Introduction
Glioblastoma (GBM) is the most common primary
malignant brain tumor in adults. High-grade GBM remains a
devastating disease despite maximal therapy with surgery, radiation
and chemotherapy with temozolomide (TMZ). GBM patients present with
only a 15- to 19-month median overall survival rate due to
chemoradiotherapy resistance and recurrence (1). Recent studies have identified several
molecular alterations involved in GBM carcinogenesis, progression,
chemotherapeutic resistance and recurrence, such as IDH1, 1p/19q,
MGMT, ATRX and PTEN, but the precise mechanisms underlying this
malignancy and its rapid recurrence have not been fully elucidated
(2–4). Targeting key genes and signaling
pathways is considered a promising approach for the diagnosis and
treatment of cancer. Therefore, finding potential biomarkers with
bioinformatic analysis contributes to a better understanding of the
occurrence and development of GBM.
Homo sapiens ZW10 interacting kinetochore
protein (ZWINT) is a known component of the kinetochore complex
required for the mitotic spindle checkpoint and plays crucial roles
in mitotic cycle maintenance (5).
The kinetochore is a highly complex structure that is central to
many essential activities during cell division. The kinetochore, a
tri-laminar plate to which microtubules attach, connects
chromosomes to the spindle to ensure the accurate segregation of
chromosomes in mitosis and meiosis (6). ZWINT encodes a protein that is
clearly involved in kinetochore function, possibly by regulating
the association between ZW10 and centromere complexes during
mitotic and mitotic prometaphase (7). It is known that abnormal mitosis is a
common feature of most malignancies. Although the exact role of the
molecular makeup of the kinetochore and how individual components
of the kinetochore interact with each other are unknown, growing
evidence shows that ZWINT is often highly expressed in a number of
human cancers and is linked with poor clinical prognosis and early
recurrence (8–10). However, its role in human GBM
remains unclear. In our research, we aimed to investigate the
expression of ZWINT and its biological significance in this primary
malignancy.
Materials and methods
Dataset processing
TCGA (The Cancer Genome Atlas, http://cancergenome.nih.gov/) is a public repository
for data storage that is freely available to users. A variety of
human cancer and tumor subtype genomic mutation profiles (11), transcriptomic data (12), and clinical data (13) have been generated, providing a
systematic characterization of methylation (14), miRNA expression (15), and oncogenic processes (16). Gene expression profiles of GBM were
downloaded from the TCGA dataset, which contains 529 GBM samples
and 10 normal samples. The data of the expression profile chip
level 3 of these samples were sorted out for analyzing the
differentially expressed genes (DEGs). However, multiple sets of
data were assessed for certain samples in practice, thus the actual
number of downloaded files was more than the original samples (548
GBM samples vs. 10 normal samples). We used P<0.05 and |FC|
(fold change) ≥2 as the criteria, and the edgeR (https://bioconductor.org/packages/release/bioc/html/edgeR.html)
(17,18) package in R 3.4.1 was used to
identify DEGs in the GBM samples compared with normal brain samples
to finally obtain the DEG list. Another gene dataset, GSE15824
(19), was downloaded from the NCBI
GEO database (https://www.ncbi.nlm.nih.gov/geo/), and GPL570 was the
platform file. Standardization data were carried out using the RMA
algorithm of the Affy (http://bioconductor.org/packages/release/bioc/html/Affy.html)
(20) package in R software, which
were used for the subsequent analysis.
GO and KEGG pathway analyses
Database for Annotation, Visualization and
Integrated Discovery (DAVID) 6.8 (http://david.abcc.ncifcrf.gov/) is an online platform
that is used for gene annotation, visualization and integrated
discovery (21,22). Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analyses were implemented with the DAVID database. Using this
comprehensive tool, we can understand the biological meaning behind
the DEGs more quickly and effectively. P<0.05 indicated a
statistically significant difference.
PPI network
The Search Tool for the Retrieval of Interacting
Genes (STRING, http://string-db.org) database was
queried to construct the protein-protein interaction (PPI) network
(23). A confidence score ≥0.9 was
set as the cutoff criterion, and disconnected nodes were excluded
from the network. CytoHubba and Molecular Complex Detection (MCODE)
in Cytoscape 3.5.1 were performed to identify hub genes and
significant modules of the PPI network (24,25).
The filter conditions were as follows: Degree cutoff=2, node score
cutoff=0.2, k-core=2, and max. depth=100.
Correlation analysis
The GSE15824 dataset contained 40 GBM samples and 5
normal brain tissues. R software was used to evaluate the
correlation between the expression of ZWINT and other
related hub genes, and the Pearson correlation coefficient (R) was
used to reflect the degree of linear correlations. The comparison
criteria were as follows: Within −1<R<1, if R>0, indicated
a positive correlation between the two genes; R<0 indicated a
negative correlation, and the greater the absolute value of R, the
more significant was the negative correlation; and R=0 indicated
nonlinear correlation.
Cell culture
The human GBM cell lines U251, U87 MG, A172, and
U373 were obtained from the American Type Culture Collection
(ATCC). The specific origin of U87 MG cells is unknown in the ATCC
version. These four cell lines were authenticated using short
tandem repeat (STR) profiling analysis following ISO 9001:2008 and
ISO/IEC 17025:2005 quality standards. The percent matches for U251,
U373, A172 and U87 MG cells were 100, 89, 100 and 100%,
respectively. They were cultured in DMEM supplemented with 10%
fetal bovine serum (FBS) (Ausbian, USA) and incubated at 37°C in a
humidified atmosphere of 5% CO2.
Immunohistochemistry microarray
A commercially available GBM tissue microarray (TMA)
slide was purchased from Shanghai GeneChem Co., Ltd., for IHC
analysis. A specific primary antibody against ZWINT was utilized
for immunohistochemistry (IHC) with a 2-step protocol. IHC scores
were calculated according to the intensity and proportion of
positive staining. Staining intensity was classified as 0
(negative), 1 (weak), 2 (moderate) and 3 (strong). The positive
cell ratio of the stained cells was scored as 0 (<1%), 1
(1–25%), 2 (26–50%), 3 (51–75%) and 4 (76–100%). The multiplied
result of the two scores represented the protein levels of ZWINT,
and an immunoreactive score (IRS) >6 was defined as a high
expression level of ZWINT, while an IRS ≤6 indicated a low level of
ZWINT.
Quantitative PCR (qPCR)
Total RNA was extracted from cells using a TRIzol
kit (cat. no. 3101-100, Pufei Biotechnology), and complementary DNA
(cDNA) was synthesized with the M-MLV reagent (Promega). The SYBR
Master Mix (Takara) and Mx3000P Real-Time PCR machine were used to
perform RT-qPCR. The PCR primers (GeneChem) were ZWINT-F,
5′-CACGTAGAGGCCATCAAAATTGG-3′ and ZWINT-R,
5′-CGGAGTTGTGTCCGTTTCCT-3′; GAPDH-F, 5′-TGACTTCAACAGCGACACCCA-3′
and GAPDH-R, 5′-CACCCTGTTGCTGTAGCCAAA-3′. Relative quantification
was calculated using the ΔΔCq method (26), normalized based on GAPDH, and
conducted in triplicate.
ZWINT shRNA design and vector
transfection
According to the cDNA sequence of ZWINT (Gene
Bank accession no. NM_007057) and the design principle of the small
hairpin RNA (shRNA), three target characteristic sequences and
nonspecific sequences were selected. shRNA against ZWINT was
transfected into 293T cells using Lipofectamine 2000™ reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), and the expression of
ZWINT at the protein level was detected by Western blot analylsis
to determine the transfection efficiency. The optimal target
sequence was selected: 5′-TTCTCCGAACGTGTCACGT-3′. ZWINT
shRNA and the control shRNA were provided by GeneChem Co., Ltd.
ZWINT shRNA was used to knock down ZWINT expression
in the U251 and U87 MG cells with Lipofectamine 2000 reagent. At 72
h after transfection, U251 and U87 cells were harvested for later
research.
Western blot assay
293T cells were lysed in radioimmunoprecipitation
assay (RIPA) buffer after transfection with shRNA for 72 h. A BCA
Protein Assay Kit was used to detect the total protein
concentration. Equal amounts of protein lysates were
electrophoresed in 10% SDS-PAGE gels and then transferred to
polyvinylidene difluoride (PVDF) membranes, which were blocked by
Tris-buffered saline with Tween (TBST) containing 5% milk. Protein
expression was probed with mouse anti-FLAG (F1804; Sigma-Aldrich;
Merck KGaA) and goat anti-mouse IgG (SC-2005; Santa Cruz
Biotechnology, Inc.). Protein expression in 293T cells that were
exogenously transfected with a plasmid encoding FLAG-tagged ZWINT
was detected by mouse anti-FLAG and mouse anti-GAPDH (SC-32233;
Santa Cruz Biotechnology, Inc.). The ECL-PLUS kit (Thermo Fisher
Scientific. Inc) was used to detect the signals on X-ray film.
GAPDH was used as the reference protein to calculate the relative
protein levels. Independent tests were performed in triplicate.
Cell growth and MTT proliferation
assay
The number of cells at each time point was counted
by a Celigo Image Cytometer (Nexcelom Bioscience). Briefly, the GBM
cells were cultured in 96-well plates at an initial density of
2×103 cells/well. Each group had three wells (10
µl/well) and was incubated at 37°C in an atmosphere of 5%
CO2. Fluorescence photomicrographs were captured, and
cells with green fluorescence were measured by a Celigo Image
Cytometer. Cell growth curves were generated for a time course of 5
days. MTT [3- (4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide] (Genview Scientific. Inc.) was used to measure cell
viability. The infected cells (2×103 cells/well) were
collected and reseeded in 96-well plates. From the first to the 5th
day, a total of 20 µl MTT solution (5 mg/ml) was added to the
cells. Subsequently, the supernatants were removed, 100 µl DMSO was
added to each well, and the plates were oscillated for 3 min.
Finally, the optical density (OD) at 490 nm was measured with a
microplate reader.
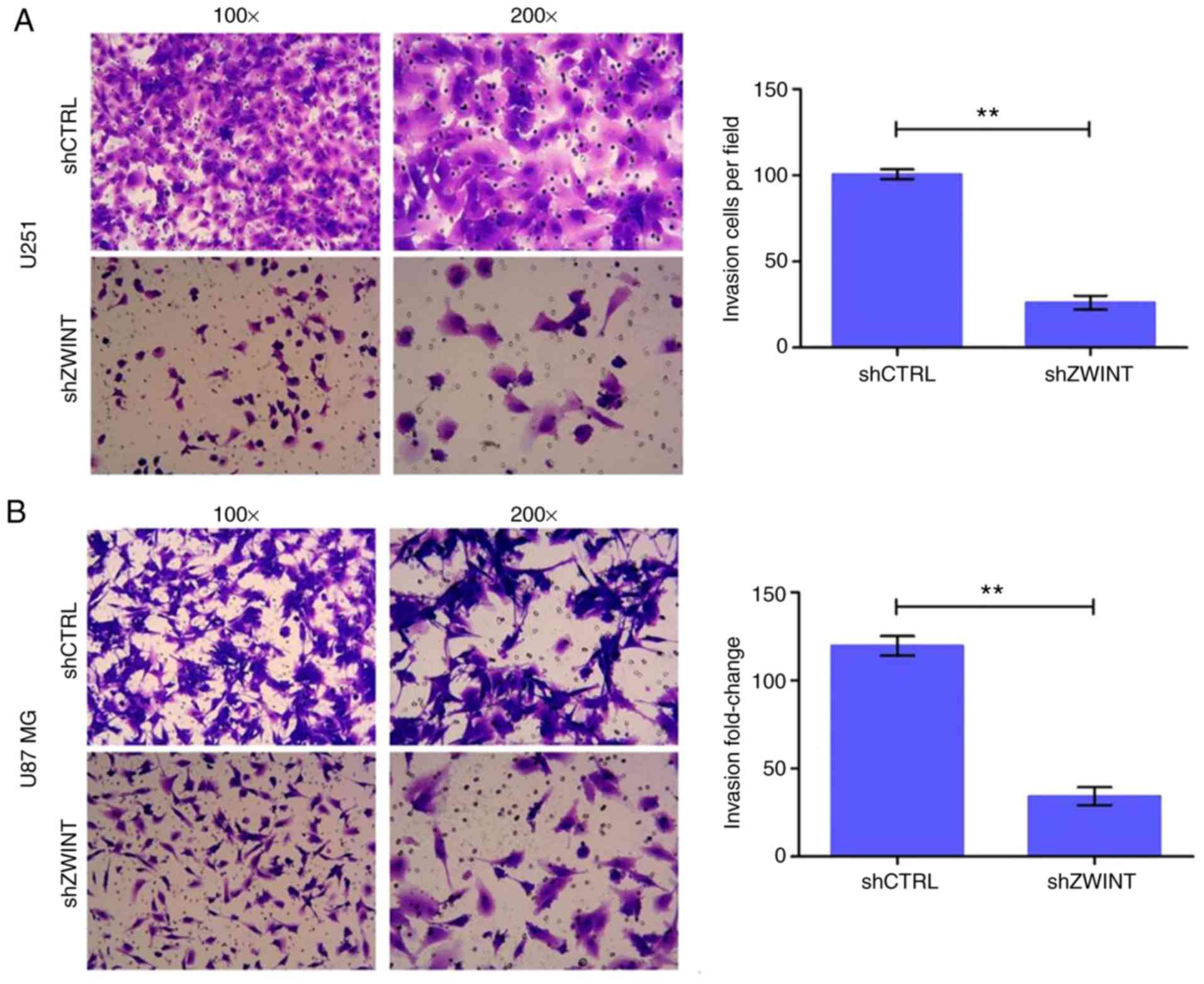
Transwell chamber assay
U251 and U87 MG cells were transfected with
ZWINT shRNA or NC, cultured for 72 h and harvested. The
invasion assay, in which the Transwell chamber was coated with
Matrigel, was conducted according to the manufacturer's
instructions for the Corning Invasion Kit (Corning Inc.). A total
of 500 µl cell suspension in FBS-free DMEM (105
cells/well in a 24-well plate) was added to the upper chamber, and
750 µl DMEM containing 30% FBS was added to the lower chamber.
Cells were removed from the upper chamber of the filter with a
cotton swab after 24 h of incubation at 37°C. Cells on the
underside were washed with PBS, stained with Giemsa, captured with
a digital camera and counted in 5 randomly selected fields of
vision at ×100 magnification and 9 randomly fields of vision at
×200 magnification under a phase contrast microscope.
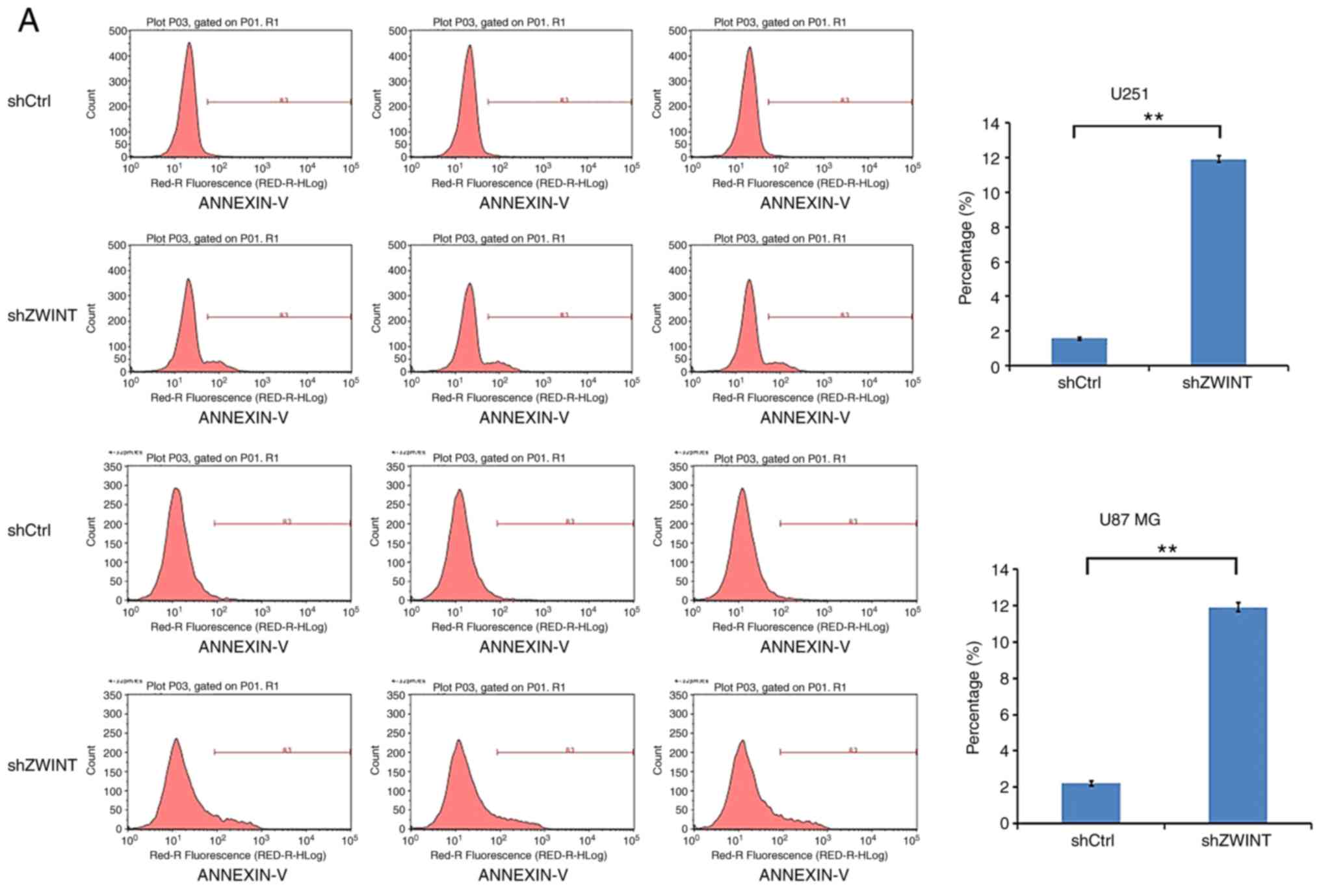
Flow cytometry and caspase-3/7
assay
Apoptotic cells were assessed using the Annexin V
apoptosis kit (88–8007; eBioscience). In short, cells were
trypsinized, washed and centrifuged at 283 × g for 5 min. Then, 1X
binding buffer was used to wash the cells again by resuspending the
cells in 200 µl. Then, the cells were incubated with 10 µl Annexin
V-APC at room temperature for 15 min in the dark. Finally, the
Annexin V-stained cells were analyzed with a flow cytometer
(Millipore) to determine the proportion of apoptotic cells.
Caspase-3/7 are central effector caspases in
apoptosis and are usually used to measure apoptotic activities. GBM
cells were first transfected with the ZWINT shRNA and NC.
Next, 1×104 infected cells/well were seeded in a 96-well
plate. Caspase-Glo 3/7 reagent (100 µl, G8091; Promega) was added
to each well, the plate was shaken for 30 min constantly and
incubation was carried out at ambient temperature for 2 h. The
luminescence signal was detected with an M2009PR (Tecan Infinite)
plate reader.
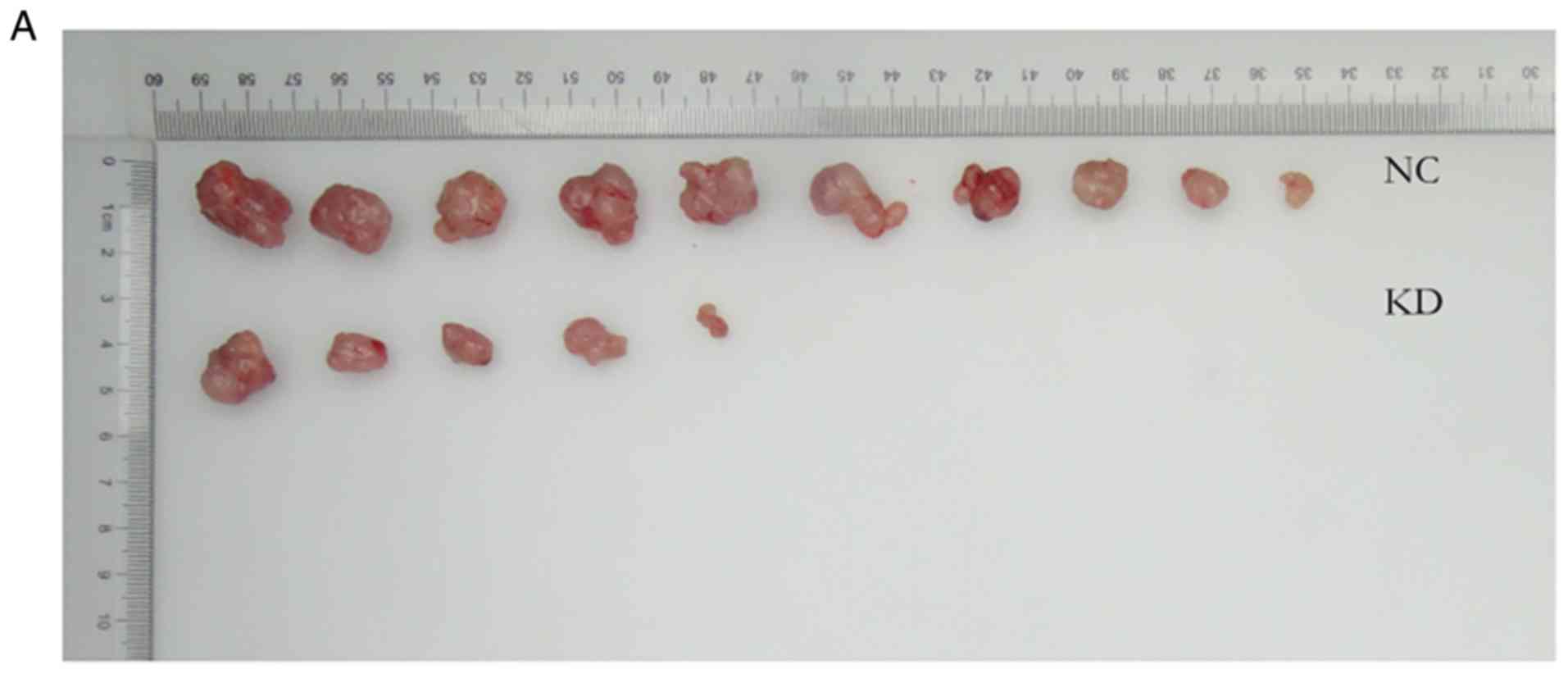
Nude mouse study
Animal experiments were approved by the Animal Care
Committee of Tongji Hospital of Huazhong University of Science and
Technology (Wuhan, Hubei, China), and strictly followed the
institutional regulations and state guidelines on experimental
animals. BALB/c athymic female nude mice (4-week-old; ~20 g) were
purchased from Shanghai Lingchang Biological Polytron Technologies
Inc. They were maintained in a constant temperature, humidity,
sterile environment and fed with food and water according to the
national regulations. Subcutaneous xenografts of human GBM were
established by injecting 5×106 ZWINT shRNA U87 MG
cells or NC cells into the right hind limbs of the mice; 10 mice in
each group. A digital caliper was used to measure tumor growth once
every 3 days. Thirty-six days after cell inoculation, the animals
were euthanized using an intraperitoneal injection of 150 mg/kg
pentobarbital sodium before collecting the tumors. Tumor tissues
were excised and weighted. The tumor volume was defined as V≈π/6 ×
L × W2, where L stands for the tumor length and W is the
tumor width.
Statistical analysis
SPSS version 20.0 (IBM Corp.) was used for data
analysis. Measurement data are expressed as the mean ± standard
deviation. Student's t-test was used to compare data between two
groups. The Pearson correlation coefficient was used for the
correlation analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
GO annotation and KEGG pathway
enrichment analysis
The downloaded gene expression profiles in the TCGA
database included 2,387 upregulated genes and 3,873 downregulated
genes. The GO functional analysis demonstrated that the upregulated
genes were mainly involved in biological processes associated with
mitotic nuclear division, cell division, sister chromatid cohesion,
protein binding and cell proliferation, and the ZWINT gene
was mainly enriched in cell division, mitotic sister chromatid
segregation, kinetochore, and mitotic cell cycle checkpoint. The
downregulated genes were mainly enriched in positive regulation of
excitatory postsynaptic potential, neurotransmitter transport, axon
terminus, and ion channel binding. Moreover, five significant
signaling pathways were overexpressed in the upregulated genes,
including the cell cycle, cellular senescence, DNA replication, Ras
and MAPK signaling pathways. Only one KEGG pathway was found for
downregulated genes when P=0.05 (Table
I).
 | Table I.Functional and pathway enrichment
analysis of DEGs in GBM. |
Table I.
Functional and pathway enrichment
analysis of DEGs in GBM.
| Category | GO/KEGG ID | Description | Count | FDR |
|---|
| Upregulated |
| BP | GO:0007067 | Mitotic nuclear
division | 64 | 3.18E-15 |
|
| GO:0051301 | Cell division | 78 | 5.45R-15 |
|
| GO:0007062 | Sister chromatid
cohesion | 34 | 5.28E-10 |
|
| GO:0008283 | Cell
proliferation | 58 | 1.90E-04 |
|
| GO:0000086 | G2/M transition of
mitotic cell cycle | 27 | 0.018169 |
| CC | GO:0015935 | Small ribosomal
subunit | 20 | 3.22E-13 |
|
| GO:0005829 | Cytosol | 343 | 9.55E-10 |
|
| GO:0005634 | Nucleus | 497 | 4.31E-07 |
|
| GO:0005737 | Cytoplasm | 478 | 0.018475 |
|
| GO:0000777 | Condensed
chromosome kinetochore | 20 | 0.018475 |
| MF | GO:0005515 | Protein
binding | 745 | 4.65E-06 |
|
| GO:0001077 | Transcriptional
activator activity | 42 | 3.17E-04 |
|
| GO:0000978 | RNA polymerase II
core promoter proximal region sequence-specific DNA binding | 54 | 9.19E-04 |
|
| GO:0003682 | Chromatin
binding | 56 | 0.003782 |
|
| GO:0043565 | Sequence-specific
DNA binding | 68 | 0.006257 |
|
| KEGG:hsa04014 | MAPK signaling
pathway | 84 | 1.02E-05 |
|
| KEGG:hsa04022 | cGMP-PKG signaling
pathway | 55 | 0.000239 |
|
| KEGG:hsa04110 | Cell cycle | 44 | 0.000364 |
|
| KEGG:hsa04014 | Ras signaling
pathway | 66 | 0.012203 |
|
| KEGG:hsa03030 | DNA
replication | 14 | 0.036848 |
| Downregulated |
| BP | GO:2000463 | Positive regulation
of excitatory postsynaptic potential | 14 | 8.31E-05 |
|
| GO:0006836 | Neurotransmitter
transport | 15 | 6.88E-04 |
|
| GO:0007214 | γ-aminobutyric acid
signaling pathway | 12 | 0.045329 |
|
| GO:0007193 | Adenylate
cyclase-inhibiting G-protein coupled receptor signaling
pathway | 18 | 0.039186 |
|
| GO:0007264 | Small GTPase
mediated signal transduction | 54 | 0.045329 |
| CC | GO:0030426 | Growth cone | 39 | 1.00E-05 |
|
| GO:0043679 | Axon terminus | 22 | 8.72E-05 |
|
| GO:0032809 | Neuronal cell body
membrane | 13 | 1.61E-04 |
|
| GO:0048471 | Perinuclear region
of cytoplasm | 118 | 0.002053 |
|
| GO:0098793 | Presynapse | 23 | 0.006006 |
| MF | GO:0005088 | Ras
guanyl-nucleotide exchange factor activity | 34 | 0.001815 |
|
| GO:0019905 | Syntaxin
binding | 26 | 0.003059 |
|
| GO:0044325 | Ion channel
binding | 32 | 0.010635 |
|
| GO:0004683 |
Calmodulin-dependent protein kinase
activity | 12 | 0.023968 |
|
| GO:0004674 | Protein
serine/threonine kinase activity | 74 | 0.032426 |
PPI network
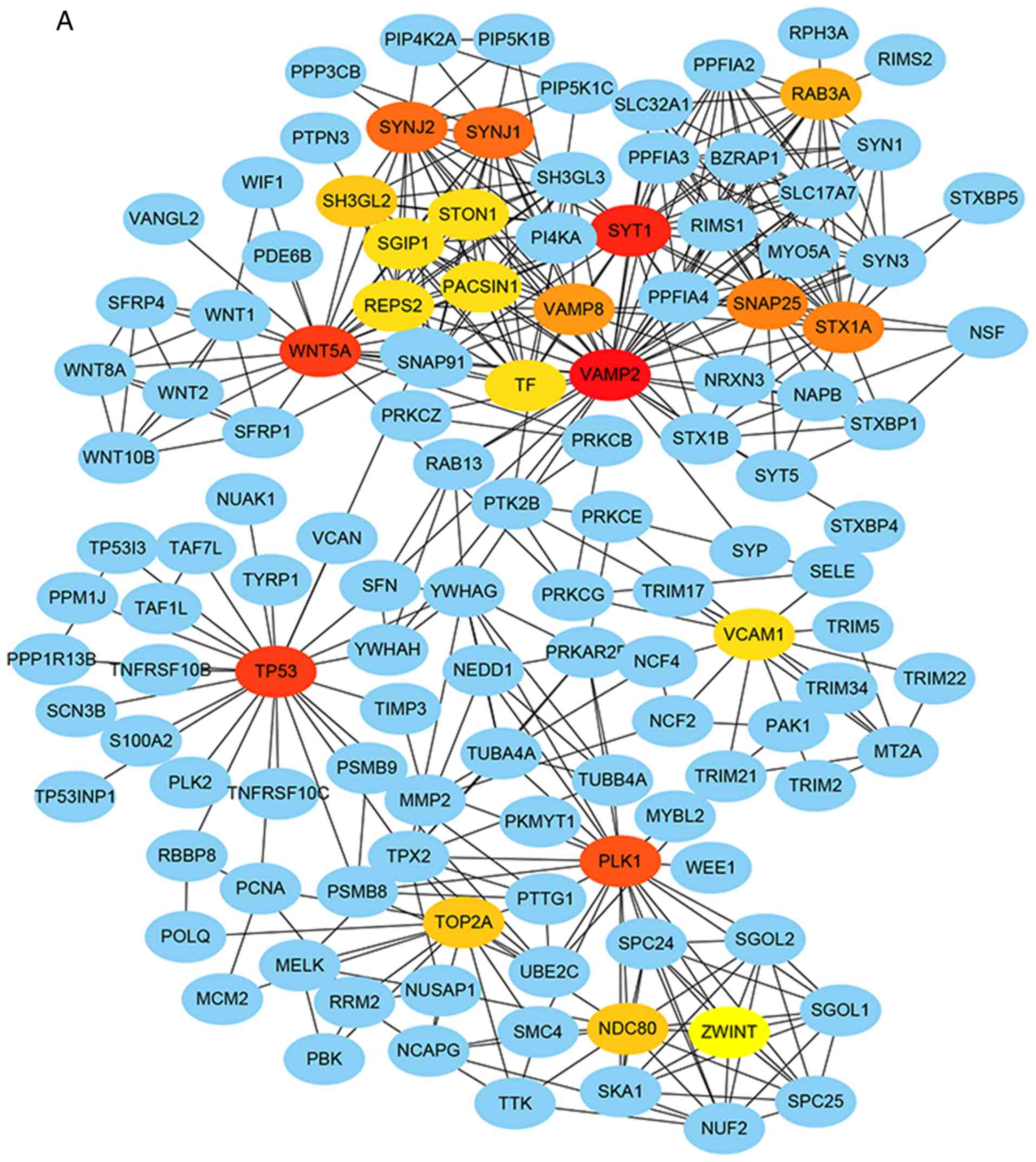
The PPI network of DEGs included 1,843 nodes and
5,116 edges, with each node representing a DEG and the edges
indicating the interactions between DEGs. Twenty genes were
selected as hub genes, which included tumor protein p53
(TP53), polo like kinase 1 (PLK1), nuclear division
cycle 80 (NDC80) and Wnt family member 5A (WNT5A).
Moreover, ZWINT interacted with kinetochore protein NDC80
homolog (NDC80), serine/threonine-protein kinase PLK1
(PLK1), spindle and kinetochore associated complex subunit 1
(SKA1) and tripartite motif containing 17
(Terf/TRIM17) (Fig. 1A). The
significant module included 9 nodes and 36 edges (Fig. 1B). GO and KEGG pathway enrichment
analyses revealed that genes in this module, such as NDC80,
SKA1 and NUF2 component of NDC80 kinetochore complex
(NUF2), were mainly associated with cell division, mitotic
cell cycle, chromosome segregation, and small GTPase-mediated
signal transduction, and they interacted with ZWINT.
Correlation between the expression of
ZWINT and other hub genes
Correlation analysis of the public dataset GSE15824
showed that ZWINT mRNA expression was significantly
positively correlated with NDC80, PLK1, NUF2, SKA1, SPC24
component of NDC80 kinetochore complex (SPC24), SPC25
component of NDC80 kinetochore complex (SPC25), shugoshin 1
(SGOL1) and shugoshin 2 (SGOL2) expression, and the
correlation coefficients were R=0.929, R=0.879, R=0.945, R=0.876,
R=0.888, R=0.965, R=0.804, and R=0.939, respectively. According to
the biological functions of ZWINT, which was enriched in
GBM, these 8 hub genes may also be correlated with cell division
and mitotic cell cycle (Fig.
2).
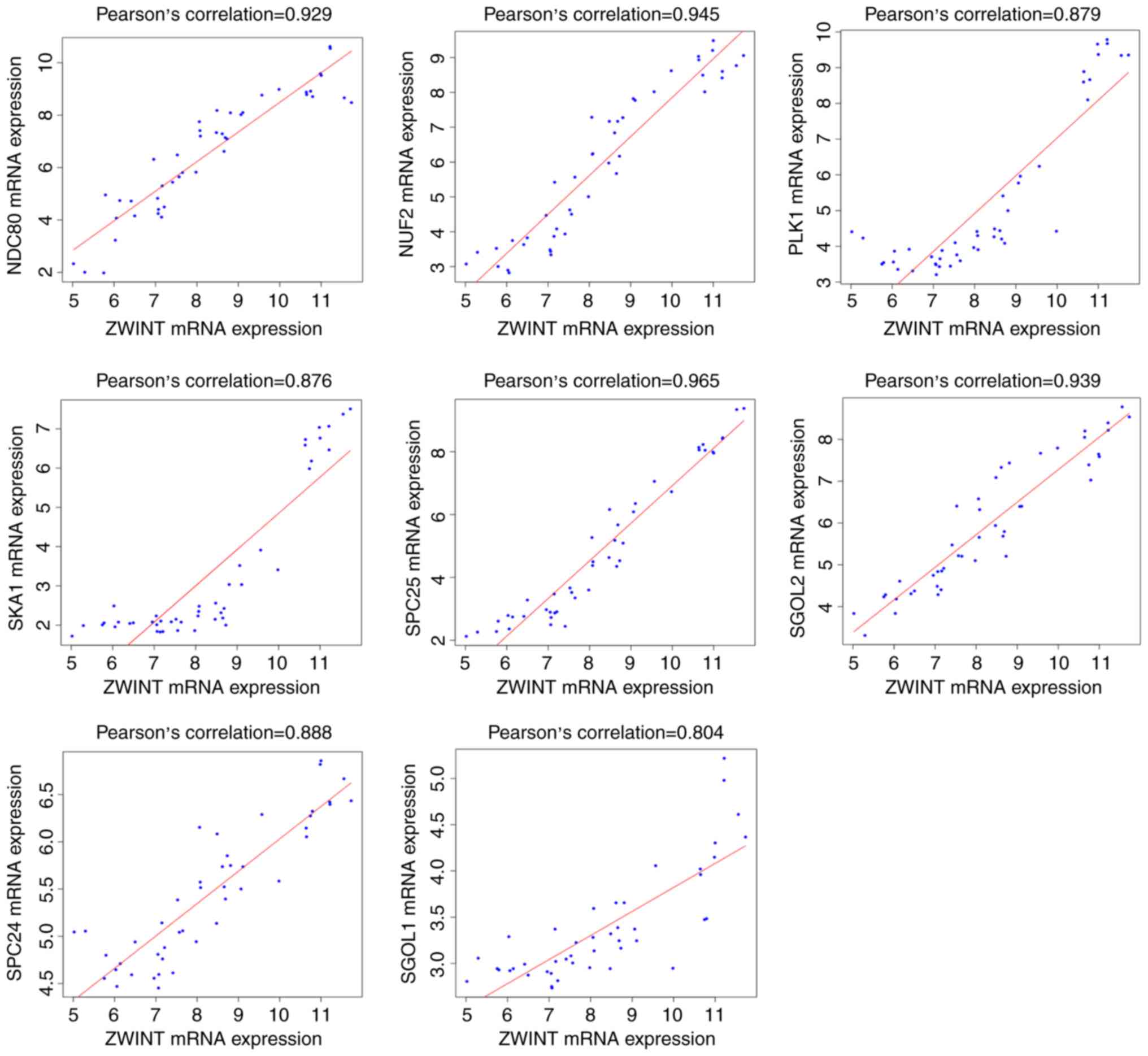 | Figure 2.Correlation between the expression of
ZWINT and the levels of other hub genes in GBM tissues.
Correlation analysis of the public dataset GSE15824 showed that
ZWINT mRNA expression was significantly positively
correlated with the cell division and mitotic cell cycle markers
NDC80, PLK1, NUF2, SKA1, SPC24, SPC25, SGOL1 and SGOL2.
ZWINT, Homo sapiens ZW10 interacting kinetochore protein;
GBM, glioblastoma; NDC80, NDC80 homolog; PLK1,
serine/threonine-protein kinase PLK1; NUF2, NUF2 component
of NDC80 kinetochore complex; SKA1, spindle and kinetochore
associated complex subunit 1; SPC24, SPC24 component of
NDC80 kinetochore complex; SPC25, SPC25 component of NDC80
kinetochore complex; SGOL1, shugoshin 1; SGOL2,
shugoshin 2. |
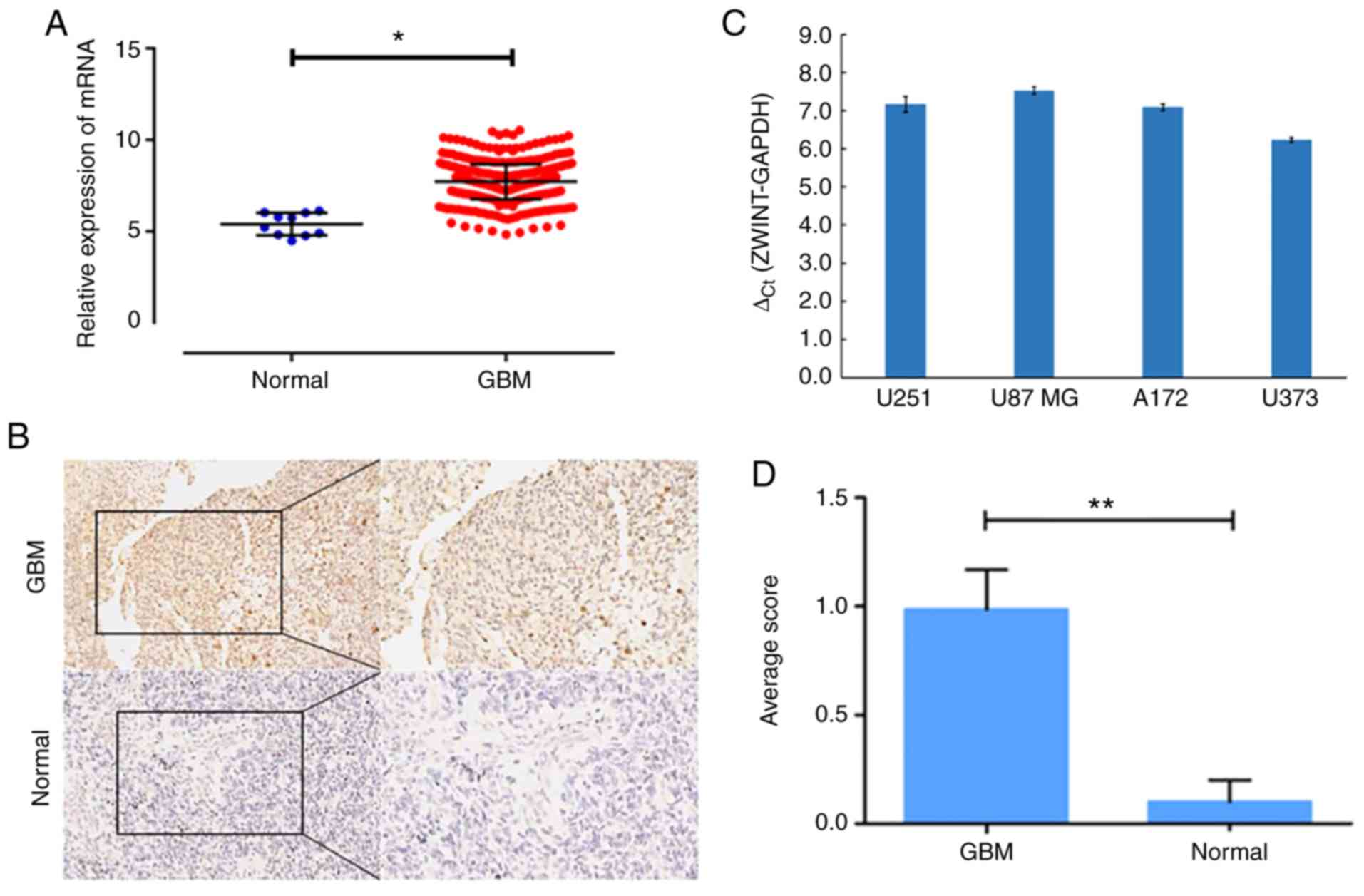
High expression of ZWINT in human GBM
tissues and cell lines
Our data confirmed that ZWINT was among the
upregulated genes, and the expression level of ZWINT mRNA in
GBM tissues was significantly higher than that in normal brain
tissues (548 GBM samples vs. 10 normal samples, Fig. 3A). In addition, IHC analysis of the
TMA was performed to examine the protein level of ZWINT, and a
significant increase was identified in GBM tissues compared with
the level noted in the paired normal brain tissues (Fig. 3B). Scoring results showed that ZWINT
was overexpressed in GBM tissues, and the difference was
statistically significant (P<0.05) (Fig. 3D). qPCR analysis revealed that ZWINT
was upregulated in all four human GBM cell lines (Fig. 3C). Therefore, U251 and U87 MG cells
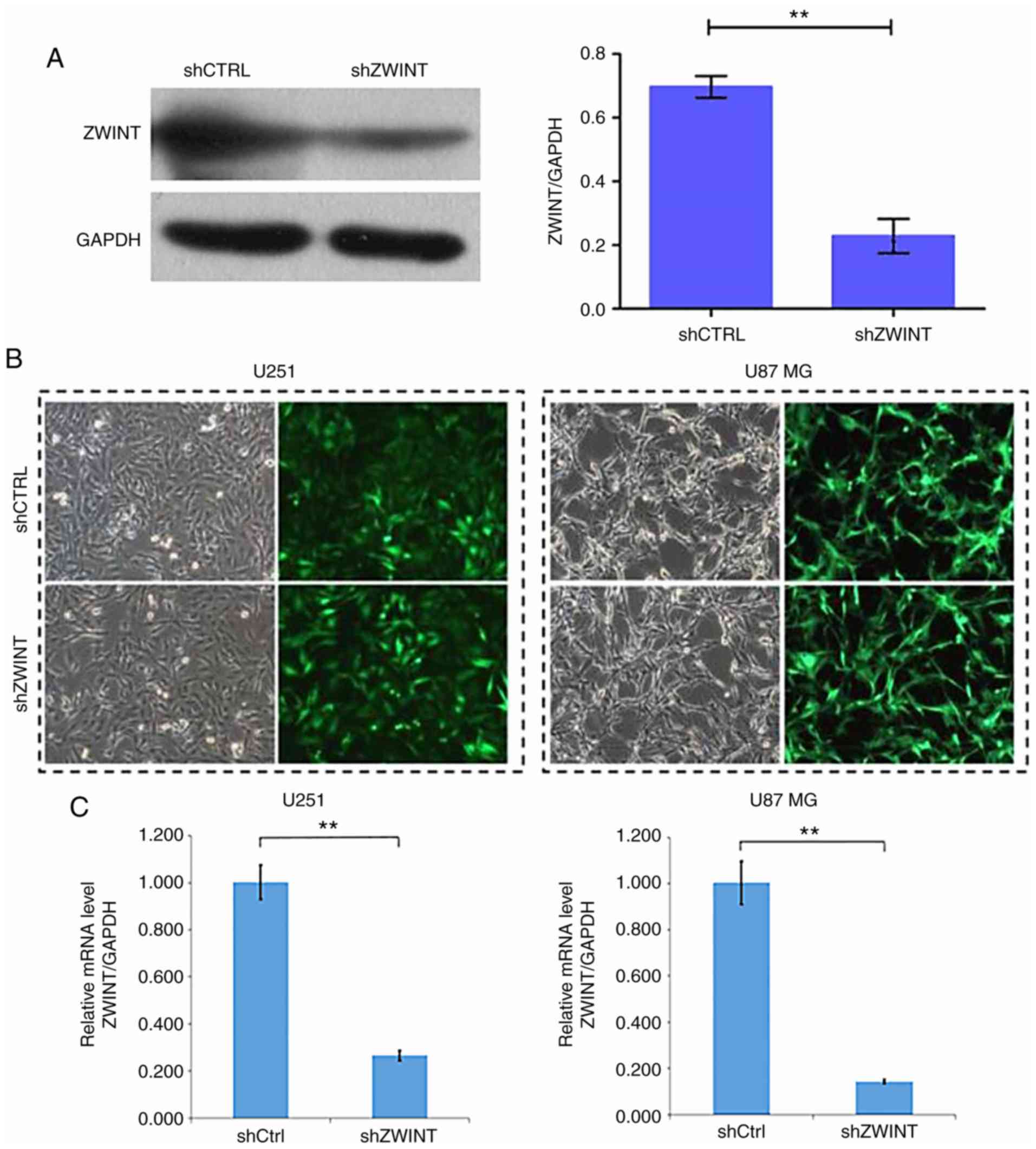
were selected for ZWINT-knockdown experiments. Western blot
analysis revealed that in the 293T cell line transfected with shRNA
targeting ZWINT (shZWINT), the expression level of ZWINT
protein was significantly reduced, indicating an effective
lentivirus-delivered shRNA sequence (Fig. 4A). Over 80% of U251 and U87 MG cells
showed green fluorescent protein expression under a fluorescence
microscope after infection with the recombinant lentiviruses
(Fig. 4B). As detected by qPCR, a
significant reduction in the mRNA level was found in U251 and U87
MG cells infected with shZWINT compared with cells infected with
shCtrl (Fig. 4C).
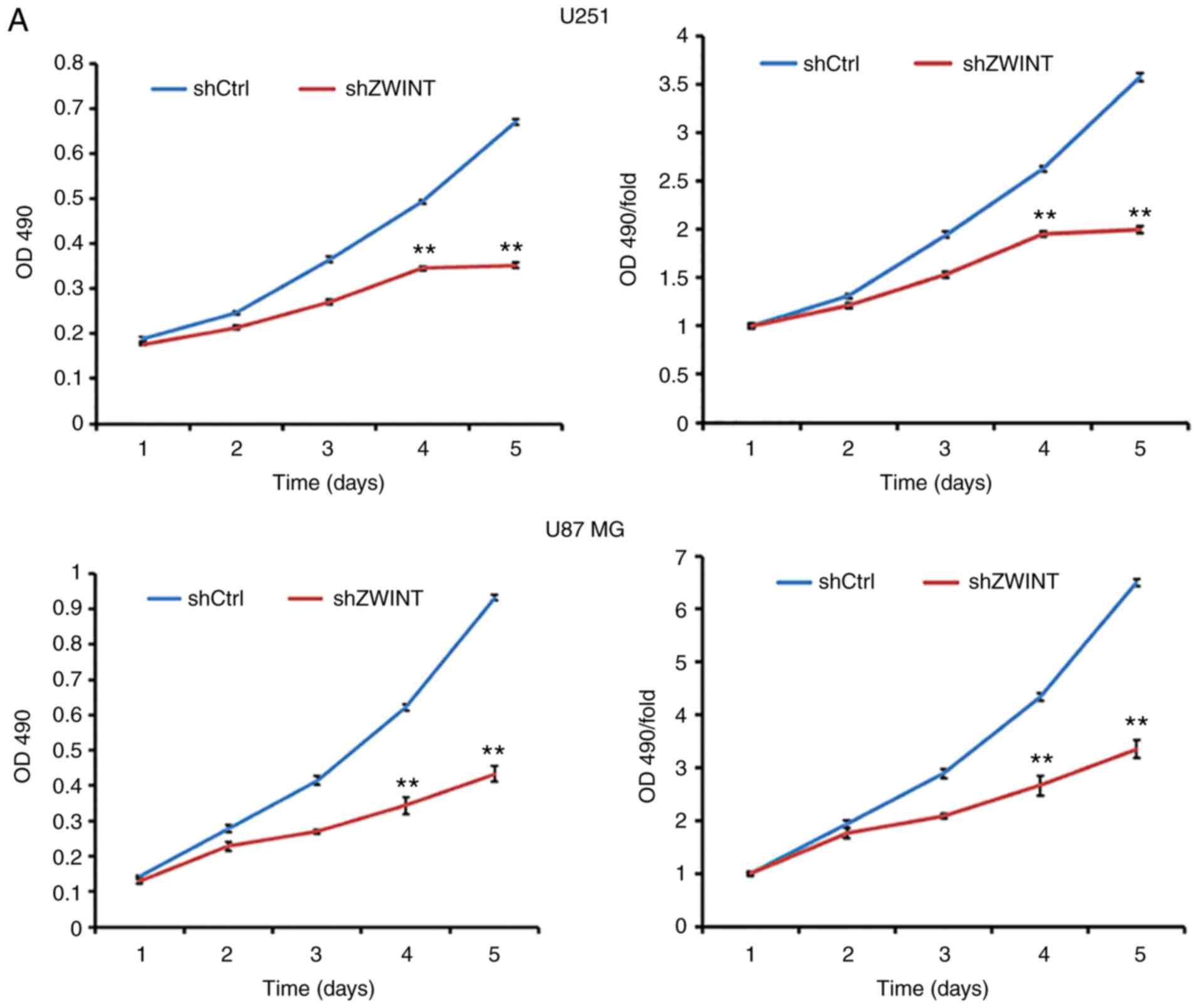
Knockdown of ZWINT inhibits GBM cell
proliferation
The proliferation activity in U251 and U87 MG
ZWINT-knockdown cells was explored by MTT assays. The results
showed that the growth rate of shZWINT U251 and U87 MG cells was
slower than that of the shCtrl group (P<0.01, Fig. 5A). Similarly, the growth curve
counted by the Celigo Image Cytometer revealed that clonogenic
survival in the shZWINT group was markedly decreased (Fig. 5B). Therefore, we speculated that
ZWINT may act as an oncogene to promote GBM cell
proliferation.
Knockdown of ZWINT suppresses GBM cell
invasion
Transwell assays were performed to investigate the
role of ZWINT in GBM cell invasion. As expected, the number of
cells in the shZWINT group that passed through the membrane into
the lower chamber was significantly lower than that in the shCtrl
group. The results showed that there were fewer invading cells in
the shZWINT group than that in the control group at 24 h after
invasion, and the invasive ability was significantly inhibited
(P<0.01, Fig. 6).
Knockdown of ZWINT increases GBM cell
apoptosis
In FACS analysis with Annexin-V, significantly
increased apoptosis was found in the ZWINT shRNA-infected U251 and
U87 MG cells from 1.57 to 11.91% (P<0.01) (Fig. 7A). Similarly, as shown in
caspase-3/7 activity analysis, the level of caspase-3/7 activity in
the cells expressing shZWINT was significantly higher than that in
cells expressing shCtrl (P<0.01) (Fig. 7B). Overall, ZWINT may have an
important role in inhibiting cell apoptosis.
Knockdown of ZWINT inhibits tumor
growth
To further demonstrate the role of ZWINT in
vivo, we performed tumor xenograft assays. The average growth
rate of the tumors in the shZWINT mouse group was significantly
slower than that of the shCtrl group, and the shZWINT group had
significantly decreased tumor volumes and weights compared to those
of the shCtrl group (Fig. 8). The
results indicated that ZWINT may act as a potent protumorigenic
factor that can accelerate GBM growth.
Discussion
Currently, the molecules involved in cell cycle
regulation have garnered wide attention due to their potential
contribution to neoplastic transformation, and targeting cell cycle
checkpoints may provide substantial improvement to malignancy
therapy (27). However, there have
been limited studies regarding these molecules in glioblastoma
(GBM). In the present study, we first analyzed the differentially
expressed genes (DEGs) between GBM and normal brain tissues from a
TCGA dataset, and then ZWINT was analyzed by high-content
screening. There are few data regarding ZWINT expression in
tumorigenesis, especially in regards to GBM progression. This
finding prompted us to perform qPCR and IHC to verify ZWINT
expression in GBM cell lines and tissues. Our data demonstrated
that ZWINT was markedly overexpressed at both the mRNA and protein
levels in GBM. We further determined that interfering with ZWINT
could inhibit the proliferation and invasion and accelerate the
apoptosis of human GBM cell lines in vitro, and it was
significantly related to the tumor growth process in vivo.
Consequently, targeting ZWINT may constitute a powerful strategy
for the development of novel therapies for GBM.
Gene polymorphism is one of the biological bases of
tumorigenesis, thus tumors can be considered genetic diseases.
Kinetochore assembly is arguably the most critical step in
determining accurate chromosome segregation, which is crucial for
maintaining genomic integrity. Kinetochores are composed of a
number of proteins (NDC80, MIS12, ZW10, Zwint-1, ZWILCH), which are
responsible for stabilizing the kinetochore-microtubule (KMT)
attachment and recruiting the components of the spindle assembly
checkpoint (SAC) (28–31). ZWINT interacts with ZW10 to ensure
correct chromosome motility and mitotic spindle checkpoint
operation (32). ZWINT
encodes a protein that is located in prophase kinetochores before
ZW10 does, even remaining detectable on the kinetochore until late
anaphase, and is distributed in the cytoplasm of interphase cells.
We speculated that only correcting erroneous
kinetochore-microtubule attachment in a timely manner and
regulating spindle checkpoint function can maintain mitotic cell
cycle integrity. ZWINT, as a part of the kinetochore, is required
for cell proliferation and growth. Therefore, increased ZWINT
causing chromosome instability can be associated with cancer
progression.
In the era of ‘big’ data, a large number of tumor
gene expression profiles have been widely used. This study made
comprehensive use of various database resources and bioinformatic
software to mine the DEGs during the occurrence and development of
GBM to screen effective molecular targets. Functional and signaling
pathway enrichment analyses were applied to identify several hub
genes that interact with ZWINT to participate in the mitosis and
regulation of the cell cycle of GBM cells. Tumorigenesis is a
complex pathogenetic process that is driven by specific genetic and
epigenetic alterations. In our present research, 6,710 DEGs were
screened in total and consisted of 2,387 upregulated genes and
3,873 downregulated genes. The upregulated genes are mainly
responsible for mitotic nuclear division, cell division, sister
chromatid cohesion and cell proliferation, and the downregulated
genes mostly participate in signal transduction, ion transport, and
regulation of synaptic potential. Twenty hub genes had high degrees
in the PPI network. We found that ZWINT was associated with NDC80
homolog (NDC80), serine/threonine-protein kinase PLK1
(PLK1), spindle and kinetochore associated complex subunit 1
(SKA1), which interact with each other and are mainly
involved in the mitotic cell cycle, cell division, signal
transduction, and AMPK signaling pathway. Abnormal gene expression
or dysfunction is closely related to neoplasia.
The mitotic regulator NDC80 is highly expressed in
various human malignancies, including hepatocellular carcinoma,
colon cancer and osteosarcoma (33–35).
NDC80 is also called Hec1, and its complexes together with NUF2
component of NDC80 kinetochore complex (NUF2), SPC24
component of NDC80 kinetochore complex (SPC24) and SPC25
component of NDC80 kinetochore complex (SPC25) participate
in the spindle assembly checkpoint and regulation of mitosis and
chromosome segregation (36–39).
Lin et al showed that Hec1 sequentially recruits Zwint-1 and
ZW10 to kinetochores during the mitotic phase of the cell cycle,
and interruption of the centromeric recruitment led to chromosomal
missegregation, incomplete activation of spindle examination
points, and ultimately cell death (31). The above findings offer a
theoretical basis for how NDC80 and ZWINT promote faithful
chromosome segregation and control the spindle checkpoint.
Tripartite motif containing 17 (Terf/TRIM17) is a
tripartite motif (TRIM) protein, and its coiled-coil domain is
required for interaction with ZWINT. Therefore, its role in
oncogenic events may be by regulating the turnover of the ZWINT
protein. Endo et al found that Terf/TRIM17 exhibits E3
ubiquitin ligase activity by stimulating the degradation of the
kinetochore protein ZWINT and negatively regulating cell
proliferation via the proteasomal pathway in mammalian cells
(5). We can use these interactions
to inhibit the growth of tumor cells, and the specific molecular
mechanism will be elucidated by future studies.
PLK1 and SKA1 are two newly discovered
genes associated with mitosis and tumorigenesis. PLK1
encodes a serine/threonine protein kinase, which is a critical
regulator of cell cycle progression, cytokinesis, mitosis and the
DNA damage response (40). The
deletion of the PLK1 protein dramatically inhibited cancer cell
proliferation and induced apoptosis (41). The SKA1 complex is a
microtubule-binding subcomplex of the outer kinetochore and is
essential for proper chromosome segregation (42). Previous data have shown that
knockdown of SKA1 inhibits cell proliferation and migration
and blocks the cell cycle; moreover, inhibition of SKA1 by
small-molecule inhibitors could restrain the activity of the AKT
and ERK signaling pathways (43,44).
These studies have shown that ZWINT, NDC80, PLK1, SKA1 and
Terf/TRIM17 participate in the pathogenesis of malignant neoplasms
by affecting mitosis and cell cycle progression, which supports our
findings.
The present research has some limitations. First, it
is not sufficient to predict target genes only based on
bioinformatics, and further mechanistic studies of the way ZWINT
affects tumor cell proliferation, invasion and apoptosis are
necessary to better understand the roles of the underlying
molecule. Second, only two GBM cell lines were used in our study,
and the function of ZWINT should be demonstrated in more cell
lines. Finally, additional clinical information is needed to
identify its prognostic value in GBM.
In summary, our preliminary research demonstrated
that ZWINT may be a promising biomarker as it promotes GBM cell
proliferation and invasion and inhibits apoptosis. Knockdown of
ZWINT inhibited tumorigenesis in a xenograft model. Data
mining and integration may be an efficient tool with which to
predict the progression of GBM. The DEGs identified by
comprehensive bioinformatic analyses may represent a valuable
resource that may predict the progression of GBM.
Acknowledgements
Not applicable.
Funding
This study was supported by a grant from the
National Natural Sciences Foundation of China (no. 81772680).
Availability of data and materials
All datasets generated during our present study are
available through data mining from public gene databases.
Authors' contributions
All the authors have made substantial contributions
to this manuscript, LY, NH, and MZ were involved in the conception
and design of the study. MZ guided the cell biology and animal
experiments and was responsible for the whole project; XZ and YZ
performed the bioinformatic data collection, integration analysis
and figure processing. LY conducted the cell biology experiments;
RC contributed to the animal experiments and project management; LY
and NH drafted and edited the manuscript; and MZ critically revised
the manuscript for important intellectual content, as well as the
collection of data. All authors confirmed and approved the final
manuscript.
Ethics approval and consent to
participate
The animal experiments in the present study were
approved by the Ethics Committee for Animal Experimentation of
Tongji Hospital, Tongji Medical College Huazhong University of
Science and Technology and in accordance to the institutional and
university guidelines on the care and use of experimental
animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liebelt BD, Shingu T, Zhou X, Ren J, Shin
SA and Hu J: Glioma stem cells: Signaling, microenvironment, and
therapy. Stem Cells Int. 2016:78498902016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Biedermann J, Preussler M, Conde M,
Peitzsch M, Richter S, Wiedemuth R, Abou-El-Ardat K, Krüger A,
Meinhardt M, Schackert G, et al: Mutant IDH1 differently affects
redox state and metabolism in glial cells of normal and tumor
origin. Cancers (Basel). 11(pii): E20282019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chai RC, Zhang KN, Chang YZ, Wu F, Liu YQ,
Zhao Z, Wang KY, Chang YH, Jiang T and Wang YZ: Systematically
characterize the clinical and biological significances of 1p19q
genes in 1p/19q non-codeletion glioma. Carcinogenesis.
40:1229–1239. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Asif S, Fatima R, Krc R, Bennett J and
Raza S: Comparative proteogenomic characterization of glioblastoma.
CNS Oncol. 8:CNS372019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Endo H, Ikeda K, Urano T, Horie-Inoue K
and Inoue S: Terf/TRIM17 stimulates degradation of kinetochore
protein ZWINT and regulates cell proliferation. J Biochem.
151:139–144. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Starr DA, Saffery R, Li Z, Simpson AE,
Choo KH, Yen TJ and Goldberg ML: HZwint-1, a novel human
kinetochore component that interacts with HZW10. J Cell Sci.
113:1939–1950. 2000.PubMed/NCBI
|
|
7
|
Famulski JK, Vos L, Sun X and Chan G:
Stable hZW10 kinetochore residency, mediated by hZwint-1
interaction, is essential for the mitotic checkpoint. J Cell Biol.
180:507–520. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peng F, Li Q, Niu SQ, Shen GP, Luo Y, Chen
M and Bao Y: ZWINT is the next potential target for lung cancer
therapy. J Cancer Res Clin Oncol. 145:661–673. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang XY, Wu B, Ma SL, Yin L, Wu MC and Li
AJ: Decreased expression of ZWINT is associated with poor prognosis
in patients with HCC after surgery. Technol Cancer Res Treat.
17:15330338187941902018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ying H, Xu Z, Chen M, Zhou S, Liang X and
Cai X: Overexpression of Zwint predicts poor prognosis and promotes
the proliferation of hepatocellular carcinoma by regulating
cell-cycle-related proteins. Onco Targets Ther. 11:689–702. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kumar S, Clarke D and Gerstein MB:
Leveraging protein dynamics to identify cancer mutational hotspots
using 3D structures. Proc Natl Acad Sci USA. 116:18962–18970. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ding J, McConechy MK, Horlings HM, Ha G,
Chun Chan F, Funnell T, Mullaly SC, Reimand J, Bashashati A, Bader
GD, et al: Systematic analysis of somatic mutations impacting gene
expression in 12 tumour types. Nat Commun. 6:85542015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deng M, Brägelmann J, Schultze JL and
Perner S: Web-TCGA: An online platform for integrated analysis of
molecular cancer data sets. BMC Bioinformatics. 17:722016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Johann PD, Jäger N, Pfister SM and Sill M:
RF_Purify: A novel tool for comprehensive analysis of tumor-purity
in methylation array data based on random forest regression. BMC
Bioinformatics. 20:4282019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shivakumar M, Lee Y, Bang L, Garg T, Sohn
KA and Kim D: Identification of epigenetic interactions between
miRNA and DNA methylation associated with gene expression as
potential prognostic markers in bladder cancer. BMC Med Genomics.
10 (Suppl 1):S302017. View Article : Google Scholar
|
|
16
|
Cooper LA, Demicco EG, Saltz JH, Powell
RT, Rao A and Lazar AJ: PanCancer insights from the cancer genome
atlas: The pathologist's perspective. J Pathol. 244:512–524. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
McCarthy DJ, Chen Y and Smyth GK:
Differential expression analysis of multifactor RNA-Seq experiments
with respect to biological variation. Nucleic Acids Res.
40:4288–4297. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Grzmil M, Morin PJ Jr, Lino MM, Merlo A,
Frank S, Wang Y, Moncayo G and Hemmings BA: MAP kinase-interacting
kinase 1 regulates SMAD2-dependent TGF-β signaling pathway in human
glioblastoma. Cancer Res. 71:2392–2402. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Z, Zhao K and Tian H: Integrated
analysis of differential expression and alternative splicing of
non-small cell lung cancer based on RNA sequencing. Oncol Lett.
14:1519–1525. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43((Database Issue)): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Franz M, Lopes CT, Huck G, Dong Y, Sumer O
and Bader GD: Cytoscape.js: A graph theory library for
visualisation and analysis. Bioinformatics. 32:309–311.
2016.PubMed/NCBI
|
|
25
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. Bmc Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Visconti R, Della Monica R and Grieco D:
Cell cycle checkpoint in cancer: A therapeutically targetable
double-edged sword. J Exp Clin Cancer Res. 35:1532016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheeseman IM and Desai A: Molecular
architecture of the kinetochore-microtubule interface. Nat Rev Mol
Cell Biol. 9:33–46. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Varma D and Salmon ED: The KMN protein
network-chief conductors of the kinetochore orchestra. J Cell Sci.
125:5927–5936. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Obuse C, Iwasaki O, Kiyomitsu T, Goshima
G, Toyoda Y and Yanagida M: A conserved Mis12 centromere complex is
linked to heterochromatic HP1 and outer kinetochore protein
Zwint-1. Nat Cell Biol. 6:1135–1141. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin YT, Chen Y, Wu G and Lee WH: Hec1
sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful
chromosome segregation and spindle checkpoint control. Oncogene.
25:6901–6914. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Woo Seo D, Yeop You S, Chung WJ, Cho DH,
Kim JS and Su Oh J: Zwint-1 is required for spindle assembly
checkpoint function and kinetochore-microtubule attachment during
oocyte meiosis. Sci Rep. 5:154312015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ju LL, Chen L, Li JH, Wang YF, Lu RJ, Bian
ZL and Shao JG: Effect of NDC80 in human hepatocellular carcinoma.
World J Gastroenterol. 23:3675–3683. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xing XK, Wu HY, Chen HL and Feng HG: NDC80
promotes proliferation and metastasis of colon cancer cells. Genet
Mol Res. 15:2016. View Article : Google Scholar :
|
|
35
|
Xu B, Wu DP, Xie RT, Liu LG and Yan XB:
Elevated NDC80 expression is associated with poor prognosis in
osteosarcoma patients. Eur Rev Med Pharmacol Sci. 21:2045–2053.
2017.PubMed/NCBI
|
|
36
|
Matsuo Y, Maurer SP, Surrey T and Toda T:
Purification and characterisation of the fission yeast Ndc80
complex. Protein Expr Purif. 135:61–69. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Valverde R, Ingram J and Harrison SC:
Conserved tetramer junction in the kinetochore Ndc80 complex. Cell
Rep. 17:1915–1922. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Suzuki A, Badger BL and Salmon ED: A
quantitative description of Ndc80 complex linkage to human
kinetochores. Nat Commun. 6:81612015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ferrara M, Sessa G, Fiore M, Bernard F,
Asteriti IA, Cundari E, Colotti G, Ferla S, Desideri M, Buglioni S,
et al: Small molecules targeted to the microtubule-Hec1 interaction
inhibit cancer cell growth through microtubule stabilization.
Oncogene. 37:231–241. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liao Y, Lin D, Cui P, Abbasi B, Chen C,
Zhang Z, Zhang Y, Dong Y, Rui R and Ju S: Polo-like kinase 1
inhibition results in misaligned chromosomes and aberrant spindles
in porcine oocytes during the first meiotic division. Reprod Domest
Anim. 53:256–265. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ma X, Wang L, Huang D, Li Y, Yang D, Li T,
Li F, Sun L, Wei H, He K, et al: Polo-like kinase 1 coordinates
biosynthesis during cell cycle progression by directly activating
pentose phosphate pathway. Nat Commun. 8:15062017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sivakumar S and Gorbsky GJ:
Phosphatase-regulated recruitment of the spindle- and
kinetochore-associated (Ska) complex to kinetochores. Biol Open.
6:1672–1679. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhao LJ, Yang HL, Li KY, Gao YH, Dong K,
Liu ZH, Wang LX and Zhang B: Knockdown of SKA1 gene inhibits cell
proliferation and metastasis in human adenoid cystic carcinoma.
Biomed Pharmacother. 90:8–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang K, Sun J, Teng J, Yu Y, Zhong D and
Fan Y: Overexpression of spindle and kinetochore-associated protein
1 contributes to the progression of prostate cancer. Tumour Biol.
39:10104283177019182017. View Article : Google Scholar : PubMed/NCBI
|