Introduction
Breast cancer is the most frequently diagnosed
malignancy in women worldwide, and is the second most common cause
of cancer-related morality among women (1). It is estimated that >1 million
people worldwide are diagnosed with breast cancer annually,
claiming 400,000 lives every year (2). In patients with breast cancer, distant
site metastases are considered the main cause of death (3,4). It
has been widely reported that highly expressed oncogenes promote
the development and progression of breast cancer (5–9).
Therefore, investigation of the roles of these genes and their
regulatory mechanism may provide a promising way to develop
therapeutic strategies for treating breast cancer.
Paired box (PAX) 6, a member of the PAX family of
proteins, is an important transcription factor in multiple
biological processes (10). PAX6 is
a key member of the retinal determination gene network (RDGN),
which is a conservative pathway required for the development of a
number of organs in mammals, including eyes, pancreas and central
nervous system (11,12). Recent studies have indicated that
aberrant expression of RDGN members is involved in cancer
initiation and progression (13–17).
Indeed, PAX6 is overexpressed in various types of human cancers,
which suggests its oncogenic roles (7,18–24).
Consistently, downregulation of PAX6 by gene silencing results in
the suppression on tumor progression in xenografted nude mice
(7). In breast cancer, specific
microRNAs inhibit cell proliferation and invasion by targeting
PAX6, which can be reversed by PAX6 overexpression (25,26).
Knockdown of PAX6 exerts significant inhibitory roles in breast
cancer cell proliferation and tumor progression in luminal type
(MCF7) and triple-negative (MDA-MB-231) breast cancer cell lines
(7). Moreover, PAX6 overexpression
is associated with a poor prognosis in patients with breast cancer
(27) and metastasis might be
affected by the methylation status of PAX6 (28). Taken together, these findings
suggested that PAX6 might be exploited as a potential target for
the treatment of breast cancer.
In breast cancer, ~90% of mortalities are associated
with metastasis of cancer cells (29). Thus, it is important to understand
the mechanism underlying the metastatic process as well as the
factors and pathways contributing to this process (30–33).
Increasing evidence supports that epithelial- mesenchymal
transition (EMT) serves a crucial role in breast tumor metastasis
(34,35). During EMT, cancer cells lose their
polarity and adhesion, and gain invasive and metastatic features
(36,37). It has been reported that the
transforming growth factor-β (TGF-β)-SMAD pathway induces EMT,
therefore promoting metastasis (38).
Although the aforementioned previous studies have
suggested an enhancive role of PAX6 in breast cancer progression,
the underlying mechanism remains unknown. The objectives of the
present study included: i) investigate the effects of PAX6 on
breast cancer cell metastasis; ii) explore the underlying mechanism
of the pro-metastatic property of PAX6. Herein, PAX6 was stably
overexpressed in breast cancer cells, revealing the pro-metastatic
property of PAX6. Additionally, analysis on the expression of PAX6
and TGF-β-SMAD signaling-associated proteins on human breast cancer
tissue array, and key factors involved in EMT revealed that
TGF-β-SMAD-mediated EMT may contribute to this biological
process.
Materials and methods
Cell culture and zebrafish models
MCF7 and MDA-MB-231 (Procell Life Science &
Technology Co., Ltd.; STR profile reports in Data S1 and S2, respectively) cell lines were cultured
in DMEM high glucose medium (Hyclone; GE Healthcare Life Sciences)
containing 10% fetal bovine serum (FBS) and 1%
penicillin/streptomycin, and incubated at 37°C in humidified air
containing 5% CO2. Adult wild-type zebrafish (AB line;
China Zebrafish Resource Center) were maintained under a 14 h
light/10 h dark cycle photoperiod according to the standard
procedures. The embryos were obtained from the natural mating of
adult zebrafish bred then maintained in bathing water. Fertilized
eggs were collected and transferred to 6-well plates (39). Plates were kept in an incubator with
the temperature maintained at 28±0.5°C and monitored daily until 2
days post-fertilization (dpf). To inhibit melanin formation, 0.003%
phenylthiocarbamide (Sigma-Aldrich; Merck KGaA) was added to the
bathing medium after 10–12 h post-fertilization (40).
Plasmid and transfection
The open reading frame of human PAX6 cDNA
(NM_000280; Shanghai Genechem Co., Ltd.) was cloned into the vector
pcDNA3.1 (Invitrogen; Thermo Fisher Scientific, Inc.) to generate
the PAX6 expression vector. The PAX6 expression vector and empty
pcDNA3.1 vector (2 µg) were stably transfected into MCF7 and
MDA-MB-231 cells using the FuGENE HD Transfection Reagent (Promega
Corporation) according to the manufacturer's protocol. After 24 h,
cells were passaged 1:15 in DMEM high glucose medium containing 600
µg/ml of G418 (Invitrogen; Thermo Fisher Scientific, Inc.). After
~2 weeks, individual clones were isolated by serial dilution.
Stably transfected cells were confirmed by western blotting, and a
total of four cell lines were established: i) PAX6-MCF7, MCF7 cells
with high expression of PAX6; ii) Ctl-MCF7, MCF7 cells with empty
pcDNA3.1; iii) PAX6-MDA-MB-231, MDA-MB-231 cells with high
expression of PAX6; iv) Ctl-MDA-MB-231, MDA-MB-231 cells lines with
empty pcDNA3.1.
Immunohistochemistry
Two human breast cancer tissue arrays with same
catalog number (cat. no. BR1504b) were obtained from Xian Alenabio
Biotechnology Co., Ltd. Each tissue array contains 70 malignant
primary breast cancer tissues and 4 paracarcinoma tissues.
Paraffin-embedded sections were stained as previously described
(41). Sections were deparaffinized
in xylene and rehydrated in graded alcohol concentrations. Sodium
citrate buffer was used for antigen retrieval. Tissues were treated
with 3% hydrogen peroxide in methanol to block endogenous
peroxidase activity, followed by 1% bovine serum albumin in PBS for
30 min at room temperature to block non-specific binding. Sections
were incubated with the primary antibody overnight at 4°C, and then
incubated with the secondary antibodies at room temperature for 30
min. The primary antibodies used were selected based on previous
studies and included PAX6 (1:100; Abcam; cat. no. ab197768)
(25) and SMAD2 [1:100; Cell
Signaling Technology, Inc. (CST); cat. no. 5339] (42); the secondary antibodies included
horseradish peroxidase-labeled anti-mouse IgG (1:1,000; Abcam; cat.
no. ab6789) and anti-rabbit IgG (1:1,000; Abcam; cat. no. ab97080).
DAB was used for chromogenic reaction, and nuclei were
counterstained with hematoxylin. One tissue array was used for PAX6
staining and another one was used for SMAD2 staining. To compare
PAX6 expression in paracarcinoma and breast cancer samples, the 4
cases of para-carcinoma and 20 randomly selected cases of malignant
primary breast cancer were used. However, to analyze the
relationship between PAX6 and SMAD2 staining patterns, all 70 cases
of malignant primary breast cancer were used. After analyzing 70
samples of PAX6 staining results, samples were divided in PAX6 low
expression and PAX6 high expression. Subsequently, the same samples
were used in the SMAD2 staining tissue array to determine SMAD2
expression in PAX6 low expression group and PAX6 high expression
group. All protein expressions were scored blindly by two
investigators. The staining intensity was rated on a 0–3 scale: 0
(negative), 1 (weak), 2 (moderate) and 3 (intense) (Fig. 1B). In addition, the percentage of
positively stained cells was also rated on a 0–3 scale: 0 (0-25%),
1 (26-50%), 2 (51-75%), and 3 (76-100%). The protein expression was
scored by multiplying the intensity score by the percentage score
of staining. PAX6 low and high expression was established based on
a staining below and above the mean expression of PAX6 staining,
respectively. The mean expression of PAX6 staining was determined
by calculating the mean value of PAX6 expression from all 70 cases
of malignant primary breast cancer.
Analysis of public microarray
datasets
All data were downloaded from http://gdac.broadinstitute.org/runs/stddata_latest/data/BRCA/20160128.
Clinical information and gene expression data [output as
RNA-sequencing by expectation-maximization (RSEM) values] were
obtained from Broad GDAC (January 2016; Firehose analysis run;
http://gdac.broadinstitute.org) to
assess the association between PAX6 expression and the overall
survival time of patients. Gene expression values were grouped into
three categories based on z-scores for PAX6 gene expression: Low
(z-score <-1.96; n=464), regular (−1.96≤ z-score ≤1.96; n=533)
and high (z-score >1.96; n=96).
Western blot analysis
Cells were seeded (1×105 cells/ml) in a
6-well plate (Corning Inc.) and cultured overnight. Cells were
harvested and collected in cold RIPA lysis buffer (Thermo Fisher
Scientific, Inc.) containing protease inhibitor cocktail. The
lysates were cleared at 20,000 × g for 10 min at 4°C, and protein
concentration was determined using the bicinchoninic acid protein
assay. Samples were boiled in 5X SDS-PAGE loading buffer and
western blotting was performed according to the Bio-Rad
electrophoresis protocol (43). The
samples were separated with 6, 10 or 12% SDS-PAGE gel (60 µg
protein per lane) by electrophoresis based on the molecular weight
of tested proteins and then electrophoretically transferred onto
PVDF membranes (Invitrogen; Thermo Fisher Scientific, Inc.).
Non-specific binding was blocked in blocking buffer TBST (TBS with
0.05% Tween-20) with 5% (w/v) skim milk for 2 h at room
temperature. The blots were subsequently incubated with primary
antibodies overnight at 4°C, washed with TBST three times, followed
by horseradish peroxidase (HRP)-conjugated secondary antibody
incubation for 2 h at room temperature. Protein expressions were
visualized by enhanced chemiluminescence (Amersham; Cytiva), and
images were captured using Image Studio System (version 5.0; LI-COR
Biosciences). Primary antibodies were selected according to
previous findings as cited and included: TGF-β (1:1,000; CST; cat.
no. 3711) (44), TGF-β receptor II
(TGF-βR II) (1:1,000; CST; cat. no. 79424) (45), SMAD2 (1:1,000; CST; cat. no. 5339)
(42), Snail zinc-finger protein
(SNAIL; 1:1,000; CST; cat. no. 3879) (42), β-actin (1:40,000; Sigma-Aldrich,
Merck KGaA; cat. no. A5441) and PAX6 (1:200; Abcam; cat. no.
ab197768) (25). Primary antibodies
of EMT-associated proteins included E-cadherin (1:1,000;
Proteintech Group, Inc.; cat. no. 20874-1-AP), N-cadherin (1:2,000;
Proteintech Group, Inc.; cat. no. 22018-1-AP), vimentin (1:1,000;
Proteintech Group, Inc.; cat. no. 10366-1-AP) and fibronectin
(1:500, Proteintech Group, Inc.; cat. no. 15613-1-AP) were selected
based on a previous literature (46). Secondary antibodies were
HRP-conjugated donkey anti-rabbit IgG (1:10,000; Zhongshan Golden
Bridge Biotechnology; cat. no. ZB5301) and HRP-conjugated donkey
anti-mouse IgG (1:10,000; Zhongshan Golden Bridge Biotechnology;
cat. no. ZB2305). The expression levels of target proteins were
normalized to β-actin and unpaired Student's t-test was applied to
statistically comparing the relative expression levels of proteins
using GraphPad Prism software 6.0 (GraphPad Software, Inc.).
Cell proliferation assay
Cell proliferation was evaluated by Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) according to
the manufacturer's protocol. Briefly, a total of 1×104
cells per well were seeded into 96-well plates in triplicate and
maintained at 37°C. After 6, 12, 24, 48 and 72 h, 10 µl CCK-8
medium was added into the wells. After incubation at 37°C for 3 h,
the absorbance was measured at a wavelength of 450 nm. The CCK-8
assays were repeated three times.
Gap closure assay
A total of 3×105 cells/ml were seeded
into each well of a Culture-Insert 2 Well in µ-Dish 35 mm plates
(Ibidi GmbH). After 24 h growth (95-100% confluent cell layer), the
Culture-Insert 2 Well was removed gently by using sterile tweezers
to generate a cell-free gap. The dish was washed with PBS to remove
cell debris and non-attached cells and filled with DMEM high
glucose medium with 10% FBS. Images were captured at 0 and 24 h
under a light microscope. Gap closure was analyzed using ImageJ
software (version 1.48; National Institutes of Health). The gap
closure assay was performed in triplicate.
Transwell migration assay
Cell migration was tested using the 24-mm Transwell
insert with 8.0 µm pore polycarbonate membrane. The top chambers
were seeded with 1×105 cells/ml in culture medium
without FBS, whereas the bottom chambers were filled with culture
medium containing 10% FBS. The cells were allowed to migrate for 24
h before being fixed with 4% paraformaldehyde for 20 min. Cells
were washed three times with PBS and stained with 0.1% crystal
violet for 40 min at room temperature. After staining, the cells
were washed three times with PBS and those remained on the top
surface of the membrane were scraped off with a cotton swab. The
migrated cells were imaged using a light microscope. Cell
quantification was performed on eight randomly selected fields
using ImageJ software (version 1.48; National Institutes of
Health). The Transwell migration assay was performed in
triplicate.
Tumor xenograft assay in
zebrafish
Pax6-MCF7 and Ctl-MCF7 stable cells were harvested
and resuspended at a density of 1×106 cells/ml, and then
treated with CM-Dil dye (Sigma-Aldrich, Merck KGaA) according to
the manufacturers' instructions. CM-Dil-labeled cells were loaded
into borosilicate glass capillary needles and injected into the
yolk sac of zebrafish embryos at 2 dpf to generate tumor
xenografts, as previously described (47). Zebrafish embryos were collected and
maintained in an incubator at 28±0.5°C; at 2 dpf, embryos were
anesthetized with 200 mg/l tricaine (0.003%; Sigma-Aldrich, Merck
KGaA) and positioned on a 10-cm petri dish coated with 1% agarose
for transplantation with CM-Dil-labeled cells at a density of
1×106/ml. After implantation, zebrafish were maintained
at 34°C for 1 day and randomly selected and photographed using a
fluorescence microscope. A total of 30 zebrafish embryos were used
in each group, and the experiments were repeated three times.
Statistical analysis
Kaplan-Meier survival curves were generated using
WinStat (version 1.1.2) for Excel (R Fitch Software, A-Prompt
Corp.) (48). PAX6 gene expression
values were grouped into three categories based on z-scores by
using R (version 3.2.2), aforementioned, and log-rank test was used
to compare patient survival curves between these groups. For the
remaining results, statistical analysis between groups was
performed using one-way ANOVA followed by Dunnett's post-hoc test
using GraphPad Prism software 6.0 (GraphPad Software, Inc.) and
expressed as mean ± SEM. P<0.05 was considered to indicate a
statistically significant difference.
Results
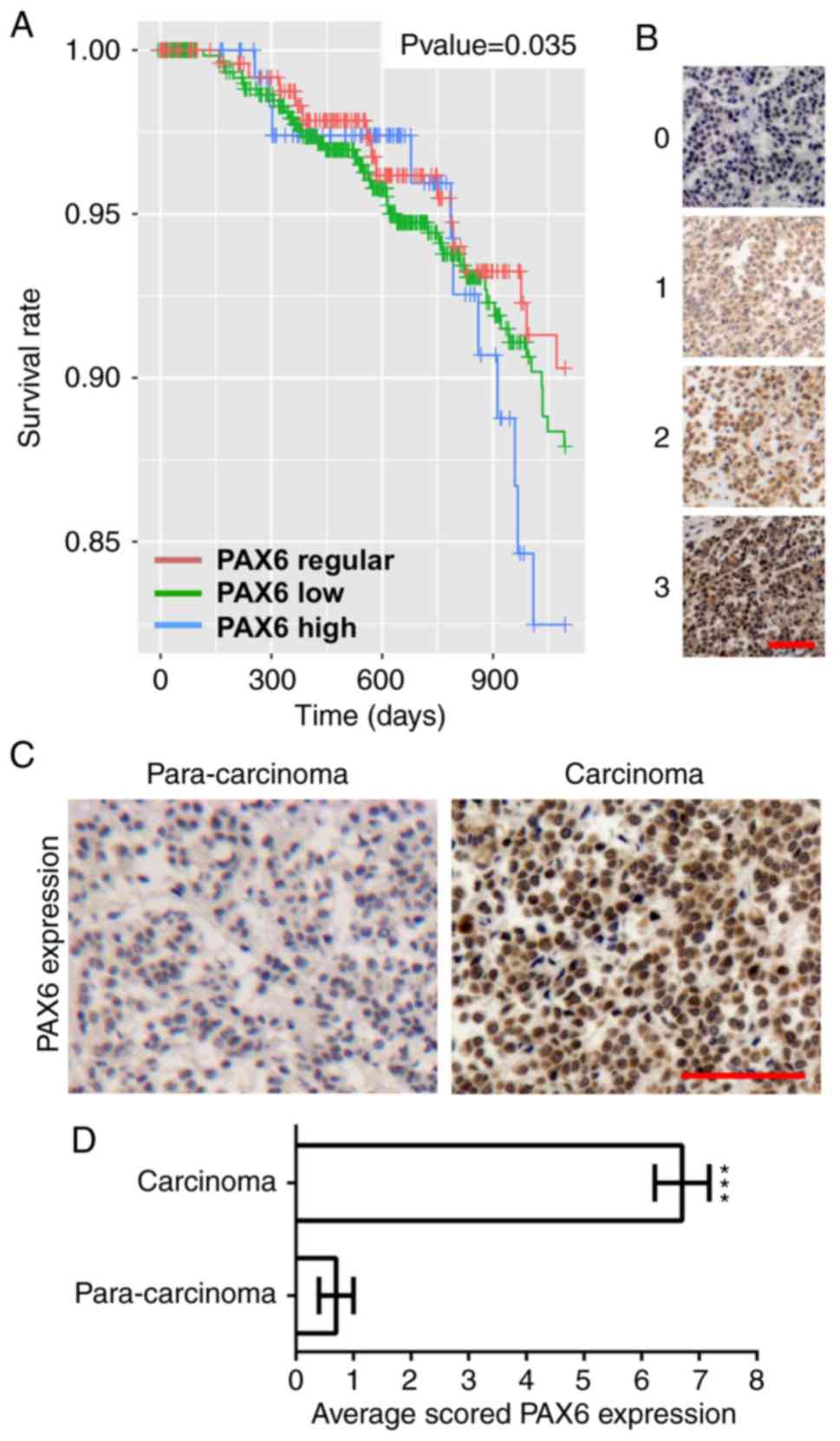
High expression of PAX6 in human
breast cancer is associated with poor prognosis
To analyze the association between the expression
levels of PAX6 and breast cancer progression, Kaplan-Meier survival
curves and PAX6 expression in human breast cancer and paracarcinoma
samples were assayed. As shown in Fig.
1A, high expression levels of PAX6 significantly associated
with poor patient prognosis, as measured by the decrease in breast
cancer survival in this group. The representative picture of each
staining intensity score was shown in Fig. 1B. To compare PAX6 expression in
paracarcinoma and breast cancer samples, the 4 cases of
para-carcinoma and 20 randomly selected cases of malignant primary
breast cancer from a human breast cancer tissue array were stained
with PAX6 and analyzed. A representative picture of high expression
of PAX6 in breast cancer is presented in Fig. 1C. PAX6 expression was scored by
multiplying the intensity score by the percentage score of PAX6
staining. As a result, PAX6 expression in breast cancer tissues was
demonstrated to be markedly increased compared with that in
paracarcinoma tissues (Fig.
1D).
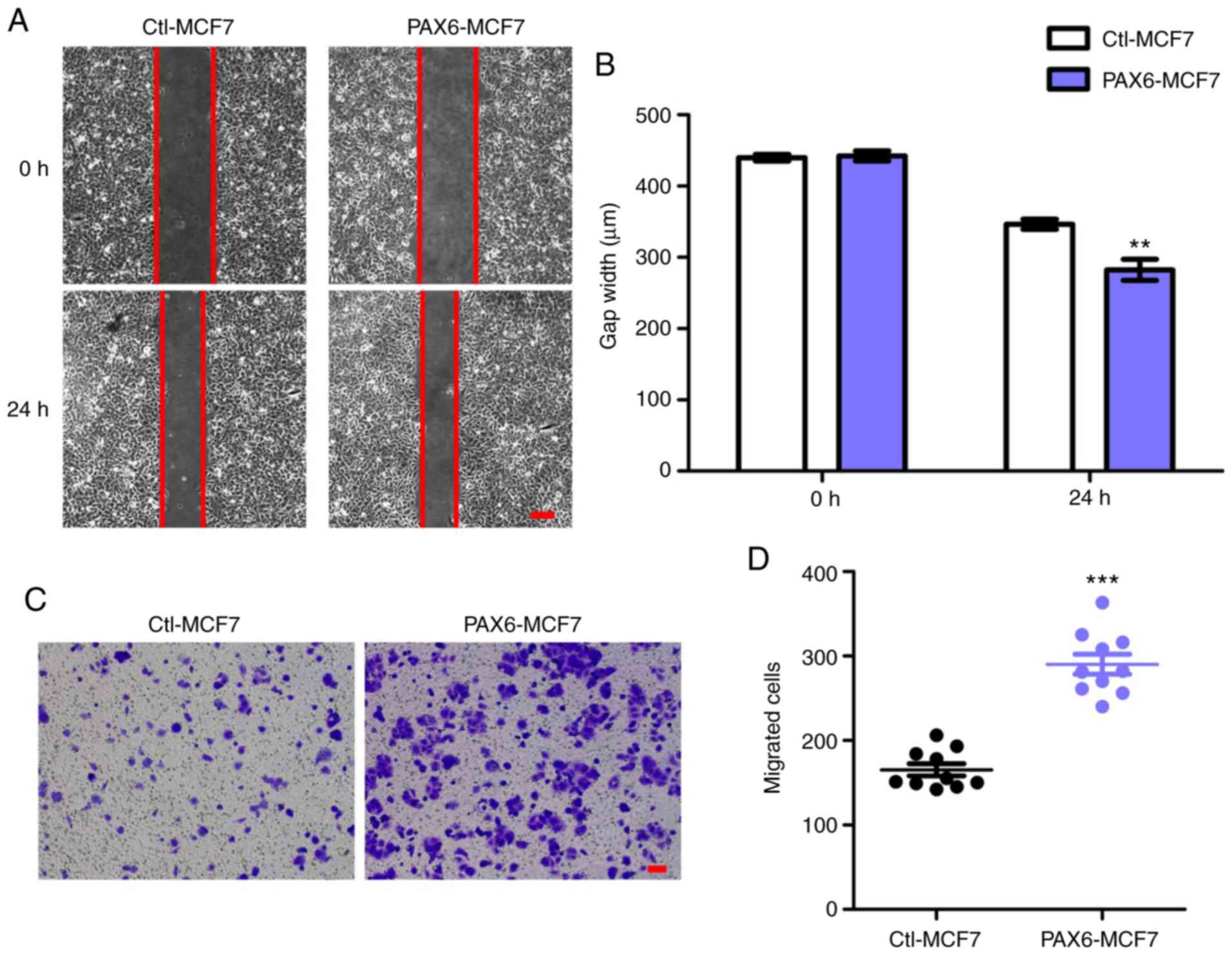
Overexpression of PAX6 facilitates
migration in the MCF7 human breast cancer cell line
Since highly expressed PAX6 associated with
decreased breast cancer survival rate (Fig. 1A) and tumor metastasis is the main
cause of death in breast cancer (49), we hypothesized that overexpression
of PAX6 may positively affect migration and metastasis. Owing to
its low metastatic potential (50,51),
the MCF7 cell line was used to generate an MCF7 cell line that
stably overexpressed PAX6 to investigate the effects of PAX6
overexpression on tumor migration and metastasis. PAX6
overexpression did not significantly affect cell proliferation
(Fig. S1). Results from the gap
closure assay, performed as a migration assay, showed that
overexpression of PAX6 promoted the migration of breast cancer
cells, significantly reducing the gap width after 24 h compared
with the Ctl-MCF7 group (Fig. 2A and
B). In addition, transwell assays also revealed an increase in
the number of migrated cells in PAX6-MCF7 cells compared with the
Ctl-MCF7 group (Fig. 2C and D).
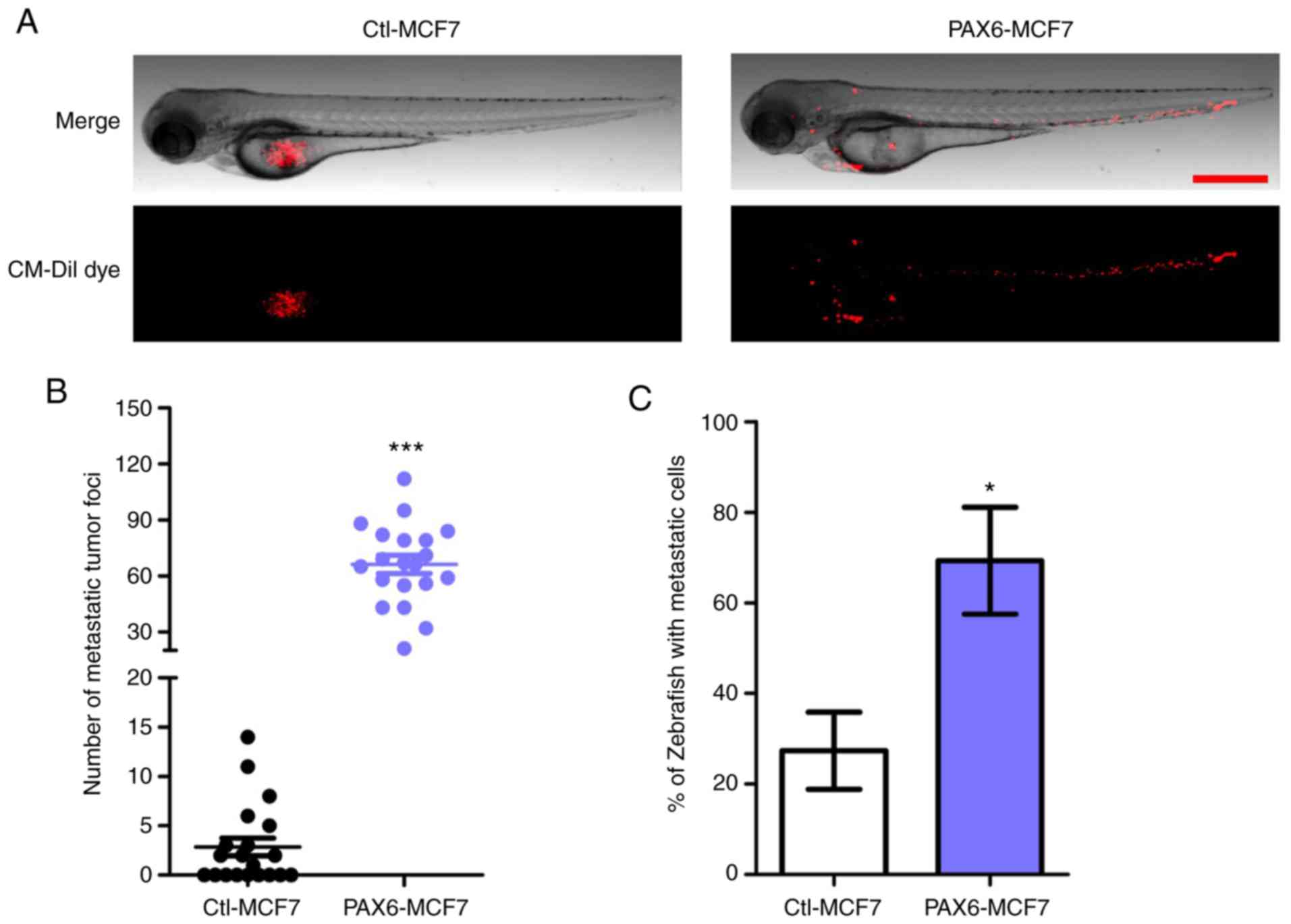
Effects of PAX6 overexpression on
tumor metastasis in vivo
To further verify the metastasis-promoting effects
of PAX6 overexpression in vivo, zebrafish tumor xenograft
assays were performed. PAX6-MCF7 cells or Ctl-MCF7 were injected
into zebrafish at 2 dpf and tumor metastasis in zebrafish were
recorded and analyzed 24 h later (Fig.
3). PAX6-MCF7 cells migrated to the head, blood vessels and the
tail regions of the zebrafish, whereas most of the Ctl-MCF7 cells
remained in the injection site (Fig. 3A
and B). Consistently, ~70% of the zebrafish injected with
PAX6-MCF7 cells displayed a tumor metastasis phenotype, which was
indicated by any foci observed outside of the injection site. By
contrast, the percentage was 30% when the zebrafish were injected
with Ctl-MCF7 cells (Fig. 3C).
High expression of PAX6 is associated
with activated TGF-β-SMAD signaling pathway in human breast
cancer
Previous studies have demonstrated that activated
TGF-β-SMAD signaling pathway mediates the formation of breast tumor
metastasis (38). To test if this
is the case for the pro-metastatic PAX6, key factors involved in
the TGF-β-SMAD signaling pathway were examined in a breast cancer
tissue array and two breast cancer cell lines highly expressing
PAX6. MCF7 is a luminal type with low metastatic potential, whereas
MDA-MB-231 is a triple-negative breast cancer cell line with high
metastatic potential (51). The
analysis of a tissue array containing 70 cases of malignant primary
breast cancer, revealed that low expression levels of PAX6 are
associated with low expression levels of SMAD2, whereas high
expression levels of PAX6 are significantly associated with high
expression levels of SMAD2 (Fig. 4A and
B). In addition, elevated protein expression levels of TGF-βR
II, SMAD2 and SNAIL were observed in PAX6-MCF7 and PAX6-MDA-MB-231
cells compared with the corresponding control (Fig. 4C and D). By contrast, PAX6
overexpression did not seem to affect TGF-β expression levels.
Moreover, no obvious changes in cell morphology were observed for
PAX6-MCF7 and PAX6-MDA-MB-231 in comparison with the control cells
(Fig. S2). Collectively, these
findings suggested that PAX6 may serve a role in the activation of
TGF-β-SMAD pathway in human breast cancer.
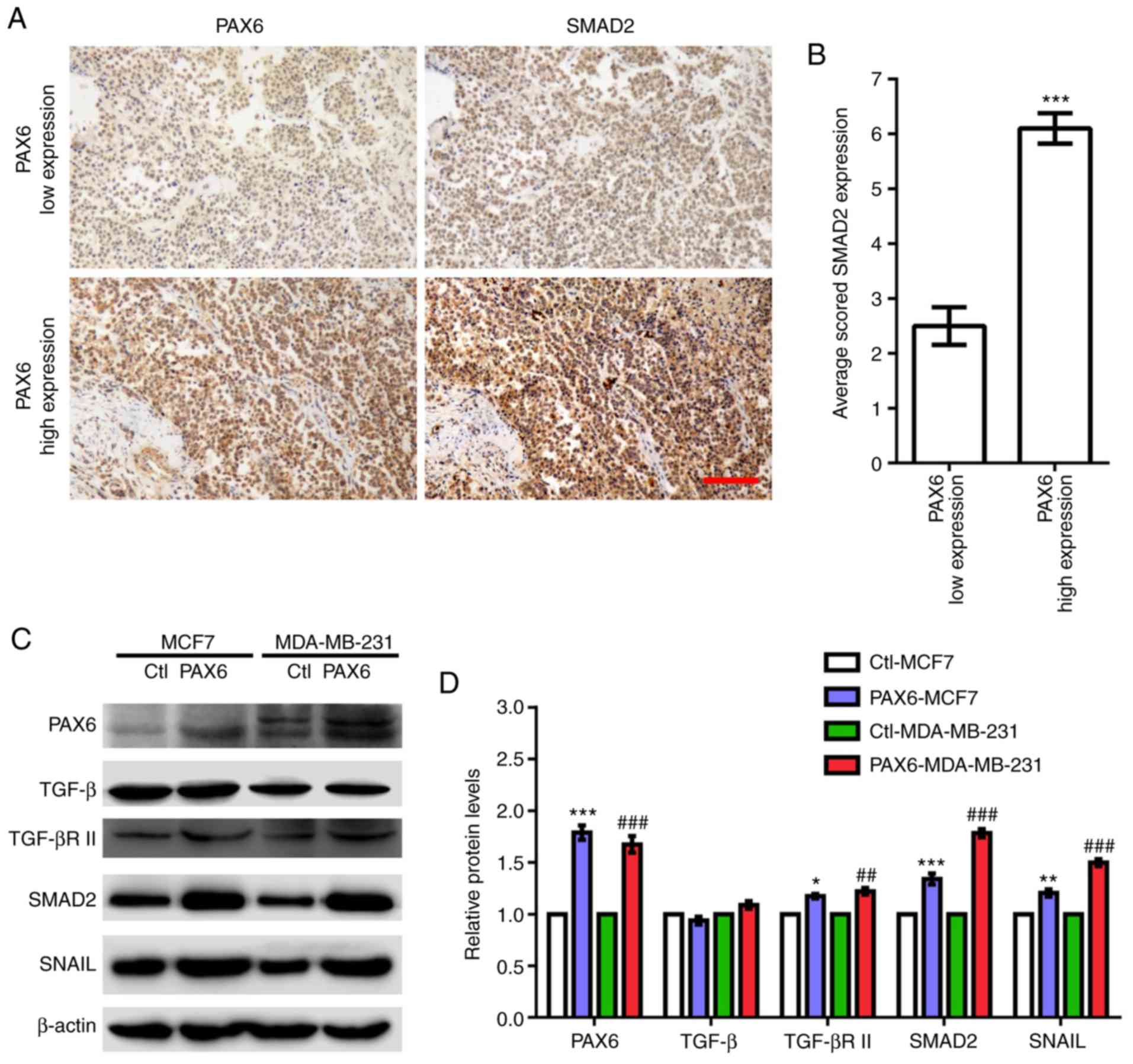 | Figure 4.Highly expressed PAX6 correlates with
activated TGF-β-SMAD signaling in human breast cancer. (A)
Representative images of human breast cancer tissue array stained
with anti-PAX6 and anti-SMAD2 antibodies. (B) Quantification of
SMAD2 staining in low and high expressing PAX6 breast cancer
samples. (C) Western blotting results of key factors involved in
TGF-β-SMAD signaling pathway including TGF-β, TGF-βR II, SMAD2 and
SNAIL in Ctl-MCF7, PAX6-MCF7, Ctl-MDA-MB-231 and PAX6-MDA-MB-231.
(D) Relative protein expression levels normalized to β-actin from
western blot in C were analyzed. Data are expressed as the mean ±
SEM; n=3; *P<0.05, **P<0.01 and ***P<0.001 vs. Ctl-MCF7;
##P<0.01 and ###P<0.001 vs.
Ctl-MDA-MB-231. Scale bar, 100 µm. Ctl, control; PAX6, paired box
6; SNAIL, Snail zinc finger protein; TGF-β, transforming growth
factor β; TGF-βR II, TGF-β receptor II. |
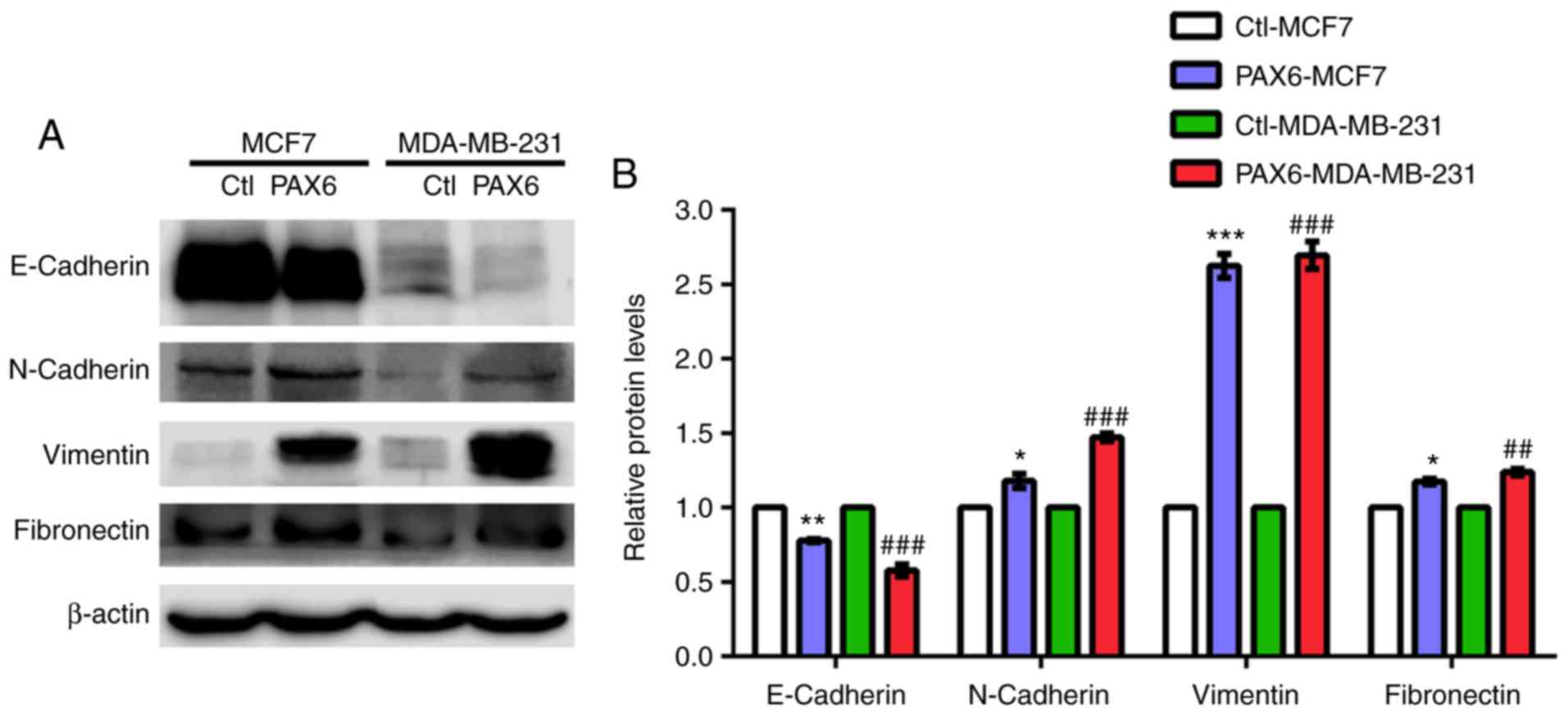
PAX6 overexpression induces EMT in
human breast cancer
It has been reported that signaling pathways such as
TGF-β, mitogen-activated protein kinase, PI3K/AKT and
extracellular-signal-regulated kinase are highly associated with
EMT-related metastasis (46,52,53).
Additionally, aforementioned results suggested a role of PAX6
overexpression in the activation of metastasis and TGF-β-SMAD
signaling pathway. Thus, we hypothesized that high expression of
PAX6 may promote metastasis through TGF-β-SMAD-mediated EMT. PAX6
overexpression in MCF7 and MDA-MB-231 cell lines resulted in a
significantly decreased expression level of the epithelial marker
E-cadherin compared with the respective controls, whereas the
protein expression levels of the mesenchymal markers N-cadherin,
vimentin and fibronectin were significantly increased (Fig. 5A and B).
Discussion
Since metastasis is the main cause of mortality in
human breast cancer, elucidating the underlying mechanism and
discovering therapeutic targets are crucial to improve breast
cancer treatment. PAX6 is known for its oncogenic role in several
types of cancers, including breast cancer (7,18–27).
However, the relationship between PAX6 and breast tumor metastasis,
as well as the underlying mechanism involved in this process need
further investigation. In the present study, overexpression of PAX6
was found to promote migration and metastasis, and
TGF-β-SMAD-mediated EMT was found to be possibly involved in this
process (Fig. 6). Specifically,
PAX6 activates TGF-β signaling and the downstream signal
transduction though SMADs, mediating EMT transcription factors
(TFs) in the nucleus. EMT TFs regulate the expression of
EMT-associated proteins, such as E-cadherin and N-cadherin, to
promote cell metastasis, ultimately affecting survival in breast
cancer patients.
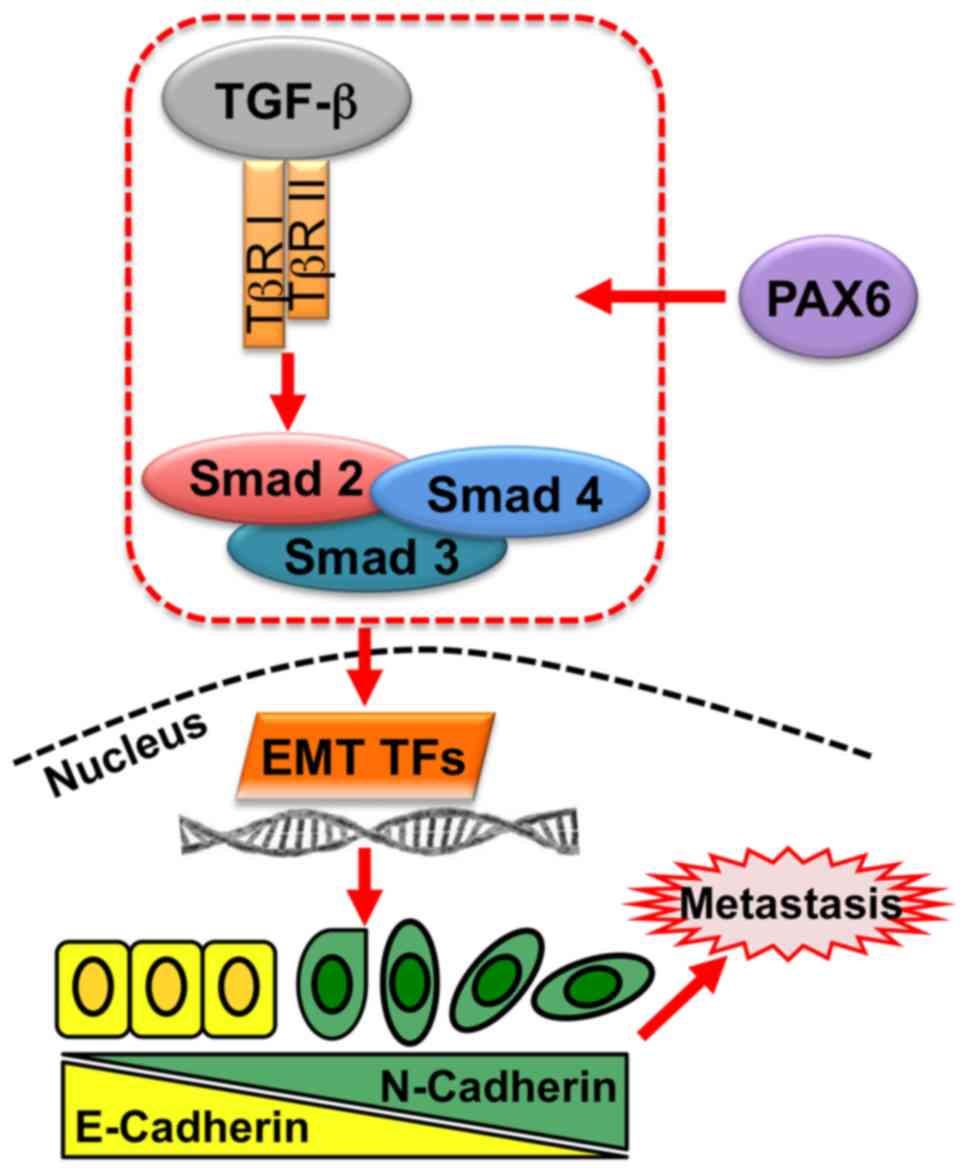 | Figure 6.Pro-metastatic activity of PAX6 in
breast cancer regulated by TGF-β-SMAD-mediated EMT. In breast
cancer, PAX6 regulates TGF-β-SMAD signaling through TGF-βR I,
TGF-βR II, and SMAD complexes, comprising SMAD2, SMAD3 and SMAD4,
which regulate EMT TFs, such as Snail zinc-finger protein, Snail
family transcriptional repressor 2, ZEB1 and ZEB2. EMT TFs
stimulate target genes involved in EMT, thus promoting metastasis
of cancer cells. EMT, epithelial-mesenchymal transition; PAX6,
paired box 6; TF, transcription factor; TGF-β, transforming growth
factor β; TβR, TGF-β receptor; ZEB, zinc-finger E-box binding
homeobox. |
Previous studies indicate that PAX6 plays important
regulatory roles in the progression of breast cancer (7,25–27).
However, these findings were derived from the downregulation of
PAX6 or from tissue microarrays. In accordance with previous
studies from tissue microarrays (27), staining results from the present
study suggested that high PAX6 expression may be associated with a
poor prognosis in breast cancer patients. In the present study,
PAX6 was stably overexpressed in human breast cancer cells to
investigate the oncogenic action of PAX6 on breast cancer. PAX6
overexpression had no significant effect on cell proliferation,
whereas it markedly promoted cell migration and metastasis. The
pro-metastatic ability of PAX6 revealed in the current study is
consistent with previous findings based on the knockdown of PAX6
(7,25,26),
further supporting the potential of PAX6 as a therapeutic target
for breast cancer treatment. Interestingly, knockdown of PAX6
remarkably inhibits cell viability, DNA synthesis and colony
formation in MCF-7 and MDA-MB-231 cells, as well as tumorigenesis
in xenograft mice models, indicating that PAX6 regulates growth of
both ER-positive and -negative breast cancer cells (7). These results contrast with the
findings of the present study, where PAX6 overexpression had no
apparent effect on cell proliferation, highlighting the importance
of studying the oncogenic function of transcription factors
utilizing multiple approached and techniques, such as knockdown and
overexpression techniques. Besides PAX6, other members of RDGN
network, including eyes absent homolog 2, homeobox protein SIX1
(SIX1) and dachshund homolog 1, have also been demonstrated to
mediate progression of breast cancer (38,50,54–58),
suggesting a possible interaction and transcriptional regulation of
RDGN components in breast cancer. PAX6 can regulate the
transcription of homeobox protein SIX1 (59,60)
and in turn, SIX1 is able to induce breast cancer metastasis via
TGF-β signaling and EMT (50,58).
One possibility is that PAX6 regulates SIX1 transcription, and
therefore promotes metastasis. Similarly, it has been reported that
PAX6 acts as a transcription factor to bind directly to the
promoter region of zinc-finger E-box binding homeobox (ZEB)2,
mediating its transcriptional activity, which promotes metastasis
in non-small cell lung cancer (NSCLC) (46).
TGF-β signaling is initiated by the interaction of
the cytokine TGF-β with TGF-βR II, which triggers the recruitment
of TGF-β receptor I to activate the downstream signal transduction
though SMADs (61,62). It has been widely reported that
aberrant regulation of TGF-β-SMAD signaling results in breast
cancer progression (63). High
levels of TGF-β-SMAD signaling is frequently found in breast
cancer, acting as a tumor promoter in the advanced stages of the
disease by activating metastasis, angiogenesis and EMT (64,65).
It has also been reported that PAX6 regulates TGF-β signaling. For
example, there is a co-localization and physical interaction
between PAX6 and TGF-β in murine eyes (66). In addition. microRNA-135b, a direct
target of PAX6, is able to inhibit TGF-β signaling (67). The present study demonstrated the
positive effects of PAX6 overexpression on the activation of
TGF-β-SMAD signaling in breast cancer cells, with the expression
levels of TGF-βR II, SMAD2 and SNAIL being significantly increased.
By contrast, overexpression of PAX6 had no obvious effect on TGF-β
expression, suggesting that the increased expression of TGF-βR II
was not due to the upregulation of TGF-β. It has been reported that
the expression levels of TGF-β receptor are not simply a passive
requirement of signaling but instead, the expression levels of
TGF-β receptors can actively modulate TGF-β responses, such as
enhancing its sensitivity to TGF-β (68). Therefore, we hypothesized that PAX6
increased TGF-β-SMAD signaling possibly through enhancing the
sensitivity to TGF-β instead of increasing expression levels of
TGF-β. Similarly, high levels of SIX1, the downstream factor of
PAX6 in the RDGN network is associated with the activation of
TGF-β-SMAD signaling during breast cancer progression (38,54,55).
Moreover, PAX6 overexpression has been reported to reverse the
inhibitory effects of SMAD3 downregulation-induced cell
proliferation and metastasis (23)
further verifying the role of PAX6 in activating TGF-β-SMAD
signaling pathway. However, the interaction between PAX6 and TGF-β
during cancer progression and the way PAX6 regulates TGF-β-SMAD
signaling pathway remains unknown, therefore it would be valuable
to explore the role of TGF-β stimulation in this context.
Since PAX6 overexpression increased breast cancer
cell metastasis, as well as the expression levels of TGF-β-SMAD
signaling and mesenchymal markers, the possible involvement of
TGF-β-SMAD mediated EMT in pro-metastatic property of PAX6 was
revealed. Numerous studies have shown that activation of TGF-β-SMAD
signaling pathway potentiates breast tumor metastasis (65,69,70).
During cancer metastasis, TGF-β-SMAD pathway can induce EMT, which
is driven by an interactive network of transcription factors
including SNAIL, Snail family transcriptional repressor 2, ZEB1 and
ZEB2 (69). EMT plays important
roles in breast cancer metastasis (34,71).
Moreover, it has been demonstrated that PAX6 overexpression
markedly promotes NSCLC metastasis by mediating EMT, affecting
survival in patients with NSCLC (46). Accordingly, results from the present
study demonstrated a significant increase in pro-migratory and
pro-metastatic activity, as well as the expression levels of
TGF-β-SMAD signaling and the mesenchymal markers N-cadherin,
vimentin and fibronectin, and a decrease in the epithelial marker
E-cadherin, induced by the overexpression of PAX6. These results
suggested that high levels of PAX6 may increase the metastasis of
breast cancer possibly through TGF-β-SMAD-mediated EMT, leading to
poor clinical outcomes.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
We appreciate the work of Mrs Ximin Wang for the
maintenance of zebrafish, and we are grateful to Dr Ming Fa for the
critical reading and editing of the manuscript.
Funding
This work was supported by The National Science
Foundation for Young Scientists of China (grant no. 81802629), for
MJ. This work was also supported by The European Union's Horizon
2020 Research and Innovation Programme (VISGEN; grant no. 734862),
The Future and Emerging Technologies Open Scheme for Research and
Innovation Actions (NEURAM; grant no. 712821) and The Higher
Education Institutional Excellence Programme of the Ministry for
Innovation and Technology in Hungary, within the thematic programme
of ‘Innovation for the sustainable life and environment’ from the
University of Pécs, for AS.
Availability of data and materials
The data generated and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
MJ and KCL conceived the project and designed the
experiments. MJ, DLG and RCW performed the experiments and analyzed
the data. AS analyzed the data and edited the manuscript. MJ wrote
the manuscript. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Akram M, Iqbal M, Daniyal M and Khan AU:
Awareness and current knowledge of breast cancer. Biol Res.
50:332017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Waks AG and Winer EP: Breast cancer
treatment. JAMA. 321:3162019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tungsukruthai S, Petpiroon N and
Chanvorachote P: Molecular mechanisms of breast cancer metastasis
and potential anti-metastatic compounds. Anticancer Res.
38:2607–2618. 2018.PubMed/NCBI
|
|
4
|
Peart O: Metastatic breast cancer. Radiol
Technol. 88:519M–539M. 2017.PubMed/NCBI
|
|
5
|
Riobo-Del Galdo NA, Lara Montero Á and
Wertheimer EV: Role of hedgehog signaling in breast cancer:
Pathogenesis and therapeutics. Cells. 8:3752019. View Article : Google Scholar
|
|
6
|
Chu PY, Hou MF, Lai JC, Chen LF and Lin
CS: Cell reprogramming in tumorigenesis and its therapeutic
implications for breast cancer. Int J Mol Sci. 20:18272019.
View Article : Google Scholar
|
|
7
|
Zong X, Yang H, Yu Y, Zou D, Ling Z, He X
and Meng X: Possible role of Pax-6 in promoting breast cancer cell
proliferation and tumorigenesis. BMB Rep. 44:595–600. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pastuszak-Lewandoska D, Kordiak J, Antczak
A, Migdalska- Sęk M, Czarnecka KH, Górski P, Nawrot E,
Kiszałkiewicz JM, Domańska-Senderowska D and Brzeziańska-Lasota E:
Expression level and methylation status of three tumor suppressor
genes, DLEC1, ITGA9 and MLH1, in non-small cell lung cancer. Med
Oncol. 33:752016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu A, Ahsanul Kabir Khan M, Chen F, Zhong
Z, Chen HC and Song Y: Overexpression of autotaxin is associated
with human renal cell carcinoma and bladder carcinoma and their
progression. Med Oncol. 33:1312016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Strachan T and Read AP: PAX genes. Curr
Opin Genet Dev. 4:427–438. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang J, Lu JP, Suter DM, Krause KH, Fini
ME, Chen B and Lu Q: Isoform- and dose-sensitive feedback
interactions between paired box 6 gene and delta-catenin in cell
differentiation and death. Exp Cell Res. 316:1070–1081. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Elso C, Lu X, Weisner PA, Thompson HL,
Skinner A, Carver E and Stubbs L: A reciprocal translocation
dissects roles of Pax6 alternative promoters and upstream
regulatory elements in the development of pancreas, brain, and eye.
Genesis. 51:630–646. 2013.PubMed/NCBI
|
|
13
|
Liu Y, Han N, Zhou S, Zhou R, Yuan X, Xu
H, Zhang C, Yin T and Wu K: The DACH/EYA/SIX gene network and its
role in tumor initiation and progression. Int J Cancer.
138:1067–1075. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Anantharajan J, Zhou H, Zhang L, Hotz T,
Vincent MY, Blevins MA, Jansson AE, Kuan JWL, Ng EY, Yeo YK, et al:
Structural and functional analyses of an allosteric EYA2
phosphatase inhibitor that has on target effects in human lung
cancer cells. Mol Cancer Ther. 18:1484–1496. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chu Y, Chen Y, Li M, Shi D, Wang B, Lian
Y, Cheng X, Wang X, Xu M, Cheng T, et al: Six1 regulates leukemia
stem cell maintenance in acute myeloid leukemia. Cancer Sci.
110:2200–2210. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kingsbury TJ, Kim M and Civin CI:
Regulation of cancer stem cell properties by SIX1, a member of the
PAX-SIX-EYA-DACH network. Adv Cancer Res. 141:1–42. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Benzina S, Beauregard AP, Guerrette R,
Jean S, Faye MD, Laflamme M, Maïcas E, Crapoulet N, Ouellette RJ
and Robichaud GA: Pax-5 is a potent regulator of E-cadherin and
breast cancer malignant processes. Oncotarget. 8:12052–12066. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luo J, Li H and Zhang C: MicroRNA-7
inhibits the malignant phenotypes of non-small cell lung cancer in
vitro by targeting Pax6. Mol Med Rep. 12:5443–5448. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Y, Li Y, Liu Y, Xie P, Li F and Li G:
PAX6, a novel target of microRNA-7, promotes cellular proliferation
and invasion in human colorectal cancer cells. Dig Dis Sci.
59:598–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shyr CR, Tsai MY, Yeh S, Kang HY, Chang
YC, Wong PL, Huang CC, Huang KE and Chang C: Tumor suppressor PAX6
functions as androgen receptor co-repressor to inhibit prostate
cancer growth. Prostate. 70:190–199. 2010.PubMed/NCBI
|
|
21
|
Muratovska A, Zhou C, He S, Goodyer P and
Eccles MR: Paired-box genes are frequently expressed in cancer and
often required for cancer cell survival. Oncogene. 22:7989–7997.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Maulbecker CC and Gruss P: The oncogenic
potential of Pax genes. EMBO J. 12:2361–2367. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qian Z, Zhang Q, Hu Y, Zhang T, Li J, Liu
Z, Zheng H, Gao Y, Jia W, Hu A, et al: Investigating the mechanism
by which SMAD3 induces PAX6 transcription to promote the
development of non-small cell lung cancer. Respir Res. 19:2622018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lai JP, Mertens RB, Mirocha J, Koo J,
Venturina M, Chung F, Mendez AB, Kahn M and Dhall D: Comparison of
PAX6 and PAX8 as immunohistochemical markers for pancreatic
neuroendocrine tumors. Endocr Pathol. 26:54–62. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zou Q, Yi W, Huang J, Fu F, Chen G and
Zhong D: MicroRNA-375 targets PAX6 and inhibits the viability,
migration and invasion of human breast cancer MCF-7 cells. Exp Ther
Med. 14:1198–1204. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Meng Y, Zou Q, Liu T, Cai X, Huang Y and
Pan J: microRNA-335 inhibits proliferation, cell-cycle progression,
colony formation, and invasion via targeting PAX6 in breast cancer
cells. Mol Med Rep. 11:379–385. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xia X, Yin W, Zhang X, Yu X, Wang C, Xu S,
Feng W and Yang H: PAX6 overexpression is associated with the poor
prognosis of invasive ductal breast cancer. Oncol Lett.
10:1501–1506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Urrutia G, Laurito S, Campoy E, Nasif D,
Branham MT and Roqué M: PAX6 promoter methylation correlates with
MDA-MB-231 cell migration, and expression of MMP2 and MMP9. Asian
Pac J Cancer Prev. 19:2859–2866. 2018.PubMed/NCBI
|
|
29
|
Wang Y and Zhou BP: Epithelial-mesenchymal
transition in breast cancer progression and metastasis. Chin J
Cancer. 30:603–611. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Valle Oseguera CA and Spencer JV: Human
cytomegalovirus interleukin-10 enhances matrigel invasion of
MDA-MB-231 breast cancer cells. Cancer Cell Int. 17:242017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He Z, Xu Q, Wang X, Wang J, Mu X, Cai Y,
Qian Y, Shao W and Shao Z: RPLP1 promotes tumor metastasis and is
associated with a poor prognosis in triple-negative breast cancer
patients. Cancer Cell Int. 18:1702018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stalker L, Pemberton J and Moorehead RA:
Inhibition of proliferation and migration of luminal and
claudin-low breast cancer cells by PDGFR inhibitors. Cancer Cell
Int. 14:892014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Borrull A, Ghislin S, Deshayes F, Lauriol
J, Alcaide-Loridan C and Middendorp S: Nanog and Oct4
overexpression increases motility and transmigration of melanoma
cells. J Cancer Res Clin Oncol. 138:1145–1154. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu Y, Sarkissyan M and Vadgama JV:
Epithelial-mesenchymal transition and breast cancer. J Clin Med.
5:132016. View Article : Google Scholar
|
|
35
|
Prieto-García E, Díaz-García CV,
García-Ruiz I and Agulló- Ortuño MT: Epithelial-to-mesenchymal
transition in tumor progression. Med Oncol. 34:1222017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Abudureheman A, Ainiwaer J, Hou Z, Niyaz
M, Turghun A, Hasim A, Zhang H, Lu X and Sheyhidin I: High MLL2
expression predicts poor prognosis and promotes tumor progression
by inducing EMT in esophageal squamous cell carcinoma. J Cancer Res
Clin Oncol. 144:1025–1035. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Farabaugh SM, Micalizzi DS, Jedlicka P,
Zhao R and Ford HL: Eya2 is required to mediate the pro-metastatic
functions of Six1 via the induction of TGF-β signaling,
epithelial-mesenchymal transition, and cancer stem cell properties.
Oncogene. 31:552–562. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jin M, He Q, Zhang S, Cui Y, Han L and Liu
K: Gastrodin suppresses pentylenetetrazole-induced seizures
progression by modulating oxidative stress in zebrafish. Neurochem
Res. 43:904–917. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Karlsson J, von Hofsten J and Olsson PE:
Generating transparent zebrafish: A refined method to improve
detection of gene expression during embryonic development. Mar
Biotechnol (NY). 3:522–527. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Harrell JC, Dye WW, Allred DC, Jedlicka P,
Spoelstra NS, Sartorius CA and Horwitz KB: Estrogen receptor
positive breast cancer metastasis: Altered hormonal sensitivity and
tumor aggressiveness in lymphatic vessels and lymph nodes. Cancer
Res. 66:9308–9315. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Singha PK, Pandeswara S, Geng H, Lan R,
Venkatachalam MA, Dobi A, Srivastava S and Saikumar P: Increased
Smad3 and reduced Smad2 levels mediate the functional switch of
TGF-β from growth suppressor to growth and metastasis promoter
through TMEPAI/PMEPA1 in triple negative breast cancer. Genes
Cancer. 10:134–149. 2019.PubMed/NCBI
|
|
43
|
Zhang B, Wang L, Ji X, Zhang S, Sik A, Liu
K and Jin M: Anti-inflammation associated protective mechanism of
berberine and its derivatives on attenuating
pentylenetetrazole-induced seizures in zebrafish. J Neuroimmune
Pharmacol. 15:309–325. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lv W, Wang J and Zhang S: Effects of
cisatracurium on epithelial-to-mesenchymal transition in esophageal
squamous cell carcinoma. Oncol Lett. 18:5325–5331. 2019.PubMed/NCBI
|
|
45
|
Wei CY, Tan QX, Zhu X, Qin QH, Zhu FB, Mo
QG and Yang WP: Expression of CDKN1A/p21 and TGFBR2 in breast
cancer and their prognostic significance. Int J Clin Exp Pathol.
8:14619–14629. 2015.PubMed/NCBI
|
|
46
|
Wu DM, Zhang T, Liu YB, Deng SH, Han R,
Liu T, Li J and Xu Y: The PAX6-ZEB2 axis promotes metastasis and
cisplatin resistance in non-small cell lung cancer through PI3K/AKT
signaling. Cell Death Dis. 10:3492019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mercatali L, La Manna F, Groenewoud A,
Casadei R, Recine F, Miserocchi G, Pieri F, Liverani C, Bongiovanni
A, Spadazzi C, et al: Development of a patient-derived xenograft
(PDX) of breast cancer bone metastasis in a zebrafish model. Int J
Mol Sci. 17:13752016. View Article : Google Scholar
|
|
48
|
Huang Z, Duan H and Li H: Identification
of gene expression pattern related to breast cancer survival using
integrated TCGA datasets and genomic tools. Biomed Res Int.
2015:8785462015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Scimeca M, Urbano N, Bonfiglio R, Duggento
A, Toschi N, Schillaci O and Bonanno E: Novel insights into breast
cancer progression and metastasis: A multidisciplinary opportunity
to transition from biology to clinical oncology. Biochim Biophys
Acta Rev Cancer. 1872:138–148. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Micalizzi DS, Christensen KL, Jedlicka P,
Coletta RD, Barón AE, Harrell JC, Horwitz KB, Billheimer D,
Heichman KA, Welm AL, et al: The Six1 homeoprotein induces human
mammary carcinoma cells to undergo epithelial-mesenchymal
transition and metastasis in mice through increasing TGF-beta
signaling. J Clin Invest. 119:2678–2690. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lacroix M and Leclercq G: Relevance of
breast cancer cell lines as models for breast tumours: An update.
Breast Cancer Res Treat. 83:249–289. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Katsuno Y, Lamouille S and Derynck R:
TGF-β signaling and epithelial-mesenchymal transition in cancer
progression. Curr Opin Oncol. 25:76–84. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sadek KW, Haik MY, Ashour AA, Baloch T,
Aboulkassim T, Yasmeen A, Vranic S, Zeidan A and Al Moustafa AE:
Water-pipe smoking promotes epithelial-mesenchymal transition and
invasion of human breast cancer cells via ERK1/ERK2 pathways.
Cancer Cell Int. 18:1802018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Patrick AN, Cabrera JH, Smith AL, Chen XS,
Ford HL and Zhao R: Structure-function analyses of the human
SIX1-EYA2 complex reveal insights into metastasis and BOR syndrome.
Nat Struct Mol Biol. 20:447–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Iwanaga R, Wang CA, Micalizzi DS, Harrell
JC, Jedlicka P, Sartorius CA, Kabos P, Farabaugh SM, Bradford AP
and Ford HL: Expression of Six1 in luminal breast cancers predicts
poor prognosis and promotes increases in tumor initiating cells by
activation of extracellular signal-regulated kinase and
transforming growth factor-beta signaling pathways. Breast Cancer
Res. 14:R1002012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
McCoy EL, Iwanaga R, Jedlicka P, Abbey NS,
Chodosh LA, Heichman KA, Welm AL and Ford HL: Six1 expands the
mouse mammary epithelial stem/progenitor cell pool and induces
mammary tumors that undergo epithelial-mesenchymal transition. J
Clin Invest. 119:2663–2677. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhao F, Wang M, Li S, Bai X, Bi H, Liu Y,
Ao X, Jia Z and Wu H: DACH1 inhibits SNAI1-mediated
epithelial-mesenchymal transition and represses breast carcinoma
metastasis. Oncogenesis. 4:e1432015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Micalizzi DS, Wang CA, Farabaugh SM,
Schiemann WP and Ford HL: Homeoprotein Six1 increases TGF-beta type
I receptor and converts TGF-beta signaling from suppressive to
supportive for tumor growth. Cancer Res. 70:10371–10380. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Pappu KS and Mardon G: Genetic control of
retinal specification and determination in Drosophila. Int J Dev
Biol. 48:913–924. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hoshiyama D, Iwabe N and Miyata T:
Evolution of the gene families forming the Pax/Six regulatory
network: Isolation of genes from primitive animals and molecular
phylogenetic analyses. FEBS Lett. 581:1639–1643. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Buck MB and Knabbe C: TGF-beta signaling
in breast cancer. Ann N Y Acad Sci. 1089:119–126. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Taylor MA, Lee YH and Schiemann WP: Role
of TGF-β and the tumor microenvironment during mammary
tumorigenesis. Gene Expr. 15:117–132. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Imamura T, Hikita A and Inoue Y: The roles
of TGF-β signaling in carcinogenesis and breast cancer metastasis.
Breast Cancer. 19:118–124. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Oft M, Heider KH and Beug H: TGFbeta
signaling is necessary for carcinoma cell invasiveness and
metastasis. Curr Biol. 8:1243–1252. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Khoshakhlagh M, Soleimani A, Binabaj MM,
Avan A, Ferns GA, Khazaei M and Hassanian SM: Therapeutic potential
of pharmacological TGF-β signaling pathway inhibitors in the
pathogenesis of breast cancer. Biochem Pharmacol. 164:17–22. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shubham K and Mishra R: Pax6 interacts
with SPARC and TGF-β in murine eyes. Mol Vis. 18:951–956.
2012.PubMed/NCBI
|
|
67
|
Bhinge A, Poschmann J, Namboori SC, Tian
X, Jia Hui Loh S, Traczyk A, Prabhakar S and Stanton LW: miR-135b
is a direct PAX6 target and specifies human neuroectoderm by
inhibiting TGF-β/BMP signaling. EMBO J. 33:1271–1283. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Rojas A, Padidam M, Cress D and Grady WM:
TGF-beta receptor levels regulate the specificity of signaling
pathway activation and biological effects of TGF-beta. Biochim
Biophys Acta. 1793:1165–1173. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Xie F, Ling L, van Dam H, Zhou F and Zhang
L: TGF-β signaling in cancer metastasis. Acta Biochim Biophys Sin
(Shanghai). 50:121–132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View Article : Google Scholar : PubMed/NCBI
|