In the past decade, the study of exosomes has become
more mature. These nano-sized vesicles can be secreted by various
types of cells. The diameter of exosomes is 30–150 nm (4,5). The
membranes of exosomes, which can fuse with cell membranes to
transport content, are similar to cell membranes (6). The exosomes contain different
biological molecules, such as nucleic acids, proteins and lipids.
Exosomes can mediate the transport of these substances to achieve
intercellular communication in physiological and pathological
states (7,8). The contents of exosomes reflect the
characteristics of parental cells. Under the protection of the
lipid bilayer structure, the contents of exosomes can avoid
degradation by various enzymes in the blood, and thus be stable in
body fluids such as blood, tissue fluid, urine, saliva and
intercellular fluid (9–12). These unique characteristics make
exosomes reliable biomarkers. Increasing evidence suggests that
exosomes play critical roles in tumorigenesis, tumor growth,
metastasis, and therapy resistance (13). Exosomal contents have begun to be
used in cancer diagnosis and prognosis. In this review, the
mechanism and clinical value of exosomes in the development of
breast cancer were examined.
Numerous types of cells can secrete exosomes under
normal and pathological conditions. Exosomes are mainly derived
from the polyvesicles formed by the intracellular lysosomal
microsome invagination, which are released into the extracellular
matrix after fusion of their extracellular membranes with those of
the polyvesicles (13). GTPases
(Rab GTPase), intercellular pH, p53 regulatory protein tumor
suppressor activation pathway 6 (TSAP6), heparanase, psoralen
(14) and other enzymes can act as
modulators of the secretion of exosomes (13).
Exosomes can be separated from cells or body fluid
supernatants by various methods, including ultracentrifugal
precipitation, ultrafiltration centrifugal precipitation, sucrose
density gradient centrifugal precipitation, immunoaffinity-based
capture, and commercial kit precipitation (13). Several new methods have recently
emerged, including nanomembrane filtration, antibody-coated
magnetic bead separation, and microfluidic immunoaffinity
separation methods. Among them, the commercial kit precipitation
can quickly separate and increase the recovery of exogenous
substances (13).
Biotechniques, which have been used to detect
isolated exosomes include transmission electron microscopy (TEM),
scanning electron microscopy (SEM), atomic force microscopy (AFM),
cryo-TEM, the nanosight system, fluorescence-activated cell sorting
(FACS), and western blotting. SEM, TEM and cryo-TEM can usually be
used to identify the size of exosomes and distinguish them from
other extracellular vesicles. NTA (Nanoparticle tracking analysis)
can monitor and quantify particle size, concentration and relative
strength in real time. Western blotting and FACS can be used to
identify exosomal surface markers. Microarray and high-throughput
sequencing techniques accurately detect the relative expression of
nucleic acid (15). QuantStudio 3D
digital polymerase chain reaction can accurately quantify the
expression of miRNAs (15).
LC-MS/MS (liquid chromatography/tandem mass spectrometry), western
blotting and enzyme linked immunosorbent assay can be used to
screen and identify exosomal proteins (16). Integrated magneto-electrochemical
analysis is a fast and on-site method for detecting the protein
profile in exosomes. The sensitivity and speed have been
demonstrated to be superior than those of traditional methods
(16).
Breast cancer-derived exosomes play numerous roles
in cell-to-cell and cell-to-microenvironment interactions
(summarized in Table I). Examples
of exosomal contents participating in the development of breast
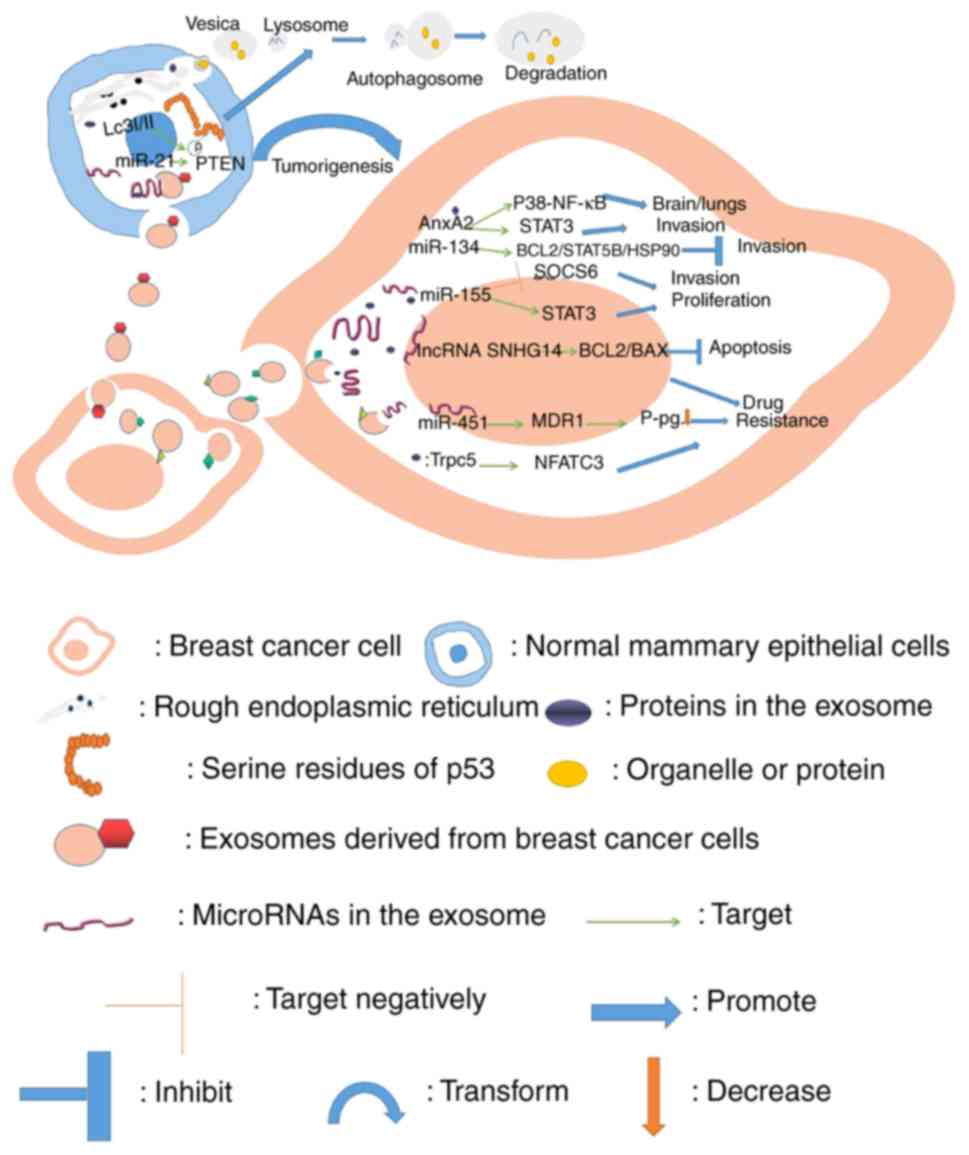
cancer and their known roles are presented in Figs. 1–3.
Exosomes contain miRNAs, lncRNAs and proteins
released from breast cancer cells into target cells, such as other
breast cancer cells or normal mammary epithelial cells. The
contents of breast cancer cells have been revealed to lead to cell
proliferation, invasion and drug resistance (38,54,63,64,67,68,72,73,75),
while in normal mammary epithelial cells, they may promote the
transformation of normal cells into breast cancer cells (34). In addition, exosomal contents may
result in autophagy of normal epithelial cells (76).
The exosomes released from breast cancer cells can
act on vascular endothelial cells, leading to endothelial cell
necrosis and migration of intravascular cancer cells (59–60).
Moreover, exosomes can also be delivered to EPCs (endothelial
progenitor cells) and prevent EPC from differentiating into normal
vascular endothelial cells, which may cause thrombosis (28). In addition, exosomes in
myeloid-derived suppressor cells (MDSCs) can also enter the
endothelial cells of blood vessels and promote the angiogenesis of
breast cancer (30).
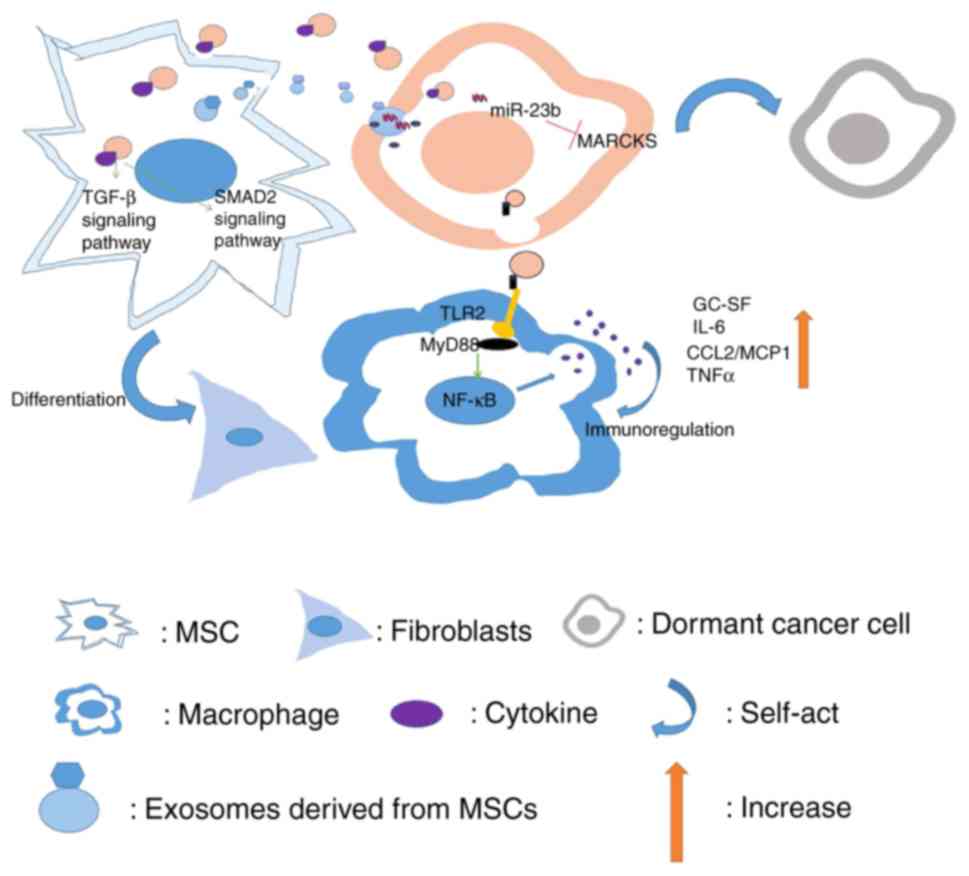
Exosomes transferred from breast cancer cells into
macrophages may activate NF-κB, release cytokines and stimulate
autoimmune regulation (29).
Exosomes from breast cancer cells also play a role in mesenchymal
stem cells (MSCs), thus promoting the differentiation into
fibroblasts (36). Conversely,
exosomes derived from MSCs can also act on breast cancer cells,
leading to dormancy of breast cancer cells (47).
Tumorigenesis is a qualitative process in tumor
development, which determines the existence and absence of tumors.
Breast cancer-derived exosomes can promote tumorigenesis in normal
mammary epithelial cells by inducing production of cancer cell
growth-promoting factors which are helpful in producing a
microenvironment that allows tumor growth (76). In addition, miRNA (miR)-21 and
miR-10b in exosomes of breast cancer cells and serum of patients
have been revealed to instigate non-tumorigenic epithelial cells to
form tumors through a dicer-dependent mechanism by targeting
phosphatase and tension homolog deleted on chromosome ten (PTEN)
and Homeobox D10 (HOXD10) respectively (34). Exosomal miR-100 has been
demonstrated to offset the effects of transcription factors that
are induced by epithelial-mesenchymal transition (EMT) and regulate
genes involved in tumorigenesis directly (27).
Tumor growth, apoptosis and dormancy play an
important role in the progression of breast cancer. For example,
breast cancer cell-derived exosomal miR-128 was revealed to promote
proliferation of MCF-7 cells by targeting the BAX gene (48). miR-10b in exosomes of breast cancer
was revealed to target HOXD10 and lead to breast tumor
proliferation (24). Additionally,
docetaxel-resistant miRNAs (miR-24, miR-26a, and miR-27a) in
exosomes are thought to activate the mitogen-activated protein
kinase (MAPK) signaling pathway, and have been revealed to be
involved in breast cancer proliferation (19). miR-1246 was also revealed to be
significantly increased in breast cancer cells, especially in
MDA-MB-231 exosomes, and could be transferred to target cyclin G2
(CCNG2) directly, leading to cell proliferation (49). Exosomal miR-155 in breast cancer may
also promote proliferation of breast cancer cells by inhibiting
suppressor of cytokine signaling 6 (SOCS6) and activating signal
transducer and activator of transcription 3 (STAT3) (75). Non-coding RNA can also promote cell
proliferation. Exosomal long non-coding RNA (lncRNA) UCA1 was
revealed to produce AKT/mammalian target of rapamycin (mTOR)
signaling (77), leading to
increased proliferation of breast cancer cells (40). While the proliferation of cancer
cells increases, some molecules can also inhibit apoptosis. For
example, exosomes derived from tamoxifen-resistant MCF-7 cells were
revealed to carry miR-222 and miR-221, act on parental MCF-7 cells,
and reduce the expression of p27 and ERa (estrogen receptor α) in
target cells. The proliferation and the cell cloning ability of
sensitive breast cancer cells was enhanced, while the apoptotic
ability was weakened (50). lncRNA
SNHG14 in exosomes was also revealed to inhibit apoptosis of breast
cancer cells via the Bcl-2/Bax pathway (63).
In addition to the exosomes involved in the
communication between cancer cells, the exosomes derived from
cancer cells can also participate in shaping the microenvironment,
thus affecting proliferation of breast cancer. Exosomes from MCF-7
breast cancer contain human telomerase reverse transcriptase
(hTERT) mRNA that can be translated into a fully active telomerase
enzyme that can promote proliferation in recipient
telomerase-negative fibroblasts (78). Exosomal miR-21 was revealed to lead
to activation of NF-κB and promote the proliferation of stromal
cells in breast phyllodes tumors (79).
Exosomes from microenvironment cells and non-tumor
cells may have a similar role in breast cancer cells. Upon exosome
transfer from tumor fibroblasts to breast cancer cells, unshielded
RN7SL1 may produce enhanced tumor growth (20). The expression of miR-21 and miR-34a
in MSC-derived exosomes has been revealed to be high. Among them,
miR-34a was demonstrated to target transcription factor E2F3. This
microenvironment was beneficial to the growth and survival of
breast cancer cells (32). In
addition, exosomal miR-34a has been revealed to target tropomyosin
alpha-1 chain (TPM1), programmed cell death protein 4 (PDCD4), and
Bcl-2 (32,33,80–82).
Exosomal miR-19a from astrocytes which leads to PTEN loss may
induce chemokine CC ligand (CCL)2 secretion, leading to brain
metastasis outgrowth by promoting metastatic breast cancer (MBC)
cells, and inhibiting apoptosis (51). Expression of lnc-Y1 and lnc-7SK
derived from MSC exosomes has been revealed to be high and this
microenvironment was beneficial to the growth of breast cancer
cells (32).
However, certain other exosomes inhibit the
proliferation of breast cancer cells. On the one hand, some of
these exosomes can act as tumor suppressor genes, thereby
inhibiting cancer cell proliferation. On the other hand, some
exosomes appear to inhibit cell proliferation, but they are
actually dormant cells, thus laying the foundation for cancer cell
resuscitation.
Bone marrow MSC-derived exosomal miR-100 may reduce
growth of breast cancer by directly targeting HOXA1 (27). Human umbilical vein endothelial
cells (HUVECs) were revealed to be capable of transmitting miR-503
to breast tumor cells via exosomes and then reduce cancer cell
growth by targeting cyclin D2 (CCND2) and cyclin D3 (CCND3)
(52). It can be inferred that
these exosomes can be used to treat cancer. Nevertheless, the bone
marrow MSC-derived miR-23b was revealed to be transferred into
breast cancer cells through exosomes, inhibit MARCKS (a protein
involved in the promotion of cell cycling) and decrease
proliferation of dormant cells, thereby laying a foundation for
tumor recurrence (47). Lim et
al reported that exosomal miR-127, miR-197, miR-222 and miR-223
released from bone marrow stromal cells negatively promoted breast
cancer cell growth by targeting CXCL12, resulting in induction of
dormancy of breast cancer cells (53).
The role of exosome-induced breast cancer metastasis
in promoting cancer progression is important. Cancer stem cells
play an important role in the recurrence of breast cancer cell
metastasis. Evidence suggests that exosomes derived from cancer
stem cells have high levels of miR-155 that mediate the loss of
CCAAT/enhancer-binding protein (C/EBP)-β, resulting in the loss of
TGF-β, inducing cells to obtain an EMT phenotype (83). Furthermore, exosomal miR-155 has
also been revealed to inhibit SOCS6, activate STAT3, and promote
metastasis of breast cancer cells (75).
Transfer of exosomes between cancer cells promotes
cancer metastasis. miR-24, miR-26a, and miR-27a in exosomes of
docetaxel-resistant breast cancer cells are involved in cancer
invasion and metastasis by activating the MAPK signaling pathway
(19). Breast cancer cell exosomal
miR-200c and miR-141 have also been demonstrated to promote breast
cancer cell metastasis via a FOXP3-KAT2B-miR200c/141 axis (55). Exosomal miR-1246 secreted by MBCs
was revealed to be transported to non-metastatic and non-malignant
breast cancer cells, thereby inhibiting their target gene cyclin-g2
and resulting in enhanced breast cancer cell survival rates and
mobility (49). Breast cancer cells
have been revealed to deliver through exosomes, miR-10b to normal
cells in which expression of miR-10b was lower than that of
MDA-MB-231 cells. In non-malignant breast cancer cell line HMLE,
miR-10b which has been revealed to promote migration of normal
cells, targeted HOXD10 and Kruppel like factor 4 (KLF4) (35,56).
In addition to miRNAs, other molecules derived from cancer cells
can also promote metastasis of cancer cells. Exosomal lncRNA UCA1
may enhance breast cancer cell activity, invasion and metastasis
(40) through AKT-mTOR signaling
(77). Maji et al revealed
that breast cancer cell-derived exosomal Annexin A2 was responsible
for brain and lung metastasis through activation of p38 MAPK
mediated by macrophages, STAT3 and nuclear factor (NF)-kB, thereby
improving levels of interleukin (IL)-6 and tumor necrosis factor
(TNF)-α (84). In addition,
Annexins A5 and A6 in breast cancer-derived exosomes may also be
involved in cancer invasion and metastasis (37,64,67,72,73).
The effect of breast cancer-derived exosomes on the
microenvironment can also alter cancer metastasis. Exosomal Wnt 5a
mRNA in MCF-7 was revealed to promote tumor invasion by activation
of β-catenin-independent Wnt signaling in macrophages, where it
could be translated to Wnt 5a protein (85). In addition, brain metastatic cancer
cell-derived miR-181c in exosomes was demonstrated to target
3-phosphoinositide-dependent protein kinase-1 in endothelial cells,
which disrupted the blood-brain barrier and caused breast cancer
cells to metastasize to the brain (60). Similarly, exosomal miR-939 was
demonstrated to directly target VE-cadherin, destroy the vascular
endothelial barrier, and finally promote breast cancer cell
migration (59). Exosomal miR-21
was also revealed to activate NF-κB and enhance invasion of stromal
cells in breast phyllodes tumors (79).
Similar to proliferation, exosomes from the
microenvironment can in turn affect cancer migration. RN7SL1 mRNA
in exosomes of breast tumor fibroblasts could promote tumor
metastasis through the NOTCH-MYC signaling pathway (86). It was reported that exosomal
miR-223, released from tumor-associated macrophages that are
activated by IL-4 derived from CD4+ T cells could
promote metastasis of breast cancer cells through the
Mef2c/β-catenin pathway (62).
miR-19a could be transported from astrocytes to breast cancer cells
by exosomes, resulting in PTEN loss and promotion of brain
metastasis of breast cancer cells (51). Doxorubicin was demonstrated to
induce the expression of exosomal miR-126a in MDSCs, leading to
breast cancer lung metastasis via an IL-33/IL-13-mediated pathway
by which the miR-126a-S100A8/9 axis was regulated (18). Moreover, cancer-associated
fibroblast (CAF)-derived exosomal miR-21, miR-143, and miR-378e
have been reported to induce EMT phenotype and promote cell
aggressiveness and metastasis by increasing organic cation
transporter octamer-binding transcription factor 3/4 (Oct3/4),
Nanog, and Sox2 expression in breast cancer cells (87). CAF-like (i.e., p85α−/−)
fibroblast-derived exosomes were also revealed to deliver Wnt10b
protein via the Wnt pathway, which resulted in enhanced migration,
and EMT of breast cancer cells (74).
As aforementioned, there are numerous molecules that
promote cancer migration. However, there are also some anticancer
molecules from exosomes that can negatively regulate cancer
migration. For example, miR-130a-3p was demonstrated to be
downregulated in exosomes of breast cancer stem cell-like cells,
and this inhibited cell invasion and metastasis by directly
targeting RAB5B (30). Breast
cancer cell exosomes carrying miR-134 were also revealed to act on
the target proteins of receptor cells such as Bcl-2, STAT5B and
heat shock protein 90 (Hsp90) to reduce tumor cell migration,
invasion (54).
Exosomes promote endothelial cell proliferation and
increase angiogenesis by transferring cancer-promoting molecules.
Hypoxic breast cancer cells have been revealed to transmit miR-210,
which has been demonstrated to inhibit Ephrin-A3 and
protein-tyrosine phosphatase 1B (PTP1B) via exosomes, and promote
angiogenesis of breast cancer (88). When exosomes derived from MDA-MB-231
were co-cultured with HUVECs in vitro, they promoted
proliferation and DNA synthesis of HUVECs in a time- and
dose-dependent manner through activation of the MAPK pathway,
resulting in changes in the vascular endothelial cell cycle and
promotion of neovascularization (26). Doxorubicin-induced exosomal miR-126a
released from MDSCs was demonstrated to contribute to induction of
Th2 T cells and angiogenesis in breast cancer (18).
Similarly, some exosomal molecules can inhibit
angiogenesis and play an anticancer role to some extent. Exosomal
transport of miR-23b and miR-320b from breast cancer cells treated
with docosahexaenoic acid reduced the expression of PLAU, AMOTL1,
NRP1 and ETS2 genes in the recipient cells and inhibited
angiogenesis in breast cancer (25). In addition, miR-16 in exosomes of
MSCs contributed to the anti-angiogenic mechanism that directly
regulates vascular endothelial growth factor (VEGF) expression
(89). Bone marrow MSC-derived
exosomes could also negatively regulate VEGF in breast cancer cells
by transporting miR-100, and the mTOR/hypoxia-inducible factor
(HIF)-1α signaling axis was revealed to be involved in regulation,
leading to the reduction of angiogenesis (90).
Breast cancer cell-derived exosomes have been
reported to induce tissue factor (TF)-independent platelet
activation and aggregation, as well as TF-dependent plasma
coagulation and platelet activation and aggregation by producing
thrombin and increasing vessel thrombosis (89). By targeting IL6 receptor, breast
cancer cell-derived exosomal miR-21 was demonstrated to inhibit the
action of endothelial progenitor cells that can be used for
thrombectomy and reduce deep vein thrombosis, thereby facilitating
thrombosis (28).
Exosomes derived from breast cancer cell lines were
revealed to induce autophagy at the phosphorylation site of the
15th serine residue of p53 in normal human primary mammary
epithelial cells. Autophagic proteins LC3 I and II were clearly
detected in lysates of human mammary epithelial cells (HMECs)
incubated with exosomes from MDA-MB-231 cells for up to 24 h
(76).
Immune killing of tumor cells cannot be ignored.
When the immune function is low or the tumor cells have the ability
to escape from immune surveillance, tumors arise quietly. Exosomes
derived from breast cancer can inhibit the activity of T cells by
stimulating the proliferation of MDSCs and enhancing the
immunosuppressive function, leading to immune evasion (29). Exosomes derived from breast cancer
cell lines (MDA-MB-231 and MCF7) have been revealed to also be
involved in the immunoregulation of macrophages. After contacting
macrophages, the exosomes were revealed to bind to the surface of
the macrophage Toll-like receptor 2 (TLR2) through its
palmitoylated protein, which activates MyD88 in macrophages. The
NF-κB signaling pathway has been revealed to release a large number
of inflammatory cytokines including granulocyte colony-stimulating
factor (GC-SF), IL-6, CCL2/monocyte chemoattractant protein 1
(MCP1) and TNFα, and immunoregulate macrophages (29). This immune response was demonstrated
to eventually enhance the rate of development of metastatic tumors.
Exosomal proteins including calreticulin (CALR) (64), MHC class I molecules (31,65–67),
and proteasome activator complex subunit 2 (PSME2) (64) have been reported to be involved in
immune evasion of breast cancer. However, dendritic cell-derived
exosomes were revealed to activate innate and acquired immune
responses, which produce specific anti-breast cancer immune effects
(26).
Drug therapy of cancer is an important part of the
anticancer process. The prognosis of patients is determined to some
extent by drug resistance in the course of cancer treatment. It has
been revealed that exosomes derived from cancer stem cells and
chemoresistant breast cancer cells have higher levels of miR-155 by
targeting forkhead box O3a (FOXO3a), and the drug resistance can be
transmitted by exosomes to sensitive breast cancer cells (83).
Exosomal miR-451 released from breast cancer cells
was revealed to bind to multidrug resistance 1 (MDR1) and reduce
the expression of P-glycoprotein (P-gp), resulting in cell
resistance to doxorubicin, along with poor therapeutic efficacy
(91). The expression levels of
miR-17, miR-30a, miR-100 and miR-222 in exosomes were also
increased and mediated drug resistance after treatment of
paclitaxel or doxorubicin in MCF-7 cells (92). In addition, upregulated exosomal
miR-1246 in breast cancer cells may lead to enhanced drug
resistance and anti-chemotherapy activity by targeting CCNG2
(49). Moreover,
docetaxel-resistant exosomal miRNAs such as miR-24 (38,39,41),
miR-23a (39), miR-222 (38), and miR-452 (42) were revealed to be capable of
targeting p27, Sprouty2, PTEN, and anaphase promoting complex 4
(APC4) mRNAs respectively in target cells, conferring resistance to
docetaxel-sensitive cells. Exosomal miR-4443 has also been revealed
to lead to cell drug resistance (43). Similarly, miR-29-enriched exosomes
have been revealed to be delivered to recipient breast cancer cells
to promote resistance to doxorubicin by targeting PTEN (44). Except for the aforementioned
molecules, tamoxifen-resistant LCC2-derived exosomes have been
revealed to have high levels of lncRNA UCA1, rendering parental
MCF-7 cells resistant to tamoxifen (45). In exosomes of SKBR-3/Tr and BT474/Tr
breast cancer cells resistant to trastuzumab, the expression level
of lncRNA-SNHG14 was significantly higher than that of the parental
cells via the Bcl-2/Bax pathway (63). It has been revealed that transient
receptor potential channel 5 (TrpC5) in exosomes of breast cancer
may confer drug resistance to breast cancer cells (68) mediated by transcription factor
nuclear factor of activated T-cell isoform c3 (NFATc3) (93,94).
It has been reported that ATP binding cassette subfamily G member 2
(ABCG2), a novel multidrug efflux transporter in exosomes in breast
cancer, was associated with resistance to mitoxantrone and
camptothecin analogs (69).
Glutathione S-transferase P1 (GSTP1)-rich exosomes were able to
confer drug resistance and were revealed to be useful in predicting
chemoresistance (70). In addition,
exosomes carrying ubiquitin carboxy-terminal hydrolase (UCH)-L1 and
P-gp from doxorubicin-resistant human breast cancer cells
(MCF7/ADM) could be integrated into cells sensitive to doxorubicin
(MCF7/WT) to transfer chemoresistance (71).
Microenvironment-derived exosomes can also transmit
drug resistance information to cancer cells. CAF-derived exosomal
mitochondrial DNA (mtDNA) from hormonal therapy-naive breast cancer
cells in patients promoted the development of hormonal
therapy-resistant disease (95).
Exosomal RNA RN7SL1 in breast tumor fibroblasts may lead to
resistance to therapy in breast cancer cells (86). Exosomal miR-155 from
tumor-associated macrophages (TAMs) was secreted into exosomes and
transported to cancer cells. There, it targeted telomeric
repeat-binding factor 1, causing cancer cells to resist cisplatin
(96).
MDA-MB-231 human breast cancer cells overexpressing
exosomal miR-940 can promote osteogenic differentiation of host
mesenchymal cells to induce extensive osteoblastic lesions in the
final tumor (17). Breast
cancer-derived exosomes were revealed to induce the differentiation
of MSCs into fibroblasts by activating the signaling pathway
mediated by TGF-β receptor (97).
They can also transform adipose tissue-derived MSCs into
myofibroblast-like cells by activating the SMAD2-mediated
intracellular signaling pathway (97). Concurrently, activation of SMAD2 was
revealed to cause upregulation of fibroblast-related functional
factors including stromal cell-derived factor 1 (SDF1), VEGF1, CCL5
and TGF-β, thereby increasing the expression of α-smooth muscle
actin (α-SMA) and presenting a phenotype of tumor-related
fibroblasts (97). Conversely,
exosomal miR-21, miR-143, and miR-378e in CAFs induced cell
dedifferentiation to a stem-like state, through increased Oct3/4,
Nanog, and Sox2 expression in breast cancer cells (87).
Exosomal content used as biomarkers has several
characteristics: i) Exogenous substances are stable and not
degraded by external enzymes, thereby inferring that the integrity
and function of the substance are unchanged (98); ii) exosomal substances can easily to
be obtained in a noninvasive way in a wide range of body fluids,
such as blood, urine, and saliva; iii) the number of proteins and
RNAs in exosomes is higher than other types of extracellular
vesicles and the content in exosomes is higher than in serum in
several tumors, this indicates exosomal content may be rich in
exosomes which could improve the detection sensitivity (99); iv) exosomal proteins and RNAs are
cell-specific which may reflect the pathological state of different
diseases (100). Therefore,
exosomal contents are valuable markers for early diagnosis,
prediction of prognosis, and evaluation of therapy in breast cancer
(101).
Complete mtDNA was found in circulating exosomes of
hormone-resistant breast cancer and may predict poor prognosis
(102). CXC receptor 4 was
revealed to be a highly expressed mRNA in the plasma exosomes of
breast cancer and an indicator of poor prognosis (23). Breast cancer serum exosomal RN7SL1
RNA was revealed to be significantly associated with invasive
breast cancer and may be a prognosis biomarker (20). Increased expression of exogenous
exosomal hTERT mRNA, which reflects tumor burden and clinical
status of patients in serum may indicate early diagnosis and
recurrence of breast cancer (21).
microRNAs have been best described in studies that
have analyzed the presence of genetic materials in exosomes.
Combination of serum exosomal miR-21 and miR-1246 may be used as
markers for early diagnosis (103,104). Exosomal miR-301 and miR-155 are
considered to be early predictors of breast cancer (105–107). Serum exosomal miR-101, miR-372 and
miR-373 can be used as early diagnostic markers of breast cancer
(3). Increased expression of
exosomal miR-126a has been found in the serum of breast cancer
patients resistant to doxorubicin therapy, which could be used as a
predictor of chemosensitivity, diagnosis and prognosis of breast
cancer (18). Serum exosomal
miR-130a-3p of breast cancer may be used as a biomarker for the
diagnosis and prognosis of breast cancer (30). Breast cancer exosomal miR-200c and
miR-141 in plasma may be used as early predictors of cancer
metastasis (55). Log-rank test
analysis has revealed that the level of exosomal miR-378 was
inversely correlated with the overall survival rate and was
associated with adverse prognosis (75). In a study by Mihelich et al,
an ideal candidate biomarker, miR-182, that contributed to disease
aggressiveness was detected in the serum exosomes from breast
cancer patients. It may be a biomarker of disease prognosis
(108). Sueta et al
assessed the level of serum exosomal RNA in breast cancer patients.
The results revealed that miR-338-3p, miR124-3p, and miR-340-5p
were upregulated rapidly and miR-29b-3p, miR-130a-3p, miR-20b-5p,
miR-18a-5p, miR-486-5p, miR-17-5p, miR-195-5p, and miR-93-5p were
downregulated in patients with breast cancer relapse (109). These results suggest that these
genes are associated with breast cancer recurrence and can be used
as prognostic indicators (109).
Breast cancer is a dynamic disease, and good
diagnostic indicators can identify the type of disease and treat it
in a timely manner. Especially for patients who have no metastasis,
appropriate interventions, such as neoadjuvant therapy is required
to prevent development of MBC. To evaluate the prognosis of breast
cancer, researchers have compared the expression of miRNAs in serum
exosomes at different stages of breast cancer. Expression of miR-21
and miR-105 in MBC was revealed to be significantly higher than
those in healthy individuals (110). However, in comparison of
expression in localized breast cancer (LBC) and MBC, only miR-21
expression was statistically significant (110). Serum exosomal miR-105 can be used
as a potential marker to distinguish healthy individuals from
patients with LBC (110).
Therefore, with false-negative results that occur during the
treatment, combination of miR-21 and miR-105 may help improve
diagnosis (110). Zhou et
al (22) found high expression
of exosomal miR-105 in the serum of breast cancer patients with
pre-metastasis, ongoing metastasis or distant metastasis. This
indicates that miR-105 may predict early metastasis and be used as
a prognostic indicator (22). The
level of exosomal miR-143 was revealed to be higher in patients
with high grade (III/IV) breast cancer, and it could be used as an
indicator of breast cancer grading (87).
Breast cancer can be divided into the following
molecular types: Luminal A, luminal B, HER2-positive and
triple-negative. miR-100 has been revealed to be downregulated in
various molecular subtypes of breast cancer, and it may be used as
a marker for diagnosis and prognosis (27). Expression of serum exosomal miR-222
in luminal type A breast cancer was revealed to be significantly
lower than in luminal type B and triple-negative breast cancer
(TNBC) (110). Serum exosomal
miR-27a was increased in patients with HER2-positive breast cancer,
and serum miR-27b in exosomes was increased in HER2-positive and
TNBC (111). The level of exosomal
miR-373 was higher in patients with TNBC than other subtypes of
breast cancer, and it has been revealed as a biological indicator
of aggressive, hormone receptor-negative breast cancer (112,113). Exosomal miR-939 was demonstrated
to play an important role in metastasis via the blood or lymph node
and was inversely related to prognosis in TNBC (59). These indicators are important to
differentiate between numerous subtypes of breast cancer and
targeted treatment should be implemented to prevent recurrence and
resistance to therapy (110).
Exosomal miRNAs can also be used for monitoring
therapy efficacy. As previously demonstrated after neoadjuvant
chemotherapy (4 weeks after doxorubicin treatment), the tumor
diameter in patients with LBC was decreased, expression of serum
exosomal miR-21 was downregulated, and the expression level of
miR-221 was decreased in patients with lymph node infection. These
indicators can thus be used as prognostic indicators for
doxorubicin treatment (110). It
has also been demonstrated that the levels of serum exosomal
miR-376a, miR-27a, miR-155, and miR-376c in breast cancer patients
are dynamic, and reflect the cancer status and return to normal
after adjuvant therapy (114). It
was revealed that breast cancer patients treated with neoadjuvant
therapy had increased expression of exosomal miR-503 (52), which may be used as an indicator of
treatment efficacy (3). Therefore,
dynamic measurement of miRNA levels in exosomes can reduce the rate
of misdiagnosis and can also monitor treatment response and effect
in real time.
Except for miRNAs, exosomes are acknowledged to
contain several other ncRNAs, such as lncRNAs, which are also
markers for breast cancer. lncRNA GAS5 in breast cancer cell
exosomes, an inhibitor of cell proliferation and apoptosis
promoter, could be used to evaluate the efficacy of chemotherapy
and radiotherapy and be a biomarker of apoptosis induction
(115). Exosomal lncRNA-SNHG14 was
revealed to be increased in serum of HER2-positive breast cancer
with resistance to trastuzumab, and it may be used as a diagnostic
marker for breast cancer. The receiver operating characteristic
(ROC) curve, which is used to evaluate diagnostic value, revealed
an area under the curve (AUC) of 0.774, along with a diagnostic
sensitivity and specificity of 80.0 and 72.5% with the established
cut-offs (3.09), respectively (63).
Several exosomal proteins are expressed in different
stages or types of breast cancer and can be used as markers for
general cancer diagnosis and prognosis. Moon et al (116,117) revealed that the development of
developmentally regulated endothelial locus 1 (Del-1) and
fibronectin derived from circulating exosomes in plasma from breast
cancer patients may be used as promising biomarkers in the early
stage of the disease. Del-1 was also revealed as a potential marker
for distinguishing breast cancer from benign breast tumors and
other diseases. Ning et al (71) have demonstrated that exosomes
containing UCH-L1 can transfer chemoresistance to recipient cells,
and these exosomes can be used as non-invasive diagnostic
biomarkers for detecting chemoresistance in patients with breast
cancer. Exosomes containing amphiregulin (AREG) which binds to cell
surface epidermal growth factor receptor (EGFR), were revealed to
increase receptor breast cancer invasive ability of cells (118). Therefore, exosomes containing AREG
may be used as prognostic and/or predictive markers (26). In a prior study it was revealed that
compared with controls without disease for 5 years, patients with
breast cancer had higher levels of survivin and its splice variants
in serum. The results indicated that exosomal survivin-2B may be
used as a diagnostic and/or prognostic indicator for early breast
cancer patients (119). In a
recent study, glypican 1 (GPC1) was identified as a novel cancer
biomarker. It was revealed that, compared with healthy controls,
75% of breast cancer patients had high expression of
GPC1+ circulating exosomes (120). In addition to tetraspanin CD9,
HSP70, epithelial cell adhesion molecule (EpCAM) (121) and Annexin-1 (122), the metalloproteinase ADAM10
(121,124) was also revealed to be uniquely
expressed in breast cancer of serum or pleural effusion-derived
exosomes from patients (123).
Potential biomarkers of breast cancer are summarized in Table II.
Via the secretion of exosomes, breast cancer cells
affect the whole systemic environment to regulate the onset and
development of cancer (124,125). Therefore, it is a superior
approach for cancer therapy to prevent exosomal formation and
secretion (124,125). In breast cancer, psoralen may
reduce exosomal formation and secretion, thereby leading to MDR
reversal (14). A novel strategy
for treating cancer is the removal of HER2-positive exosomes from
the whole circulatory system by application of affinity plasma
exchange, established by the biotechnology company Aethlon Medical,
Inc. This new strategy is helpful to inhibit the progression of
HER2-positive breast cancer (126).
The important roles of exosomes in breast cancer
suggest that they can be exploited as a therapeutic target.
Increased mRNA expression levels of CD9, CD63, CD81 and tumor
susceptibility gene 101 (TSG101) were revealed in breast cancer
stem cell exosomes, which may serve as useful biomarkers to provide
new strategies for cancer treatment and prevention (127). Exosomal CCNG2 mRNA and miR-1246
could be used as therapeutic targets for breast cancer by inducing
cell proliferation, migration, invasion, metastasis and drug
resistance (49). miR-660 in serum
exosomes is considered to regulate breast cancer cell
proliferation, migration, and invasion, thereby serving as a
therapeutic target for breast cancer (128). lncRNA MALAT1 was also revealed to
be highly expressed in breast cancer cell exosomes and promoted
cell proliferation, and thus may be used as a target gene for
breast cancer treatment (129).
Exosomal lncRNA-SNHG14 could be used as a therapeutic target for
HER2-positive breast cancer with resistance to trastuzumab which
was revealed to lead to distant metastasis (63).
Exosomes derived from stem cells are an efficient
mode of drug delivery. The advantages are as follows: i) Stem cells
can produce more exosomes (130);
ii) exosomes derived from stem cells have improved ability for
expansion in vivo (130);
iii) stem cells harness strong differentiation ability (130); and iv) stem cells are free from
activating the immune system, which renders stem cell exosomes a
safe drug delivery tool (131).
Numerous strategies may be used to treat breast
cancer by exosomes: Transfection of anti-RNA substances or
anti-oncogenes into exosomes, drugs that interfere with loading or
delivery of exosomal contents that promote tumors, or removal of
specific exosomal compounds from circulation. Natural or synthetic
exosomes in a specific cell type can utilize affinity to deliver
activity-modulating compounds to other cancer cells or
microenvironments to promote tumor suppressor contents or inhibit
tumor promoter contents (3).
Exosomes derived from bone MSCs were demonstrated to
effectively transport anti-miR-142-3p, thereby inhibiting the
expression of miR-142-3p and miR-150 and increasing the level of
APC mRNA and P2X purinoceptor 7 mRNA, thus leading to antitumor
effects including decrease of cell viability, increase of
apoptosis, significantly reduced tumor volume and weight, and
extended survival (6). The let-7
miRNA from HCC70 cell-derived exosomes in human breast ductal
carcinoma could inhibit EGFR-positive breast cancer in vivo
(132). In addition, 293
cell-derived exosomes could deliver surface-modified with anti-EGFR
peptides through cell engineering to inhibit the proliferation of
breast cancer cells (133).
The microenvironment plays an important role in
tumor development. Exosomes released from pre-adipocytes in the
microenvironment of breast cancer can promote tumor invasion and
metastasis (134). It has been
reported that shikonin could inhibit tumor growth and increase the
sensitivity of breast cancer cells to endocrine therapy (135). The differentiation of
pre-adipocytes treated with shikonin was revealed to be reduced,
and the content of miR-140 in the exosomes was increased, so that
the SOX9/miR-140 signaling pathway in target cells was enhanced,
thereby inhibiting the stem cell formation, tumor migration, and
tumor growth of ductal carcinoma in situ (DCIS) in
vivo (58). Therefore, drugs
targeting the microenvironment are important for the prevention of
invasive breast cancer.
Breast cancer cell exosomes carrying miR-134 were
revealed to increase sensitivity to anticancer drugs (54). During breast cancer neoadjuvant
chemotherapy, HUVECs were capable of transmitting miR-503 to breast
tumor cells via exosomes, which was helpful to the antitumor
response (52). Tumor-derived
exosomes are composed of various cytosol membranous tumor antigens,
molecules involved in antigen presentation, which may be promising
candidates for cancer vaccines (136,137).
In addition to breast cancer, exosomes have been
reported in several other tumors and benign diseases. Thus far,
they have been recognized as potential biomarkers for cancer
diagnostics due to their characteristics (139–142). Since exosomes can be secreted by
all cells and circulate in the body fluids, specific detection and
isolation of cancer cell-derived exosomes are urgently required to
identify the different diseases. GPC1+ circulating
exosomes were detected in the serum of patients with pancreatic
ductal adenocarcinoma (PDCA) with absolute specificity and
sensitivity (120). CLDN4, EPCAM
as well as other exosome ‘surfaceome’ were revealed to be
PDAC-specific biomarker candidates (143). Surface-enhanced Raman scattering
technology for rapid and label-free exosomal detection (Exo-SERS)
and multiplex proximity extension assays (PEA) are helpful in the
identification of various exosomes from different sources (144,145). Furthermore, due to the
heterogeneity of exosomes, it is desirable to investigate them
individually by using different techniques such as antibody-DNA
conjugates and next-generation sequencing which combines a
proximity-dependent barcoding assay or a sequential quantification
analysis utilizing DNA-PAINT. In the future, profiling of multiple
exosomal surface biomarkers at the single-exosome level may be
detected to distinguish exosomes from different diseases (146,147). An increasing number of specific
exosomal molecules may be established for the identification of
different diseases.
Substances in the exosomes, such as DNAs, mRNAs,
proteins, miRNAs and lncRNAs, play important roles in tumor cell
communication and remodeling of the tumor microenvironment. The
study of exosomes provides a new perspective for the diagnosis and
prognosis of tumors. Furthermore, exosomes and their contents have
become new targets for cancer treatment.
Although exosome research is promising, some issues
affect research of exosomes. (1)
In vivo, the potential effect of exosomes from cancer cells
on recipient cells may rely on some environmental parameters and
accessibility barriers (34). It is
impossible to guarantee whether the exosomal content in the in
vitro co-culture technique is within the physiological range
(148). (2) All cells can release exosomes. Cancer
cells or CAF-derived exosomes account for a small proportion, thus
there are several research obstacles in finding markers to identify
exosomes of cancer cell origin and increasing the number of
exosomes in order to extract and detect abundant contents (149). (3)
An endogenous control used to normalize the expression of exosomal
contents is lacking. Without a reference content, different
publications may report conflicting data on the level of exosomal
contents. Furthermore, standardization of the approaches are
important in order to obtain common concepts for exosome assays and
reporting results, with international cooperation among the
scientists (150). (4) When the patient has other diseases, the
expression of exosomal contents is variable and not specific, thus
the appropriate statistical tests should be used when analyzing the
data (112). (5) As aforementioned, when exosomes are
used to treat breast cancer, stem cell-derived exosomes can be
excellent drug carriers, however, they also have several
shortcomings. For example, their biodistribution and in vivo
clearance functions have not been elucidated (151–154), which may have adverse side effects
in humans, such as acute renal injury (155). In addition, stem cell-derived
exosomes have some cytotoxicity and tumor tropism (6). Therefore, a method is required to
control the particle size of exosomes, allowing them to fully
function in the body, and be effectively removed in a timely manner
(6). This will provide more
constructive insights into the diagnosis and treatment of breast
cancer exosomes after these issues are resolved (43).
Not applicable.
This study was supported by the National Natural
Science Foundation of China (grant no. 81702078), the Natural
Science Foundation of Jiangsu Province (grant no. BK20170356), the
Natural Science Fund for Colleges and Universities of Jiangsu
Province (grant no. 17KJB320016), the Suzhou Science and Technology
Project (grant no. SYS201728), the Postgraduate Research and
Practice Innovation Program of Jiangsu Province (grant no.
KYCX18_2535), the Young Stuff Pre-research Fund Project of The
Second Affiliated Hospital of Soochow University (grant no.
SDFEYQN1718).
Not applicable.
HY conceived the study. LW and BW collected the
literature and wrote the review. HW, JM and YR prepared the tables
and participated in the writing of the manuscript. All authors
read, reviewed, and approved the final manuscript. LW and BW
contributed equally to this work.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sempere LF, Keto J and Fabbri M: Exosomal
MicroRNAs in breast cancer towards diagnostic and therapeutic
applications. Cancers (Basel). 9(pii): E712017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bardelli A and Pantel K: Liquid biopsies,
what we do not know (Yet). Cancer Cell. 31:172–179. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kalra H, Drummen GP and Mathivanan S:
Focus on extracellular vesicles: Introducing the next small big
thing. Int J Mol Sci. 17:1702016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Naseri Z, Oskuee RK, Jaafari MR and
Forouzandeh Moghadam M: Exosome-mediated delivery of functionally
active miRNA-142-3p inhibitor reduces tumorigenicity of breast
cancer in vitro and in vivo. Int J Nanomedicine. 13:7727–7747.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tkach M and Théry C: Communication by
extracellular vesicles: Where we are and where we need to go. Cell.
164:1226–1232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang X, Shi H, Yuan X, Jiang P, Qian H
and Xu W: Tumor-derived exosomes induce N2 polarization of
neutrophils to promote gastric cancer cell migration. Mol Cancer.
17:1462018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jin Y, Chen K, Wang Z, Wang Y, Liu J, Lin
L, Shao Y, Gao L, Yin H, Cui C, et al: DNA in serum extracellular
vesicles is stable under different storage conditions. BMC Cancer.
16:7532016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Q, Shao Y, Zhang X, Zheng T, Miao M,
Qin L, Wang B, Ye G, Xiao B and Guo J: Plasma long noncoding RNA
protected by exosomes as a potential stable biomarker for gastric
cancer. Tumour Biol. 36:2007–2012. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Street JM, Koritzinsky EH, Glispie DM,
Star RA and Yuen PS: Urine exosomes: An emerging trove of
biomarkers. Adv Clin Chem. 78:103–122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nair S, Tang KD, Kenny L and Punyadeera C:
Salivary exosomes as potential biomarkers in cancer. Oral Oncol.
84:31–40. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang H, Fu H, Xu W and Zhang X: Exosomal
non-coding RNAs: A promising cancer biomarker. Clin Chem Lab Med.
54:1871–1879. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang X, Xu C, Hua Y, Sun L, Cheng K, Jia
Z, Han Y, Dong J, Cui Y and Yang Z: Exosomes play an important role
in the process of psoralen reverse multidrug resistance of breast
cancer. J Exp Clin Cancer Res. 35:1862016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Conte D, Verri C, Borzi C, Suatoni P,
Pastorino U, Sozzi G and Fortunato O: Novel method to detect
microRNAs using chip-based QuantStudio 3D digital PCR. BMC
Genomics. 16:8492015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jeong S, Park J, Pathania D, Castro CM,
Weissleder R and Lee H: Integrated magneto-electrochemical sensor
for exosome analysis. ACS Nano. 10:1802–1809. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Singh CK, Kumar A and Roy SS: Quantitative
analysis of the methane gas emissions from municipal solid waste in
India. Sci Rep. 8:29132018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deng Z, Rong Y, Teng Y, Zhuang X,
Samykutty A, Mu J, Zhang L, Cao P, Yan J, Miller D and Zhang HG:
Exosomes miR-126a released from MDSC induced by DOX treatment
promotes lung metastasis. Oncogene. 36:639–651. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sebolt-Leopold JS and Herrera R: Targeting
the mitogen-activated protein kinase cascade to treat cancer. Nat
Rev Cancer. 4:937–947. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou R, Chen KK, Zhang J, Xiao B, Huang Z,
Ju C, Sun J, Zhang F, Lv XB and Huang G: The decade of exosomal
long RNA species: An emerging cancer antagonist. Mol Cancer.
17:752018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Goldvaser H, Gutkin A, Beery E, Edel Y,
Nordenberg J, Wolach O, Rabizadeh E, Uziel O and Lahav M:
Characterisation of blood-derived exosomal hTERT mRNA secretion in
cancer patients: A potential pan-cancer marker. Br J Cancer.
117:353–357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou W, Fong MY, Min Y, Somlo G, Liu L,
Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR, et al:
Cancer-secreted miR-105 destroys vascular endothelial barriers to
promote metastasis. Cancer Cell. 25:501–515. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rodriguez M, Silva J, Herrera A, Herrera
M, Peña C, Martín P, Gil-Calderón B, Larriba MJ, Coronado MJ,
Soldevilla B, et al: Exosomes enriched in
stemness/metastatic-related mRNAS promote oncogenic potential in
breast cancer. Oncotarget. 6:40575–40587. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ,
Hou JH, Fu J, Zeng MS, Yun JP, Wu QL, et al: Knockdown of miR-21 in
human breast cancer cell lines inhibits proliferation, in vitro
migration and in vivo tumor growth. Breast Cancer Res. 13:R22011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hannafon BN, Carpenter KJ, Berry WL,
Janknecht R, Dooley WC and Ding WQ: Exosome-mediated microRNA
signaling from breast cancer cells is altered by the
anti-angiogenesis agent docosahexaenoic acid (DHA). Mol Cancer.
14:1332015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bi TL, Sun JJ, Tian YZ and Zhou YF:
Research progress of relationship between exosomes and breast
cancer. Sheng Li Xue Bao. 68:352–358. 2016.(In Chinese). PubMed/NCBI
|
|
27
|
Chen D, Sun Y, Yuan Y, Han Z, Zhang P,
Zhang J, You MJ, Teruya-Feldstein J, Wang M, Gupta S, et al:
miR-100 induces epithelial-mesenchymal transition but suppresses
tumorigenesis, migration and invasion. PLoS Genet. 10:e10041772014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang W, Yuan X, Xu A, Zhu X, Zhan Y, Wang
S and Liu M: Human cancer cells suppress behaviors of endothelial
progenitor cells through miR-21 targeting IL6R. Microvasc Res.
120:21–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chow A, Zhou W, Liu L, Fong MY, Champer J,
Van Haute D, Chin AR, Ren X, Gugiu BG, Meng Z, et al: Macrophage
immunomodulation by breast cancer-derived exosomes requires
Toll-like receptor 2-mediated activation of NF-κB. Sci Rep.
4:57502014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kong X, Zhang J, Li J, Shao J and Fang L:
MiR-130a-3p inhibits migration and invasion by regulating RAB5B in
human breast cancer stem cell-like cells. Biochem Biophys Res
Commun. 501:486–493. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Clayton A and Tabi Z: Exosomes and the
MICA-NKG2D system in cancer. Blood Cells Mol Dis. 34:206–213. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vallabhaneni KC, Penfornis P, Dhule S,
Guillonneau F, Adams KV, Mo YY, Xu R, Liu Y, Watabe K, Vemuri MC
and Pochampally R: Extracellular vesicles from bone marrow
mesenchymal stem/stromal cells transport tumor regulatory microRNA,
proteins, and metabolites. Oncotarget. 6:4953–4967. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Melo SA, Sugimoto H, O'Connell JT, Kato N,
Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, et
al: Cancer exosomes perform cell-independent microRNA biogenesis
and promote tumorigenesis. Cancer Cell. 26:707–721. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ochieng J, Pratap S, Khatua AK and Sakwe
AM: Anchorage-independent growth of breast carcinoma cells is
mediated by serum exosomes. Exp Cell Res. 315:1875–1888. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brandes AA, Franceschi E, Tosoni A, Hegi
ME and Stupp R: Epidermal growth factor receptor inhibitors in
neuro-oncology: Hopes and disappointments. Clin Cancer Res.
14:957–960. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhong S, Li W, Chen Z, Xu J and Zhao J:
MiR-222 and miR-29a contribute to the drug-resistance of breast
cancer cells. Gene. 531:8–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li X, Liu X, Xu W, Zhou P, Gao P, Jiang S,
Lobie PE and Zhu T: c-MYC-regulated miR-23a/24-2/27a cluster
promotes mammary carcinoma cell invasion and hepatic metastasis by
targeting Sprouty2. J Biol Chem. 288:18121–18133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li X, Wu Y, Liu A and Tang X: Long
non-coding RNA UCA1 enhances tamoxifen resistance in breast cancer
cells through a miR-18a-HIF1α feedback regulatory loop. Tumour
Biol. 37:14733–14743. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Giglio S, Cirombella R, Amodeo R, Portaro
L, Lavra L and Vecchione A: MicroRNA miR-24 promotes cell
proliferation by targeting the CDKs inhibitors p27Kip1 and
p16INK4a. J Cell Physiol. 228:2015–2023. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen WX, Cai YQ, Lv MM, Chen L, Zhong SL,
Ma TF, Zhao JH and Tang JH: Exosomes from docetaxel-resistant
breast cancer cells alter chemosensitivity by delivering microRNAs.
Tumour Biol. 35:9649–9659. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He Y, Deng F, Yang S, Wang D, Chen X,
Zhong S, Zhao J and Tang J: Exosomal microRNA: A novel biomarker
for breast cancer. Biomark Med. 12:177–188. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shen H, Li L, Yang S, Wang D, Zhong S,
Zhao J and Tang J: MicroRNA-29a contributes to drug-resistance of
breast cancer cells to adriamycin through PTEN/AKT/GSK3β signaling
pathway. Gene. 593:84–90. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xu CG, Yang MF, Ren YQ, Wu CH and Wang LQ:
Exosomes mediated transfer of lncRNA UCA1 results in increased
tamoxifen resistance in breast cancer cells. Eur Rev Med Pharmacol
Sci. 20:4362–4368. 2016.PubMed/NCBI
|
|
46
|
Matula Z, Németh A, Lőrincz P, Szepesi Á,
Brózik A, Buzás EI, Lőw P, Német K, Uher F and Urbán VS: The role
of extracellular vesicle and tunneling nanotube-mediated
intercellular cross-talk between mesenchymal stem cells and human
peripheral T cells. Stem Cells Dev. 25:1818–1832. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ono M, Kosaka N, Tominaga N, Yoshioka Y,
Takeshita F, Takahashi RU, Yoshida M, Tsuda H, Tamura K and Ochiya
T: Exosomes from bone marrow mesenchymal stem cells contain a
microRNA that promotes dormancy in metastatic breast cancer cells.
Sci Signal. 7:ra632014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wei Y, Li M, Cui S, Wang D, Zhang CY, Zen
K and Li L: Shikonin inhibits the proliferation of human breast
cancer cells by reducing tumor-derived exosomes. Molecules.
21(pii): E7772016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gorczynski RM, Zhu F, Chen Z, Kos O and
Khatri I: A comparison of serum miRNAs influencing metastatic
growth of EMT6 vs 4THM tumor cells in wild-type and CD200R1KO mice.
Breast Cancer Res Treat. 162:255–266. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wei Y, Lai X, Yu S, Chen S, Ma Y, Zhang Y,
Li H, Zhu X, Yao L and Zhang J: Exosomal miR-221/222 enhances
tamoxifen resistance in recipient ER-positive breast cancer cells.
Breast Cancer Res Treat. 147:423–431. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang L, Zhang S, Yao J, Lowery FJ, Zhang
Q, Huang WC, Li P, Li M, Wang X, Zhang C, et al:
Microenvironment-induced PTEN loss by exosomal microRNA primes
brain metastasis outgrowth. Nature. 527:100–104. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bovy N, Blomme B, Frères P, Dederen S,
Nivelles O, Lion M, Carnet O, Martial JA, Noël A, Thiry M, et al:
Endothelial exosomes contribute to the antitumor response during
breast cancer neoadjuvant chemotherapy via microRNA transfer.
Oncotarget. 6:10253–10266. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lim PK, Bliss SA, Patel SA, Taborga M,
Dave MA, Gregory LA, Greco SJ, Bryan M, Patel PS and Rameshwar P:
Gap junction-mediated import of microRNA from bone marrow stromal
cells can elicit cell cycle quiescence in breast cancer cells.
Cancer Res. 71:1550–1560. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
O'Brien K, Lowry MC, Corcoran C, Martinez
VG, Daly M, Rani S, Gallagher WM, Radomski MW, MacLeod RA and
O'Driscoll L: miR-134 in extracellular vesicles reduces
triple-negative breast cancer aggression and increases drug
sensitivity. Oncotarget. 6:32774–32789. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang G, Zhang W, Li B, Stringer-Reasor E,
Chu C, Sun L, Bae S, Chen D, Wei S, Jiao K, et al: MicroRNA-200c
and microRNA- 141 are regulated by a FOXP3-KAT2B axis and
associated with tumor metastasis in breast cancer. Breast Cancer
Res. 19:732017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Singh R, Pochampally R, Watabe K, Lu Z and
Mo YY: Exosome-mediated transfer of miR-10b promotes cell invasion
in breast cancer. Mol Cancer. 13:2562014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Fong MY, Zhou W, Liu L, Alontaga AY,
Chandra M, Ashby J, Chow A, O'Connor ST, Li S, Chin AR, et al:
Breast-cancer-secreted miR-122 reprograms glucose metabolism in
premetastatic niche to promote metastasis. Nat Cell Biol.
17:183–194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gernapudi R, Yao Y, Zhang Y, Wolfson B,
Roy S, Duru N, Eades G, Yang P and Zhou Q: Targeting exosomes from
preadipocytes inhibits preadipocyte to cancer stem cell signaling
in early-stage breast cancer. Breast Cancer Res Treat. 150:685–695.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Di Modica M, Regondi V, Sandri M, Iorio
MV, Zanetti A, Tagliabue E, Casalini P and Triulzi T: Breast
cancer-secreted miR-939 downregulates VE-cadherin and destroys the
barrier function of endothelial monolayers. Cancer Lett.
384:94–100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Tominaga N, Kosaka N, Ono M, Katsuda T,
Yoshioka Y, Tamura K, Lötvall J, Nakagama H and Ochiya T: Brain
metastatic cancer cells release microRNA-181c-containing
extracellular vesicles capable of destructing blood-brain barrier.
Nat Commun. 6:67162015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Baroni S, Romero-Cordoba S, Plantamura I,
Dugo M, D'Ippolito E, Cataldo A, Cosentino G, Angeloni V, Rossini
A, Daidone MG and Iorio MV: Exosome-mediated delivery of miR-9
induces cancer-associated fibroblast-like properties in human
breast fibroblasts. Cell Death Dis. 7:e23122016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yang M, Chen J, Su F, Yu B, Su F, Lin L,
Liu Y, Huang JD and Song E: Microvesicles secreted by macrophages
shuttle invasion-potentiating microRNAs into breast cancer cells.
Mol Cancer. 10:1172011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Dong H, Wang W, Chen R, Zhang Y, Zou K, Ye
M, He X, Zhang F and Han J: Exosome-mediated transfer of
lncRNA-SNHG14 promotes trastuzumab chemoresistance in breast
cancer. Int J Oncol. 53:1013–1026. 2018.PubMed/NCBI
|
|
64
|
Klinke DJ II, Kulkarni YM, Wu Y and
Byrne-Hoffman C: Inferring alterations in cell-to-cell
communication in HER2+ breast cancer using secretome
profiling of three cell models. Biotechnol Bioeng. 111:1853–1863.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zheng Y, Campbell EC, Lucocq J, Riches A
and Powis SJ: Monitoring the Rab27 associated exosome pathway using
nanoparticle tracking analysis. Exp Cell Res. 319:1706–1713. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Bard MP, Hegmans JP, Hemmes A, Luider TM,
Willemsen R, Severijnen LA, van Meerbeeck JP, Burgers SA,
Hoogsteden HC and Lambrecht BN: Proteomic analysis of exosomes
isolated from human malignant pleural effusions. Am J Respir Cell
Mol Biol. 31:114–121. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Palazzolo G, Albanese NN, DI Cara G, Gygax
D, Vittorelli ML and Pucci-Minafra I: Proteomic analysis of
exosome-like vesicles derived from breast cancer cells. Anticancer
Res. 32:847–860. 2012.PubMed/NCBI
|
|
68
|
Ma X, Chen Z, Hua D, He D, Wang L, Zhang
P, Wang J, Cai Y, Gao C, Zhang X, et al: Essential role for
TrpC5-containing extracellular vesicles in breast cancer with
chemotherapeutic resistance. Proc Natl Acad Sci USA. 111:6389–6394.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kong JN, He Q, Wang G, Dasgupta S, Dinkins
MB, Zhu G, Kim A, Spassieva S and Bieberich E: Guggulsterone and
bexarotene induce secretion of exosome-associated breast cancer
resistance protein and reduce doxorubicin resistance in MDA-MB-231
cells. Int J Cancer. 137:1610–1620. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yang SJ, Wang DD, Li J, Xu HZ, Shen HY,
Chen X, Zhou SY, Zhong SL, Zhao JH and Tang JH: Predictive role of
GSTP1-containing exosomes in chemotherapy-resistant breast cancer.
Gene. 623:5–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ning K, Wang T, Sun X, Zhang P, Chen Y,
Jin J and Hua D: UCH-L1-containing exosomes mediate
chemotherapeutic resistance transfer in breast cancer. J Surg
Oncol. 115:932–940. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kesimer M, Scull M, Brighton B, DeMaria G,
Burns K, O'Neal W, Pickles RJ and Sheehan JK: Characterization of
exosome-like vesicles released from human tracheobronchial ciliated
epithelium: A possible role in innate defense. FASEB J.
23:1858–1868. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Villarreal L, Méndez O, Salvans C, Gregori
J, Baselga J and Villanueva J: Unconventional secretion is a major
contributor of cancer cell line secretomes. Mol Cell Proteomics.
12:1046–1060. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chen Y, Zeng C, Zhan Y, Wang H, Jiang X
and Li W: Aberrant low expression of p85α in stromal fibroblasts
promotes breast cancer cell metastasis through exosome-mediated
paracrine Wnt10b. Oncogene. 36:4692–4705. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Muluhngwi P and Klinge CM: Identification
of miRNAs as biomarkers for acquired endocrine resistance in breast
cancer. Mol Cell Endocrinol. 456:76–86. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Dutta S, Warshall C, Bandyopadhyay C,
Dutta D and Chandran B: Interactions between exosomes from breast
cancer cells and primary mammary epithelial cells leads to
generation of reactive oxygen species which induce DNA damage
response, stabilization of p53 and autophagy in epithelial cells.
PLoS One. 9:e975802014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wu C and Luo J: Long non-coding RNA
(lncRNA) urothelial carcinoma-associated 1 (UCA1) enhances
tamoxifen resistance in breast cancer cells via inhibiting mTOR
signaling pathway. Med Sci Monit. 22:3860–3867. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Gutkin A, Uziel O, Beery E, Nordenberg J,
Pinchasi M, Goldvaser H, Henick S, Goldberg M and Lahav M: Tumor
cells derived exosomes contain hTERT mRNA and transform
nonmalignant fibroblasts into telomerase positive cells.
Oncotarget. 7:59173–59188. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Gong C, Nie Y, Qu S, Liao JY, Cui X, Yao
H, Zeng Y, Su F, Song E and Liu Q: miR-21 induces myofibroblast
differentiation and promotes the malignant progression of breast
phyllodes tumors. Cancer Res. 74:4341–4352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Baglio SR, Rooijers K, Koppers-Lalic D,
Verweij FJ, Pérez Lanzón M, Zini N, Naaijkens B, Perut F, Niessen
HW, Baldini N and Pegtel DM: Human bone marrow- and
adipose-mesenchymal stem cells secrete exosomes enriched in
distinctive miRNA and tRNA species. Stem Cell Res Ther. 6:1272015.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Frankel LB, Christoffersen NR, Jacobsen A,
Lindow M, Krogh A and Lund AH: Programmed cell death 4 (PDCD4) is
an important functional target of the microRNA miR-21 in breast
cancer cells. J Biol Chem. 283:1026–1033. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
miR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Yu DD, Lv MM, Chen WX, Zhong SL, Zhang XH,
Chen L, Ma TF, Tang JH and Zhao JH: Role of miR-155 in drug
resistance of breast cancer. Tumour Biol. 36:1395–1401. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Maji S, Chaudhary P, Akopova I, Nguyen PM,
Hare RJ, Gryczynski I and Vishwanatha JK: Exosomal annexin II
promotes angiogenesis and breast cancer metastasis. Mol Cancer Res.
15:93–105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Menck K, Klemm F, Gross JC, Pukrop T,
Wenzel D and Binder C: Induction and transport of Wnt 5a during
macrophage-induced malignant invasion is mediated by two types of
extracellular vesicles. Oncotarget. 4:2057–2066. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon
T, Kim BC, Benci JL, DeMichele AM, Tchou J, Marcotrigiano J and
Minn AJ: Exosome RNA unshielding couples stromal activation to
pattern recognition receptor signaling in cancer. Cell.
170:352–366.e13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Donnarumma E, Fiore D, Nappa M, Roscigno
G, Adamo A, Iaboni M, Russo V, Affinito A, Puoti I, Quintavalle C,
et al: Cancer-associated fibroblasts release exosomal microRNAs
that dictate an aggressive phenotype in breast cancer. Oncotarget.
8:19592–19608. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Jung KO, Youn H, Lee CH, Kang KW and Chung
JK: Visualization of exosome-mediated miR-210 transfer from hypoxic
tumor cells. Oncotarget. 8:9899–9910. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Lee JK, Park SR, Jung BK, Jeon YK, Lee YS,
Kim MK, Kim YG, Jang JY and Kim CW: Exosomes derived from
mesenchymal stem cells suppress angiogenesis by down-regulating
VEGF expression in breast cancer cells. PLoS One. 8:e842562013.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Pakravan K, Babashah S, Sadeghizadeh M,
Mowla SJ, Mossahebi-Mohammadi M, Ataei F, Dana N and Javan M:
MicroRNA-100 shuttled by mesenchymal stem cell-derived exosomes
suppresses in vitro angiogenesis through modulating the
mTOR/HIF-1α/VEGF signaling axis in breast cancer cells. Cell Oncol
(Dordr). 40:457–470. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Chen WX, Zhong SL, Ji MH, Pan M, Hu Q, Lv
MM, Luo Z, Zhao JH and Tang JH: MicroRNAs delivered by
extracellular vesicles: An emerging resistance mechanism for breast
cancer. Tumour Biol. 35:2883–2892. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Chen WX, Liu XM, Lv MM, Chen L, Zhao JH,
Zhong SL, Ji MH, Hu Q, Luo Z, Wu JZ and Tang JH: Exosomes from
drug-resistant breast cancer cells transmit chemoresistance by a
horizontal transfer of microRNAs. PLoS One. 9:e952402014.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Dong Y, Pan Q, Jiang L, Chen Z, Zhang F,
Liu Y, Xing H, Shi M, Li J, Li X, et al: Tumor endothelial
expression of P-glycoprotein upon microvesicular transfer of TrpC5
derived from adriamycin-resistant breast cancer cells. Biochem
Biophys Res Commun. 446:85–90. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Ma X, Cai Y, He D, Zou C, Zhang P, Lo CY,
Xu Z, Chan FL, Yu S, Chen Y, et al: Transient receptor potential
channel TRPC5 is essential for P-glycoprotein induction in
drug-resistant cancer cells. Proc Natl Acad Sci USA.
109:16282–16287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Sansone P, Savini C, Kurelac I, Chang Q,
Amato LB, Strillacci A, Stepanova A, Iommarini L, Mastroleo C, Daly
L, et al: Packaging and transfer of mitochondrial DNA via exosomes
regulate escape from dormancy in hormonal therapy-resistant breast
cancer. Proc Natl Acad Sci USA. 114:E9066–E9075. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Challagundla KB, Wise PM, Neviani P, Chava
H, Murtadha M, Xu T, Kennedy R, Ivan C, Zhang X, Vannini I, et al:
Exosome-mediated transfer of microRNAs within the tumor
microenvironment and neuroblastoma resistance to chemotherapy. J
Natl Cancer Inst. 107(pii): djv1352015.PubMed/NCBI
|
|
97
|
Cho JA, Park H, Lim EH and Lee KW:
Exosomes from breast cancer cells can convert adipose
tissue-derived mesenchymal stem cells into myofibroblast-like
cells. Int J Oncol. 40:130–138. 2012.PubMed/NCBI
|
|
98
|
Berrondo C, Flax J, Kucherov V, Siebert A,
Osinski T, Rosenberg A, Fucile C, Richheimer S and Beckham CJ:
Expression of the long non-coding RNA HOTAIR correlates with
disease progression in bladder cancer and is contained in bladder
cancer patient urinary exosomes. PLoS One. 11:e01472362016.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Dong L, Lin W, Qi P, Xu MD, Wu X, Ni S,
Huang D, Weng WW, Tan C, Sheng W, et al: Circulating long RNAs in
serum extracellular vesicles: Their characterization and potential
application as biomarkers for diagnosis of colorectal cancer.
Cancer Epidemiol Biomarkers Prev. 25:1158–1166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Dragomir M, Chen B and Calin GA: Exosomal
lncRNAs as new players in cell-to-cell communication. Transl Cancer
Res. 7 (Suppl 2):S243–S252. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Momen-Heravi F, Getting SJ and Moschos SA:
Extracellular vesicles and their nucleic acids for biomarker
discovery. Pharmacol Ther. 192:170–187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Halvaei S, Daryani S, Eslami-S Z, Samadi
T, Jafarbeik-Iravani N, Bakhshayesh TO, Majidzadeh-A K and Esmaeili
R: Exosomes in cancer liquid biopsy: A focus on breast cancer. Mol
Ther Nucleic Acids. 10:131–141. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Hannafon BN, Trigoso YD, Calloway CL, Zhao
YD, Lum DH, Welm AL, Zhao ZJ, Blick KE, Dooley WC and Ding WQ:
Plasma exosome microRNAs are indicative of breast cancer. Breast
Cancer Res. 18:902016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Shi W, Gerster K, Alajez NM, Tsang J,
Waldron L, Pintilie M, Hui AB, Sykes J, P'ng C, Miller N, et al:
MicroRNA-301 mediates proliferation and invasion in human breast
cancer. Cancer Res. 71:2926–2937. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Ma F, Zhang J, Zhong L, Wang L, Liu Y,
Wang Y, Peng L and Guo B: Upregulated microRNA-301a in breast
cancer promotes tumor metastasis by targeting PTEN and activating
Wnt/β-catenin signaling. Gene. 535:191–197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Lettlova S, Brynychova V, Blecha J, Vrana
D, Vondrusova M, Soucek P and Truksa J: MiR-301a-3p suppresses
estrogen signaling by directly inhibiting ESR1 in ERα positive
breast cancer. Cell Physiol Biochem. 46:2601–2615. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Mihelich BL, Dambal S, Lin S and Nonn L:
miR-182, of the miR-183 cluster family, is packaged in exosomes and
is detected in human exosomes from serum, breast cells and prostate
cells. Oncol Lett. 12:1197–1203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Sueta A, Yamamoto Y, Tomiguchi M,
Takeshita T, Yamamoto-Ibusuki M and Iwase H: Differential
expression of exosomal miRNAs between breast cancer patients with
and without recurrence. Oncotarget. 8:69934–69944. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Rodriguez-Martinez A, de Miguel-Pérez D,
Ortega FG, García-Puche JL, Robles-Fernández I, Exposito J,
Martorell-Marugan J, Carmona-Sáez P, Garrido-Navas MDC, Rolfo C, et
al: Exosomal miRNA profile as complementary tool in the diagnostic
and prediction of treatment response in localized breast cancer
under neoadjuvant chemotherapy. Breast Cancer Res. 21:212019.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Stevic I, Müller V, Weber K, Fasching PA,
Karn T, Marmé F, Schem C, Stickeler E, Denkert C, van Mackelenbergh
M, et al: Specific microRNA signatures in exosomes of
triple-negative and HER2-positive breast cancer patients undergoing
neoadjuvant therapy within the GeparSixto trial. BMC Med.
16:1792018. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Joyce DP, Kerin MJ and Dwyer RM:
Exosome-encapsulated microRNAs as circulating biomarkers for breast
cancer. Int J Cancer. 139:1443–1448. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Eichelser C, Stückrath I, Müller V,
Milde-Langosch K, Wikman H, Pantel K and Schwarzenbach H: Increased
serum levels of circulating exosomal microRNA-373 in
receptor-negative breast cancer patients. Oncotarget. 5:9650–9663.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Cuk K, Zucknick M, Madhavan D, Schott S,
Golatta M, Heil J, Marmé F, Turchinovich A, Sinn P, Sohn C, et al:
Plasma microRNA panel for minimally invasive detection of breast
cancer. PLoS One. 8:e767292013. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Koldemir O, Özgür E and Gezer U:
Accumulation of GAS5 in exosomes is a marker of apoptosis
induction. Biomed Rep. 6:358–362. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Moon PG, Lee JE, Cho YE, Lee SJ, Jung JH,
Chae YS, Bae HI, Kim YB, Kim IS, Park HY and Baek MC:
Identification of developmental endothelial locus-1 on circulating
extracellular vesicles as a novel biomarker for early breast cancer
detection. Clin Cancer Res. 22:1757–1766. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Moon PG, Lee JE, Cho YE, Lee SJ, Chae YS,
Jung JH, Kim IS, Park HY and Baek MC: Fibronectin on circulating
extracellular vesicles as a liquid biopsy to detect breast cancer.
Oncotarget. 7:40189–40199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Higginbotham JN, Demory Beckler M, Gephart
JD, Franklin JL, Bogatcheva G, Kremers GJ, Piston DW, Ayers GD,
McConnell RE, Tyska MJ and Coffey RJ: Amphiregulin exosomes
increase cancer cell invasion. Curr Biol. 21:779–786. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Khan S, Bennit HF, Turay D, Perez M,
Mirshahidi S, Yuan Y and Wall NR: Early diagnostic value of
survivin and its alternative splice variants in breast cancer. BMC
Cancer. 14:1762014. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Melo SA, Luecke LB, Kahlert C, Fernandez
AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari
N, et al: Glypican-1 identifies cancer exosomes and detects early
pancreatic cancer. Nature. 523:177–182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Rupp AK, Rupp C, Keller S, Brase JC,
Ehehalt R, Fogel M, Moldenhauer G, Marmé F, Sültmann H and Altevogt
P: Loss of EpCAM expression in breast cancer derived serum
exosomes: Role of proteolytic cleavage. Gynecol Oncol. 122:437–446.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Lim LH and Pervaiz S: Annexin 1: The new
face of an old molecule. FASEB J. 21:968–975. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Galindo-Hernandez O, Villegas-Comonfort S,
Candanedo F, González-Vázquez MC, Chavez-Ocaña S,
Jimenez-Villanueva X, Sierra-Martinez M and Salazar EP: Elevated
concentration of microvesicles isolated from peripheral blood in
breast cancer patients. Arch Med Res. 44:208–214. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Kooijmans SA, Vader P, van Dommelen SM,
van Solinge WW and Schiffelers RM: Exosome mimetics: A novel class
of drug delivery systems. Int J Nanomedicine. 7:1525–1541.
2012.PubMed/NCBI
|
|
125
|
Kahlert C and Kalluri R: Exosomes in tumor
microenvironment influence cancer progression and metastasis. J Mol
Med (Berl). 91:431–437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Marleau AM, Chen CS, Joyce JA and Tullis
RH: Exosome removal as a therapeutic adjuvant in cancer. J Transl
Med. 10:1342012. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Kumar D, Gupta D, Shankar S and Srivastava
RK: Biomolecular characterization of exosomes released from cancer
stem cells: Possible implications for biomarker and treatment of
cancer. Oncotarget. 6:3280–3291. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Shen Y, Ye YF, Ruan LW, Bao L, Wu MW and
Zhou Y: Inhibition of miR-660-5p expression suppresses tumor
development and metastasis in human breast cancer. Genet Mol Res.
16:2017. View Article : Google Scholar
|
|
129
|
Zhang P, Zhou H, Lu K, Lu Y, Wang Y and
Feng T: Exosome-mediated delivery of MALAT1 induces cell
proliferation in breast cancer. Onco Targets Ther. 11:291–299.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Yeo RW, Lai RC, Zhang B, Tan SS, Yin Y,
The BJ and Lim SK: Mesenchymal stem cell: An efficient mass
producer of exosomes for drug delivery. Adv Drug Deliv Rev.
65:336–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Rani S, Ryan AE, Griffin MD and Ritter T:
Mesenchymal stem cell-derived extracellular vesicles: Toward
cell-free therapeutic applications. Mol Ther. 23:812–823. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Ohno S, Takanashi M, Sudo K, Ueda S,
Ishikawa A, Matsuyama N, Fujita K, Mizutani T, Ohgi T, Ochiya T, et
al: Systemically injected exosomes targeted to EGFR deliver
antitumor microRNA to breast cancer cells. Mol Ther. 21:185–191.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
de Jong OG, Kooijmans SAA, Murphy DE,
Jiang L, Evers MJW, Sluijter JPG, Vader P and Schiffelers RM: Drug
delivery with extracellular vesicles: From imagination to
innovation. Acc Chem Res. 52:1761–1770. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Iyengar P, Combs TP, Shah SJ, Gouon-Evans
V, Pollard JW, Albanese C, Flanagan L, Tenniswood MP, Guha C,
Lisanti MP, et al: Adipocyte-secreted factors synergistically
promote mammary tumorigenesis through induction of anti-apoptotic
transcriptional programs and proto-oncogene stabilization.
Oncogene. 22:6408–6423. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Yao Y and Zhou Q: A novel antiestrogen
agent Shikonin inhibits estrogen-dependent gene transcription in
human breast cancer cells. Breast Cancer Res Treat. 121:233–240.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Cho JA, Yeo DJ, Son HY, Kim HW, Jung DS,
Ko JK, Koh JS, Kim YN and Kim CW: Exosomes: A new delivery system
for tumor antigens in cancer immunotherapy. Int J Cancer.
114:613–622. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Tan A, De La Peña H and Seifalian AM: The
application of exosomes as a nanoscale cancer vaccine. Int J
Nanomedicine. 5:889–900. 2010.PubMed/NCBI
|
|
138
|
Wu CY, Du SL, Zhang J, Liang AL and Liu
YJ: Exosomes and breast cancer: A comprehensive review of novel
therapeutic strategies from diagnosis to treatment. Cancer Gene
Ther. 24:6–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Kalluri R: The biology and function of
exosomes in cancer. J Clin Invest. 126:1208–1215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Skog J, Würdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Tanaka Y, Kamohara H, Kinoshita K,
Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M and Baba H:
Clinical impact of serum exosomal microRNA-21 as a clinical
biomarker in human esophageal squamous cell carcinoma. Cancer.
119:1159–1167. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Costa-Silva B, Aiello NM, Ocean AJ, Singh
S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, et
al: Pancreatic cancer exosomes initiate pre-metastatic niche
formation in the liver. Nat Cell Biol. 17:816–826. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Castillo J, Bernard V, San Lucas FA,
Allenson K, Capello M, Kim DU, Gascoyne P, Mulu FC, Stephens BM,
Huang J, et al: Surfaceome profiling enables isolation of
cancer-specific exosomal cargo in liquid biopsies from pancreatic
cancer patients. Ann Oncol. 29:223–229. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Zhang P, Wang L, Fang Y, Zheng D, Lin T
and Wang H: Label-free exosomal detection and classification in
rapid discriminating different cancer types based on specific Raman
phenotypes and multivariate statistical analysis. Molecules.
24(pii): E29472019. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Larssen P, Wik L, Czarnewski P, Eldh M,
Löf L, Ronquist KG, Dubois L, Freyhult E, Gallant CJ, Oelrich J, et
al: Tracing cellular origin of human exosomes using multiplex
proximity extension assays. Mol Cell Proteomics. 16:502–511. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Wu D, Yan J, Shen X, Sun Y, Thulin M, Cai
Y, Wik L, Shen Q, Oelrich J1, Qian X, et al: Profiling surface
proteins on individual exosomes using a proximity barcoding assay.
Nat Commun. 10:38542019. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Chen C, Zong S, Liu Y, Wang Z, Zhang Y,
Chen B and Cui Y: Profiling of exosomal biomarkers for accurate
cancer identification: Combining DNA-PAINT with
machine-learning-based classification. Small. 15:e19010142019.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Shimomura A, Shiino S, Kawauchi J,
Takizawa S, Sakamoto H, Matsuzaki J, Ono M, Takeshita F, Niida S,
Shimizu C, et al: Novel combination of serum microRNA for detecting
breast cancer in the early stage. Cancer Sci. 107:326–334. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Roccaro AM, Sacco A, Maiso P, Azab AK, Tai
YT, Reagan M, Azab F, Flores LM, Campigotto F, Weller E, et al: BM
mesenchymal stromal cell-derived exosomes facilitate multiple
myeloma progression. J Clin Invest. 123:1542–1555. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Schwarzenbach H: The clinical relevance of
circulating, exosomal miRNAs as biomarkers for cancer. Expert Rev
Mol Diagn. 15:1159–1169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Takahashi Y, Nishikawa M, Shinotsuka H,
Matsui Y, Ohara S, Imai T and Takakura Y: Visualization and in vivo
tracking of the exosomes of murine melanoma B16-BL6 cells in mice
after intravenous injection. J Biotechnol. 165:77–84. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Smyth T, Kullberg M, Malik N, Smith-Jones
P, Graner MW and Anchordoquy TJ: Biodistribution and delivery
efficiency of unmodified tumor-derived exosomes. J Control Release.
199:145–155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Bala S, Csak T, Momen-Heravi F, Lippai D,
Kodys K, Catalano D, Satishchandran A, Ambros V and Szabo G:
Biodistribution and function of extracellular miRNA-155 in mice.
Sci Rep. 5:107212015. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Tian T, Wang Y, Wang H, Zhu Z and Xiao Z:
Visualizing of the cellular uptake and intracellular trafficking of
exosomes by live-cell microscopy. J Cell Biochem. 111:488–496.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Grange C, Tapparo M, Bruno S, Chatterjee
D, Quesenberry PJ, Tetta C and Camussi G: Biodistribution of
mesenchymal stem cell-derived extracellular vesicles in a model of
acute kidney injury monitored by optical imaging. Int J Mol Med.
33:1055–1063. 2014. View Article : Google Scholar : PubMed/NCBI
|