Introduction
Liver cancer is one of the most common malignant
tumor types, with an incidence rate ranked sixth, and a mortality
rate ranked third worldwide (1).
Liver cancer is characterized by a rapid development, poor curative
effect and high recurrence rate, and the five-year survival rate is
<30% (2,3). Surgery is the only available method to
treat developed cases, but most patients are diagnosed at an
advanced stage and have already lost the opportunity for surgery.
At present, drug treatment including chemotherapy and targeted
therapy is not effective for the treatment of liver cancer
(3–5), and the prognosis of liver cancer
patients is still poor (4,6,7).
Therefore, it is crucial to develop a more efficient and safer drug
for the treatment of liver cancer.
Autophagy, a highly conserved biological process in
eukaryotic cells, is a lysosomal-mediated metabolic process that
degrades damaged proteins or organelles to maintain cell metabolism
and cell renewal, which plays a key role in maintaining homeostasis
in the cellular environment (8–10).
Dysregulation of autophagy is associated with a variety of diseases
including multiple forms of cancer (11,12).
Autophagy plays a ‘double-edged sword’ role, promotion and
suppression, in tumor cells, which depends on the type and stage of
tumor development (13). In the
early stages of cancer cell expansion, autophagy provides energy
and nutrients to normal cells, which can inhibit the proliferation
of cancer cells (14). During tumor
progression, autophagy can enhance the tolerance of cancer cells to
low-nutrient environments to promote cancer cell proliferation, and
also increase the resistance of cancer cells to anticancer drugs
(15). A previous study has
revealed that deletion of the autophagy marker gene ATG6/Beclin-1
often occurs in a variety of cancer types (16). In addition, in animal models,
Beclin-1 gene-deficient mice were more susceptible to developing
cancer types (17). The
aforementioned studies indicated that Beclin-1 is a tumor
suppressor gene, and that autophagy is closely related to the
occurrence and development of tumors. Generally, autophagy is a
non-specific degradation of intracellular components, but autophagy
can also specifically degrade some target proteins (9,10). If
specific cytoprotective factors are degraded, autophagic cell death
will occur (18). Autophagy and
apoptosis, two different forms of programmed cell death, often
occur simultaneously when cells are under stress (19). The relationship between autophagy
and apoptosis is a research hotspot. Studies have revealed that
there are four relationships between autophagy and apoptosis
(19,20): i) Autophagy precedes apoptosis and
maintains cell homeostasis; ii) apoptosis inhibits autophagic cell
death and inhibition of apoptosis stimulates autophagic cell death;
iii) autophagy degrades damaged organelles and proteins as well as
attenuating DNA damage, thereby inhibiting apoptosis; iv) autophagy
and apoptosis exist independently. The gene regulatory network
behind autophagy and apoptosis is highly linked. The anti-apoptotic
protein B-cell lymphoma 2 (Bcl-2) inhibits the formation of
autophagosomes by binding to the pro-autophagic protein, Beclin-1
(21,22). Notably, caspases, a highly studied
group of essential apoptotic proteins, can cause the inhibition of
autophagy by breaking down autophagy-associated proteins, including
p62, Beclin-1, ATG5 and ATG3 (23–25).
In addition, multiple autophagy-related proteins are also involved
in apoptosis when cells are under stress (26,27).
Mammalian target of rapamycin (mTOR), a serine/threonine protein
kinase, is involved in various physiological functions, such as
cell proliferation, apoptosis, autophagy and the cell cycle
(28,29). mTOR is primarily activated by the
PI3K/AKT/mTOR signaling pathway (28). The activated PI3K/AKT/mTOR signaling
pathway plays an important role in the occurrence and development
of various tumor types (30).
Inhibition of this pathway has become an important target for
cancer therapy, and multiple mTOR inhibitors have shown
satisfactory efficacy (31–33).
Veratramine, a known natural steroidal alkaloid, is
found in a variety of plants of the lily family. Previous studies
have revealed that veratramine lowers blood pressure, antagonizes
sodium ion channels and functions as an analgesic. A recent study
revealed that veratramine inhibited tumor cell proliferation in
vitro (34). Bai et al
(35) also revealed that
veratramine inhibited the downstream signaling pathway of
transcription factor activator protein-1 (AP-1) that regulates
multiple cell functions including proliferation, apoptosis and
epithelial-mesenchymal transition (EMT). These results indicated
that veratramine may have antitumor activity, but the effect of
veratramine on liver cancer and the mechanism behind its antitumor
activity are unclear. In the present study, the aim was to
investigate the effects of veratramine on liver cancer in
vitro and in vivo. In addition, as part of the present
work, a focus was applied to investigate whether veratramine can
induce autophagic cell death in HepG2 cells, in order to clarify
the method of action behind its antitumor activity.
Materials and methods
Cell lines and reagents
Veratramine (with a purity ≥98%) was extracted and
isolated from the roots and rhizomes of Veratrum nigrum L.,
and provided by Nanjing Spring & Autumn Biological Engineering
Co., Ltd. The chemical structure of veratramine is presented in
Fig. 1A. The human liver cancer
HepG2 cell line was purchased from the American Type Culture
Collection. FBS, penicillin streptomycin and cell culture media
were purchased from Hangzhou Sijiqing Biological Engineering
Materials Co., Ltd. The Cell Counting Kit-8 (CCK-8) assay kit was
obtained from Nanjing KeyGen Biotech Co., Ltd. The apoptosis
detection kit and antibodies used for western blotting were
purchased from Beyotime Institute of Biotechnology. The reverse
transcription kit and PCR kit were purchased from Vazyme Biotech
Co., Ltd. Acridine orange (AO) and 3-methyladenine (3-MA) were
obtained from Sigma-Aldrich; Merck KGaA. All other reagents and
solvents (AR grade) were purchased from Sinopharm Chemical Reagent
Co., Ltd. Ten BALB/c nude mice, aged 4–6 weeks (16–18 g), were
obtained from Shanghai Slack Laboratory Animals Co., Ltd. These
mice were housed in specialized mouse cages at ~20°C, humidity
50–60%, specific pathogen-free, dark conditions, and fed with
normal food and clean water.
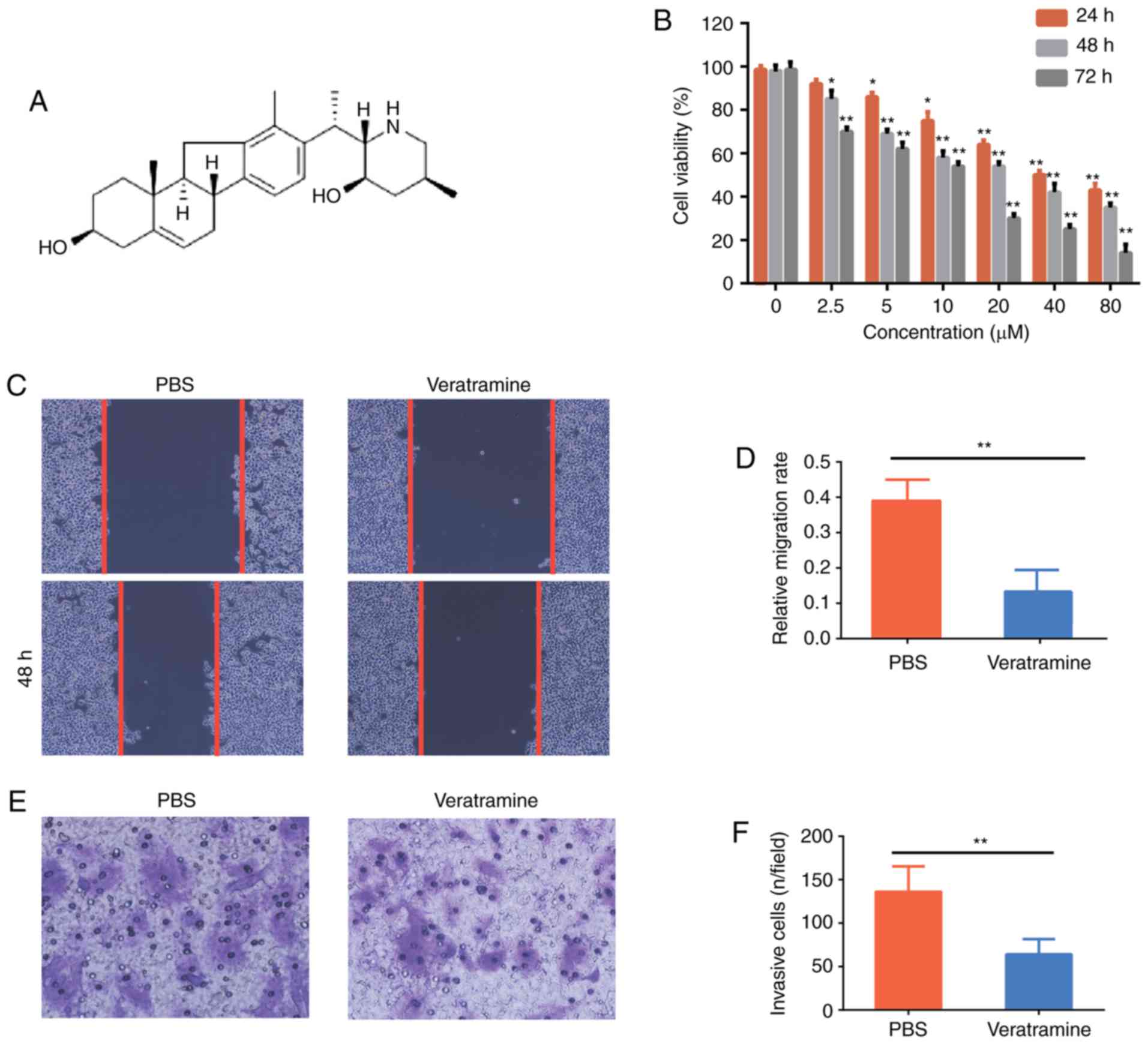 | Figure 1.Veratramine inhibits HepG2 cell
proliferation, migration and invasion in vitro. (A)
Structure of veratramine. (B) HepG2 cells were treated with various
concentrations of veratramine (0, 2.5, 5, 10, 20, 40 and 80 µM) for
24, 48 and 72 h. The cell viability was determined using CCK-8
assays. (C) HepG2 cells were treated with veratramine (40 µM) for
48 h. Cell migration was analyzed using wound scratch assays.
Representative images are presented (magnification, ×200). (D)
Statistical data for the wound scratch assays. (E) Cells were
treated as described and cell invasion was detected using Transwell
assays. Representative images are presented (magnification, ×200).
(F) Quantitative analysis of invading cells. Data are presented as
the mean ± SD of three independent experiments. *P<0.05,
**P<0.01 vs. the control group (PBS). |
Cell culture
The human liver cancer HepG2 cells were cultured in
RPMI-1640 media containing 10% FBS, penicillin (100 units/ml) and
streptomycin (100 units/ml). The cells were maintained in a humid
incubator (Sanyo XD-101; Sanyo) with 5% CO2 at 37°C.
Cell viability assay
Cell viability was assessed using a CCK-8 assay.
HepG2 cells (5×103 cells/well) were seeded in 96 well
plates for 24 h, and then treated with various concentrations of
veratramine (0, 2.5, 5, 10, 20, 40 and 80 µM) for an additional 24,
48 or 72 h. Subsequently, 10 µl of CCK-8 solution was added to each
well and the cells were further incubated for 4 h. The absorbance
was measured at 450 nm using a microplate reader (Bio-Rad
Laboratories, Inc.). IC50 values were calculated using
the cell survival data curve.
Cell migration and invasion assay
The effect of veratramine on HepG2 cell migration
and invasion was assessed using a wound healing assay and Transwell
assay, respectively. For the wound healing assay, HepG2 cells were
seeded in 6 well plates and cultured to 90% confluence. The cell
monolayer was scratched using a 10-µl pipette tip. Subsequently,
the cells were treated with veratramine (40 µM) for 48 h. After
drug treatment, the cells were observed through an inverted
microscope (Olympus Corporation), and the width of the scratch was
measured.
For the Transwell assay, Transwell culture inserts
were placed into the wells of 6-well plates and coated with a layer
of Matrigel™. Subsequently, RPMI-1640 medium containing 10% FBS was
added to the lower chamber and HepG2 cells were seeded in the upper
chamber. After veratramine (40 µM) treatment for 48 h, the invaded
cells in the lower chamber were fixed with 1% formaldehyde for 10
min at 25°C and stained with 0.5% crystal violet for another 5 min.
Finally, the cells were observed under an inverted microscope
(magnification, ×200).
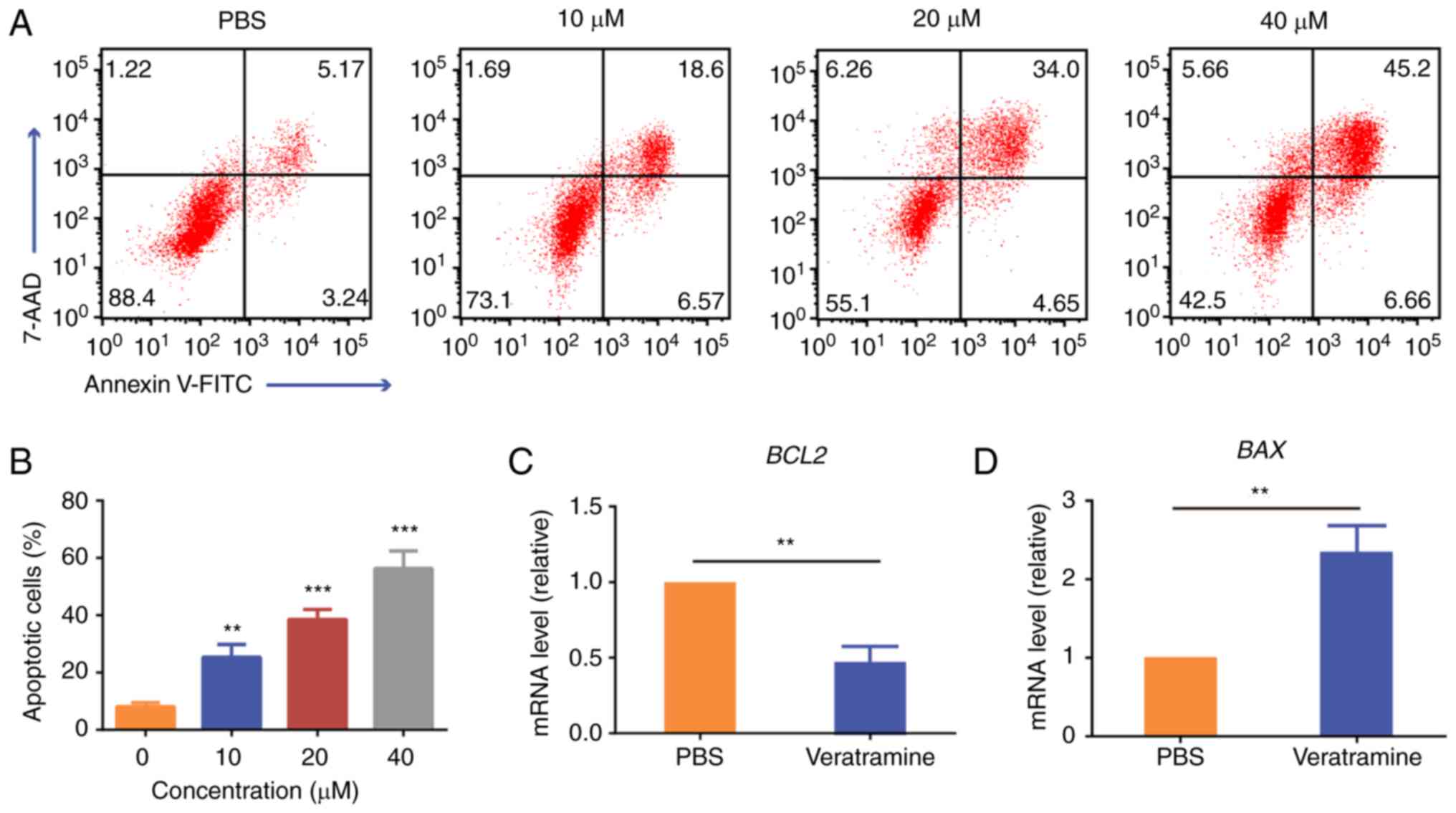
Detection of apoptosis
Cell apoptosis was evaluated using the Annexin
V-FITC/7AAD double staining assay. Briefly, HepG2 cells
(3×105 cells/well) were seeded in 6-well plates for 24
h. Subsequently, the cells were treated with increasing
concentrations of veratramine (10, 20 and 40 µM) for 48 h. After
incubation, the cells were harvested and stained with Annexin
V-FITC and 7AAD according to the manufacturer's instructions.
Apoptotic cells were quantified using flow cytometry (FACSCalibur;
BD Biosciences) and the data were analyzed using FlowJo software
(FlowJo V10; Tree Star, Inc.).
Total RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
RT-qPCR was used to evaluate the expression levels
of the key apoptotic genes, BCL2 and BAX, and
AP-1 downstream gene ETS1. After phosphate-buffered
saline (PBS) or veratramine treatment for 48 h, HepG2 cells were
subjected to total RNA isolation using TRIzol® reagent.
Subsequently, cDNA was synthesized using the HiScript III 1st
Strand cDNA Synthesis kit and qPCR was performed using the ChamQ
SYBR qPCR Master Mix according to the manufacturer's instructions.
Thermocycling parameters were optimized as 5 min at 95°C, followed
by 40 cycles at 95°C (15 sec), 60°C (30 sec); melt curve: 95°C (15
sec), 60°C (30 sec) and 95°C (15 sec). PCR primers were provided by
Guangzhou RiboBio Co., Ltd., and β-actin was used as the internal
reference. Quantitative analysis of relative mRNA expression levels
was performed according to the 2−ΔΔCq method (36). Primer sequences were as follows:
BCL2 forward, 5′GTGGAGGAGCTCTTCAGGGA3′ and reverse,
5′AGGCACCCAGGGTGATGCAA3′); BAX forward,
5′GGCCCACCAGCTCTGAGCAGA3′ and reverse, 5′GCCACGTGGGCGTCCCAAAGT3′;
ETS1 forward, 5′GCGCGCTAGCAACTTGCTACCATCCCGT3′ and reverse,
5′GCGCAAGCTTTGCCATCACTCGTCGGC3′; β-actin forward,
5′ACAAAGTGGTCATTGAGGGC3′and reverse, 5′GCCGTCAGGCAGCTCGTAGC3′.
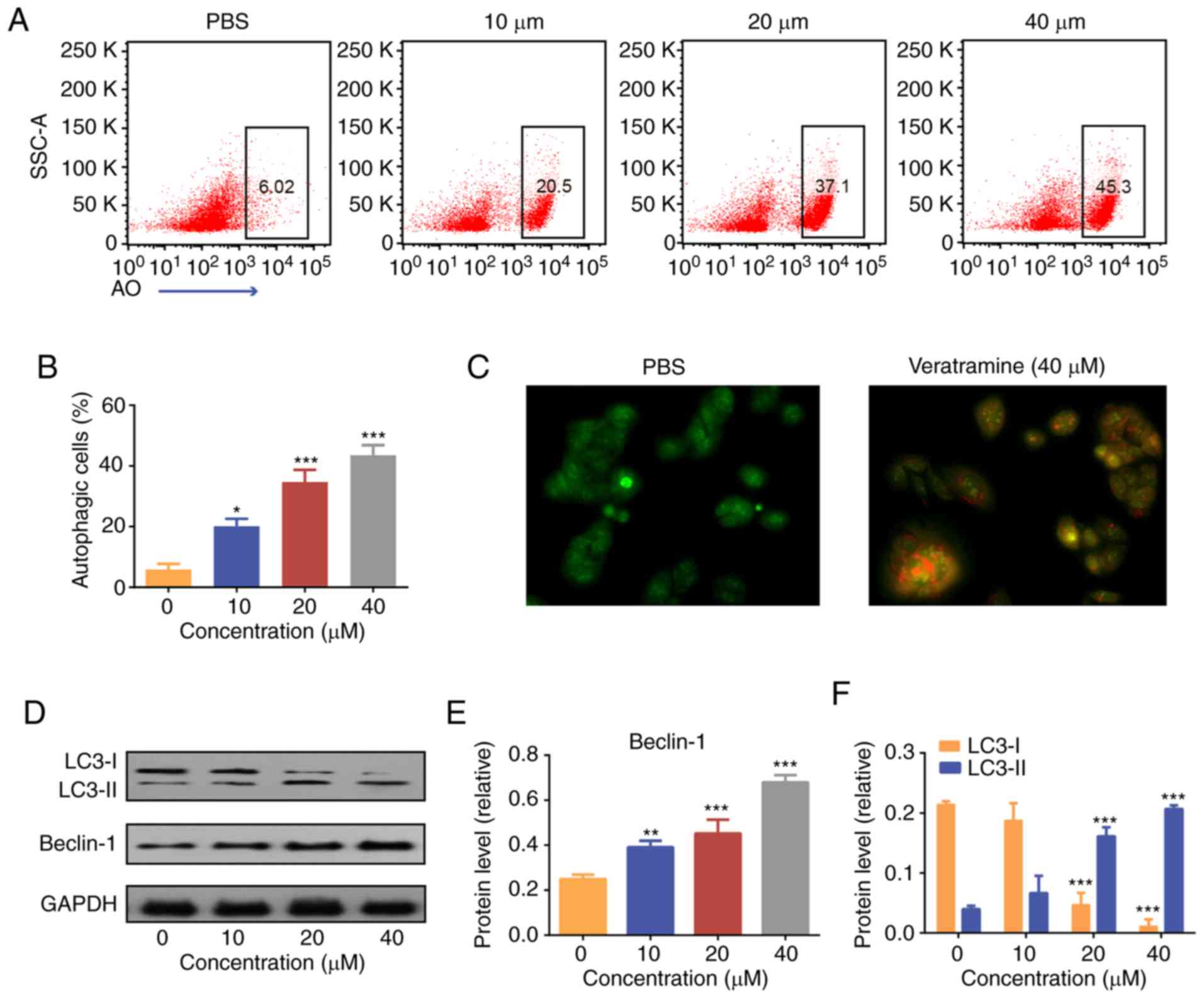
Acridine orange staining
Formation of acidic vesicular organelles (AVO) is a
hallmark of autophagic cells. Acridine orange (AO) dye readily
penetrates cell membranes and visualizes the occurrence of the
acidic vesicular organelles (AVOs). Therefore, AO staining was used
to detect autophagy. HepG2 cells were cultured overnight and
treated with varying concentrations of veratramine (10, 20 and 40
µM) for 48 h. Subsequently, the cells were stained with 10 µg/ml of
AO dye for 20 min at 37°C in the dark. AVO formation (Red) was
observed using fluorescence microscopy (magnification, ×200). The
percentage of autophagic cells was measured using flow
cytometry.
Western blotting
HepG2 cells were cultured overnight and then treated
with varying concentrations of veratramine (10, 20 and 40 µM) for
48 h. An equal volume of PBS was used as a control. After drug
treatment, the cells were harvested and lysed in RIPA lysis buffer
supplemented with a protease inhibitor cocktail. Cell debris were
removed by centrifugation at 14,000 × g at 4°C for 15 min and the
protein concentration was determined using BCA assays.
Subsequently, 30 µg of total protein was separated on a 10%
SDS-PAGE gel and electroblotted onto PVDF membranes (EMD
Millipore). The membranes were blocked with 5% skim milk in
TBS-Tween-20 (TBST; 20 mmol/l Tris-HCl, 150 mmol/l NaCl and 0.1%
Tween-20) for 1 h at room temperature (RT), and then incubated with
anti-Beclin1 (product no. 3495), anti-LC3 (product no. 3868),
anti-PI3K (product no. 4249), anti-phospho-PI3K (product no.
17366), anti-Akt (product no. dilution 4685), anti-phospho-Akt
(product no. 9614), anti-mTOR (product no. 2983), anti-phospho-mTOR
(product no. 5536) or anti-GAPDH (product no. 5174; all with a
dilution 1:1,000; all from Cell Signaling Technology, Inc.) primary
antibodies at 4°C, overnight. After washing 3×10 min with TBST, the
membranes were incubated with horseradish peroxidase-conjugated
secondary antibodies (product no. 7074; dilution 1:2,000; Cell
Signaling Technology, Inc.) for 1 h at RT. The protein bands were
visualized using ECL reagent and the G:BOX chemiXR5 Imaging System
(Syngene). Band intensities were quantified using Image-Pro Plus
6.0 software, with GAPDH used as an internal control.
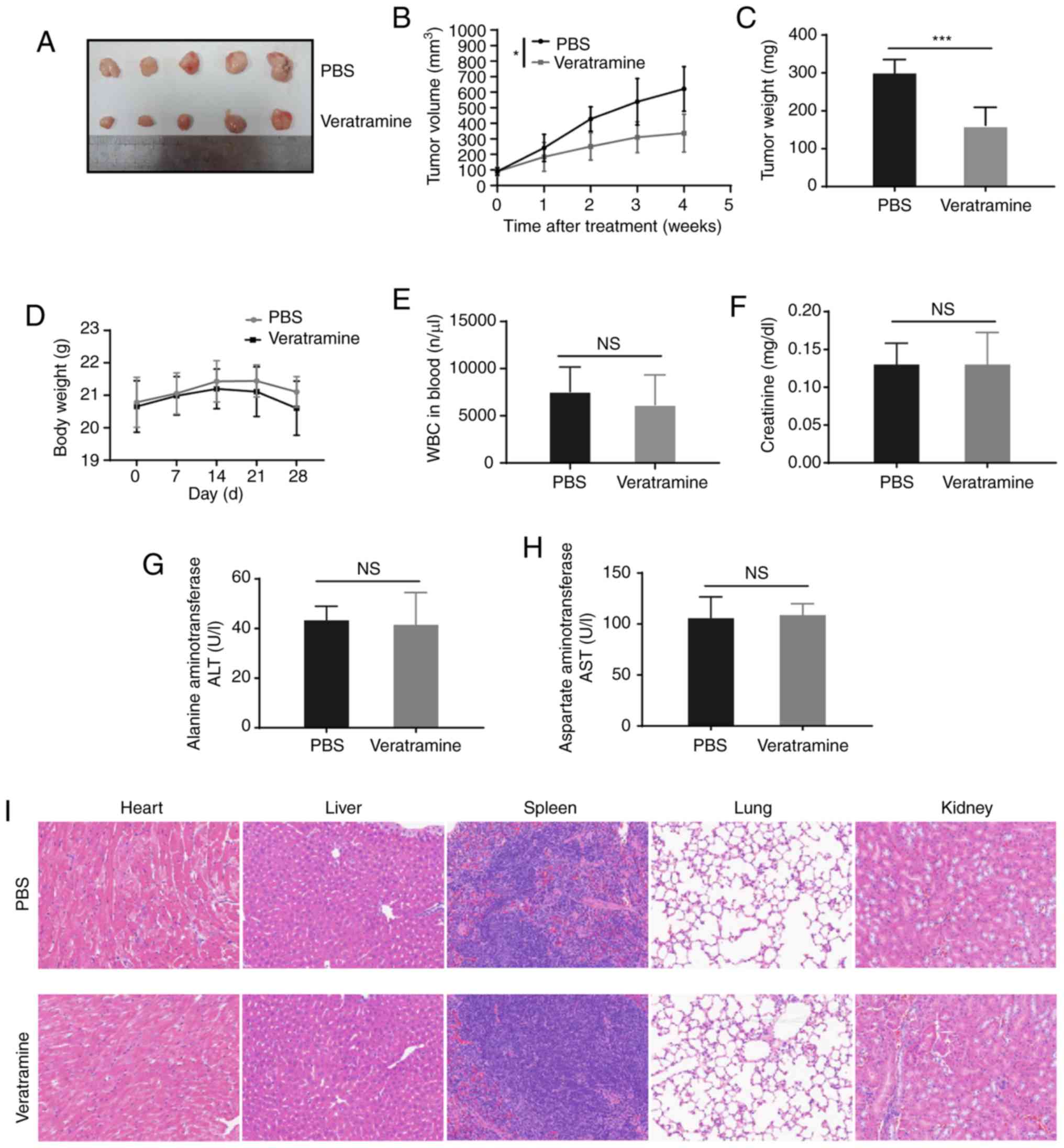
Anti-tumor growth effects in vivo
The liver cancer xenograft model was established by
subcutaneous injection of human HepG2 cells (1×106 cells
suspended in 0.1 ml PBS for each mouse) into the armpits of nude
female BALB/c mice (6–8 weeks old). Once the tumors reached a
volume of approximately 75–100 mm3, the mice were
randomly divided into two groups, with five mice per group, a PBS
group and veratramine group. PBS (200 µl) or veratramine (2 mg/kg)
were injected into the mice via the tail vein. Based on the
IC50 value of veratramine on HepG2 cells in
vitro, and previous studies (37–39), 2
mg/kg of veratramine was used for the animal experiment. The mice
were treated three times a week for 4 weeks. During the treatment,
the weights of the mice and the volume of the tumors were measured
every week. The volume of the tumors was calculated according to
the following formula: V = 0.5 × length × width2. Three
days after the last treatment, blood was collected from the mice
and the mice were sacrificed. Subcutaneous tumors from the mice
were excised and weighed. The number of white blood cells,
neutrophils and platelets, as well as the concentrations of alanine
aminotransferase, aspartate aminotransferase and creatinine were
examined. The heart, liver, spleen, lungs and kidneys were excised
and then fixed with 4% paraformaldehyde. The paraffin sections of
the organs were performed for pathological evaluation with
hematoxylin-eosin (H&E) staining (hematoxylin, 5 min; eosin, 2
min) at room temperature. The results of H&E staining were
evaluated by pathologist in a blinded fashion. The animal
experiments were approved by the Animal Care and Use Committee of
Yijishan Hospital of Wannan Medical College.
Statistical analysis
The experiments were performed in triplicate and the
data are presented as the mean ± SD. Data were analyzed using
GraphPad Prism 6.0 software (GraphPad Software, Inc.). Student
t-tests or ANOVAs with Tukey's post hoc test were used to evaluate
the data, and P<0.05 was considered to indicate a statistically
significant difference.
Results
Veratramine inhibits HepG2 cell
proliferation, migration and invasion
CCK-8, wound healing and Transwell assays were
performed to examine the effects of veratramine on HepG2 cell
proliferation, migration and invasion, respectively. As revealed in
Fig. 1B, veratramine significantly
inhibited HepG2 cell proliferation in a time- and dose-dependent
manner, and the IC50 values of veratramine on HepG2
cells for 24, 48, and 72 h were 26.54 µM, 19.81 and 9.12 µM,
respectively. Furthermore, the data revealed that 40 µM of
veratramine treatment reduced the cell migration rate compared to
the PBS control (Fig. 1C and D).
Consistently, the number of invasive cells was also decreased after
veratramine treatment for 48 h. Collectively, these results
indicated that veratramine not only suppressed cell growth but also
reduced cell migration and invasion in HepG2 cells. This
demonstrated the potential for veratramine to treat liver
cancer.
Veratramine induces apoptosis in HepG2
cells
The effect of veratramine on apoptosis was
investigated using Annexin V-FITC/7AAD double staining and RT-qPCR.
The cells were treated with the indicated dose of veratramine for
48 h and the same volume of PBS was used as a negative control. As
indicated in Fig. 2A and B, flow
cytometric data revealed that veratramine induced apoptosis in
HepG2 cells, and the proportion of apoptotic cells (Annexin
V+) increased from 8.41% (PBS control) to 51.86% (40 µM
of veratramine). Although multiple signaling pathways are involved
in apoptosis, the majority of them are regulated by the Bcl-2
family (40). BCL2 and
BAX are representative genes in the Bcl-2 family. Among
them, BCL2 is an anti-apoptotic gene, whereas BAX is
a pro-apoptotic gene. Studies have indicated that the ratio of
BCL2 to BAX is considered to be a decisive factor as
to whether a cell will commit to apoptosis. Therefore, in order to
further verify whether veratramine can induce apoptosis, the mRNA
expression levels of BCL2 and BAX were detected using
RT-qPCR. RT-qPCR results revealed that veratramine significantly
upregulated the expression level of BAX and downregulated
the expression level of BCL2, which further demonstrated
that veratramine induced apoptosis in HepG2 cells (Fig. 2C and D).
Veratramine induces autophagy in HepG2
cells
To explore the mechanism of action behind the
antitumor activity, the effect of veratramine on autophagy in HepG2
cells was investigated. AO staining was performed to label
autophagosomes. Flow cytometric results revealed that the number of
autophagic cells (AO+) increased with increasing
veratramine concentrations. After 48 h of veratramine treatment (40
µM), the proportion of autophagic cells reached 45.3% (Fig. 3A and B). Moreover, fluorescence
microscopy images further confirmed that veratramine induced
autophagy in HepG2 cells (Fig. 3C).
Autophagy is strictly regulated by gene expression, with Beclin-1
and LC3 proteins being markers of autophagy (41,42).
During autophagy, LC3-I is converted to LC3-II and then localized
to the autophagosome membrane (41). The expression levels of LC3-II are
positively correlated with the degree of autophagy, and the ratio
of LC3-II/I can be used to estimate the level of autophagy
(41,42). In the present study, western blot
analysis revealed that Beclin-1 and LC3-II proteins were
significantly upregulated, while LC3-I protein was significantly
downregulated, in the veratramine-treated cells. The Beclin-1
protein expression levels and the ratio of LC3-II/I were the
highest in the high concentration group (40 µM) (Fig. 3D-F). Overall, the aforementioned
results demonstrated that veratramine treatment could induce
autophagy in HepG2 cells.
Veratramine induces autophagy by
inhibiting the PI3K/Akt/mTOR signaling pathway
The PI3K/Akt/mTOR signaling pathway is an
intracellular signaling pathway that plays an important role in the
pathogenesis of cancer (30). As a
downstream protein of PI3K and Akt, phosphorylated mTOR protein
promotes cancer cell proliferation and angiogenesis (28,29). A
previous study has demonstrated that the PI3K/Akt/mTOR pathway is
associated with autophagy (43).
Therefore, it was hypothesized that veratramine may inhibit the
PI3K/Akt/mTOR pathway to promote autophagic cell death. In order to
verify this hypothesis, the phosphorylation levels of PI3K, Akt and
mTOR proteins were assessed using western blotting. The results
revealed that the levels of p-PI3K, p-Akt and p-mTOR proteins were
significantly lower in veratramine-treated cells than in the
control cells, and the changes were achieved in a dose-dependent
manner (Fig. 4A and B). Moreover, a
previous study demonstrated that veratramine modulated
AP-1-dependent gene transcription by directly binding to
programmable DNA (35). ETS
proto-oncogene 1, transcription factor (ETS1), an
AP-1 downstream gene (35),
has been revealed to be predominantly expressed in various tumor
subtypes and can mediate activation of the PI3K/Akt/mTOR pathway
(44,45). As anticipated, the mRNA level of
ETS1 was significantly downregulated in veratramine-treated
HepG2 cells (Fig. 4C). Based on
these results, it was concluded that veratramine induced autophagy
by blocking the PI3K/Akt/mTOR signaling pathway in HepG2 cells.
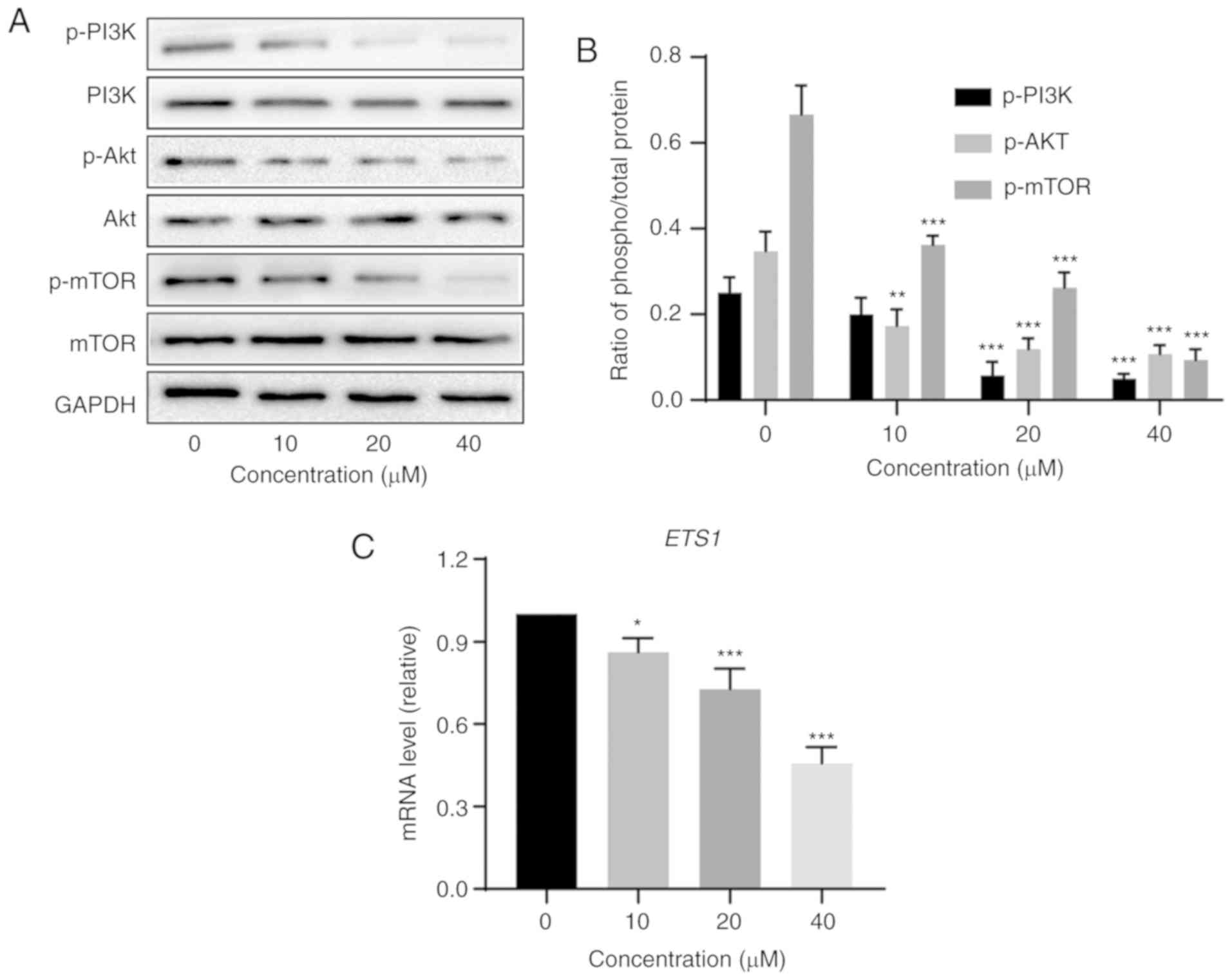 | Figure 4.Veratramine inhibits the
PI3K/Akt/mTOR signaling pathway in HepG2 cells. (A) Western blot
analysis of PI3K, Akt and mTOR protein expression levels in HepG2
cells treated with various concentrations of veratramine (0, 10, 20
and 40 µM) for 48 h. (B) Quantitative data of p-PI3K, p-Akt, p-mTOR
phospho/total proteins. (C) The qPCR analysis of ETS1 mRNA
level in HepG2 cells treated with various concentrations of
veratramine (0, 10, 20 and 40 µM) for 48 h. The data are presented
as the mean ± SD of three independent experiments. *P<0.05,
**P<0.01, ***P<0.001 vs. the control group. ETS1, ETS
proto-oncogene 1, transcription factor. |
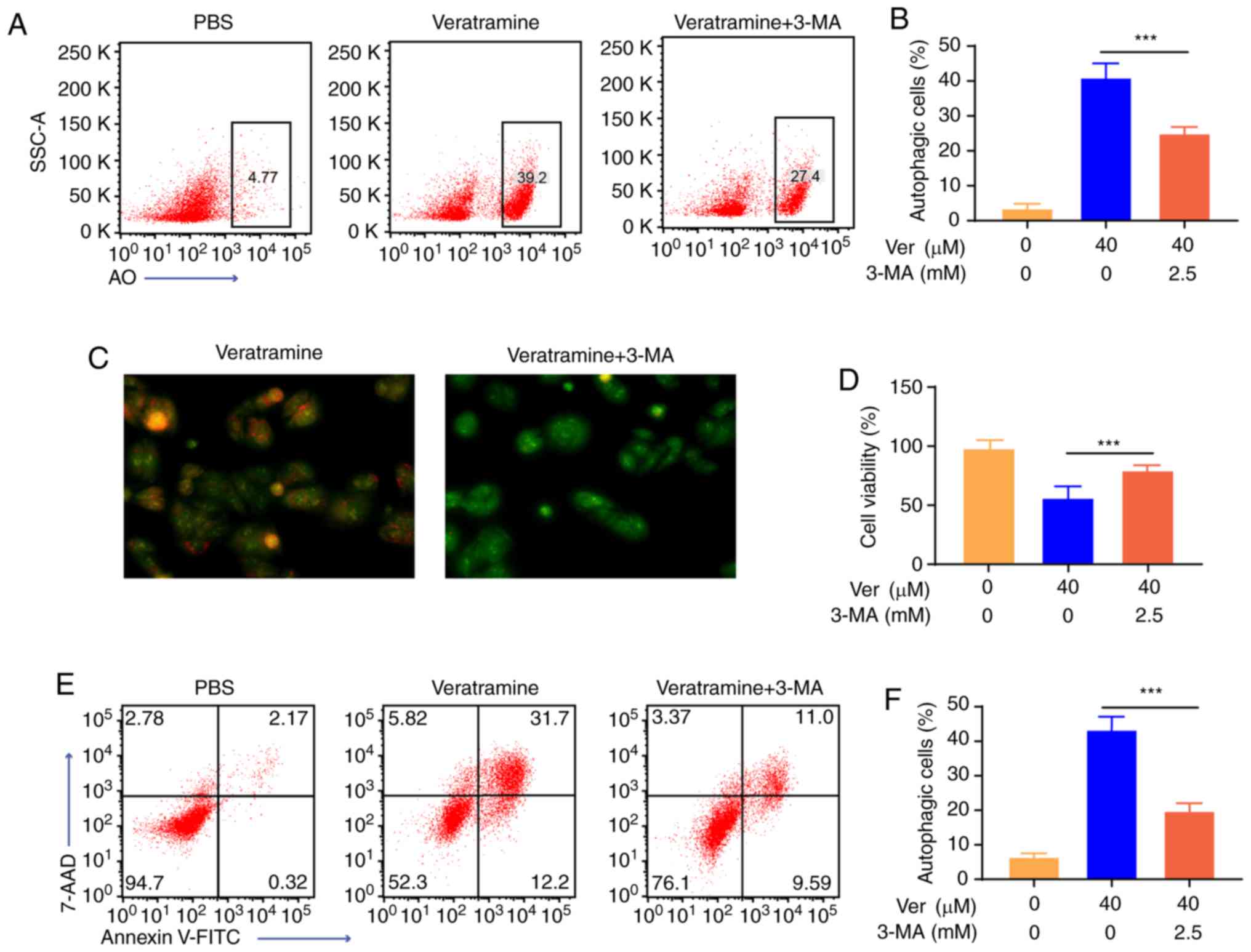
Veratramine induces autophagy-mediated
apoptosis in HepG2 cells
To evaluate the effect of autophagy on
veratramine-induced cell death, autophagy inhibitor 3-MA was used
and the proportion of apoptotic cells and the cell viability was
examined. Cells were co-incubated with veratramine (40 µM) for 48 h
in the presence or absence of 3-MA (2.5 mM). As revealed in
Fig. 5A and B, after the addition
of 3-MA, the number of autophagosomes was significantly decreased,
demonstrating that autophagy induced by veratramine was
significantly inhibited. Fluorescence microscopy results further
confirmed that autophagic vacuoles were reduced after 3-MA
co-treatment (Fig. 5C). Moreover,
cell viability in cells treated with a combination of veratramine
and 3-MA, was higher than that in cells treated with veratramine
alone, which indicated that 3-MA attenuated the cytotoxicity of
veratramine on HepG2 cells (Fig.
5D). Similarly, the proportion of apoptotic cells in the
co-treatment group was lower compared with that of the veratramine
alone treatment group (Fig. 5E and
F). Collectively, these results indicated that veratramine
induced-autophagy promoted apoptosis in HepG2 cells.
Veratramine suppresses liver cancer
xenografted tumor growth with a low systemic toxicity
Based on the findings of the present in vitro
experiments, the antitumor effect of veratramine was further
verified in the HepG2 subcutaneous xenograft mouse model. No mice
died or acted abnormally before the end of the animal experiment.
By monitoring the tumor volume, it was observed that veratramine
treatment significantly inhibited tumor growth compared with the
PBS control (Fig. 6A-B). Consistent
with the tumor volume, tumor weight in the veratramine group was
significantly lower than the tumor weight in the PBS group
(Fig. 6C). In addition, the
systemic toxicity of veratramine was also evaluated in vivo.
The weight of the mice in the veratramine-treated group was not
significantly different from that in the PBS group (Fig. 6D). Furthermore, veratramine
treatment had no significant effect on the number of blood cells as
well as the liver and kidney function compared with the PBS control
(Fig. 6E-H). Moreover, the results
of H&E staining revealed that no obvious tissue damage, such as
inflammation (infiltration of inflammatory cells) and/or necrosis
(tissue structural destruction), was observed in tissue sections of
the heart, liver, spleen, lung and kidney (Fig. 6I), which indicated that veratramine
had no obvious organ toxicity. Collectively, these results
demonstrated that veratramine not only suppressed tumor growth
in vivo, but also had no obvious systemic toxicity.
Discussion
Natural products are invaluable as resources for the
discovery and development of various drugs, especially antitumor
drugs (46–48). Numerous chemotherapeutic drugs
derived from natural products, such as taxol and
hydroxycamptothecin, have exhibited favorable clinical efficacy
(47,48). Veratramine, a steroidal alkaloid
isolated from plants of the lily family, has been reported to
possess multiple pharmacological activities, such as lowering blood
pressure and acting as an antithrombotic agent (49,50).
Recent studies demonstrated that veratramine may have antitumor
activities (34). Liver cancer is a
malignant tumor that is insensitive to existing drug treatments
(4), thus, it is extremely urgent
to find a safe and effective drug to treat liver cancer. In the
present study, veratramine was revealed to significantly inhibit
cell proliferation, invasion and metastasis in HepG2 cells, and the
IC50 value of veratramine on HepG2 cells at 48 h was
only 19.81 µM, which demonstrated that veratramine had a strong
antitumor effect. Notably, it was also observed that veratramine
significantly inhibited tumor growth in a liver cancer subcutaneous
xenograft model. In addition, common toxic side effects associated
with chemotherapeutic drugs were also evaluated. It was revealed
that veratramine had no significant effect on the number of blood
cells, weight, and liver/kidney function in mice. Based on these
findings, it was inferred that veratramine had characteristics such
as a high efficiency and low systemic toxicity and was a promising
antitumor agent.
Autophagy is a cellular metabolic process possessed
by eukaryotes in which cytoplasmic cargo is delivered to lysosomes
and intracellular components are degraded and recycled (10). Autophagy often occurs at low levels
in cells and is rapidly upregulated when cells are under stress
(8). Evidence suggests that
induction of autophagic cell death is a promising strategy for the
treatment of cancer (51). In the
present study, it was revealed that veratramine could concurrently
induce autophagy and apoptosis in HepG2 cells. To confirm the
relationship between autophagy and apoptosis in veratramine-induced
HepG2 cell death, 3-MA was used to inhibit autophagy. Notably,
after autophagy was inhibited, the proportion of apoptotic cells
was reduced, and cell viability was also reduced, which indicated
that veratramine induced autophagy-mediated apoptosis in HepG2
cells.
mTOR kinase can act directly on autophagy-related
proteins to regulate the formation of autophagosomes (52). The PI3/Akt/mTOR signaling pathway is
one of the main pathways involved in autophagy, in which activated
PI3K promotes the phosphorylation of Akt, leading to
phosphorylation of mTOR, which inhibits autophagy (30,43).
In this present study, the data demonstrated that veratramine
downregulated the phosphorylation levels of PI3K, AKT and mTOR in a
concentration-dependent manner. Furthermore, ETS1, an
AP-1-dependent gene, was also downregulated, consistent with
a previous study (35). Previous
studies have demonstrated that ETS1 mediates activation of
the PI3K/Akt/mTOR pathway (44,45).
Therefore, it was concluded that veratramine inhibited the
PI3K/AKT/mTOR signaling pathway by targeting ETS1, thereby
inducing autophagic cell death in HepG2 cells. However, a
limitation of the present study, is that whether the AP-1
gene itself and AP-1-dependent genes are involved in the
antitumor effect of veratramine has not been explored.
In conclusion, the present data demonstrated that
veratramine inhibited human liver cancer HepG2 cell proliferation,
invasion and infiltration in vitro, and veratramine could
also significantly suppress tumor growth with a low systemic
toxicity in vivo. In addition, veratramine induced
autophagy-mediated apoptosis in HepG2 cells by blocking the
activation of the PI3K/Akt/mTOR signaling pathway. Therefore, the
present study suggested that veratramine may be a potential agent
for the treatment of liver cancer.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81241102) and the
fund of the Traditional Chinese Medicine Scientific Research
Project of Anhui Provincial Health Department (2012zy59).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LY and YX participated in its design and performed
experiments, analyzed the data and wrote the draft manuscript. PX,
WZ and YG interpreted the results, performed the animal experiments
and reviewed the manuscript. FX and ZJ conceived the study,
performed data analysis and manuscript editing and improvement. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The animal experiments were approved by the Animal
Care and Use Committee of Yijishan Hospital of Wannan Medical
College.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kudo M: Systemic therapy for
hepatocellular carcinoma: Latest advances. Cancers (Basel).
10:E4122018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li D, Sedano S, Allen R, Gong J, Cho M and
Sharma S: Current treatment landscape for advanced hepatocellular
carcinoma: Patient outcomes and the impact on quality of life.
Cancers (Basel). 11:E8412019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Colagrande S, Inghilesi AL, Aburas S,
Taliani GG, Nardi C and Marra F: Challenges of advanced
hepatocellular carcinoma. World J Gastroenterol. 22:7645–7659.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ikeda K: Recent advances in medical
management of hepatocellular carcinoma. Hepatol Res. 49:14–32.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kawaguchi T, Nakano D, Okamura S, Shimose
S, Hayakawa M, Niizeki T, Koga H and Torimura T: Spontaneous
regression of hepatocellular carcinoma with reduction in
angiogenesis-related cytokines after treatment with sodium-glucose
cotransporter 2 inhibitor in a cirrhotic patient with diabetes
mellitus. Hepatol Res. 49:479–486. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shimose S, Tanaka M, Iwamoto H, Niizeki T,
Shirono T, Aino H, Noda Y, Kamachi N, Okamura S, Nakano M, et al:
Prognostic impact of transcatheter arterial chemoembolization
(TACE) combined with radiofrequency ablation in patients with
unresectable hepatocellular carcinoma: Comparison with TACE alone
using decision-tree analysis after propensity score matching.
Hepatol Res. 49:919–928. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Doherty J and Baehrecke EH: Life, death
and autophagy. Nat Cell Biol. 20:1110–1117. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dikic I: Proteasomal and autophagic
degradation systems. Annu Rev Biochem. 86:193–224. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mizushima N and Komatsu M: Autophagy:
Renovation of cells and tissues. Cell. 147:728–741. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Levine B and Kroemer G: Autophagy in the
pathogenesis of disease. Cell. 132:27–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Retnakumar SV and Muller S:
Pharmacological autophagy regulators as therapeutic agents for
inflammatory bowel diseases. Trends Mol Med. 25:516–537. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Long M and McWilliams TG: Monitoring
autophagy in cancer: From bench to bedside. Semin Cancer Biol. Jul
15–2019.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yazdani HO, Huang H and Tsung A:
Autophagy: Dual response in the development of hepatocellular
carcinoma. Cells. 8:E912019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye
WC, Zhang DM and Chen ZS: Autophagy and multidrug resistance in
cancer. Chin J Cancer. 36:522017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Elgendy M, Ciro M, Abdel-Aziz AK, Belmonte
G, Dal Zuffo R, Mercurio C, Miracco C, Lanfrancone L, Foiani M and
Minucci S: Beclin 1 restrains tumorigenesis through Mcl-1
destabilization in an autophagy-independent reciprocal manner. Nat
Commun. 5:56372014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Edinger AL and Thompson CB: Defective
autophagy leads to cancer. Cancer Cell. 4:422–424. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shimizu S, Yoshida T, Tsujioka M and
Arakawa S: Autophagic cell death and cancer. Int J Mol Sci.
15:3145–3153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nikoletopoulou V, Markaki M, Palikaras K
and Tavernarakis N: Crosstalk between apoptosis, necrosis and
autophagy. Biochim Biophys Acta. 1833:3448–3459. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gordy C and He YW: The crosstalk between
autophagy and apoptosis: Where does this lead? Protein Cell.
3:17–27. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee EF, Smith NA, Soares da Costa TP,
Meftahi N, Yao S, Harris TJ, Tran S, Pettikiriarachchi S, Perugini
MA, Keizer DW, et al: Structural insights into BCL2 pro-survival
protein interactions with the key autophagy regulator BECN1
following phosphorylation by STK4/MST1. Autophagy. 15:785–795.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Levine B, Sinha SC and Kroemer G: Bcl-2
family members: Dual regulators of apoptosis and autophagy.
Autophagy. 4:600–606. 2008. View Article : Google Scholar
|
|
23
|
Wu H, Che X, Zheng Q, Wu A, Pan K, Shao A,
Wu Q, Zhang J and Hong Y: Caspases: A molecular switch node in the
crosstalk between autophagy and apoptosis. Int J Biol Sci.
10:1072–1083. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hou W, Han J, Lu C, Goldstein LA and
Rabinowich H: Autophagic degradation of active caspase-8: A
crosstalk mechanism between autophagy and apoptosis. Autophagy.
6:891–900. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Djavaheri-Mergny M, Maiuri MC and Kroemer
G: Cross talk between apoptosis and autophagy by caspase-mediated
cleavage of Beclin 1. Oncogene. 29:1717–1719. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Salminen A, Kaarniranta K and Kauppinen A:
Beclin 1 interactome controls the crosstalk between apoptosis,
autophagy and inflammasome activation: Impact on the aging process.
Ageing Res Rev. 12:520–534. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng
P, Hogan RN, Gilpin C and Levine B: Autophagy gene-dependent
clearance of apoptotic cells during embryonic development. Cell.
128:931–946. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Paquette M, El-Houjeiri L and Pause A:
mTOR pathways in cancer and autophagy. Cancers (Basel). 10:E182018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Boutouja F, Stiehm CM and Platta HW: mTOR:
A cellular regulator interface in health and disease. Cells.
8:E182019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Alzahrani AS: PI3K/Akt/mTOR inhibitors in
cancer: At the bench and bedside. Semin Cancer Biol. 59:125–132.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wagle N, Grabiner BC, Van Allen EM, Hodis
E, Jacobus S, Supko JG, Stewart M, Choueiri TM, Gandhi L, Cleary
JM, et al: Activating mTOR mutations in a patient with an
extraordinary response on a phase I trial of everolimus and
pazopanib. Cancer Discov. 4:546–553. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jovanović B, Mayer IA, Mayer EL, Abramson
VG, Bardia A, Sanders ME, Kuba MG, Estrada MV, Beeler JS, Shaver
TM, et al: A randomized phase II neoadjuvant study of cisplatin,
paclitaxel with or without everolimus in patients with stage II/III
triple-negative breast cancer (TNBC): Responses and long-term
outcome correlated with increased frequency of DNA damage response
gene mutations, TNBC subtype, AR status, and Ki67. Clin Cancer Res.
23:4035–4045. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li J, Kim SG and Blenis J: Rapamycin: One
drug, many effects. Cell Metab. 19:373–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang J, Li HL, Shen YH, Jin HZ, Yan SK,
Liu RH and Zhang WD: Antitumor activity of extracts and compounds
from the rhizomes of veratrum dahuricum. Phytother Res.
22:1093–1096. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bai F, Liu K, Li H, Wang J, Zhu J, Hao P,
Zhu L, Zhang S, Shan L, Ma W, et al: Veratramine modulates
AP-1-dependent gene transcription by directly binding to
programmable DNA. Nucleic Acids Res. 46:546–557. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang J, Li HL, Shen YH, Jin HZ, Yan SK,
Liu XH, Zeng HW, Liu RH, Tan YX and Zhang WD: Antitumor and
antiplatelet activity of alkaloids from veratrum dahuricum.
Phytother Res. 24:821–826. 2010.PubMed/NCBI
|
|
38
|
Cong Y, Guo J, Tang Z, Lin S, Zhang Q, Li
J and Cai Z: Metabolism study of veratramine associated with
neurotoxicity by using HPLC-MSn. J Chromatogr Sci. 53:1092–1099.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lyu C, Zhang Y, Zhou W, Zhang S, Kou F,
Wei H, Zhang N and Zuo Z: Gender-dependent pharmacokinetics of
veratramine in rats: In vivo and in vitro evidence. AAPS J.
18:432–444. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gross A, McDonnell JM and Korsmeyer SJ:
BCL-2 family members and the mitochondria in apoptosis. Genes Dev.
13:1899–1911. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yorimitsu T and Klionsky DJ: Autophagy:
Molecular machinery for self-eating. Cell Death Differ. 12 (Suppl
2):S1542–S1552. 2005. View Article : Google Scholar
|
|
42
|
Cao Y and Klionsky DJ: Physiological
functions of Atg6/Beclin 1: A unique autophagy-related protein.
Cell Res. 17:839–849. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Porta C, Paglino C and Mosca A: Targeting
PI3K/Akt/mTOR signaling in cancer. Front Oncol. 4:642014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xu S, Ge J, Zhang Z and Zhou W: MiR-129
inhibits cell proliferation and metastasis by targeting ETS1 via
PI3K/AKT/mTOR pathway in prostate cancer. Biomed Pharmacother.
96:634–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Meng S, Jian Z, Yan X, Li J and Zhang R:
LncRNA SNHG6 inhibits cell proliferation and metastasis by
targeting ETS1 via the PI3K/AKT/mTOR pathway in colorectal cancer.
Mol Med Rep. 20:2541–2548. 2019.PubMed/NCBI
|
|
46
|
Tu Y, Zhu S, Wang J, Burstein E and Jia D:
Natural compounds in the chemoprevention of alcoholic liver
disease. Phytother Res. 33:2192–2212. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Annovazzi L, Caldera V, Mellai M, Riganti
C, Battaglia L, Chirio D, Melcarne A and Schiffer D: The DNA
damage/repair cascade in glioblastoma cell lines after
chemotherapeutic agent treatment. Int J Oncol. 46:2299–2308. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dotan E, Cohen SJ, Starodub AN, Lieu CH,
Messersmith WA, Simpson PS, Guarino MJ, Marshall JL, Goldberg RM,
Hecht JR, et al: Phase I/II trial of labetuzumab govitecan
(anti-CEACAM5/SN-38 antibody-drug conjugate) in patients with
refractory or relapsing metastatic colorectal cancer. J Clin Oncol.
35:3338–3346. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li Q, Zhao YL, Long CB, Zhu PF, Liu YP and
Luo XD: Seven new veratramine-type alkaloids with potent analgesic
effect from Veratrum taliense. J Ethnopharmacol. 244:1121372019.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chandler CM and McDougal OM: Medicinal
history of North American. Veratrum. Phytochem Rev. 13:671–694.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Deng S, Shanmugam MK, Kumar AP, Yap CT,
Sethi G and Bishayee A: Targeting autophagy using natural compounds
for cancer prevention and therapy. Cancer. 125:1228–1246. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jung CH, Ro SH, Cao J, Otto NM and Kim DH:
mTOR regulation of autophagy. FEBS Lett. 584:1287–1295. 2010.
View Article : Google Scholar : PubMed/NCBI
|