Introduction
Breast cancer (BC) is the most frequently diagnosed
cancer and the leading cause of cancer-related mortality among
women worldwide (1,2). Despite the improved prognosis of
individuals with BC and advances in radical surgery along with
adjuvant therapy (3,4), BC is associated with common recurrence
and persistently high mortality rates. BC frequently leads to
multi-organ distant metastasis, such as in the lung, bone, liver,
and brain (5). Studies are underway
to identify therapeutic targets for BC, but more research in this
area is needed.
Posttranscriptional modifications can influence the
activity and stability of cellular RNAs, and the most abundant of
these modifications is N6-methyladenosine (m6A) (6,7). In
many species including higher eukaryotes, m6A modification is
reported to be involved in mRNA splicing, translation, and
stability; these activities also occur in rRNA and miRNA (8). m6A has been implicated in the
development of numerous human diseases, including tumor formation
(9). m6A methylation is controlled
by the RNA methyltransferase complex (m6A ‘writers’), RNA
demethylases (m6A ‘erasers’), and m6A readers. The m6A ‘writer’ is
composed of Wilms tumor 1-associated protein (WTAP) (10), methyltransferase-like 3 and 14
(METTL3 and METTL14) (11), and protein virilizer homologue
(KIAA1429), while heterogeneous nuclear ribonucleoprotein
and YTH domain-containing RNA binding protein function as m6A
‘readers’ (12). AlkB homolog 5
(ALKBH5) and fat mass and obesity-associated protein (FTO) work as
m6A ‘erasers’ to remove it from the target region (13,14).
Cross-talk and dynamic equilibrium among m6A writers, readers, and
erasers play important roles in cancer development (15,16).
Current efforts focusing on m6A and tumor
development utilize advanced transcriptome-wide approaches for m6A
sequencing. It has been shown that the METTL3 level is
increased in lung carcinoma (LC) and modulates LC cell
proliferation, survival, and invasion by regulating translation of
target mRNA (17). Other research
has revealed that FTO regulates the progression of cervical
squamous cell carcinoma (CSCC) through mRNA demethylation (18). Additionally, ALKBH5 has been found
to maintain the glioblastoma stem-like cell tumorigenicity through
modulating FOXM1 (19). Both
METTL3 and METTL14 dramatically promote glioblastoma
stem cell growth, self-renewal, and tumorigenesis (20). All of this evidence indicates a role
for m6A and its enzyme system in tumor progression, but its
downstream factors and signaling pathways are still unclear.
MicroRNAs (miRNAs) are a group of small non-coding
RNAs, 21–24 nucleotides long, that are found in diverse organisms
and regulate mRNA expression (21).
Evidence indicates that m6A modification affects miRNA processing
and expression: altered METTL3/m6A may contribute to
abnormal miRNA expression in many biological processes, especially
in cancers (22). METTL3 is
reported to be modulated by HBXIP by inhibiting miRNA let-7g
in breast cancer (23). It has also
been shown that METTL14 suppresses the metastasis of
hepatocellular carcinoma by regulating m6A-dependent primary
microRNA processing (8). In the
present study, we aimed to identify the mechanism and function of
METTL14/m6A in BC cells, as well as describe the potential
effect of METTL14 on the miRNA expression profile and the
effect of hsa-miR-146a-5p modulated by METTL14 on the
function in BC cells.
Materials and methods
Study subjects and blood samples
We obtained preserved BC tissue samples and their
adjacent normal breast tissues from Nanjing Hospital (Nanjing,
Jiangsu, China); the BC patients underwent surgical treatment
between January 2017 and January 2018. Demographic and
clinicopathological information for these patients is documented in
Table I. This study was approved by
the Human Research Ethics Committee of Nanjing Medical University
(Nanjing, Jiangsu, China). All patients gave their written informed
consent according to the Declaration of Helsinki, and all
experimental procedures were conducted following its
guidelines.
 | Table I.Demographic and clinicopathological
information of the 58 BC patients. |
Table I.
Demographic and clinicopathological
information of the 58 BC patients.
|
|
| METTL14
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | No. of
patients | Low n (%) | High n (%) | P-value |
|---|
| Age (years) |
|
|
| 0.5992 |
|
<60 | 28 | 15 (53.5) | 13 (46.5) |
|
|
≥60 | 30 | 14 (46.7) | 16 (53.3) |
|
| Typing |
|
|
| 0.9389 |
| Luminal
A | 4 | 2 (50) | 2 (50) |
|
| Luminal
B | 33 | 16 (39.4) | 17 (60.6) |
|
|
HER-2+ | 9 | 5 (55.6) | 4 (44.4) |
|
|
Basal-like | 12 | 5 (41.7) | 7 (58.3) |
|
| Microvascular
invasion |
|
|
| 0.4297 |
|
Yes | 27 | 12 (44.4) | 15 (55.6) |
|
| No | 31 | 17 (54.8) | 14 (45.2) |
|
| Nerve invasion |
|
|
| 0.6489 |
|
Yes | 15 | 7 (46.7) | 8 (53.3) |
|
| No | 43 | 23 (53.5) | 20 (46.5) |
|
| TNM |
|
|
| 0.0483a |
|
T1+T2 | 54 | 30 (55.6) | 24 (44.4) |
|
| T3 | 4 | 0 | 4 (100) |
|
| Pathological
stage |
|
|
| 0.0827 |
|
I+II | 47 | 22 (46.8) | 25 (53.2) |
|
|
III | 11 | 2 (18.2) | 9 (81.8) |
|
| WHO |
|
|
| 0.0768 |
|
I+II | 27 | 12 (44.4) | 15 (55.6) |
|
|
III | 31 | 7 (22.6) | 24 (77.4) |
|
| Metastasis |
|
|
| 0.2389 |
|
pN0+pN1 | 47 | 22 (46.8) | 25 (53.2) |
|
|
pN2+pN3 | 11 | 3 (27.3) | 8 (72.7) |
|
RNA m6A quantification
Total RNA was extracted from tissues using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's manual. RNA quality was analyzed by NanoDrop (Thermo
Fisher Scientific, Inc.) and 1% agarose gel electrophoresis. The
EpiQuik™ m6A RNA Methylation Quantification Kit (P-9005-48,
Colorimetric, Epigentek, USA) was applied to analyze the m6A
content in high-quality RNAs. Briefly, 100–300 ng RNA was loaded
into test wells. Capture and detection antibody solutions were then
added to each test well separately in an appropriately diluted
concentration following the manufacturer's protocol. m6A levels
were determined according to the absorbance of each sample at 450
nm, and a standard curve was then used to calculate relative m6A
content.
Western blot analysis
Protein samples were taken from the cells lysed
using SDS (Beyotime). Total protein was determined using the Pierce
BCA assay kit (Thermo Fisher Scientific, Inc.). SDS-PAGE gels (10%)
were used to separate the different proteins, and 15 µg total
protein was loaded per lane. Then, all protein bands were visible
on polyvinylidene fluoride (PVDF) membranes (Millipore). The
membranes were immersed in primary antibodies, anti-METTL14
(dilution 1:1,000; cat. no. Ab220030; Abcam) and anti-GAPDH
(dilution 1:1,000; cat. no. AF0006; Beyotime Institute of
Biotechnology) for 1 h at room temperature. Next, the membranes
were washed and incubated with HRP-conjugated goat anti-rabbit IgG
(dilution 1:1,000; cat. no. A0216; Beyotime Institute of
Biotechnology) for 2 h. Protein bands were visualized using ECL
chemiluminescence reagent (Thermo Fisher Scientific, Inc.) and
images were captured using the ChemiDoc MP system (Bio-Rad
Laboratories, Inc.). Immunoblot data were normalized to GAPDH
levels using ImageJ software (version 1.8.0; National Institutes of
Health, Bethesda, MD, USA).
Cell culture and transfection
BC cell lines including MCF-7 and MDA-MB-231 were
obtained from Zhongqiao Xinzhou Biotechnology Co. Ltd. Cells were
sub-cultured in Dulbecco's modified Eagle's medium (DMEM)
(Zhongqiao Xinzhou Biotechnology Co. Ltd.) mixed with 10% Gibco
fetal bovine serum (FBS) (Thermo Fisher Scientific, Inc.), 100
mg/ml streptomycin (Procell), and 100 U/ml penicillin at 37°C in a
humidified atmosphere with 5% CO2. When the cell density
reached 80–90%, siRNAs (listed in Table II), hsa-miR-146a-5p inhibitor and
plasmids, purchased from GenePharma, were transfected using
Lipofectamine 3000 reagent (Thermo Fisher Scientific, Inc.) into
MCF-7 and MDA-MB-231 cells, respectively. In addition,
3-deazaadenosine (m6A inhibitor) was purchased from Sigma-Aldrich
(Merck KGaA). All procedures followed the manufacturer's
instructions.
 | Table II.Primers and sequences. |
Table II.
Primers and sequences.
| Primers | Sequences
(5′-3′) |
|---|
| METTL14-F |
AAATGCTGGACTTGGGATGATA |
| METTL14-R |
CCCATTTTCGTAAACACACTCTT |
| METTL3-F |
GAACACTGCTTGGTTGGTGTCA |
| METTL3-R |
TGGAACGAACCTCAGCTACGAT |
| WTAP-F |
TTCCCAAGAAGGTTCGATTGAG |
| WTAP-R |
AGACTCCTGCTGTTGTTGCTTTAG |
| KIAA1429-F |
CGATAACTTGATGACCCCAGAA |
| KIAA1429-R |
ATAACGGCAAGATTCCATTTC |
|
hsa-miR-146a-5p-F |
GGCGGTGAGAACTGAATTCC |
|
hsa-miR-29b-3p-F |
CGCAGTAGCACCATTTGAAAT |
|
hsa-miR-3691-5p-F |
CGCAGAGTGGATGATGGAGAC |
|
hsa-miR-221-3p-F |
GGCGGAGCTACATTGTCTGC |
| hsa-miR-4326-F |
GGGCGGTGTTCCTCTGTCT |
| all-miR-R |
AGTGCGTGTCGTGGAGTCG |
| B2M-F |
CTCTTTCTGGCCTGGAGGCTAT |
| B2M-R |
AGTCAACTTCAATGTCGGATGGAT |
| U6-F |
CGATACAGAGAAGATTAGCATGGC |
| U6-R |
AACGCTTCACGAATTTGCGT |
| Si-METTL14 |
UCUUAUCCAACCUUUCUUCCG |
Cell function as assessed by Transwell
assay
For the cell migration assay, 1×105 MCF-7
or MDA-MB-231 cells were loaded into the upper chamber inserts
(Millipore) with 500 µl of serum-free DMEM. The lower chambers
contained 700 µl DMEM with 10% FBS. After 24–48 h, the cells
underneath the membrane were stained with 800 µl crystal violet
(cat. no. C0121; Beyotime Institute of Biotechnology) at room
temperature for 30 min and counted using a light microscope in 3
random high-power fields. For the invasion experiment, chambers
precoated with matrix adhesive (Corning, USA) were applied. All
experiments were repeated three times.
CCK-8 assay
Cell proliferation abilities of the MCF-7 or
MDA-MB-231 cells were evaluated by Cell Counting Kit-8 assays.
Cells were transfected as mentioned above. Post-transfection at 48
h, the cells were digested, resuspended into single-cell
suspension, seeded into 96-well plates with 100 µl of cells per
well (2×104 cell/ml), and incubated at 37°C with 5%
CO2. CCK-8 reagent (10 µl) was added to per well after
incubation at 0, 24, 48, 72 and 96 h, and incubated for 1 h for
each time point. Absorbance was measured via a microplate reader
(Bio-Rad Laboratories) at 450 nm.
Small RNA library construction and
sequencing
Total RNA was extracted from the MCF-7 and
MDA-MB-231 cells, and then quantitated and purified. RNAs of high
quality were used to establish small RNA libraries with the
NEBNext® Multiplex Small RNA Library Prep Kit following
the manufacturer's instructions (New England Biolabs). A Hiseq2500
(Illumina, USA) was then used for small RNA sequencing.
Data filtering and mapping
The raw data were filtered and optimized with FastQC
(http://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
(24) to discard short (<15 nt)
and low-quality reads. The small RNA clean reads were then matched
to the miRBase21.0 (http://www.mirbase.org/) (25) to screen out known miRNAs (20–25 bp),
and unmapped reads were further scheduled to piRNAcluster
(http://www.smallrnagroup.uni-mainz.de/piRNAclusterDB.html).
The matched reads were then compared with the National Center for
Biotechnology Information database (https://www.ncbi.nlm.nih.gov/) to find known piRNAs.
Unmapped reads were then assigned to Genomic tRNA Database
(GtRNAdb, http://gtrnadb.ucsc.edu/) and tRFdb
(http://genome.bioch.virginia.edu/trfdb/) to define
tRNA derived fragments (tRFs). Finally, the remaining unmatched
reads were allocated to Rfam (http://rfam.xfam.org/) to identify small nucleolar RNA
(snoRNA) as well as small nuclear RNA (snRNA) sequences. All
identifiable small RNAs were separated and defined.
Differentially expressed miRNA
identification and target prediction
Differentially expressed (DE) miRNAs were filtered
with Ebseq2.0 packages (26).
|Log2(fold change) | >1 and false discovery rate
(FDR) <0.05 were considered to be significantly different.
Miranda and RNAhybrid were applied to determine DE miRNAs' target
mRNAs (27). mRNAs that met both
Miranda (score ≥150 and energy <-20) and RNAhybrid (energy
<-25) cutoffs were considered to be the assumed DE mRNAs.
Finally, the miRNA/mRNA interaction network was diagrammed using
Cytoscape software (28) (version
3.5.1).
Functional enrichment analysis of
target genes
Target genes screened using the database for
Annotation, Visualization, and Integrated Discovery 6.8
Bioinformatics Tool (DAVID 6.8) (29) were then submitted for Gene Ontology
(GO, http://www.geneontology.org/) (30) analysis and Kyoto Encyclopedia of
Genes and Genomes (KEGG, http://www.kegg.jp/) pathway enrichment analysis
(31). GO analyses were divided
into cellular component (CC), biological process (BP), and
molecular function (MF) analysis. KEGG pathway enrichment analysis
was conducted to explore the biological pathways in which the DE
miRNA target mRNAs were potentially involved. The cutoff threshold
was set to P-value <0.05.
METTL14 and miRNA expression as
verified by real-time quantitative PCR (RT-qPCR)
Total RNA was reverse-transcribed into complementary
DNA (cDNA) following the manufacturer's instructions
(ThermoScientific™ RevertAidAM First Strand cDNA
Synthesis kit, cat. #K1622; Thermo Fisher Scientific, Inc.). qPCR
was conducted using the ABI Q6 detection system (Applied Biosystems
Inc.; Thermo Fisher Scientific, Inc.). The delta-delta Cq method
(2−ΔΔCq) (32) was used
to measure the relative content of the miRNAs. Primers were ordered
from Sangon Biotech, and primer sequences are displayed in Table II. B2M and U6 were used as the
internal control for Mettl14 and miRNA, respectively.
Statistical analysis
SPSS 19.0 (IBM Corp.) was used to perform
statistical analysis. The P-values of the clinical indexes were
calculated using Chi-square test. The paired Student's t- test was
used for METTL14 level in 20 paired BC and control tissues.
The unpaired Student's t-test was applied for two-group comparison
of the other assays, and one-way analysis of variance (ANOVA)
followed by Tukey's multiple comparison test was used when more
than two groups were evaluated. P-value <0.05 was considered as
indicative of a statistically significant difference.
Results
m6A methyltransferase METTL14 (also
known as methyltransferase like 14 and
N6-adenosine-methyltransferase non-catalytic subunit) is aberrantly
expressed in BC tissues
Evidence indicates that there is an aberrant
N6-methyladenosine level in BC (23). We assumed that abnormal m6A
modification in BC was due to abnormal expression of m6A
methyltransferases. To test this hypothesis, the mRNA levels of
METTL3, METTL14, WTAP, and KIAA1429 in 20 paired BC
and paracancerous tissue samples were determined by qPCR. Results
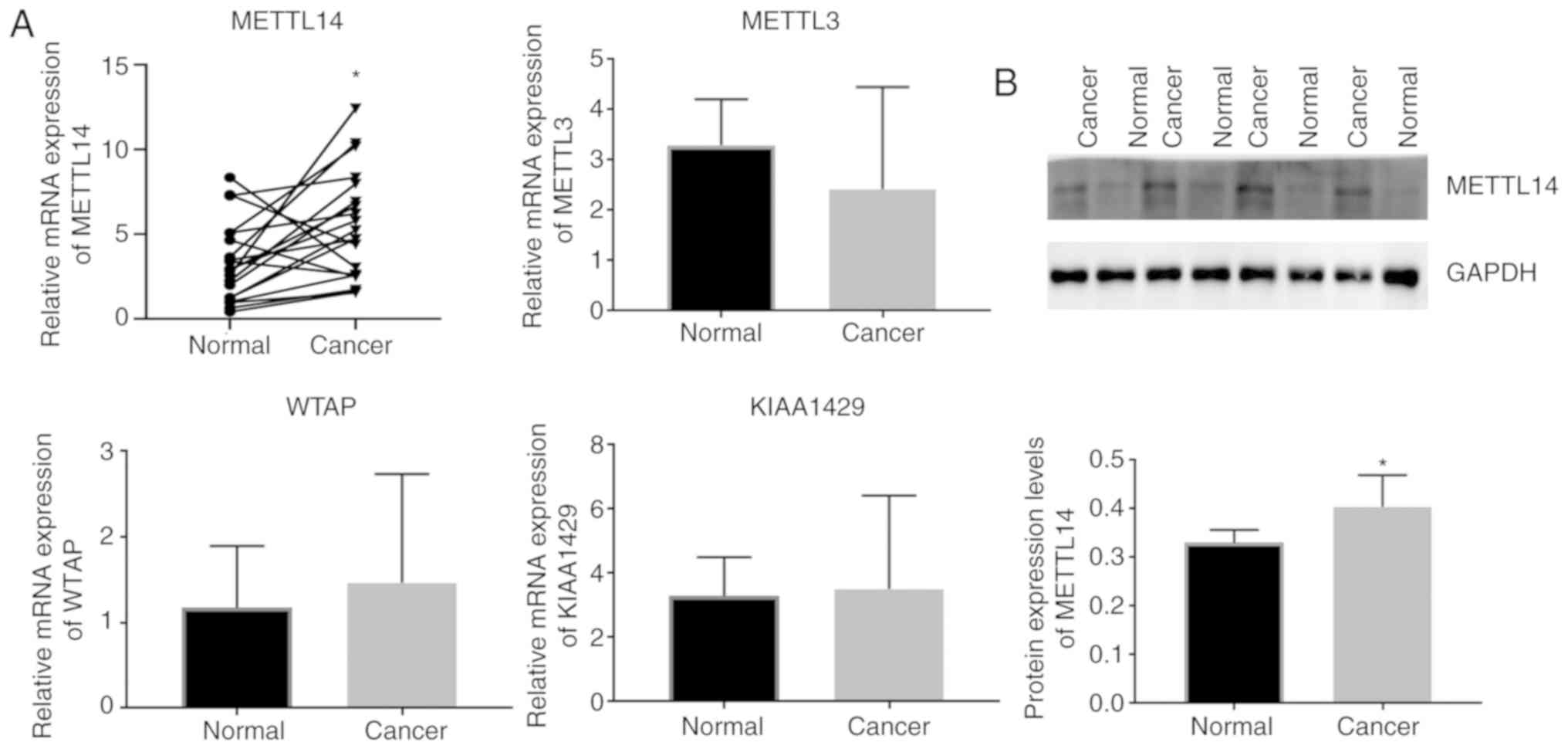
revealed that METTL14 expression was significantly increased in BC,
while no significant difference was observed in the expression of
METTL3, WTAP and KIAA1429 (Fig. 1A). Moreover, western blot analysis
showed that the METTL14 protein level was significantly increased
in BC tissues compared to that in the normal tissue (Fig. 1B). After we analyzed the demographic
and clinicopathological information of the BC patients in Table I, we found that there was no
difference between the METTL14 low and high expression
samples with regard to age, typing, microvascular invasion, nerve
invasion status, pathological stage, WHO stage or metastasis
status, while METTL14 expression was related to TNM
grade.
METTL14 modulates the m6A modification
in BC cells
METTL14 has been reported to regulate m6A
formation; therefore, we inferred that aberrant METTL14
expression may play an important role in m6A modifications in BC
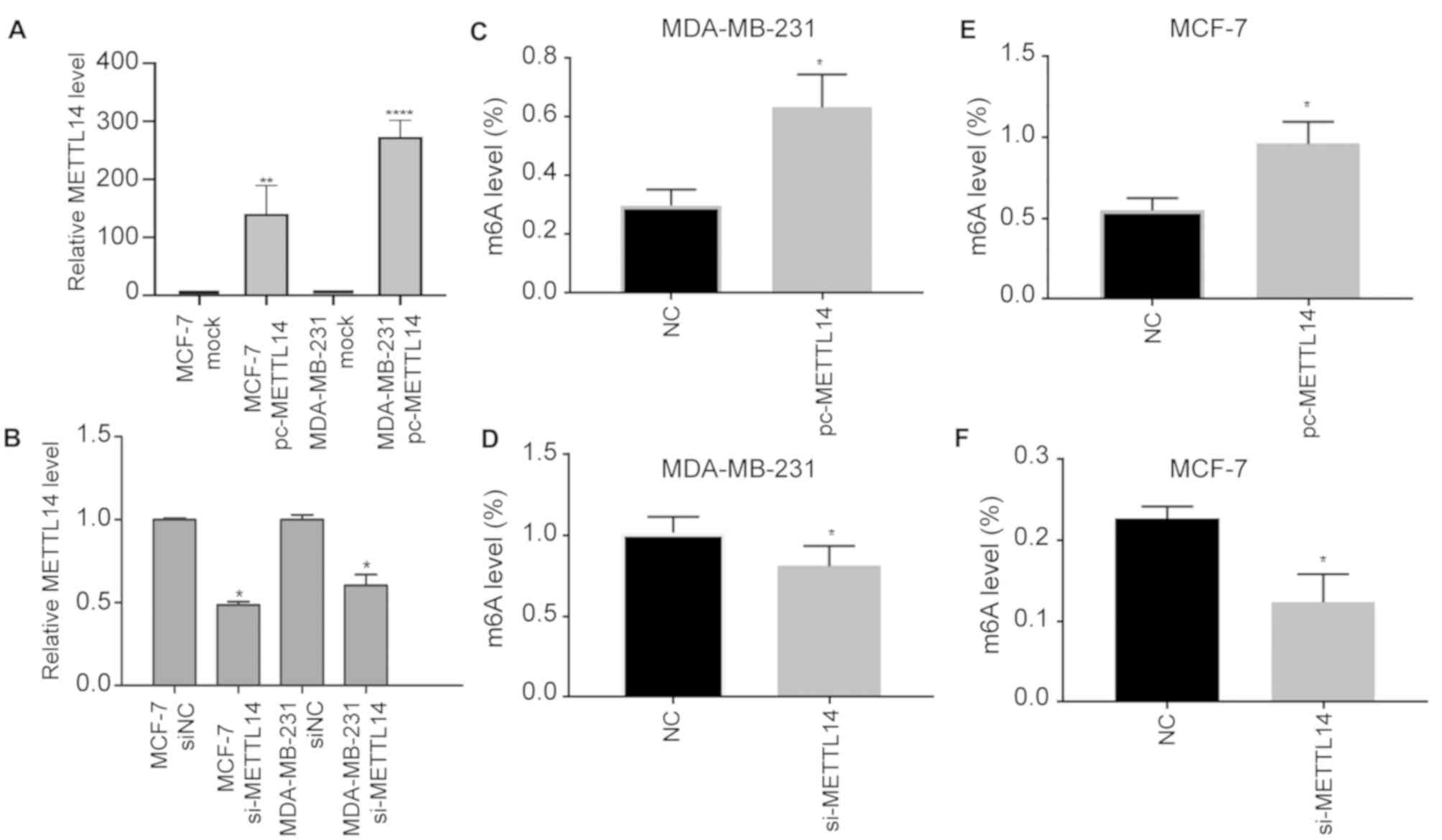
cells. We transfected METTL14 siRNA into MCF-7 and
MDA-MB-231 cells to create METTL14 knockdown cell lines
(MCF-7 si-METTL14 and MDA-MB-231 si-METTL14; Fig. 2A), and we cloned METTL14 into
the pcNDA3.1 vector to achieve overexpressing cell lines (MCF-7
pc-METTL14 and MDA-MB-231 pc-METTL14; Fig. 2B). As expected, METTL14
overexpression increased m6A levels, and knockdown resulted in
decreased m6A levels in the MDA-MB-231 (Fig. 2C and D). Consistently,
METTL14 overexpression increased m6A levels, and knockdown
resulted in decreased m6A levels in the MCF7 cells (Fig. 2E and F). After METTL14
overexpression and knockdown, the obtained results revealed that
METTL14 upregulated the m6A levels in MDA-MB-231 and MCF7
cells. Therefore, METTL14 modulates the m6A modification in
BC.
Through regulation of m6A modification
METTL14 enhances migration and invasion capacity of BC cells
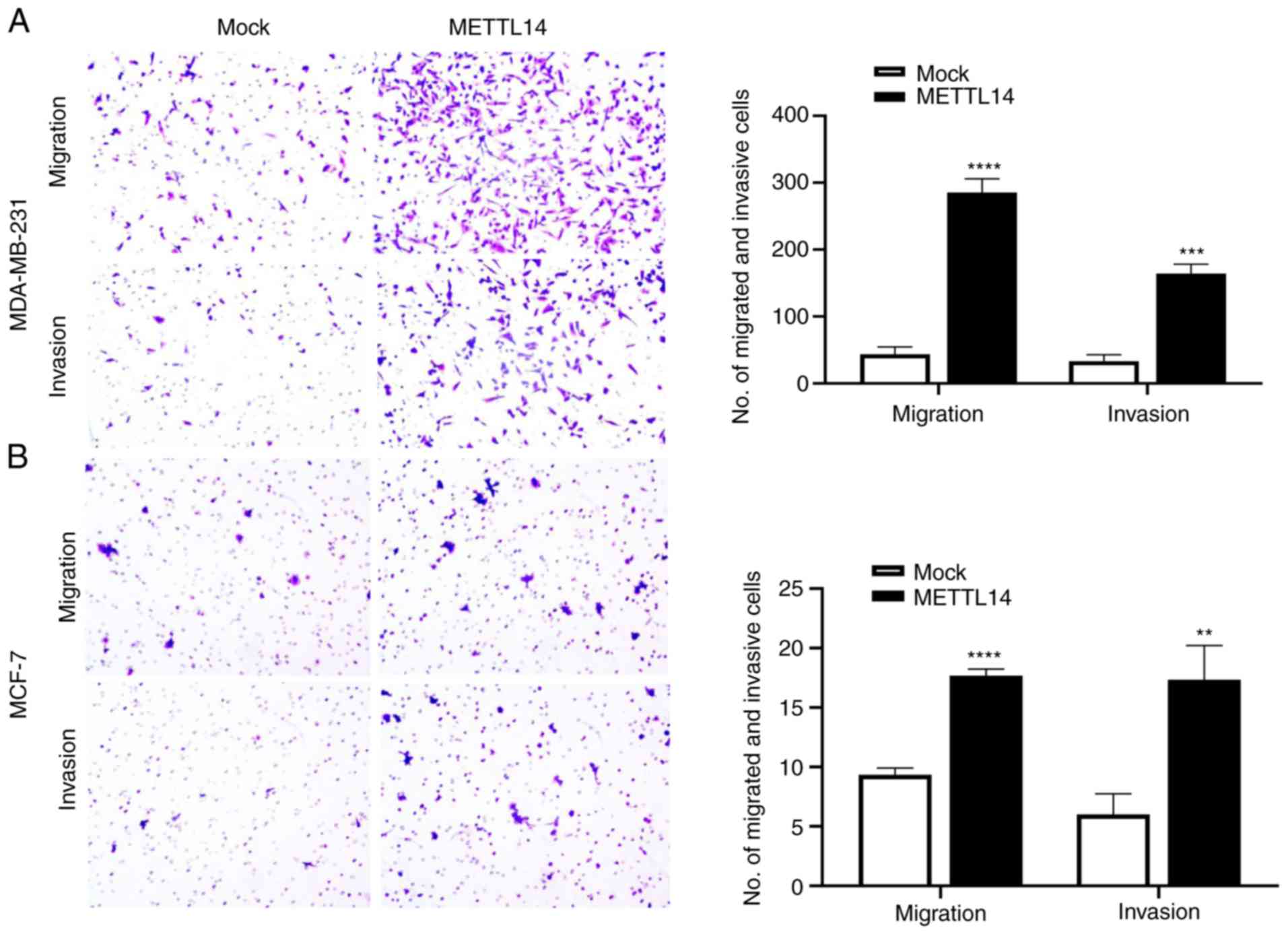
We next assessed the effect of the aberrant
expression of METTL14 on BC cell migration and invasion. The
BC cells were subsequently transfected with METTL14
overexpression or control vector for functional assays. METTL14
overexpression promoted the migration and invasion of the
MDA-MB-231 and MCF-7 cells (Fig.
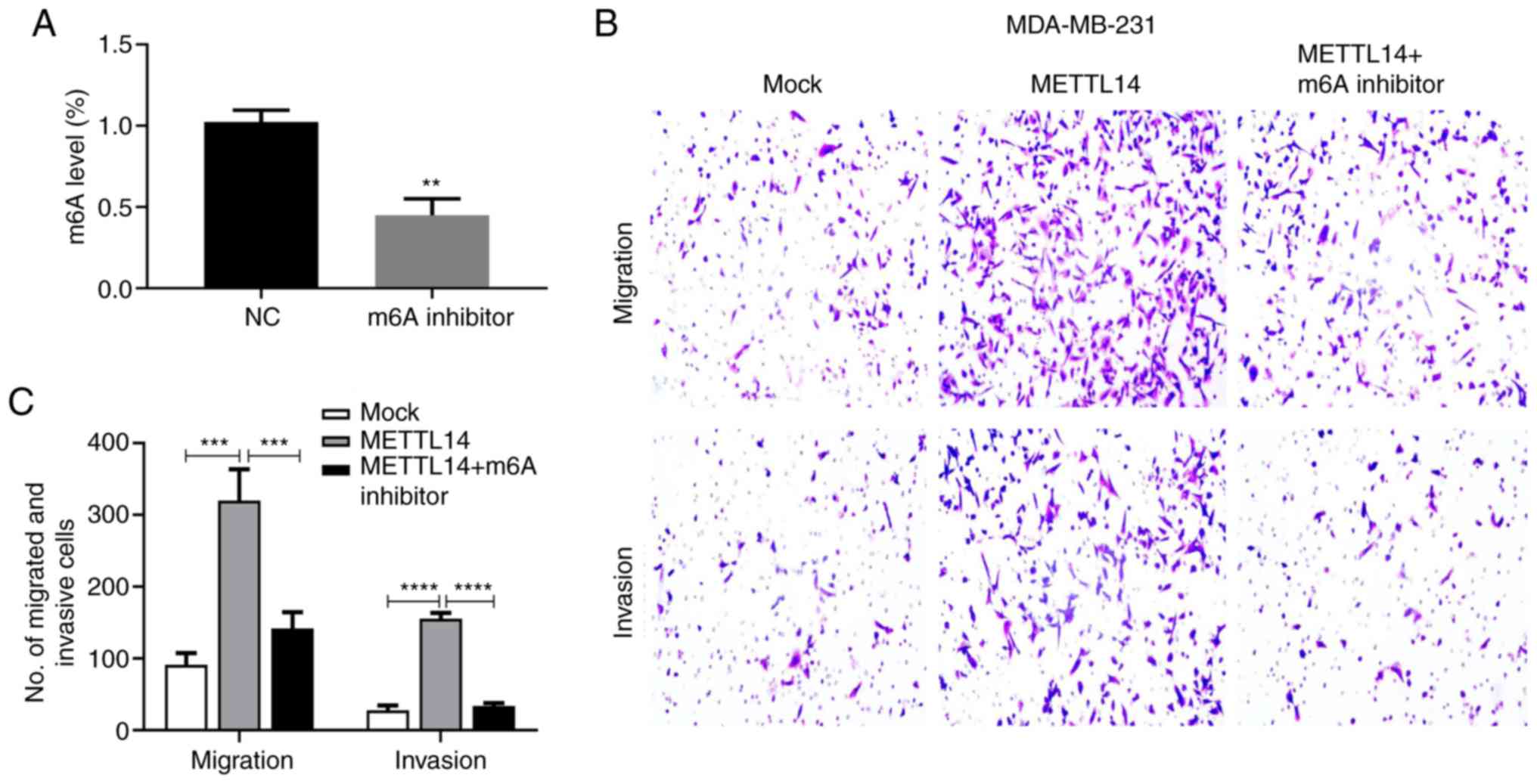
3). Moreover, it was confirmed that the m6A inhibitor
significantly inhibited the m6A level in the MDA-MB-231 cells
(Fig. 4A). With the m6A inhibitor
treatment, the enhanced effect of METTL14 on cell migration and
invasion was suppressed (Fig. 4B and
C), indicating that METTL14 may act as a positive
regulator of BC function. We also tested the effect of
METTL14 on cell proliferation using the CCK-8 assay. Results
revealed that METTL14 gain-of-expression had little effect
on cell proliferation in the MCF-7 and MDA-MB-231 cells (Fig. S1).
METTL14 overexpression reshapes the
miRNA profiling of BC cells
To explore the effect of abnormal expression of
METTL14 on miRNA processing and expression, we conducted
miRNA sequencing on METTL14 overexpression and mock groups
of MCF-7 and MDA-MB-231 cells. In cells overexpressing
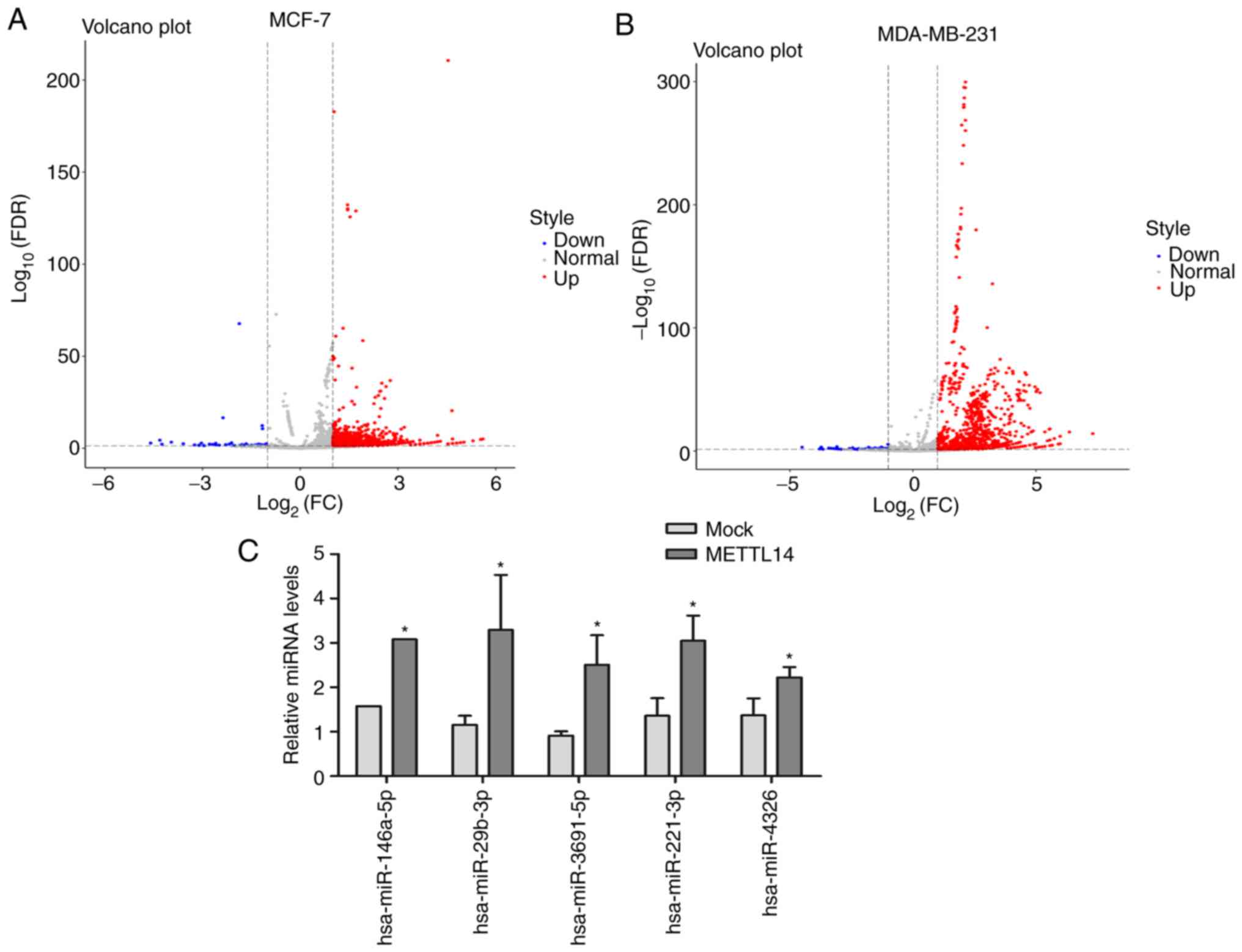
METTL14, the number of upregulated miRNAs was higher than
that of the downregulated miRNAs in both cell lines (Fig. 5A and B). Specifically, 685 miRNAs
were upregulated and only 65 were downregulated in the MDA-MB-231
cells, and 52 miRNAs were upregulated and 14 were downregulated in
the MCF-7 cells. Moreover, 34 DE miRNAs showed the same
upregulated/downregulated tendency between the MCF-7 and MDA-MB-231
cell lines with METTL14 overexpression; of these, 33 miRNAs
were upregulated and 1 miRNA was downregulated. Expression of 5
miRNAs including hsa-miR-146a-5p, hsa-miR-29b-3p, hsa-miR-3691-5p,
hsa-miR-221-3p, and hsa-miR-4326 was verified by RT-qPCR (Fig. 5C), and the results of these
expression levels of miRNAs were in accordance with miRNA
sequencing data.
Functional enrichment and miRNA-mRNA
interaction network analysis reveal the DE miRNAs targeting BC
development
To evaluate the DE miRNA targeting mRNAs in the
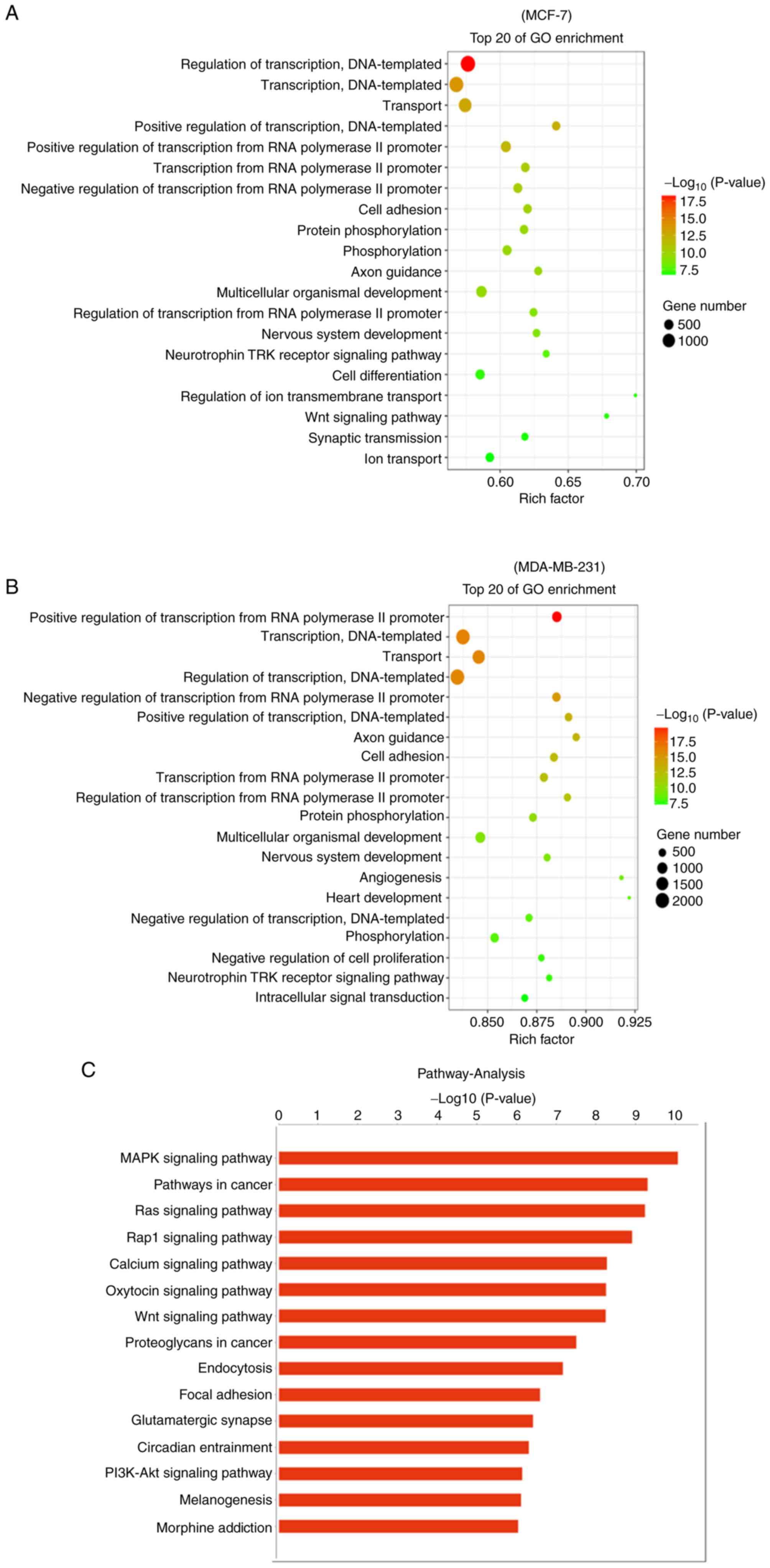
MCF-7 and MDA-MB-231 cells during METTL14 overexpression, GO and
KEGG pathway enrichment analyses were performed. The target genes
of DE miRNAs were mainly enriched in ‘regulation of transcription’,
‘cell adhesion’, ‘protein phosphorylation’, ‘phosphorylation’,
‘axon guidance’, ‘nervous system development’, and ‘neurotrophin
TRK receptor signaling pathway’ in both the MCF-7 and MDA-231 cell
lines. In addition, ‘angiogenesis’, ‘cell differentiation’ and
‘negative regulation of cell proliferation’, which are related to
BC development, were also enriched in the TOP20 GO terms (Fig. 6A and B). Moreover, KEGG pathway
enrichment analysis revealed that the DE miRNAs target mRNAs
enriched in KEGG terms such as ‘MAPK signaling pathway’, ‘Pathways
in cancer’, ‘Ras signaling pathway’, ‘PI3K-Akt signaling pathway’,
‘Focal adhesion’, ‘Wnt signaling pathway’, and ‘Rap1 signaling
pathway’ were also closely related to the occurrence and
development of cancer (Fig. 6C and
D). All GO and KEGG analyses indicated that aberrant
METTL14 expression altered the miRNA expression profile,
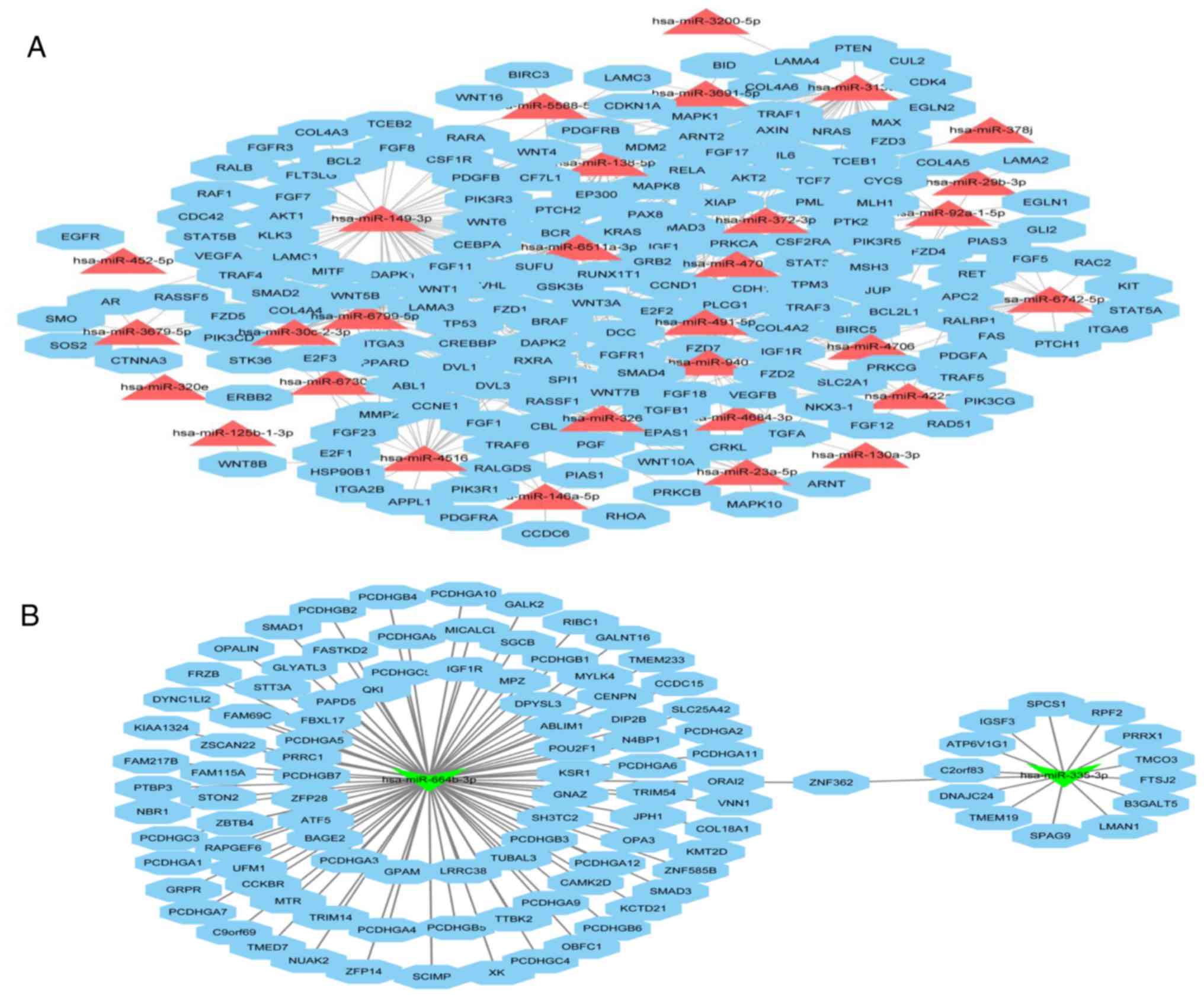
adjusting and controlling BC cell activities. We utilized Cytoscape
software to depict an integrated mRNA/miRNA interaction network
that included 30 upregulated miRNAs (Fig. 7A) and 2 downregulated (Fig. 7B) miRNAs and their targets involved
in ‘Pathways in cancer’. In this complex network, upregulated
hsa-miR-149-3p and downregulated hsa-554b-3p were shown to have
more targets that related to the cancer pathway. In addition,
hsa-miR-146a-5p, hsa-miR-29b-3p and hsa-miR-3691-5p appeared in the
network, and their expression was confirmed by RT-qPCR.
hsa-miR-146a-5p modulated by METTL14
promotes migration and invasion capacity of BC cells
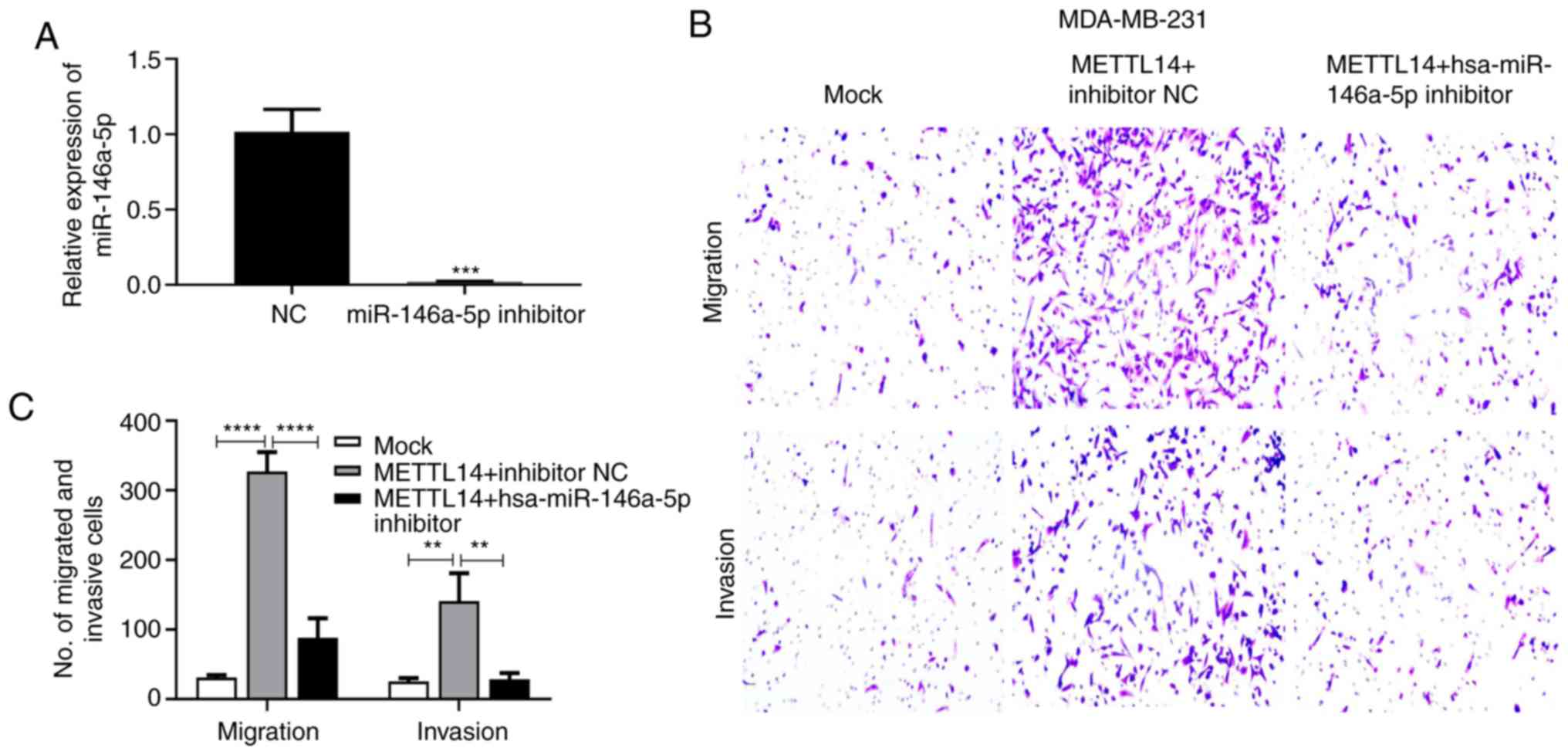
We next evaluated the effect of hsa-miR-146a-5p
modulated by METTL14 on migration and invasion of BC cells.
First, we confirmed that the hsa-miR-146a-5p inhibitor
significantly inhibited hsa-miR-146a-5p expression in the
MDA-MB-231 cells (Fig. 8A). The
METTL14 overexpression vector and hsa-miR-146a-5p inhibitor
as well as the negative control were subsequently transfected into
MDA-MB-231 cells. Overexpression of METTL14 promoted the
migration and invasion capacity of MDA-MB-231 cells, while
treatment with the hsa-miR-146a-5p inhibitor suppressed this
enhanced cell migration and invasion ability (Fig. 8B and C).
Discussion
Early studies have shown that the epigenetic
regulation of N6-methyladenosine (m6A) is involved in many
post-transcriptional regulatory processes, similarly to DNA histone
modifications. Aberrant m6A is usually associated with the
progression of disease and cancer. The present study is the first
to show that aberrant expression of RNA methyltransferase
METTL14 (methyltransferase-like 14) modulates the
development and the level of m6A in breast cancer (BC), and to
reveal that microRNA processing and expression is determined in a
N6-methyladenosine-dependent manner. This introduces the idea that
the relationship between m6A modification and miRNA processing is
important in BC development.
With the development of whole-transcriptome m6A
sequencing approaches, the functions of m6A RNA modification in
cancer biology have been rapidly elucidated. RNA methyltransferase
METTL3 or METTL14 and RNA demethylase fat mass and
obesity-associated protein (FTO) are critical for glioblastoma stem
cell (GSC) self-renewal and tumorigenesis (20). METTL3 promotes bladder cancer
proliferation and metastasis via AFF4/NF-κB/MYC signaling (33). Additionally, 70% of endometrial
tumors exhibit downregulated m6A methylation that may be due to
METTL14 mutation or reduced METTL3 expression; these
abnormalities in RNA methyltransferases promote proliferation and
tumorigenicity of endometrial cancer via AKT pathway
activation (16). Reductions in m6A
methylation lead to decreased expression of the negative AKT
regulator PHLPP2 and increased expression of the positive
AKT regulator mTORC2. However, the expression of RNA
methyltransferase in BC, as well as its role and mechanism in
cancer development, remain unclear. Our study revealed for the
first that aberrant METTL14 expression is related to BC cell
migration and invasion. We also tested the overexpression of
METTL14 on BC cell growth using the CCK-8 assay, but
METTL14 gain-of-expression had little effect on cell
proliferation in MCF-7 cells. As shown in the Gene Ontology (GO)
functional analysis, differentially expressed (DE) miRNAs were
enriched in cell adhesion terms in both MCF-7 and MDA-MB-231 cells.
Metastasis formation is a complex process, and requires the
epithelial-mesenchymal transition (EMT) (34). During EMT, cell-to-cell and
cell-to-matrix adhesion molecules are crucial for cells to break
free from the primary tumor mass and enter the bloodstream to cause
metastasis formation (35). Here,
this evidence indicates that METTL14 may control BC cell
meta-stasis by regulating miRNA levels that affect these adhesion
molecules, and should be further verified in vitro and in
vivo. This will be our future research direction.
Studies have shown that m6A modification is involved
in transcriptome regulation in disease and cancer states. m6A
modification has potential implications for miRNA biogenesis, as
well as for mRNA-miRNA interactions (36). For instance, METTL14 is
associated with hepatocellular carcinoma metastasis, and its
depletion was found to inhibit miR-126 expression by modulating
DGCR8 binding in an m6A-dependent manner (37). In our results, we were surprised to
find that METTL14 overexpression led to increased expression
of most miRNAs, especially in the cancer-related
pathway-miRNA-mRNAs interaction network: 30 miRNAs were
upregulated, and 2 miRNAs were downregulated. We hypothesize that
increased m6A expression may promote pri-miRNA processing, thus
leading to increased miRNA expression; this lays the groundwork for
future studies.
The miRNA-mRNA interaction network that we
investigated is related to the cancer pathway. hsa-miR-146a-5p,
hsa-miR-29b-3p, and hsa-miR-3691-5p were present in the network and
were expressed as confirmed by RT-qPCR. miR-146a-5p functions as a
tumor suppressor in a variety of types of cancer (38,39),
its overexpression promotes BC cell proliferation, and it could
serve as a new prognostic marker for HER2-positive BC (40). In the present study, miR-146a-5p
increased with METTL14 overexpression, which is in
accordance with previous reports. Here, overexpression of
METTL14 and hsa-miR-146a-5p expression modulated by
METTL14 promoted BC cell migration and invasion in
vitro. Whether hsa-miR-146a-5p modulated by METTL14
promotes the metastatic potential of BC cells need to be further
identified in vivo, which will be the focus of our future
research. hsa-miR-146a-5p was enriched in cell adhesion terms
through GO functional analysis, suggesting that hsa-miR-146a-5p
modulated by METTL14 is associated with metastasis.
In conclusion, abnormal m6A modification in BC was
found to be due to the abnormal expression of the m6A
methyltransferase METTL14. METTL14 gain- and
loss-of-expression was found to regulate m6A levels, and
METTL14 overexpression regulated m6A levels to enhance the
migration and invasion capacity of MCF-7 and MDA-MB-231 cells. The
miRNA/mRNA network was most enriched in the cancer-related
pathways, and DE miRNAs were found to be enriched in cell adhesion
terms. Moreover, hsa-miR-146a-5p modulated by METTL14
promoted cell migration and invasion. Thus, METTL14
modulates m6A modification and hsa-miR-146a-5p expression, and
further promotes cell migration and invasion of breast cancer
cells. Taken together, these results indicate that METTL14
may modulate the metastatic potential of breast cancer cells.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Nanjing Medical
Science and Technology Development Project (YKK16093) and the
Fundamental Research Funds for the Central Universities
(14380389).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
Conceptualization of the study and funding
acquisition was achieved by JS. Data collection was carried out by
DY, RW and XS. Investigation and analysis of the research data was
conducted by DY, RW, XS, LX and YY. Preparation of the original
draft was carried out by DY, RW, XS and LX. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
This study was approved by the Human Research Ethics
Committee of Nanjing Medical University (Nanjing, Jiangsu, China).
All the patients gave written informed consent according to the
Declaration of Helsinki, and all experimental methods were
conducted following relevant guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nechuta SJ, Caan BJ, Chen WY, Lu W, Chen
Z, Kwan ML, Flatt SW, Zheng Y, Zheng W, Pierce JP and Shu XO: Soy
food intake after diagnosis of breast cancer and survival: An
in-depth analysis of combined evidence from cohort studies of US
and Chinese women. Am J Clin Nutr. 23:123–132. 2012. View Article : Google Scholar
|
|
3
|
Zhang T, Li J, He Y, Yang F, Hao Y, Jin W,
Wu J, Sun Z, Li Y, Chen Y, et al: A small molecule targeting
myoferlin exerts promising anti-tumor effects on breast cancer. Nat
Commun. 9:37262018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Turner NC, Finn RS, Martin M, Im SA,
DeMichele A, Ettl J, Diéras V, Moulder S, Lipatov O, Colleoni M, et
al: Clinical considerations of the role of palbociclib in the
management of advanced breast cancer patients with and without
visceral metastases. Ann Oncol. 29:669–680. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Echeverria GV, Powell E, Seth S, Ge Z,
Carugo A, Bristow C, Peoples M, Robinson F, Qiu H, Shao J, et al:
High-resolution clonal mapping of multi-organ metastasis in triple
negative breast cancer. Nat Commun. 9:50792018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Machnicka MA, Milanowska K, Osman Oglou O,
Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S,
Dunin-Horkawicz S, Rother KM, et al: MODOMICS: A database of RNA
modification pathways-2013 update. Nucleic Acids Res. 41 (Database
issue):D262–D267. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Frye M, Harada BT, Behm M and He C: RNA
modifications modulate gene expression during development. Science.
361:1346–1349. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee M, Kim B and Kim VN: Emerging roles of
RNA modification: m(6)A and U-tail. Cell. 158:980–987. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao X, Chen Y, Mao Q, Jiang X, Jiang W,
Chen J, Xu W, Zhong L and Sun X: Overexpression of YTHDF1 is
associated with poor prognosis in patients with hepatocellular
carcinoma. Cancer Biomark. 21:859–868. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Li Y, Toth JI, Petroski MD, Zhang
Z and Zhao JC: N6-methyladenosine modification destabilizes
developmental regulators in embryonic stem cells. Nat Cell Biol.
16:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu Y, Dominissini D, Rechavi G and He C:
Gene expression regulation mediated through reversible
m6A RNA methylation. Nat Rev Genet. 15:293–306. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mathiyalagan P, Adamiak M, Mayourian J,
Sassi Y, Liang Y, Agarwal N, Jha D, Zhang S, Kohlbrenner E,
Chepurko E, et al: FTO-Dependent m6A Regulates Cardiac Function
During Remodeling and Repair. Circulation. 139:518–532. 2018.
View Article : Google Scholar
|
|
14
|
Shen DD, Suo FZ, Song QM, Chang J, Zhang
T, Hong JJ, Zheng YC and Liu HM: Development of formaldehyde
dehydrogenase-coupled assay and antibody-based assays for ALKBH5
activity evaluation. J Pharm Biomed Anal. 162:9–15. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Panneerdoss S, Eedunuri VK, Yadav P,
Timilsina S, Rajamanickam S, Viswanadhapalli S, Abdelfattah N,
Onyeagucha BC, Cui X, Lai Z, et al: Cross-talk among writers,
readers, and erasers of m6A regulates cancer growth and
progression. Sci Adv. 4:eaar82632018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu J, Eckert MA, Harada BT, Liu SM, Lu Z,
Yu K, Tienda SM, Chryplewicz A, Zhu AC, Yang Y, et al:
m6A mRNA methylation regulates AKT activity to promote
the proliferation and tumorigenicity of endometrial cancer. Nat
Cell Biol. 20:1074–1083. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin S, Choe J, Du P, Triboulet R and
Gregory R: The m(6)A methyltransferase METTL3 promotes translation
in human cancer cells. Mol Cell. 62:335–345. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R,
Wang YY and Zhe H: FTO regulates the chemo-radiotherapy resistance
of cervical squamous cell carcinoma (CSCC) by targeting
beta-catenin through mRNA demethylation. Mol Carcinog. 57:590–597.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S,
Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al: m6A
Demethylase ALKBH5 maintains tumorigenicity of glioblastoma
stem-like cells by sustaining FOXM1 expression and cell
proliferation program. Cancer Cell. 31:591–606.e596. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun
G, Lu Z, Huang Y, Yang CG, et al: m6A RNA methylation
regulates the self-renewal and tumorigenesis of glioblastoma stem
cells. Cell Rep. 18:2622–2634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hiro-Oki I and Yukihide T: The functions
of MicroRNAs: mRNA decay and translational repression. Trends Cell
Biol. 25:651–665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Alarcón CR, Lee H, Goodarzi H, Halberg N
and Tavazoie SF: N6-methyladenosine marks primary microRNAs for
processing. Nature. 519:482–485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang
Z, Liu Y, Zhang X, Zhang W and Ye L: HBXIP-elevated
methyltransferase METTL3 promotes the progression of breast cancer
via inhibiting tumor suppressor let-7g. Cancer Lett.
28:15652017.
|
|
24
|
Ramayo-Caldas Y, Mach N, Esteve-Codina A,
Corominas J, Castelló A, Ballester M, Estellé J, Ibáñez-Escriche N,
Fernández AI, Pérez-Enciso M and Folch JM: Liver transcriptome
profile in pigs with extreme phenotypes of intramuscular fatty acid
composition. BMC Genomics. 13:5472012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42((Database issue)): D68–D73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wright GW and Simon RM: A random variance
model for detection of differential gene expression in small
microarray experiments. Bioinformatics. 19:2448–2455. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar :
|
|
28
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hulsegge I, Kommadath A and Smits MA:
Globaltest and GOEAST: Two different approaches for Gene Ontology
analysis. BMC Proc. 3 (Suppl 4):S102009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H,
Wu M, Liang Y, Zhu F, Zhang Y, Zhang X, et al: The mA
methyltransferase METTL3 promotes bladder cancer progression via
AFF4/NF-κB/MYC signaling network. Oncogene. 38:3667–3680. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sökeland G and Schumacher U: The
functional role of integrins during intra- and extravasation within
the metastatic cascade. Mol Cancer. 18:122019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Massalha S and Weihs D: Metastatic breast
cancer cells adhere strongly on varying stiffness substrates,
initially without adjusting their morphology. Biomech Model
Mechanobiol. 16:961–970. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Erson-Bensan AE and Begik O: m6A
Modification and Implications for microRNAs. Microrna. 6:97–101.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH,
Wang F, Wang TT, Xu QG, Zhou WP and Sun SH: METTL14 suppresses the
metastatic potential of hepatocellular carcinoma by modulating N
-methyladenosine-dependent primary MicroRNA processing. Hepatology.
65:529–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang C, Zhang W, Zhang L, Chen X, Liu F,
Zhang J, Guan S, Sun Y, Chen P, Wang D, et al: miR-146a-5p mediates
epithelial-mesenchymal transition of oesophageal squamous cell
carcinoma via targeting Notch2. Br J Cancer. 118:e122018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hsieh JY, Huang TS, Cheng SM, Lin WS, Tsai
TN, Lee OK and Wang HW: miR-146a-5p circuitry uncouples cell
proliferation and migration, but not differentiation, in human
mesenchymal stem cells. Nucleic Acids Res. 41:9753–9763. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gao W, Hua J, Jia Z, Ding J, Han Z, Dong
Y, Lin Q and Yao Y: Expression of miR-146a-5p in breast cancer and
its role in proliferation of breast cancer cells. Oncol Lett.
15:9884–9888. 2018.PubMed/NCBI
|