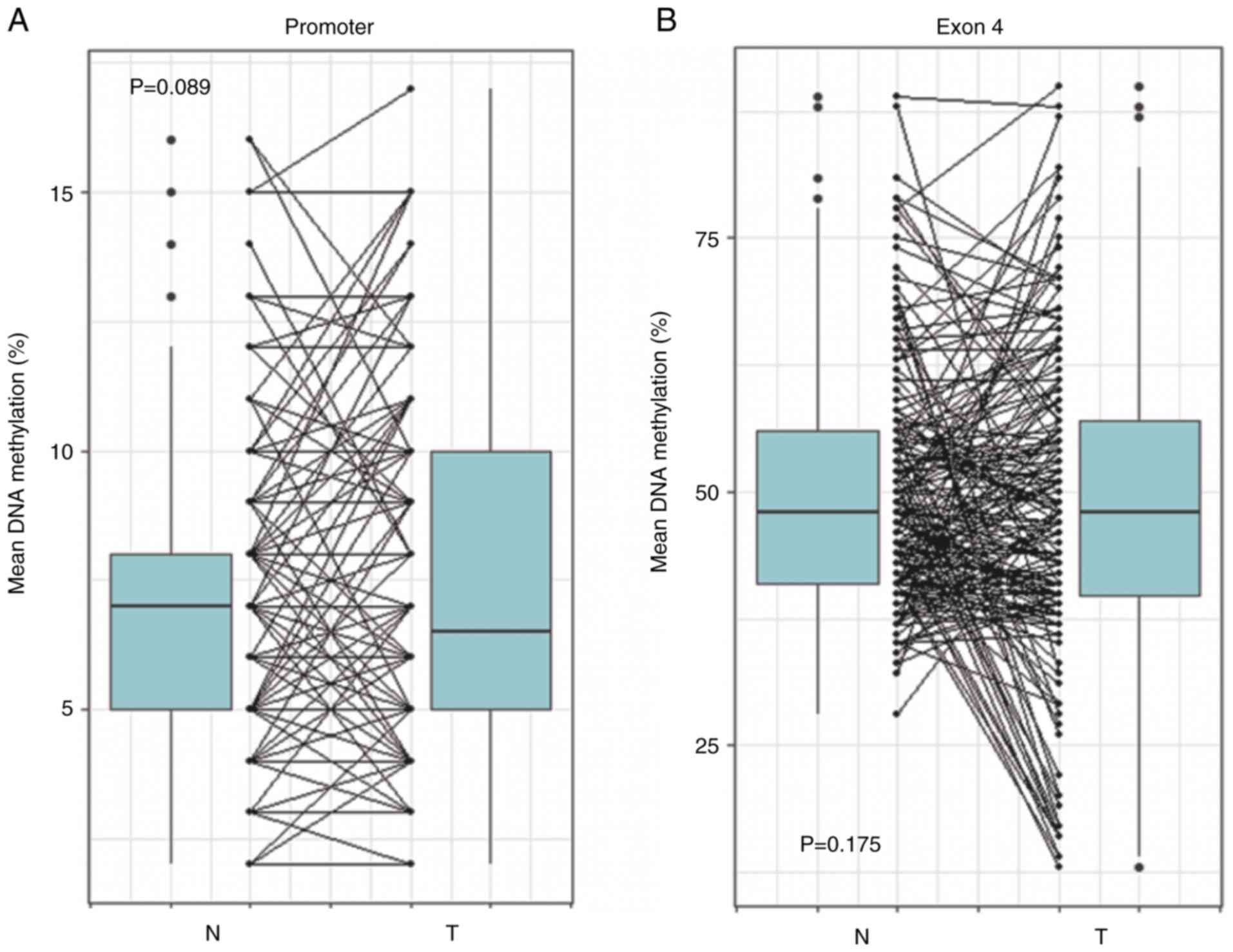
bisulfite treatment. The present results revealed that there were
no statistically significant differences of the mean DNA
methylation level of FOSB promoter (P=0.089) and exon 4
(P=0.175) between tumor and matched normal tissues (Fig. 2). The CGI of the FOSB
promoter region was completely unmethylated in all nonmalignant and
malignant lung tissues. However, compared to the MI of normal
tissues, the DNA methylation level of the FOSB exon 4 region
was markedly (>2-folds) decreased in 18 (10.2% of the total)
tumor tissues, indicating that the unmethylation of FOSB
exon 4 may be a tumor-associated event during NSCLC
tumorigenesis.
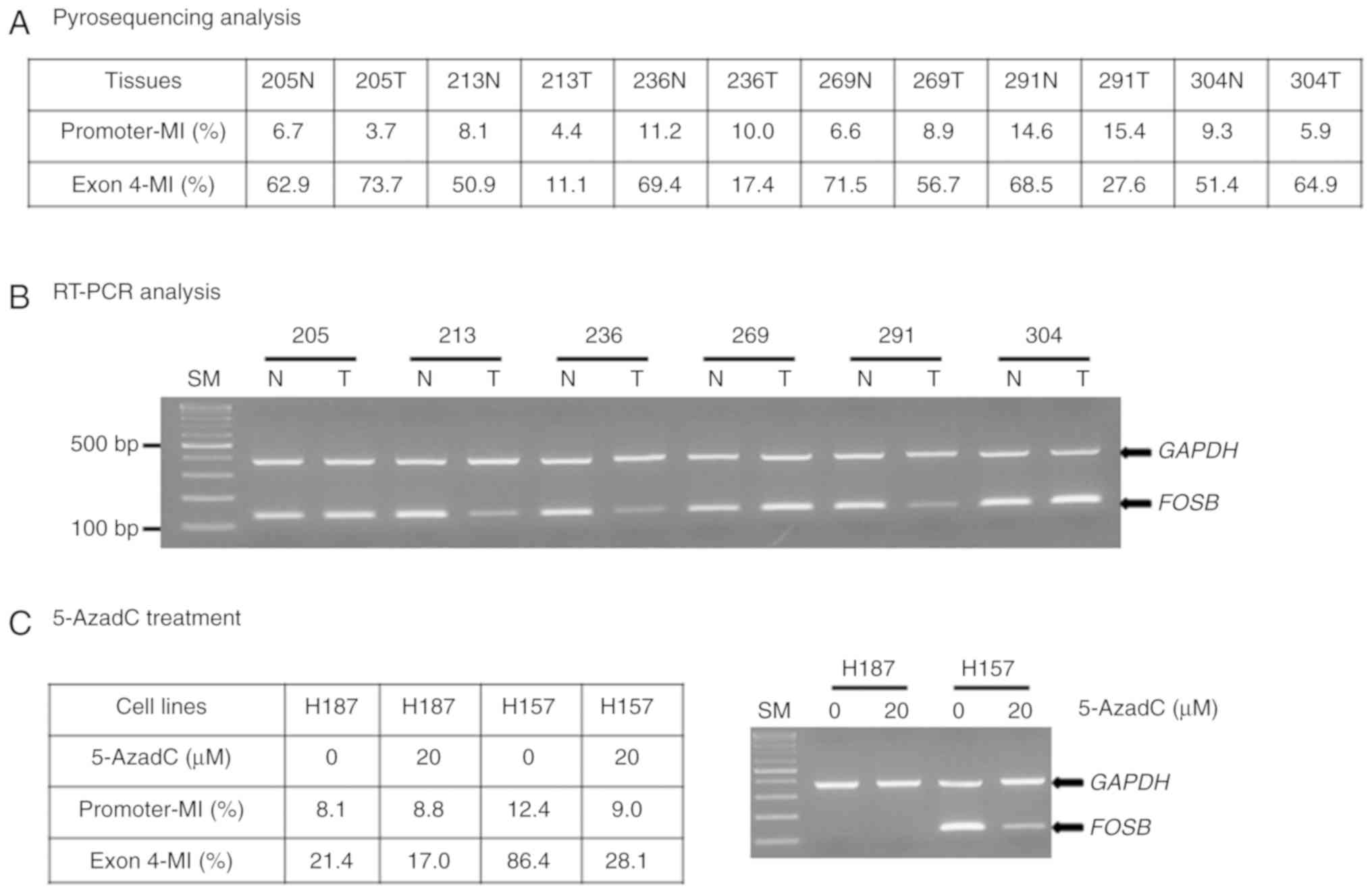
It was then investigated whether the unmethylation
of FOSB exon 4 could regulate its mRNA expression in
representative lung tissue specimens. Methylation status combined
with RT-PCR results revealed low or undetectable levels of
FOSB transcripts in tumor tissues with unmethylated alleles
(213T, 236T and 291T), and high FOSB levels in tumor and
non-tumor lung tissues with methylated alleles (Fig. 3A and B). This observation was
further substantiated by 5-AzadC treatment of NSCLC cell lines.
FOSB mRNA was absent in H187 cells with unmethylated exon 4,
and present in H157 cells with methylated alleles; notably,
FOSB expression decreased upon 5-AzadC treatment (Fig. 3C). Collectively, these results
indicated that exon 4 hypomethylation may be indicative of
FOSB gene silencing.
Association of FOSB methylation status
with clinicopathological parameters and survival outcome
Unmethylated FOSB exon 4 was more frequent in
patients with lymph node metastasis than in those without
metastasis with a borderline significance (17.4 vs. 7.7%, P=0.062;
Table I). However, no significant
association was revealed in other clinicopathological factors, such
as age, sex, smoking status, histology, and pathologic stage
(Table I). The methylation status
of the FOSB exon 4 in adenocarcinoma and squamous cell
carcinoma, respectively, was specifically analyzed (Table SII).
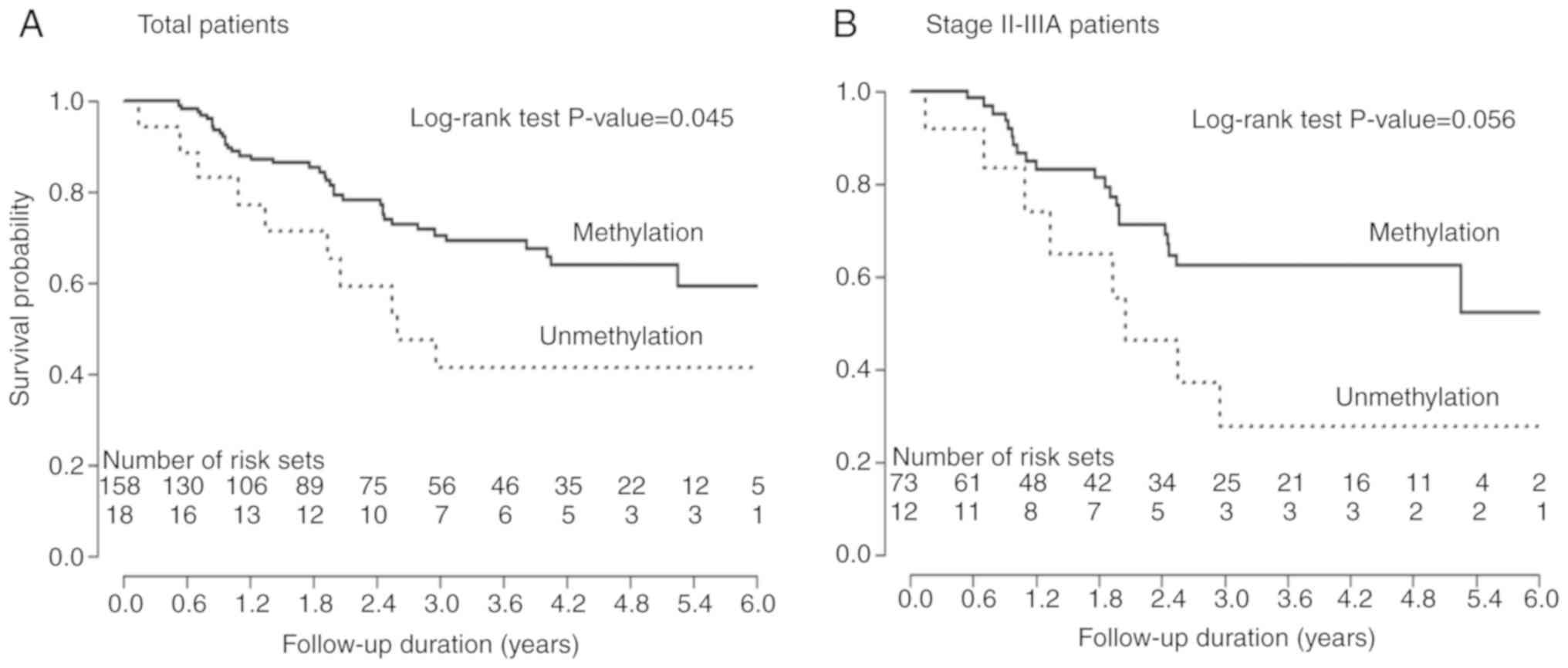
Next, Kaplan-Meier survival analysis was performed
to determine the prognostic potential of FOSB methylation
status. The patients with unmethylated FOSB had
significantly shorter OS than those with FOSB methylation
(PL-R=0.045, Fig. 4A).
After stratification, according to the clinicopathological
features, FOSB unmethylation was markedly associated with an
unfavorable OS in patients with stage II–IIIA
(PL-R=0.056; Fig. 4B).
To evaluate whether FOSB unmethylation is an independent
prognostic predictor in NSCLC, the data was further analyzed using
the Cox proportional hazards regression model after adjusting for
possible cofounding variables of survival. Multivariate analysis
revealed that FOSB unmethylation was associated with poor
survival in patients with stage II–IIIA [adjusted hazard ratio
(HR)=2.43, 95% confidence interval (CI)=1.04–5.68, P=0.040], but
not in all subjects (Table
II).
 | Table II.Overall survival according to
methylation status of FOSB genes in NSCLC patients. |
Table II.
Overall survival according to
methylation status of FOSB genes in NSCLC patients.
| Variables | No. of cases | No. of deaths
(%)a | 5-year survival
rate (%)b |
PLRc | Adjusted HR (95%
CI) | P-value |
|---|
| All subjects | 176 | 47 (26.7) | 61.2 |
|
|
|
|
Methylation | 158 | 37 (23.4) | 64.2 | 0.045 | 1 | 0.250 |
|
Unmethylation | 18 | 10 (55.6) | 41.7 |
| 1.53
(0.74–3.16)d |
|
| Stage I |
|
|
|
|
|
|
|
Methylation | 85 | 16 (18.8) | 65.8 | 0.812 | 1 | 0.905 |
|
Unmethylation | 6 | 2 (33.3) | 66.7 |
| 0.91
(0.20–4.12)e |
|
| Stage II–IIIA |
|
|
|
|
|
|
|
Methylation | 73 | 21 (28.8) | 62.5 | 0.056 | 1 | 0.040 |
|
Unmethylation | 12 | 8 (66.7) | 27.8 |
| 2.43
(1.04–5.68)e |
|
Discussion
The present study revealed that unmethylation of
FOSB exon 4 was detected in 18 (10.2%) out of the 176 NSCLC
cases and its unmethylation was related to loss of FOSB mRNA
expression. The genomes of cancer cells tend to show widespread
gene body hypomethylation alongside hypermethylation of gene
promoter (25). Numerous studies
have investigated the diagnostic and prognostic values of promoter
methylation (5). However, data on
the role and nature of gene body methylation in specific genes are
currently limited. Thus, the present results represent the first
demonstration that the gene body (exon 4) region of the FOSB
gene can undergo a DNA methylation alteration in NSCLC patients.
Similarly, Suzuki et al revealed that increased DNA
methylation in the gene body led to the upregulation of FOSB
in liver tumors of arsenic-exposed mice (24). DNA methylation in promoter sequences
is well known to silence genes and is the presumed therapeutic
target of methylation inhibition (26). Notably, DNA methylation is more
prevalent within gene bodies than appears for promoters (27,28),
and gene body methylation appears to be actively involved in
multiple gene regulation processes including alternative promoter
usage, regulation of short and long non-coding RNAs, alternative
splicing and enhancer activity (29,30),
indicating a contrasting association between DNA methylation and
expression across the promoter (negative association) vs. gene body
(positive association).
One notable finding of the present study is the
association of FOSB methylation changes with survival
outcomes in a subset of patients with NSCLC, indicating that
FOSB may be a new biomarker for the prognosis of NSCLC.
Moreover, the present results provide evidence that FOSB may
possess tumor-suppressive properties in NSCLC. These observations
are in line with a study revealing that FOSB is an anti-metastatic
protein in lung cancer because it negatively regulates MMP9
expression, which induces cell migration and invasion (21). Notably, the present data revealed
that the unmethylation of FOSB exon 4 tends to occur in
patients with lymph node metastases. FOSB expression was
significantly downregulated in gastric and pancreatic cancer
tissues and its downregulation was associated with reduced survival
(18,19). FOSB overexpression also
triggered cell death by regulating the expression of MMP9 in MCF
breast cancer cells (31).
Collectively, the present investigation indicated that FOSB exerts
a tumor-suppressive function in NSCLC. The mechanism of its action
requires future investigations. However, FOSB may be used as a
novel biomarker or a specific therapeutic target for lung
cancer.
Some of the limitations of this study include its
retrospective design and the relatively small sample size,
indicating that the results may be influenced by selection bias.
Thus, further large-scale studies and longer follow-up periods are
required to verify the clinical significance of FOSB
expression. Collectively, it was revealed that the decrease in DNA
methylation levels of the FOSB exon 4 was associated with
decreased expression of FOSB mRNA in tumor tissues and with
unfavorable outcomes of patients with later stages of lung cancer.
It is anticipated that accumulating knowledge on gene body
methylation will provide valuable information in the future,
especially to develop effective tools for DNA methylation-targeted
therapy.
Supplementary Material
Supporting Data
Acknowledgements
We would like to thank the National Biobank of
Korea, KNUH for providing patient material and data.
Funding
The present study was supported by the Basic Science
Research Program through the National Research Foundation of Korea
(NRF) (No. NRF-2016R1D1A1B03931462).
Availability of data and materials
The accompanying data underlying our findings are
available upon request to corresponding author.
Authors' contributions
DSK and JYP drafted the manuscript, conceived and
coordinated the study, and were responsible for interpretation of
the data. WKL performed the statistical analyses. All authors have
read and approved the final manuscript and agree to be accountable
for all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
The present study was conducted with the approval of
IRB, KNUH (approval no. 2014-04-210). Participants provided their
written informed consent to participate in this study and for the
publication of all associated data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Spiro SG and Silvestri GA: One hundred
years of lung cancer. Am J Respir Crit Care Med. 172:523–529. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yano T, Haro A, Shikada Y, Maruyama R and
Maehara Y: Non-Small cell lung cancer in never smokers as a
representative ‘non-smoking-associated lung cancer’: Epidemiology
and clinical features. Int J Clin Oncol. 16:287–293. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bird A: DNA methylation patterns and
epigenetic memory. Genes Dev. 16:6–21. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jankowska AM, Millward CL and Caldwell CW:
The potential of DNA modifications as biomarkers and therapeutic
targets in oncology. Expert Rev Mol Diagn. 15:1325–1337. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu F and Zhang HT: DNA methylation and
nonsmall cell lung cancer. Anat Rec (Hoboken). 294:1787–1795. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Seok Y, Lee WK, Park JY and Kim DS: TGFBI
promoter methylation is associated with poor prognosis in lung
adenocarcinoma patients. Mol Cells. 42:161–165. 2019.PubMed/NCBI
|
|
8
|
Tessema M, Yingling CM, Liu Y, Tellez CS,
Van Neste L, Baylin SS and Belinsky SA: Genome-Wide unmasking of
epigenetically silenced genes in lung adenocarcinoma from smokers
and never smokers. Carcinogenesis. 35:1248–1257. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shaulian E and Karin M: AP-1 as a
regulator of cell life and death. Nat Cell Biol. 4:E131–E136. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ting CH, Chen YC, Wu CJ and Chen JY:
Targeting FOSB with a cationic antimicrobial peptide, TP4, for
treatment of triple-negative breast cancer. Oncotarget.
7:40329–40347. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Na HH, Noh HJ, Cheong HM, Kang Y and Kim
KC: SETDB1 mediated FosB expression increasing the cell
proliferation rate during anticancer drug therapy. BMP Rep.
49:238–243. 2016. View Article : Google Scholar
|
|
12
|
Milde-Langosch K: The fos family of
transcription factors and their role in tumorigenesis. Eur J
Cancer. 41:2449–2461. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barrett CS, Millena AC and Khan SA: TGF-β
effects on prostate cancer cell migration and invasion require
FosB. Prostate. 77:72–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cervantes-Madrid DL, Nagi S and Asting
Gustafsson A: FosB transcription factor regulates COX-2 expression
in colorectal cancer cells without affecting PGE2 expression. Oncol
Lett. 13:1411–1416. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shahzad MMK, Arevalo JM, Armaiz-Pena GN,
Lu C, Stone RL, Moreno-Smith M, Nishimura M, Lee JW, Jennings NB,
Bottsford-Miller J, et al: Stress effects on FosB- and
interleukin-8 (IL8)-driven ovarian cancer growth and metastasis. J
Biol Chem. 285:35462–35470. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Renaud SJ, Kubota K, Rumi MA and Soares
MJ: The FOS transcription factor family differentially controls
trophoblast migration and invasion. J Biol Chem. 289:5025–5039.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rowther FB, Wei W, Dawson TP, Ashton A,
Singh A, Madiesse-Timchou MP, Thomas DG, Darling JL and Warr T:
Cyclic nucleotide phosphodiesterase-1C (PDE1C) drives cell
proliferation, migration and invasion in glioblastoma multiform
cells in vitro. Mol Carcinog. 55:268–279. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang C, Jiang Y, Shao W, Shi W, Gao X, Qin
W, Jiang T, Wang F and Feng S: Abnormal expression of FOSB
correlates with tumor progression and poor survival in patients
with gastric cancer. Int J Oncol. 49:1489–1496. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim JH, Lee JY, Lee KT, Lee JK, Lee KH,
Jang KT, Heo JS, Choi SH and Rhee JC: RGS16 and fosB underexpressed
in pancreatic cancer with lymph node metastasis promote tumor
progression. Tumour Biol. 31:541–548. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Milde-Langosch K, Kappes H, Riethdorf S,
Loning T and Bamberger AM: FosB is highly expressed in normal
mammary epithelia, but down-regulated in poorly differentiated
breast carcinomas. Breast Cancer Res Treat. 77:265–275. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang LQ, Zhao LH and Qiao YZ:
Identification of potential therapeutic targets for lung cancer by
bioinformatics analysis. Mol Med Rep. 13:1975–1982. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim DS, Lee WK and Park JY:
Hypermethylation of normal mucosa of esophagus-specific 1 is
associated with an unfavorable prognosis in patients with non-small
cell lung cancer. Oncol Lett. 16:2409–2415. 2018.PubMed/NCBI
|
|
23
|
Anier K, Malinovskaja K, Aonurm-Helm A,
Zharkovsky A and Kalda A: DNA methylation regulates cocaine-induced
behavioral sensitization in mice. Neuropsychopharmacol.
35:2450–2461. 2010. View Article : Google Scholar
|
|
24
|
Suzuki T, Yamashita S, Ushijima T, Takumi
S, Sano T, Michikawa T and Nohara K: Genome-Wide analysis of DNA
methylation changes induced by gestational arsenic exposure in
liver tumors. Cancer Sci. 104:1575–1585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen ZX and Riggs AD: DNA methylation and
demethylation in mammals. J Biol Chem. 286:18347–18353. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kulis M, Heath S, Bibikova M, Queiros AC,
Navarro A, Clot G, Martinez-Trillos A, Castellano G, Brun-Health I,
Pinyol M, et al: Epigenomic analysis detects widespread gene-body
DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet.
44:1236–1242. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang X, Han H, De Carvalho DD, Lay FD,
Jones PA and Liang G: Gene body methylation can alter gene
expression and is a therapeutic target in cancer. Cancer Cell.
26:577–590. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kulis M, Queiros AC, Beekman R and
Martin-Subero JI: Intragenic DNA methylation in transcriptional
regulation, normal differentiation and cancer. Biochim Biophys
Acta. 1829:1161–1174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee SM, Choi WY, Lee J and Kim YJ: The
regulatory mechanisms of intragenic DNA methylation. Epigenomics.
7:527–531. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Esteller M: Dormant hypermethylated tumor
suppressor genes: Questions and answers. J Pathol. 205:172–180.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Milde-Langosch K, Roder H, Andritzky B,
Aslan B, Hemminger G, Brinkmann A, Bamberger CM, Loning T and
Bamberger AM: The role of the AP-1 transcription factors c-Fos,
FosB, Fra-1 and Fra-2 in the invasion process of mammary
carcinomas. Breast Cancer Res Treat. 86:139–152. 2004. View Article : Google Scholar : PubMed/NCBI
|