Introduction
The ability to invade tissues and metastasize is the
major cause of cancer mortality (1–3).
Although advances in understanding cancer invasion and metastasis
have revealed several important potential molecular targets,
treatment of cancer metastasis is still limited. Metastasis occurs
through a series of interrelated steps of cell detachment,
migration, invasion, and adhesion, and is controlled by multiple
biochemical and signaling pathways. Metastatic processes have
recently been suggested to be significantly influenced by
microenvironmental factors, such as oxygen levels or pH.
Recent evidence has revealed that the pH of the
microenvironment of solid tumors is relatively acidic due to the
accumulation of lactic acid as a result of active aerobic and
anaerobic glycolysis (4). Such
environmental acidity has been reported to enhance malignant
transformation, invasion, and metastasis of several cancer cells.
Exposure of melanoma cells to acidic pH induced acid-resistant and
invasive phenotypes (5). Acidic
environments also enhanced local invasion and metastasis of human
colon cancer cells (6,7). In addition, acidity promoted the
malignant progression, induction of cancer stem cell phenotypes,
and the epithelial-mesenchymal transition in glioma and melanoma
cells (8,9). Although recent studies have revealed
that acidity upregulate transcription factors such as NF-κB and
Twist1 and is associated with alterations in adherence junction
proteins, including E-cadherin, p120-catenin, and β-catenin, the
mechanism by which acidity drives cancer cell invasion is not yet
fully understood (8,9).
Currently, multiple lines of evidence have revealed
that proteases are involved in cancer invasion and metastasis due
to their ability to cleave extracellular matrix components,
adhesion molecules, and cytokines (10). Kallikrein-related peptidases (KLKs)
are a subgroup of chymotrypsin-like serine proteases composed of
fifteen homologous members, KLKs 1-15. KLKs regulate diverse
essential physiological processes, and certain KLK members act as
signaling regulators that control cell functions through specific
membrane receptors, the protease-activated receptors (PARs)
(11–14). Notably, several members have been
reported to be overexpressed, and therefore, could serve as
potential biomarkers for diagnosis and prognosis, in various
cancers including ovarian, prostate, pancreatic and cervical cancer
(15,16). Therefore, KLKs have been extensively
studied for their use as cancer therapeutic targets (17). In the present study, it was revealed
that extracellular acidity markedly increased the expression of
KLK7 and KLK8 in gastric cancer cell lines and the role of these
enzymes in the acidity-mediated invasiveness in these cells was
investigated.
Materials and methods
Cell culture and pH adjustment
SNU-601 and AGS human gastric cancer (GC) cells were
obtained from the Korean Cell Line Bank and the American Type
Culture Collection, respectively. Cells were cultured in RPMI-1640
medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented
with 10% (v/v) fetal bovine serum and 1% PS at 37°C in an
atmosphere containing 5% CO2. For preparation of acidic
pH medium, pH 7.4 RPMI-1640 medium containing 1% FBS medium was
prepared, and then adjusted to pH 6.5 or 6.2 by adding 5M HCl
solution. Sulindac and NS398 were purchased from Calbiochem; EMD
Millipore. For Cox inhibition, cells were exposed to pH 7.4, 6.5
and 6.2 in the presence of 20 µM NS398 or 100 µM sulindac for 72 h,
and the treated cells were harvested for western blot analysis or
real-time PCR assay.
Invasion assay
To assay cell invasiveness, Matrigel-coated
Transwell chambers (Costar; Corning Incorporated) were used. Cells
in the logarithmic phase were incubated in RPMI-1640 medium
containing 1% FBS, adjusted to pH 7.4, 6.5 or 6.2, for 72 h, after
which the number of viable cells that corresponded to the equal
absorbance detected in a standard curve for a viability assay were
obtained from each pH condition and suspended in 1% FBS/RPMI-1640
medium adjusted to each pH condition (pH 7.4, 6.5 or 6.2). Then
~200 µl of the cell suspension was added to the upper portion of
the insert, and medium containing 5% FBS was added to the lower
portion of the inset. After 8 h (for AGS) and 18 h (for SNU601) of
incubation at 37°C in 5% CO2, noninvasive cells were
removed from the upper surface of the Transwell membrane with a
cotton swab, and the invaded cells on the lower layer surface were
fixed by 4% formaldehyde for 15 at RT and stained with 0.5% crystal
violet solution for 10 min RT. The numbers of invaded cells were
counted under high-power magnification (×200) or the images were
obtained by image analysis software (analysis FIVE; Olympus
Corporation).
Before the invasion assay, a standard curve for
viability in each pH medium was prepared since exposure to acidic
pH medium decreased the number of viable cells. The number of
viable cells was determined as follows: Cells cultured for 3 days
at each pH were trypsinized, resuspended in normal pH medium and
further incubated for the time required for invasion measurement,
and a standard curve for viability was prepared using an MTT assay.
Following MTT incubation, the purple formazan crystals were
dissolved using dimethyl sulfoxide and viability was subsequently
analyzed at a wavelength of 595 nm. Based on these survival curves,
the number of cells corresponding to the same absorbance was
calculated and used for this assay.
Western blot analysis
Treated cells were lysed in either a whole-cell
lysis buffer (50 mM Hepes, 150 mM NaCl, 1% Triton X-100, 5 mM EGTA,
protease inhibitor cocktail). Protein determination was performed
by BCA method. Equal amounts of protein extracts (50 µg of protein
per lane) were electrophoretically separated using 10–12% SDS-PAGE
and transferred to a nitrocellulose membrane using a standard
technique. Blocking was performed by incubation in 3% skim
milk/0.1%Tween Tris-buffered saline for 30 min at 4°C. Antibodies
were used to probe for KLK7 (dilution 1:500; cat. no. sc-514447;
Santa Cruz Biotechnology, Inc.), cyclooxygenase (COX)2 (dilution
1:200; cat. no. 160112; Cayman Chemical), KLK8 (dilution 1:1,000;
cat. no. ab150395; Abcam), COX1 (dilution 1:500; cat. no. ab695;
Abcam) and α-tubulin (dilution 1:2,000; clone no. B-5-1-2; cat. no.
32-2500; Invitrogen; Thermo Fisher Scientific, Inc.). The secondary
antibodies used were: Anti-rabbit HRP (dilution 1:2,500; cat. no.
sc-2030; Santa Cruz Biotechnology, Inc.), anti-mouse HRP (dilution
1:2,500; cat. no. sc-2031; Santa Cruz Biotechnology, Inc.) were
used. Anti-a-tubulin was used as a loading control. Protein bands
were visualized with the Super Signal West Pico chemiluminescence
kit (Pierce Biotechnology; Thermo Fisher Scientific, Inc.). Protein
blots were quantified using Alpha Ease FC software version 4.0
(Alpha Innotech). Signals were acquired using an Image Station
4000MM image analyzer (Kodak).
RNA interference (RNAi)
For the RNAi experiment, siRNAs of KLK7, KLK8, COX1
and COX2, and a scrambled siRNA control were purchased from Bioneer
Corporation. Cells were individually transfected with siRNA
oligonucleotides using an Amaxa™ Transfection System (Lonza
Bioscience) and grown for 24 h prior to exposure to acidic pH
medium.
Real-time reverse
transcription-polymerase chain reaction
Real-time PCR was performed with the Light Cycler
2.0 (Roche Diagnostics) using the Fast Start DNA Master SYBR-Green
I Kit (Roche Diagnostics). For verification of the correct
amplification product, PCR products were analyzed on a 2% agarose
gel stained with ethidium bromide. The sequences of the primers
were designed as follows: For β-actin, 5′-GACTATGACTTAGTTGCGTTA-3′
and 5′-GCCTTCATACATCTCAAGTTG-3′, for COX1,
5′-GGCGGGTACATTTCTCCATC-3′ and 5′-CCTCATGTTTGCCTTCTTTGC-3′, for
COX2, 5′-AACACAACAGAGTATGCGA-3′ and 5′-GTGTTAAATTCAGCAGCAATACG-3′.
Primers of KLK7 (product no. P106933), KLK8 (product no. P178536),
matrix metalloproteinase 7 (MMP7) (product no. P310408) and matrix
metalloproteinase 9 (MMP9) (product no. P323207) were purchased
from Bioneer Corporation. PCR was conducted at 95°C for 10 min,
followed by 45 cycles of 95°C for 15 sec, 60°C for 5 sec, and 72°C
for 7 sec. Melt curve analysis was performed to confirm that a
single product was present. Negative controls without template were
included in each run. Data were analyzed using Light Cycler
software version 4.0 (Roche Diagnostics). The 2−ΔΔCq
method was used for analysis of relative gene expression (18).
Detection of KLKs in cell culture
supernatants
Cells were incubated in RPMI-1640 medium containing
1% FBS, adjusted to pH 7.4, 6.5 or 6.2, for 72 h, after which
culture medium was obtained and release of KLK7 and KLK8 from cells
was assessed using a specific sandwich-type ELISA assay according
to established procedures with some modifications (19). Briefly, 96-well polystyrene plates
were coated with 500 ng/well of a specific capture antibody. After
overnight incubation, the plates were washed and 50 µl of culture
supernatant or standard and an equal volume of assay buffer were
added and incubated for 2 h. Plates were washed and biotinylated
antibodies were subsequently added. After incubation and washing,
alkaline phosphatase-conjugated streptavidin was added. Then,
diflunisal phosphate stock solution and terbium-based detection
solution were added, and fluorescence was assessed with a
fluorometer (Perkin-Elmer). Calibration and data reduction were
performed automatically.
Statistical analysis
All numerical data are presented as the mean ± SE of
at least three independent experiments. Student's t-test was used
for simple comparisons, and one-way ANOVA with Tukey's post hoc
test was applied for multiple comparisons. P<0.05 was considered
to indicate a statistically significant difference.
Results
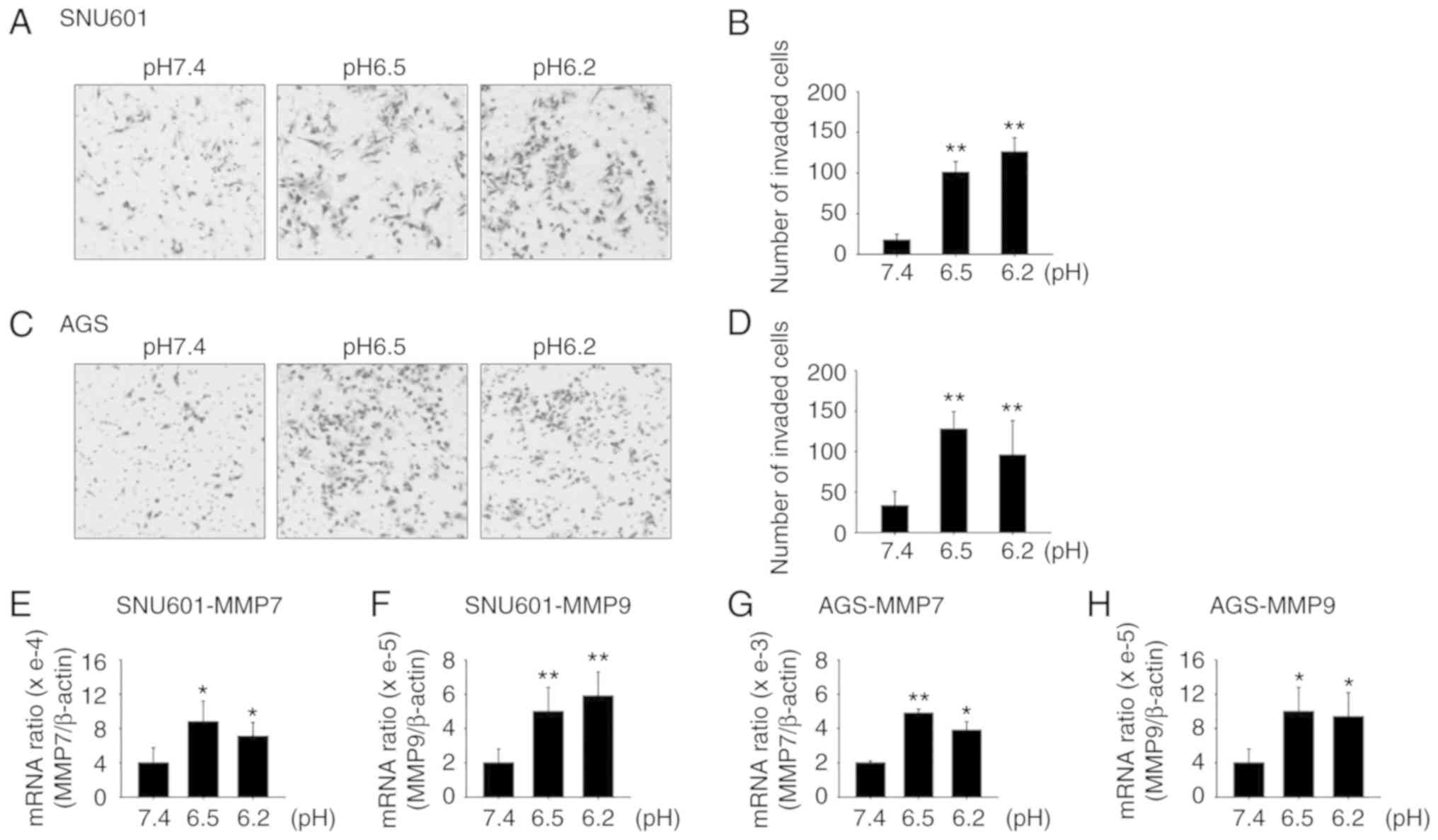
Incubation in acidic culture medium
increases GC cell invasion
Previous research has revealed that extracellular
acidity induces tumor growth, invasion, and metastasis. It was
observed that incubation of gastric cancer cell lines AGS and
SNU601 in an acidic pH medium triggered increased invasion of the
cells compared to those cultured in normal-pH medium. To detect
invasiveness, cells were cultured in culture medium adjusted to pH
7.4, 6.5, and 6.2 for 72 h, then the same number of viable cells
were obtained and added onto the upper portion of Transwell plates
which were coated with Matrigel. After 8 h for AGS and 24 h for
SNU601 cells, the number of cells that invaded to the lower chamber
was counted. The results revealed that the number of cells invading
to the lower portion of the Transwell plates increased in the
acidic environment (Fig. 1A-D). In
addition, the mRNA levels of proteolytic enzymes MMP7 and MMP9,
which are known to be involved in cancer cell invasion, were
increased, as assessed by real-time PCR (Fig. 1E-H).
Cells cultured in acidic medium
increase expression of KLK7 and KLK8
Proteases play crucial roles in cancer invasion and
metastasis due to their proteolytic activity to cleave
extracellular matrix and adhesion molecules. Although MMPs are
known to be essential proteases mediating cell invasion, a possible
involvement of other proteases in cancer cell invasion under an
acidic environment was explored. Notably, it was revealed that the
levels of KLK7 and KLK8 were significantly increased in acidic
culture conditions. Acidic pH culture conditions significantly
increased the mRNA levels of KLK7 and KLK8 in both gastric cancer
cell lines (Fig. 2A-H). To assess
whether increased mRNAs of KLK7 and KLK8 led to increased protein
levels, immunoblot assays were performed using cell lysates. When
the cells were incubated in acidic medium, the protein levels of
KLK8 increased but those of KLK7 decreased (Fig. 2I and J). Since KLKs are proteases
mainly secreted into the extracellular space, the protein levels of
KLK7 and KLK8 released into the medium were also examined using an
ELISA assay. As the pH decreased, the amounts of KLK7 and KLK8
proteins released into the medium significantly increased (Fig. 2K and L). Thus, acidity induced the
expression and secretion of KLK7 and KLK8.
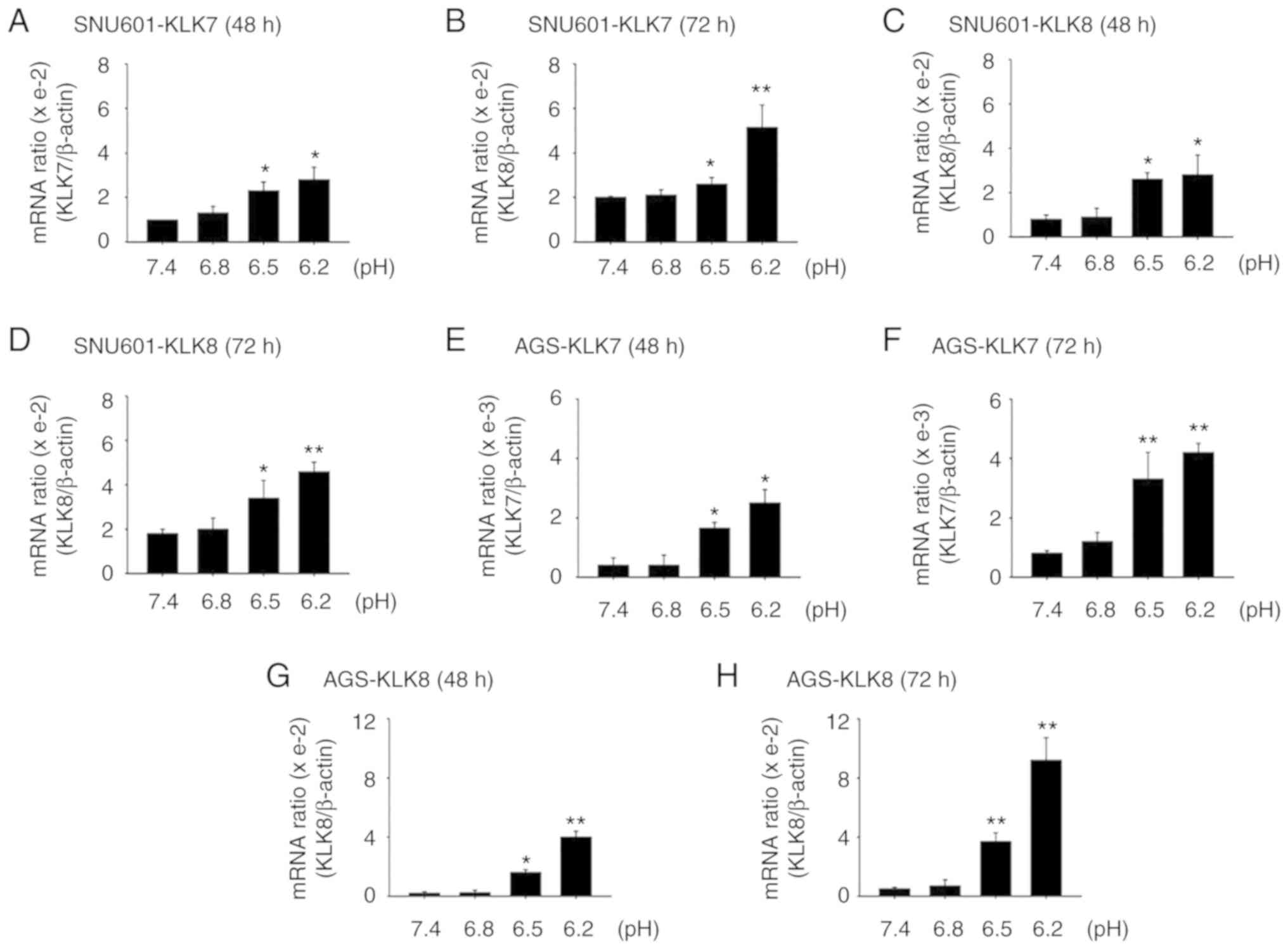 | Figure 2.Acidic culture conditions increase
expression of KLK7 and KLK8. (A-D) SNU601 and (E-H) AGS cells
exposed to normal (pH 7.4) or acidic (pH 6.8, 6.5 and 6.2) medium
for (A, C, E and G) 48 h or (B, D, F and H) 72 h were harvested and
the mRNA expression of the genes encoding (A, B, E and F) KLK7 and
(C, D, G and H) KLK8 was analyzed by real-time PCR. SNU601 and AGS
cells exposed to pH 7.4, 6.8, 6.5 and 6.2 for 72 h were collected
and total protein extracts were prepared by cell lysis and analyzed
by (I and J) immunoblotting with antibodies against KLK7 and KLK8
or medium was subjected to (K and L) ELISA assay to assess secreted
KLK7 and KLK8. *P<0.05, **P<0.01 vs. pH 7.4. KLK7,
kallikrein-related peptidase 7; KLK8, kallikrein-related peptidase
8. |
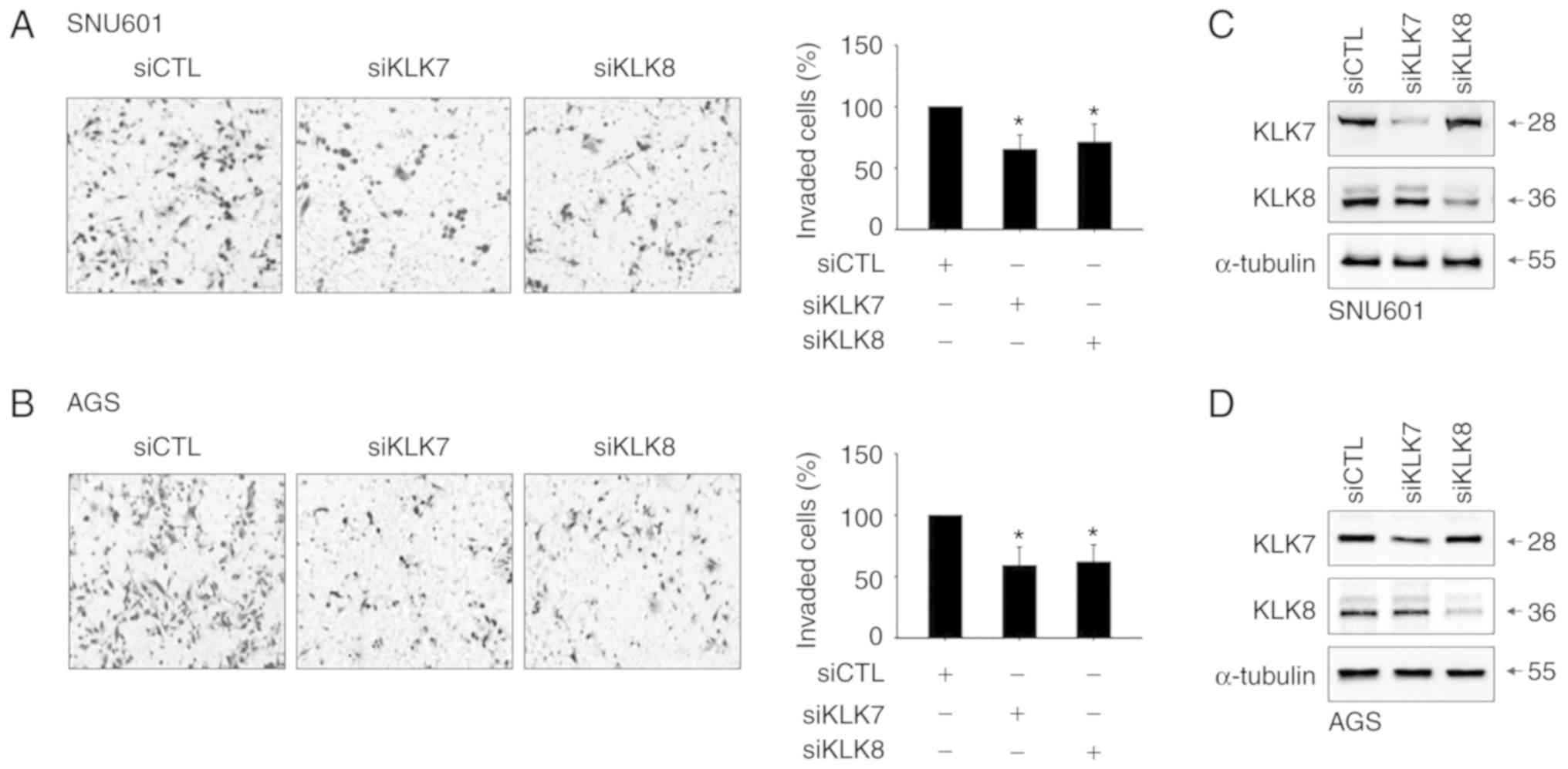
Elevation of KLK7 and KLK8 is linked
to acidity-induced invasion
Since KLK family members have been revealed to be
involved in malignant phenotypes of certain cancers, it was
hypothesized that increased KLK7 and KLK8 in acidic environments
would play a role in the acidity-mediated invasion of these cancer
cells. To examine this, cells treated with siRNAs specifically
silencing KLK7 or KLK8 were exposed to acidic medium. Cells in
which KLK7 or KLK8 were silenced exhibited low invasiveness after
incubation in acidic (pH 6.5) culture medium (Fig. 3A and B). The silencing efficiency of
the KLK7 and KLK8 siRNAs was confirmed by decreased protein levels
(Fig. 3C and D). Thus, increased
expression of KLK7 and KLK8 appears to be linked to the enhanced
invasiveness of gastric cancer cells.
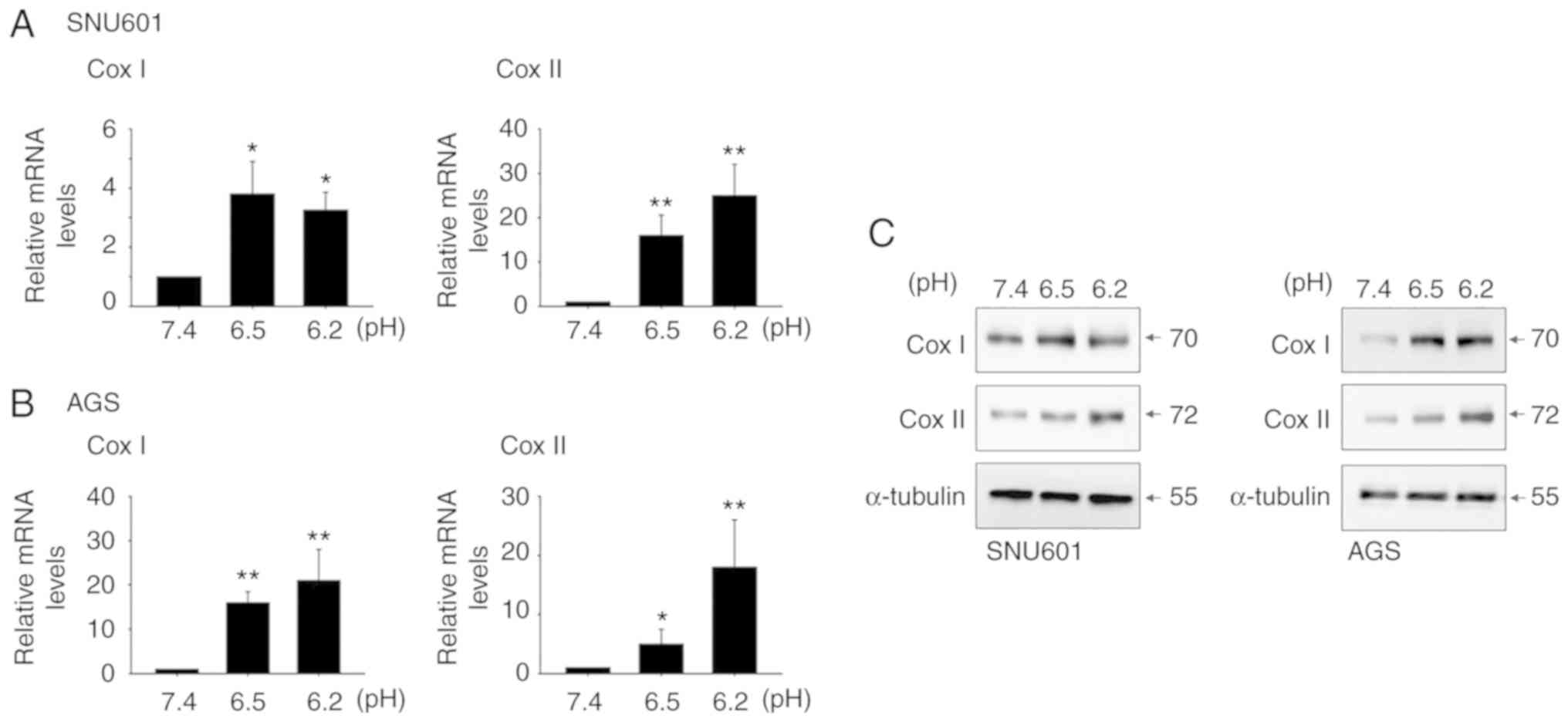
Induction of COX1 and COX2 is involved
in acidity-mediated KLK7 and KLK8 expression
To understand the regulatory mechanisms involved in
acidity-mediated KLK7 and KLK8 expression, potential upstream
regulators were examined. Previously, acidity was revealed to
stimulate inflammatory responses via the cyclooxygenase (COX)
pathway. Consistent with a previous study, acidic culture
conditions elevated mRNA and protein levels of COX2 in both GC cell
lines (20). In addition, the mRNA
and protein expression of COX1, a constitutive isoform, increased
(Fig. 4). Thus, the role of COXs in
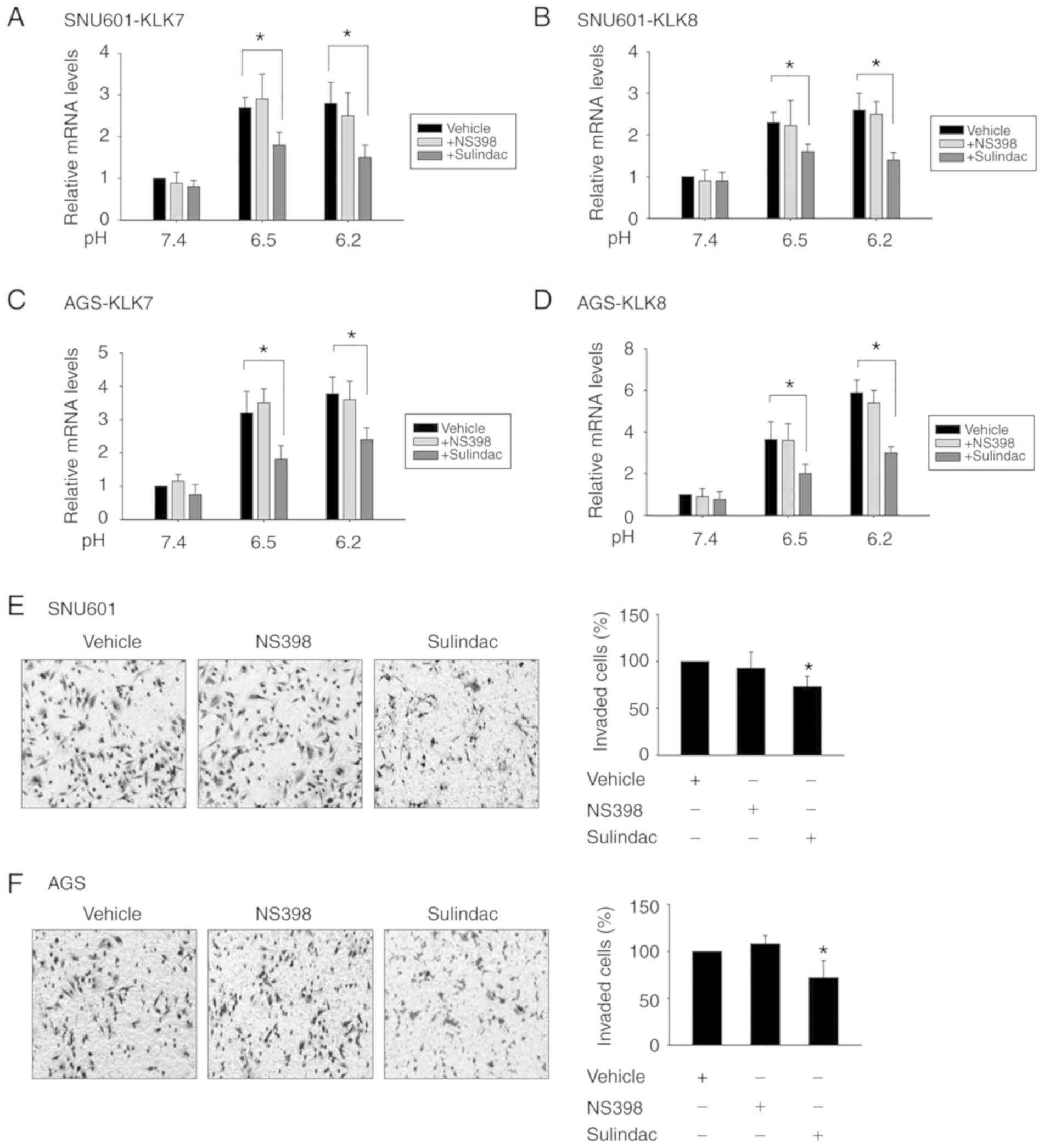
KLK induction was examined by assessing the effect of COX
inhibitors on the expression of KLK7 and KLK8. Upon exposure to
sulindac, a broad COX inhibitor, acidity-induced mRNA expression of
KLK7 and KLK8 was reduced. In contrast, exposure to selective COX2
inhibitor NS398 had little effect on the level of these peptidases
(Fig. 5A-D). Likewise, the effect
on cellular invasion at acidic pH was partially reduced by
sulindac, but not by NS398 (Fig. 5E and
F). This indicated that the COX pathway is involved in
acidity-induced KLK7 and KLK8 expression, and that COX1 plays a
more important role than COX2 in the expression of KLK7 and KLK8.
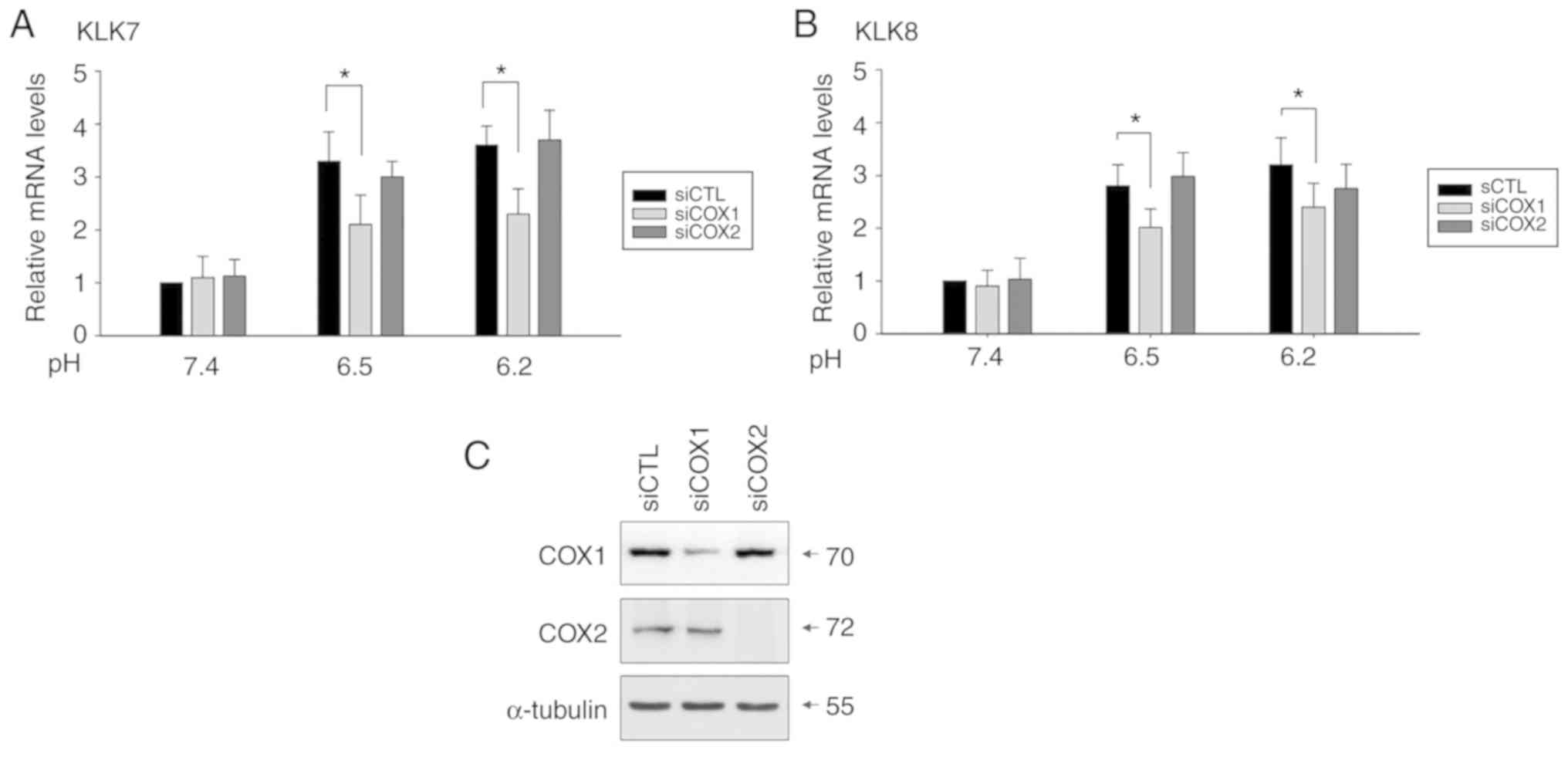
To confirm this, cells transfected with siRNAs silencing COX1 or
COX2 were exposed to acidic culture medium, and the silencing
efficiency was confirmed by decreased protein levels (Fig. 6C). Knockdown of COX1, but not COX2
partially reduced the mRNA expression of KLK7 and KLK8 (Fig. 6A and B).
Discussion
Previously, we reported that acidic culture
conditions are associated with a decrease in cell growth and an
increase in chemoresistance of gastric cancer cells (20). In the present study, it was
demonstrated that gastric cancer cells surviving in low-pH
environments (pH 6.5 and 6.2) were more invasive than cells
cultured at normal pH, in line with previous studies revealing that
extracellular acidity can enhance carcinogenesis and cancer
progression by promoting local invasion and metastasis of the tumor
(6,7). Although, mild acidic pH ranges were
selected to focus on the acidic pH that is formed inside solid
cancer tissue in this study, in future studies the effect of lower
pH, such as that in the stomach, will be investigated.
An acidic environment can affect the expression of
multiple genes in tumor cells. In searching for factors
contributing to acidity-mediated invasion, it was revealed that the
expression of certain proteases was induced by acidity. Evidence
collected over the past several years has revealed that proteases
play an essential role in invasion and metastasis of tumor cells
because they break bonds with surrounding cells or tissues and
degrade the extracellular matrix, thereby allowing tumor cells to
penetrate through physical barriers (21). Thus far, various proteases,
including cathepsin B, cathepsin D, the urokinase-type plasminogen
activator, and several matrix metalloproteases, have been revealed
to be involved in tumor invasion and metastasis (22,23).
In the present study, it was revealed that GC cells exposed to
acidic culture medium had markedly increased expression of KLK
family members KLK7 and KLK8, and a considerable amount of these
peptidases was secreted into extracellular space. Through
siRNA-based gene silencing, it was revealed that these peptidases
are involved in the promotion of acidity-mediated migration and
invasion.
KLK family members are known to be involved in the
regulation of various physiological and pathological processes in
human tissues, and certain KLKs play roles in releasing cell-cell
adhesion through proteolytic cleavage of junctional proteins, such
as desmosomal cadherins (24).
Alteration of KLK7 levels is known to be linked to several skin
disorders, including dermatitis, psoriasis, and Netherton syndrome
(25–27). Notably, overexpression of KLKs is
often linked to aggressive phenotypes in multiple cancers. A recent
study revealed that an increased KLK7 level was linked to increased
proliferation of colorectal cancer cells and associated with poor
prognosis or short survival in colon, ovarian and pancreatic
cancers (28–30). Furthermore, KLK7 has been described
as being potentially involved in metastasis in colorectal and
pancreatic cancer, and melanoma (31–34).
KLK8, found in numerous normal tissues as well as in body fluids,
is also implicated in a variety of malignant tumors, including
ovarian, cervical, salivary gland, and lung cancers when aberrantly
expressed (35–39). In addition, overexpression of KLK8
is considered to be a potential independent prognostic indicator
for CRC. The study on KLK7 and KLK8 is still in the beginning step,
and to the best of our knowledge, this is the first study revealing
that KLK7 and KLK8 are involved in acidity-mediated gastric cancer
cell invasion. Although the link between gastric cancer and KLK7
and KLK8 has not been reported, a previous study indirectly
supported the tumor promoting role of KLK7 in gastric cancer, in
which inhibition of the growth of transplanted gastric cancer by
cisplatin was accompanied by reduction of KLK7 expression (40).
Nevertheless, some studies have demonstrated the
opposite effect of KLK7 and KLK8 on cancer. For example, high
levels of KLK7 were linked to decreased proliferation in melanoma
and a favorable prognosis in melanoma patients (41,42).
Increased KLK8 was associated with either a favorable prognosis or
no effect on patient outcomes in ovarian cancer (43,44).
Since the invasion process requires protease activity to cleave
tumor cells from surrounding cells or the extracellular matrix,
regulation of cancer cell invasion by KLK may be directly linked to
the action of the downstream substrates of KLKs, such as
E-cadherin, fibronectin, laminin, IGFBP3, and midkine. Thus, these
discrepancies are likely explained by the dominant substrates
available for KLK7 or KLK8 in specific environments, although the
exact roles of KLK7 and KLK8 in specific cancer tissues and pH
environments requires further investigation through in vivo
studies.
Although various important functions of KLKs have
been identified, regulatory mechanisms involving KLK expression
have not yet been elucidated. In the present study, it was observed
that acidic pH conditions increased the expression of COX1 and COX2
in GC cells. It was hypothesized that the acidity would regulate
KLK7 and KLK8-mediated invasiveness through COX induction.
Previously, acidity-induced COX2 expression was also observed in
oral cells (45). COX is well
established to play a crucial role in a wide range of inflammatory
responses, in which acidity also plays roles. Extracellular acidity
triggers activation of immune responses in multiple immune cells,
endothelial cells, and fibroblasts (46–48),
and many chronic inflammatory diseases involve acidic pH regions
(49). Notably, the inflammatory
response acts as a critical factor affecting carcinogenesis and
cancer progression. Therefore, the present study may provide an
explanation for the induction of inflammatory responses and the
increase in tumor invasion in acidic environments, and in further
study, a more specific link between acidity-mediated inflammation
and tumor invasiveness is under investigation by evaluating the
expression of inflammatory-related genes including interleukin 8
and c-Myc.
In the present study, it was demonstrated that an
acidic environment could increase COX1-regulated KLK7 and KLK8
expression, and this event contributed to the acidity-induced
promotion of GC cell invasion. This study indicates KLK7 and KLK8
as potential targets for anticancer therapies in acidic tumor
microenvironments.
Acknowledgements
We would like to thank Ms Jeong-Eun Choi and Dr
Mi-Rae Lee for their excellent technical assistance.
Funding
This research was supported by the Basic Science
Research Program through the National Research Foundation of Korea
(NRF) funded by the Ministry of Education (NRF-2015R1D1A1A01060533
and NRF-2018R1D1A1B07046430).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
SIH designed and wrote the manuscript. SCL, KHK and
RH performed the experiments and analyzed the data. SIH, MJL and
SCL reviewed and edited the manuscript. All authors approved the
final manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lazebnik Y: What are the hallmarks of
cancer? Nat Rev Cancer. 10:232–233. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Seyfried TN and Huysentruyt LC: On the
origin of cancer metastasis. Crit Rev Oncog. 18:43–73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tannock IF and Rotin D: Acid pH in tumors
and its potential for therapeutic exploitation. Cancer Res.
49:4373–4384. 1989.PubMed/NCBI
|
|
5
|
Moellering RE, Black KC, Krishnamurty C,
Baggett BK, Stafford P, Rain M, Gatenby RA and Gillies RJ: Acid
treatment of melanoma cells selects for invasive phenotypes. Clin
Exp Metastasis. 25:411–425. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Estrella V, Chen T, Lloyd M, Wojtkowiak J,
Cornnell HH, Ibrahim-Hashim A, Bailey K, Balagurunathan Y, Rothberg
JM, Sloane BF, et al: Acidity generated by the tumor
microenvironment drives local invasion. Cancer Res. 73:1524–1535.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gatenby RA and Gillies RJ: A
microenvironmental model of carcinogenesis. Nat Rev Cancer.
8:56–61. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peppicelli S, Bianchini F, Torre E and
Calorini L: Contribution of acidic melanoma cells undergoing
epithelial-to-mesenchymal transition to aggressiveness of
non-acidic melanoma cells. Clin Exp Metastasis. 31:423–433. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hjelmeland AB, Wu Q, Heddleston JM,
Choudhary GS, MacSwords J, Lathia JD, McLendon R, Lindner D, Sloan
A and Rich JN: Acidic stress promotes a glioma stem cell phenotype.
Cell Death Differ. 18:829–840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sevenich L and Joyce JA: Pericellular
proteolysis in cancer. Genes Dev. 28:2331–2347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Darmoul D, Gratio V, Devaud H, Peiretti F
and Laburthe M: Activation of proteinase-activated receptor 1
promotes human colon cancer cell proliferation through epidermal
growth factor receptor transactivation. Mol Cancer Res. 2:514–522.
2004.PubMed/NCBI
|
|
12
|
Darmoul D, Marie JC, Devaud H, Gratio V
and Laburthe M: Initiation of human colon cancer cell proliferation
by trypsin acting at protease-activated receptor-2. Br J Cancer.
85:772–779. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gratio V, Loriot C, Virca GD,
Oikonomopoulou K, Walker F, Diamandis EP, Hollenberg MD and Darmoul
D: Kallikrein-related peptidase 14 acts on proteinase-activated
receptor 2 to induce signaling pathway in colon cancer cells. Am J
Pathol. 179:2625–2636. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ramsay AJ, Reid JC, Adams MN, Samaratunga
H, Dong Y, Clements JA and Hooper JD: Prostatic trypsin-like
kallikrein-related peptidases (KLKs) and other prostate-expressed
tryptic proteinases as regulators of signalling via
proteinase-activated receptors (PARs). Biol Chem. 389:653–668.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Loessner D, Goettig P, Preis S, Felber J,
Bronger H, Clements JA, Dorn J and Magdolen V: Kallikrein-related
peptidases represent attractive therapeutic targets for ovarian
cancer. Expert Opin Ther Targets. 22:745–763. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Avgeris M and Scorilas A:
Kallikrein-related peptidases (KLKs) as emerging therapeutic
targets: Focus on prostate cancer and skin pathologies. Expert Opin
Ther Targets. 20:801–818. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kryza T, Silva ML, Loessner D,
Heuze-Vourc'h N and Clements JA: The kallikrein-related peptidase
family: Dysregulation and functions during cancer progression.
Biochimie. 122:283–299. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shaw JL and Diamandis EP: Distribution of
15 human kallikreins in tissues and biological fluids. Clin Chem.
53:1423–1432. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hong R and Han SI: Extracellular acidity
enhances tumor necrosis factor-related apoptosis-inducing ligand
(TRAIL)-mediated apoptosis via DR5 in gastric cancer cells. Korean
J Physiol Pharmacol. 22:513–523. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chambers AF and Matrisian LM: Changing
views of the role of matrix metalloproteinases in metastasis. J
Natl Cancer Inst. 89:1260–1270. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang L and Han X: The urokinase
plasminogen activator system in breast cancer invasion and
metastasis. Biomed Pharmacother. 67:179–182. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Duffy MJ: The role of proteolytic enzymes
in cancer invasion and metastasis. Clin Exp Metastasis. 10:145–155.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Caubet C, Jonca N, Brattsand M, Guerrin M,
Bernard D, Schmidt R, Egelrud T, Simon M and Serre G: Degradation
of corneodesmosome proteins by two serine proteases of the
kallikrein family, SCTE/KLK5/hK5 and SCCE/KLK7/hK7. J Invest
Dermatol. 122:1235–1244. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Komatsu N, Saijoh K, Kuk C, Liu AC, Khan
S, Shirasaki F, Takehara K and Diamandis EP: Human tissue
kallikrein expression in the stratum corneum and serum of atopic
dermatitis patients. Exp Dermatol. 16:513–519. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Descargues P, Deraison C, Bonnart C, Kreft
M, Kishibe M, Ishida-Yamamoto A, Elias P, Barrandon Y, Zambruno G,
Sonnenberg A and Hovnanian A: Spink5-deficient mice mimic Netherton
syndrome through degradation of desmoglein 1 by epidermal protease
hyperactivity. Nat Genet. 37:56–65. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ekholm E and Egelrud T: Stratum corneum
chymotryptic enzyme in psoriasis. Arch Dermatol Res. 291:195–200.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dorn J, Gkazepis A, Kotzsch M, Kremer M,
Propping C, Mayer K, Mengele K, Diamandis EP, Kiechle M, Magdolen V
and Schmitt M: Clinical value of protein expression of
kallikrein-related peptidase 7 (KLK7) in ovarian cancer. Biol Chem.
395:95–107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Devetzi M, Trangas T, Scorilas A,
Xynopoulos D and Talieri M: Parallel overexpression and clinical
significance of kallikrein-related peptidases 7 and 14 (KLK7KLK14)
in colon cancer. Thromb Haemost. 109:716–725. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Iakovlev V, Siegel ER, Tsao MS and Haun
RS: Expression of kallikrein-related peptidase 7 predicts poor
prognosis in patients with unresectable pancreatic ductal
adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 21:1135–1142.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rezze GG, Fregnani JH, Duprat J and
Landman G: Cell adhesion and communication proteins are
differentially expressed in melanoma progression model. Hum Pathol.
42:409–418. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Talieri M, Mathioudaki K, Prezas P,
Alexopoulou DK, Diamandis EP, Xynopoulos D, Ardavanis A,
Arnogiannaki N and Scorilas A: Clinical significance of
kallikrein-related peptidase 7 (KLK7) in colorectal cancer. Thromb
Haemost. 101:741–747. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Johnson SK, Ramani VC, Hennings L and Haun
RS: Kallikrein 7 enhances pancreatic cancer cell invasion by
shedding E-cadherin. Cancer. 109:1811–1820. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Delaunay T, Deschamps L, Haddada M, Walker
F, Soosaipillai A, Soualmia F, El Amri C, Diamandis EP, Brattsand
M, Magdolen V and Darmoul D: Aberrant expression of
kallikrein-related peptidase 7 is correlated with human melanoma
aggressiveness by stimulating cell migration and invasion. Mol
Oncol. 11:1330–1347. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kishi T, Grass L, Soosaipillai A,
Shimizu-Okabe C and Diamandis EP: Human kallikrein 8: Immunoassay
development and identification in tissue extracts and biological
fluids. Clin Chem. 49:87–96. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Planque C, Choi YH, Guyetant S,
Heuze-Vourc'h N, Briollais L and Courty Y: Alternative splicing
variant of kallikrein-related peptidase 8 as an independent
predictor of unfavorable prognosis in lung cancer. Clin Chem.
56:987–997. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Darling MR, Tsai S, Jackson-Boeters L,
Daley TD and Diamandis EP: Human kallikrein 8 expression in
salivary gland tumors. Head Neck Pathol. 2:169–174. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cane S, Bignotti E, Bellone S, Palmieri M,
De las Casas L, Roman JJ, Pecorelli S, Cannon MJ, O'brien T and
Santin AD: The novel serine protease tumor-associated
differentially expressed gene-14 (KLK8/Neuropsin/Ovasin) is highly
overexpressed in cervical cancer. Am J Obstet Gynecol. 190:60–66.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Magklara A, Scorilas A, Katsaros D,
Massobrio M, Yousef GM, Fracchioli S, Danese S and Diamandis EP:
The human KLK8 (neuropsin/ovasin) gene: Identification of two novel
splice variants and its prognostic value in ovarian cancer. Clin
Cancer Res. 7:806–811. 2001.PubMed/NCBI
|
|
40
|
Zhuang GF, Tan Y, Yang YZ, Zhang JW and
Tang J: Experiment research of cisplatin implants inhibiting
transplantation tumor growth and regulating the expression of KLK7
and E-cad of tumor-bearing mice with gastric cancer. Asian Pac J
Trop Med. 9:606–609. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu Y, Prassas I, Dimitromanolakis A and
Diamandis EP: Novel biological substrates of human kallikrein 7
identified through degradomics. J Biol Chem. 290:17762–17775. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Martins WK, Esteves GH, Almeida OM, Rezze
GG, Landman G, Marques SM, Carvalho AF, L Reis LF, Duprat JP and
Stolf BS: Gene network analyses point to the importance of human
tissue kallikreins in melanoma progression. BMC Med Genomics.
4:762011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Borgono CA, Kishi T, Scorilas A, Harbeck
N, Dorn J, Schmalfeldt B, Schmitt M and Diamandis EP: Human
kallikrein 8 protein is a favorable prognostic marker in ovarian
cancer. Clin Cancer Res. 12:1487–1493. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ahmed N, Dorn J, Napieralski R, Drecoll E,
Kotzsch M, Goettig P, Zein E, Avril S, Kiechle M, Diamandis EP, et
al: Clinical relevance of kallikrein-related peptidase 6 (KLK6) and
8 (KLK8) mRNA expression in advanced serous ovarian cancer. Biol
Chem. 397:1265–1276. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cha SH, Park JE, Kwak JO, Kim HW, Kim JB,
Lee KY and Cha YN: Attenuation of extracellular acidic pH-induced
cyclooxygenase-2 expression by nitric oxide. Mol Cells. 19:232–238.
2005.PubMed/NCBI
|
|
46
|
Riemann A, Ihling A, Thomas J, Schneider
B, Thews O and Gekle M: Acidic environment activates inflammatory
programs in fibroblasts via a cAMP-MAPK pathway. Biochim Biophys
Acta. 1853:299–307. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dong L, Li Z, Leffler NR, Asch AS, Chi JT
and Yang LV: Acidosis activation of the proton-sensing GPR4
receptor stimulates vascular endothelial cell inflammatory
responses revealed by transcriptome analysis. PLoS One.
8:e619912013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Etulain J, Negrotto S, Carestia A, Pozner
RG, Romaniuk MA, D'Atri LP, Klement GL and Schattner M: Acidosis
downregulates platelet haemostatic functions and promotes
neutrophil proinflammatory responses mediated by platelets. Thromb
Haemost. 107:99–110. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Naghavi M, John R, Naguib S, Siadaty MS,
Grasu R, Kurian KC, van Winkle WB, Soller B, Litovsky S, Madjid M,
et al: pH Heterogeneity of human and rabbit atherosclerotic
plaques; a new insight into detection of vulnerable plaque.
Atherosclerosis. 164:27–35. 2002. View Article : Google Scholar : PubMed/NCBI
|