Introduction
Breast cancer (BC) is a common type of cancer that
seriously affects women's health (1). Despite significant advances in early
diagnosis, surgical treatment, and targeted therapy for BC, the
5-year overall survival rate remains poor owing to a high
metastatic rate (2). Therefore, a
better understanding of the underlying molecular and biological
mechanisms involved in the carcinogenesis and development of BC may
lead to novel therapies to treat this challenging disease.
Long noncoding RNAs (lncRNAs) are transcripts longer
than 200 nucleotides without protein-coding potential (3). Next-generation sequencing has shown
that lncRNAs are widely transcribed in the genome (4,5).
Although they were previously considered to represent
transcriptional ‘noise’, emerging evidence shows that lncRNAs play
critical roles in various biological processes, including cellular
development and determination of cell fate (6–8).
Notably, abnormal expression of lncRNAs has also been observed in
the development and progression of cancer, where it leads to the
dysregulation of cell proliferation, apoptosis, migration, and
invasion (9–11). For example, it was found that
androgen receptor negatively induced lncRNA (ARNILA) promoted
invasion and metastasis in triple-negative BC (12). Therefore, identification of key
oncogenic BC-related lncRNAs and their molecular mechanisms is
important to develop novel therapeutic strategies.
lncRNA proliferating cell nuclear antigen (PCNA) has
multiple roles in DNA replication and repair, and maintains genomic
integrity at the genetic and epigenetic levels by interacting with
chaperone proteins. It comprises at least four effective
pseudogenes, proliferating cell nuclear antigen pseudogene 1
(PCNAP1), PCNAP2, PCNAP3, and PCNAP4 (13,14).
In addition, lncRNA PCNAP can act as an endogenous RNA via microRNA
(miRNA)-dependent crosstalk. The high sequence homology of the
pseudogene with its ancestral gene means they can share a common
miRNA, leading to the regulation of ancestral genes. However, the
relationships among lncRNA PCNAP1, miRNAs, and BC require further
investigation.
Here, we investigated the expression, biological
function, and potential molecular mechanisms of lncRNA PCNAP1 in
the pathogenesis of BC. We found that lncRNA PCNAP1 was highly
expressed in tumor samples, and high lncRNA PCNAP1 levels predicted
poor prognosis. Function assays showed that knockdown of lncRNA
PCNAP1 suppressed the migration and invasion of breast cancer
cells. Regarding the mechanism, lncRNA PCNAP1 promoted the
metastasis of BC by binding and downregulating miRNA-340-5p, thus
promoting the upregulation of SRY-box transcription factor 4 (SOX4)
in BC cells. In conclusion, our results suggest that upregulation
of PCNAP1 promotes BC metastasis by modulating the
miRNA-340-5P/SOX4 axis, which may represent an alternative means of
inhibiting metastasis in BC patients.
Materials and methods
Clinical samples
Human specimens including 70 BC tissues and paired
adjacent normal tissues were collected from patients from 1 March
2013 to 31 November 2014 at the Henan Provincial People's Hospital.
The mean age of the patients was 47.90 years [standard deviation
(SD) 6.84] with a range of 36 to 72 years. The fresh tissue
specimens were collected and immediately placed in liquid nitrogen
until use. None of the patients recruited in this study had
undergone preoperative chemotherapy or radiotherapy. Follow-up was
conducted and ended on May 31, 2019. Death dates was verified by
phone contact with the patient's relatives or by hospital records.
Overall survival (OS) time were defined according to the time after
treatment. The clinicopathological variables of the patients are
shown in Table I. The clinical data
collection and research procedures were reviewed and approved by
the Medical Ethics Committee of Henan Provincial People's Hospital.
In addition, BC donors participating in the study provided written
informed consent for their tissue samples to be used for scientific
research.
 | Table I.Association between the lncRNA PCNAP1
expression level and clinicopathologic features of the BC cases
(N=70). |
Table I.
Association between the lncRNA PCNAP1
expression level and clinicopathologic features of the BC cases
(N=70).
|
|
| lncRNA PCNAP1
expression [n (%)] |
|
|---|
|
|
|
|
|
|---|
| Variables | n | Low (n=35) (%) | High (n=35)
(%) | P-value |
|---|
| Age (years) |
|
<50 | 30 | 16 (53.3) | 14 (46.7) | 0.629 |
|
≥50 | 40 | 19 (47.5) | 21 (52.5) |
|
| Tumor size
(cm) |
|
<2.5 | 47 | 25 (53.2) | 22 (46.8) | 0.445 |
|
≥2.5 | 23 | 10 (43.5) | 13 (56.5) |
|
| ER |
|
Positive | 45 | 21 (46.7) | 24 (53.3) | 0.454 |
|
Negative | 25 | 14 (56) | 11 (44) |
|
| PR |
|
Positive | 47 | 22 (46.8) | 25 (53.2) | 0.445 |
|
Negative | 23 | 13 (56.5) | 10 (43.5) |
|
| HER-2 |
|
Positive | 35 | 15 (42.9) | 20 (57.1) | 0.232 |
|
Negative | 35 | 20 (57.1) | 15 (42.9) |
|
| Differentiation
grade |
|
G1/G2 | 47 | 29 (61.7) | 18 (38.3) | 0.005 |
| G3 | 23 | 6 (26.1) | 17 (73.9) |
|
| TNM stage |
|
I/II | 40 | 28 (70) | 12 (30) |
<0.001 |
|
III | 30 | 7 (23.3) | 23 (76.7) |
|
| Lymph node
metastasis |
| No | 56 | 32 (57.1) | 24 (42.9) | 0.017 |
|
Yes | 14 | 3 (21.4) | 11 (78.6) |
|
Cell lines and culture
The human BC cell lines MCF7 and MDA-MB-231 were
provided by the Cell Bank of the Chinese Academy of Science
(Shanghai, China) and were cultivated at 37°C in a humidified
atmosphere of 5% CO2 using DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal calf serum
(Invitrogen; Thermo Fisher Scientific, Inc.).
Cell transfection
A short interfering RNA (siRNA) negative control
(NC), si-PCNAP1, miR-340-5p mimics, miR-340-5p mimics negative
control (miR-Ctrl), miR-340-5p inhibitor, miR-340-5p inhibitor
negative control (inhibitor NC), pcDNA3.1-SOX4 and pcDNA3.1 empty
vector (vector) were obtained from RiboBio (Guangzhou, China).
Using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) protocols, the siRNA NC, si-PCNAP1, miR-340-5p mimics,
miR-340-5p inhibitor and NC, pcDNA3.1-SOX4 and pcDNA3.1 empty
vector were introduced into MDA-MB-231 and MCF7 cells. Before
plasmid transfection, the MDA-MB-231 and MCF7 cells were suspended
and seeded in 6-well culture plates at 37°C. When the cell
confluence rate reached 80–90%, the cell culture medium was
replaced by serum-free fresh medium 3 h before transfection.
In vivo mouse model for analysis of
metastatic capability
All animal experiments were performed at the Animal
Laboratory Center of Henan Provincial People's Hospital. All
experimental procedures and protocols were approved by the
Institutional Animal Care and Use Committee at Henan Provincial
People's Hospital, in accordance with the Guide for the Care and
Use of Laboratory Animals (15).
Nude mice were purchased from the Animal Husbandry Center of the
Shanghai Institute of Cell Biology, Academia Sinica, Shanghai,
China. Twenty-one female athymic BALB/c nude mice (weight, 19–20 g;
4–5 weeks of age) were randomized into three groups of 7 nude mice
per group. Nude mice were anesthetized with 1% isoflurane, and
MDA-MB-231 cells transfected with siPCNAP1 or the corresponding
vector control cells (5×105 cells/100 µl of complete
medium) were intravenously injected into nude mice respectively via
the tail vein. Nude mice were housed in the animal research
facility according to institutional guidelines, with 12-h light,
and food and water ad libitum. Nude mice behavior was
observed every day, and body weights were monitored every 3 days.
The nude mice were euthanized as soon as the following symptoms
were detected: i) Severe cachexia (weight loss approaching 25%);
ii) inability to obtain food or water; general lack of moving
activities; iii) pale appearance, body coat looking unhealthy and
scruffy; iv) breathing problem; v) infection at the injection site.
Nude mice were anesthetized with 1% isoflurane and sacrificed by
decapitation on day 16 after cancer cell injection. Their lungs
were then resected for metastatic nodule counting. Tissues were
fixed and embedded in paraffin according to standard procedures. A
sampling of sections was taken across each lung in the following
manner. Two consecutive 4-µm sections were taken. Subsequently, a
number of consecutive 4-µm sections were discarded (approximately
30). This process was repeated along the entire length of each lung
lobe. The sections were then stained using hematoxylin and eosin
(H&E) (scale bar, 500 µm). Acctording to the images of H&E
staining, the lung nodule numbers were quantified microscopically
by three observers. According to Salmon et al (16), the adapted ‘Prolate Spheroid’ model
was used to calculate the tumor volume. The volume of any given
lung tumor nodule was determined, on the basis of measurements of
the radii of two sections and known separation between the two
sections. The diameter (2 × R1) of the largest tumor
nodule was 6.83 mm and the average diameter (2 × R1) was
2.59 mm, which complies with the requirements of animal welfare.
Then by Salmon's equation (16),
the volume of each lung tumor nodule was determined.
Tumor volume (V)=(4/3) × π × R1 ×
R2;
R1≥R2;
R1=[(a22 -
a12 + D2)2/(4 ×
D2) + a12]1/2;
R2=[(b22-b12
+ D2)2/(4 × D2) +
b12]1/2;
where a1 is the length (longest
dimension) of section 1 and b1 is the width (shortest
dimension) of section 1, a2 is the length (longest
dimension) of section 1 and b2 is the width (shortest
dimension) of section 2; D is the distance between section 1 and
section 2.
Bioinformatics analysis
TargetScan (http://www.targetscan.org/vert_/72) (17), EV-miRNA (http://bioinfo.life.hust.edu.cn/EVmiRNA) (18) and miRTarBase (http://miRTarBase.mbc.nctu.edu.tw/) (19) were used to predict the associations
between miRNAs and mRNA, and miRcode (http://www.mircode.org/index) (20) was used to predict the binding sites
shared by miRNAs and lncRNAs.
RNA extraction and quantitative
real-time PCR (qRT-PCR)
The miRNAs were extracted using the miRNeasy Mini
kit (cat. no. 217004; Qiagen). Poly(A) was added, and 1 µg of RNA
containing miRNA was reverse transcribed into cDNA to detect
miR-340-5p. Primers for miR-340-5p and U6 were obtained from
GeneCopoeia. To detect expression of SOX4 and lncRNA PCNAP1, total
RNA was extracted using TRIzol (Invitrogen; Thermo Fisher
Scientific, Inc.) and further reverse transcribed into cDNA using
ReverTra Ace qPCR RT-Kit (cat. no. FSK-100; Toyobo). Expression of
miRNA or SOX4 was determined in a StepOnePlus system (Invitrogen;
Thermo Fisher Scientific, Inc.) using SYBR Premix Ex Taq from
Takara. RT-qPCR was performed under the conditions at 94°C for 5
min, followed by 40 cycles at 94°C for 30 sec, 55°C for 30 sec and
72°C for 90 sec. Cq value of each sample was recorded and analyzed
utilizing the 2−ΔΔCq method according to Livak and
Schmittgen (21). U6 or GAPDH was
used as an endogenous control. The RT-qPCR primers used were as
follows: lncRNA PCNAP1-forward, 5′-CACTCCACTCTCTCTTC-3′ and lncRNA
PCNAP1-reverse, 5′-CAGAAAACCGCATCTACC-3′; U6-forward,
5′-CTCGCTTCGCA-3′ and U6-reverse, 5′-AACGCTTCACGAATTTGCGT-3′;
SOX4-forward, 5′-GACCTGCTCGACCTGAACC-3′ and SOX4-reverse,
5′-CCGGGCTCGAAGTTAAAATCC-3′; GAPDH-forward,
5′-GGTGGTCTCCTCTGACTTCAACA-3′ and GAPDH-reverse
5′-GTTGCTGTAGCCAAATTCGTTGT-3′.
Nucleo-cytoplasmic separation
For each group, MCF7 cells were cultured in a 100-mm
culture dish (~5×106 cells) with 0.5 ml buffer A [10 mM
HEPES (pH 7.9) (Hushi, China), 10 mM KCl (Hushi), 1.5 mM
MgCl2 (Hushi), 0.34 M sucrose (Hushi), 10% glycerol
(Hushi), 1 mM DTT (Biosharp, China), 0.1% Triton X-100 (Biosharp),
and protease inhibitor mixture (Roche, USA)]. A protease inhibitor
cocktail (Sigma-Aldrich; Merck KGaA), PMSF (BBI, China),
phosphatase inhibitors NaF (Hushi), Na3VO4
(Hushi, China), and Na4P2O7
(Hushi) were added. Cells were then placed on ice and washed twice
with 1X pre-cooled phosphate-buffered saline (PBS). Then, 0.5 ml
buffer A was added. Cells were scraped and transferred to labeled
EP tubes and placed on ice for 10 min, before being centrifuged at
1,500 × g and 4°C for 10 min. The supernatants were transferred to
new EP tubes and centrifuged at 6,000 × g for 10 min at 4°C. The
resulting supernatant was the cytoplasmic component, and the
precipitate obtained by low-speed centrifugation was the nuclear
component. The precipitate was washed with 500 ml buffer A, and
then centrifuged at 1,500 × g and 4°C for 10 min. This was repeated
3–4 times. The appropriate amount of buffer B [3 mM EDTA (Hushi),
0.2 mM EGTA (Hushi), 1 mM DTT, and protease inhibitor mixture] was
determined, and cocktail and NaF were also added (~100 µl per
sample). The precipitate was cleaned with buffer B, transferred to
a tissue grinder, and ground 20–30 times. It was then transferred
to a new EP tube, placed on ice for 30 min, and centrifuged at
6,000 × g for 10 min at 4°C. The resulting supernatant was the
nuclear component.
Western blot analysis
Cells were collected 48 h after transfection and
lysed in 2X sodium dodecyl sulfate (SDS) sample buffer [100 mM
Tris-HCl (pH 6.8), 10 mM EDTA, 4% SDS, and 10% glycine] to extract
total protein. The protein was quantified by bicinchoninic acid
(BCA) analysis. Protein extractions were separated by 10% SDS-PAGE
(20 µg per lane), and transferred onto polyvinylidene fluoride
(PVDF) membranes. Membranes were blocked in 5% milk at room
temperature, incubated with primary antibody in TBST with 5% BSA
overnight at 4°C, then incubated with fluorescent-tagged secondary
antibody at room temperature for 1 h, and read using enhanced
chemiluminescence (Santa Cruz Biotechnology, Dallas, USA).
anti-E-cadherin (dilution 1:2,000; product code ab76055),
anti-N-cadherin (dilution 1:2,000, product code ab18203) and
anti-SOX4 (dilution 1:5,000; product code ab134107) were purchased
from Abcam, anti-GAPDH (dilution 1:1,000; cat. no. D16H11) were
purchased from Cell Signaling Technology. Secondary antibodies were
mouse anti-rabbit IgG-HRP (dilution 1:10,000; cat. no. sc-2357) and
bovine anti-mouse IgG-HRP (dilution 1:10,000, cat. no. sc-2371),
both purchased from Santa Cruz Biotechnology.
In vitro scratch assays
A horizontal line was drawn on the back of each
6-well plate using a marker pen before the in vitro scratch
assays. After digestion with 0.25% trypsin, 5×105 cells
were seeded into each well to form a cell monolayer. A 20-µl
pipette tip was used to scrape a straight line perpendicular to the
horizontal line on the flat cell monolayer, and the cells were
washed three times with PBS to remove floating cell debris. Then,
serum-free medium was added, and the cells were cultured in a 37°C
incubator containing 5% CO2. Each plate was photographed
at two time points (0 and 48 h) to observe the healing of the
scratches, and the cell healing index was calculated as follows:
(Initial scratch width-scratch width at the time of
experiment)/initial scratch width ×100%. The experiment was
repeated three times (Magnification, ×200, scale bar, 100 µm).
Transwell assays
Transwell assays were used to assess the
aggressiveness of BC cells. We used Transwell chambers (Corning)
coated with Matrigel (BD Biosciences). The trypsin-treated
transfected cells (containing 1×105 cells per 100 µl of
serum-free DMEM) were inoculated into the upper chamber. DMEM (500
µl) supplemented with 20% fetal calf serum was added to the lower
chamber. After incubation for 72 h at 37°C in a humidified
environment containing 5% CO2, the BC cells invading the
lower chamber were fixed with methanol and stained with crystal
violet. The invading cells were photographed using an inverted
microscope (magnification, ×200, scale bar, 100 µm).
Luciferase reporter assays
The wild-type (WT) or mutant human SOX4 3′
untranslated region (UTR) sequences with many potential binding
sites were expanded and cloned into the pGL3-Basic vector (Promega
Corp.). 293T cells were transfected together with a mixture of 0.02
µg of pGL3-Basic-SOX4 and 150 nM of miRNA-340-5p mimetic.
Forty-eight hours after transfection, firefly luciferase activity
and Renilla reniformis luciferase activity were detected
using Dual-Luciferase reporter assay system (Promega Corp.), and
Renilla luciferase activity was normalized to firefly
luciferase activity.
MS2-RIP
MCF7 cells were co-transfected with pSL-MS2,
pSL-MS2-PCNAP1, and pSLMS2-mut (miRNA-340-5p) with pMS2-GFP
(AddGene, USA). After 48 h, cells were used to perform RNA
immunoprecipitation (RIP) experiments as previously described
(22). The RNA fraction isolated by
RIP was analyzed by RT-qPCR.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 6 software (GraphPad Software, Inc.). Data are presented as
mean ± SEM. The Student's t-test was used to analyze differences
between groups. Differences among more than two groups were
evaluated by one-way analysis of variance, followed by post hoc
multiple comparison with the Tukey test. The Chi-square test was
performed to analyze the count data. Pearson correlation analysis
was performed to investigate associations. The Kaplan-Meier method
was used to analyze survival rates, and the log-rank test was
performed to compare the differences. A multivariate analysis of
all the variables that were found to be significantly correlated in
the univariate analysis was performed using a Cox
proportional-hazards regression model. Two-sided P-values <0.05
were considered to indicate a statistically significant
difference.
Results
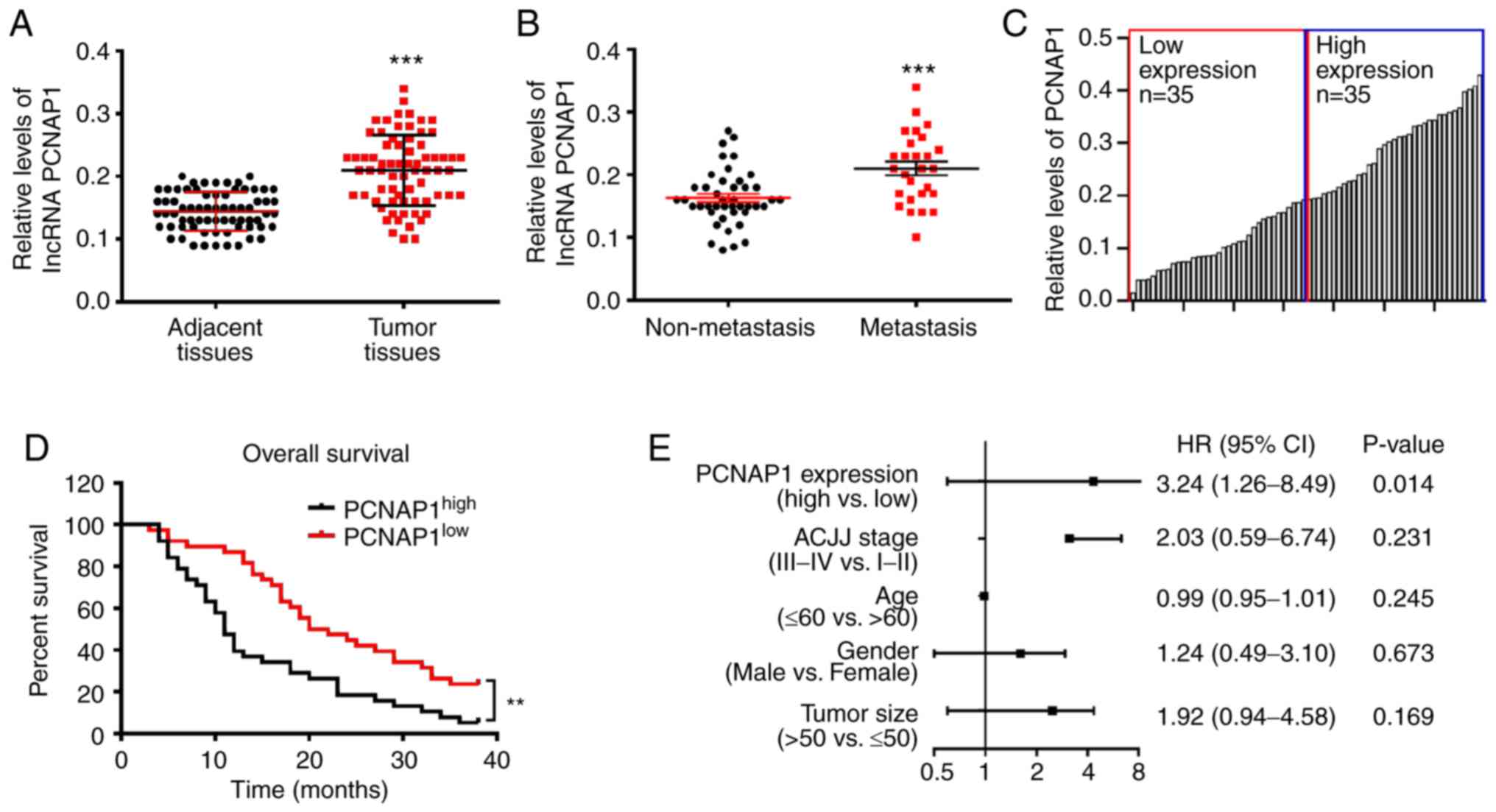
lncRNA PCNAP1 is upregulated in human
BC tissues and high lncRNA PCNAP1 levels predict poor overall
survival
To determine the involvement of lncRNA PCNAP1 in the
development of BC, we collected 70 pairs of BC tumor tissues and
paired adjacent normal tissues. The correlations of lncRNA PCNAP1
expression with clinicopathological features of BC patients are
shown in Table I. Based on
statistical analyses of our results, high lncRNA PCNAP1 expression
was significantly correlated with differentiation grade, TNM stage,
and lymph node metastases in BC patients (P<0.05). However, the
expression of lncRNA PCNAP1 was not associated with other
clinicopathological factors of BC patients, including age, tumor
size, estrogen receptor (ER), progesterone receptor (PR), and human
epidermal growth factor receptor 2 (HER-2) status (P>0.05).
These data indicate that upregulation of lncRNA PCNAP1 may have a
critical role in BC progression. We then performed RT-qPCR to check
the relative expression levels of lncRNA PCNAP1. We found that the
basal expression level of lncRNA PCNAP1 in BC was 0.217, which was
significantly higher than that in corresponding adjacent healthy
tissues (P<0.001) (Fig. 1A).
Intriguingly, patients with metastasis had higher lncRNA PCNAP1
levels compared with those without metastasis (P<0.001)
(Fig. 1B). Kaplan-Meier survival
analysis and log-rank tests were performed to further assess the
clinical significance of lncRNA PCNAP1 in the prognosis of BC
patients. We divided the samples into low (below the median, n=35)
and high (above the median, n=35) lncRNA PCNAP1 expression groups
based on to the mean level of lncRNA PCNAP1; the cut-off value for
determining low expression/high expression was 0.190, and the mean
expression level was 0.202 (Fig.
1C). An increased level of lncRNA PCNAP1 was significantly
associated with poor overall survival (OS) [hazard ratio (HR),
3.24; 95% confidence interval (CI), 1.26–8.49; P=0.014] (Fig. 1D and E). Taken together, these data
suggest that increased lncRNA PCNAP1 expression may have an
important role in the development and progression of BC.
Knockdown of lncRNA PCNAP1 suppresses
the migration and invasion of breast cancer cells in vitro and in
vivo
To explore the role of lncRNA PCNAP1 in BC
progression, loss-of-function experiments using siRNAs (siPCNAP1#a
and siPCNAP1#b) and CCK-8 assays were performed in breast cancer
cell lines MDA-MB-231 and MCF7. Levels of lncRNA PCNAP1 expression
were markedly decreased in both cell types; by ~80% or ~70%,
respectively, compared with the scrambled controls (P<0.001 in
MDA-MB-231; P<0.01 in MCF7) (Fig. 2A
and B). However, no significant effect of lncRNA PCNAP1
knockdown on BC cell viability was observed, consistent with the
results of the CCK-8 assays (Fig. 2C
and D). Given that lncRNA PCNAP1 levels were increased in
metastatic samples, we conducted cell migration, invasion, and
wound healing assays to determine whether there was an effect on BC
cell metastasis. Loss of lncRNA PCNAP1 significantly decreased the
speed of scratch closure in the two BC cell lines: By ~64%
(P<0.01) in MDA-MB-231 and by ~82% (P<0.001) in MCF7
(Fig. 2E-H). In addition,
inhibition of lncRNA PCNAP1 decreased invasion dramatically, as
shown by the Transwell invasion assay results (P<0.01) (Fig. 2I-K). Cells with lncRNA PCNAP1
knockdown showed prominently increased protein levels of epithelial
marker E-cadherin, and decreased levels of mesenchymal marker
N-cadherin (Fig. 2L). Furthermore,
we explored the role of lncRNA PCNAP1 in metastasis of BC in
vivo by tail vein injection with highly aggressive siCtrl and
siPCNAP1 MDA-MB-231 cells. As shown in Fig. 2M and N, histological examination
confirmed that the injection of siCtrl cells led to lung metastasis
in BALB/c nude mice; these metastases were obviously inhibited in
the nude mice injected with siPCNAP1 cells. Furthermore, lncRNA
PCNAP1 knockdown also obviously descreased the volume of tumor
nodules in the lung (Fig. 2O).
However, the weight of mice did not change after injection of
siPCNAP1 cells (Fig. 2P). Taken
together, these data suggest that lncRNA PCNAP1 is a positive
regulator of migration, invasion, and epithelial-mesenchymal
transition both in vitro and in vivo.
 | Figure 2.Knockdown of lncRNA PCNAP1 suppresses
the migration and invasion of BC cells in vitro and in
vivo. (A and B) Loss-of-function experiments showed that lncRNA
PCNAP1 expression levels were markedly decreased following
transfection with siPCNAP1#a and siPCNAP1#b compared with scrambled
controls (siCtrl) in BC cells (by ~80 and ~70% in MDA-MB-231 and
MCF7 cells, respectively). (C and D) Knockdown of lncRNA PCNAP1 had
no significant effect on BC cell viability in MDA-MB-231 and MCF7
cells, as confirmed by CCK-8 assay results. (E-H) Loss of lncRNA
PCNAP1 significantly decreased the speed of wound closure in BC
cells (by ~64 and ~82% in MDA-MB-231 and MCF7 cells, respectively).
(I-K) Transwell invasion assays showed that lncRNA PCNAP1
inhibition significantly reduced invasion. (L) Cells with lncRNA
PCNAP1 knockdown (siPCNAP1#a and siPCNAP1#b) showed observably
increased protein levels of epithelial marker E-cadherin, and
decreased levels of mesenchymal marker N-cadherin. (M and N)
Injection of siCtrl cells led to lung metastasis in BALB/c nude
mice; these metastases were inhibited in nude mice injected with
siPCNAP1-transfected MDA-MB-231 cells. (O) Lung metasasis tumor
volume of nude mice. Results are representative of three
independent experiments. (P) Changes in nude mouse body weight.
Data are shown as mean ± SD. *P<0.05, **P<0.01,
***P<0.001, compared with the siCtrl group. BC, breast cancer;
lncRNA, long noncoding RNA; PCNAP1, proliferating cell nuclear
antigen pseudogene 1. |
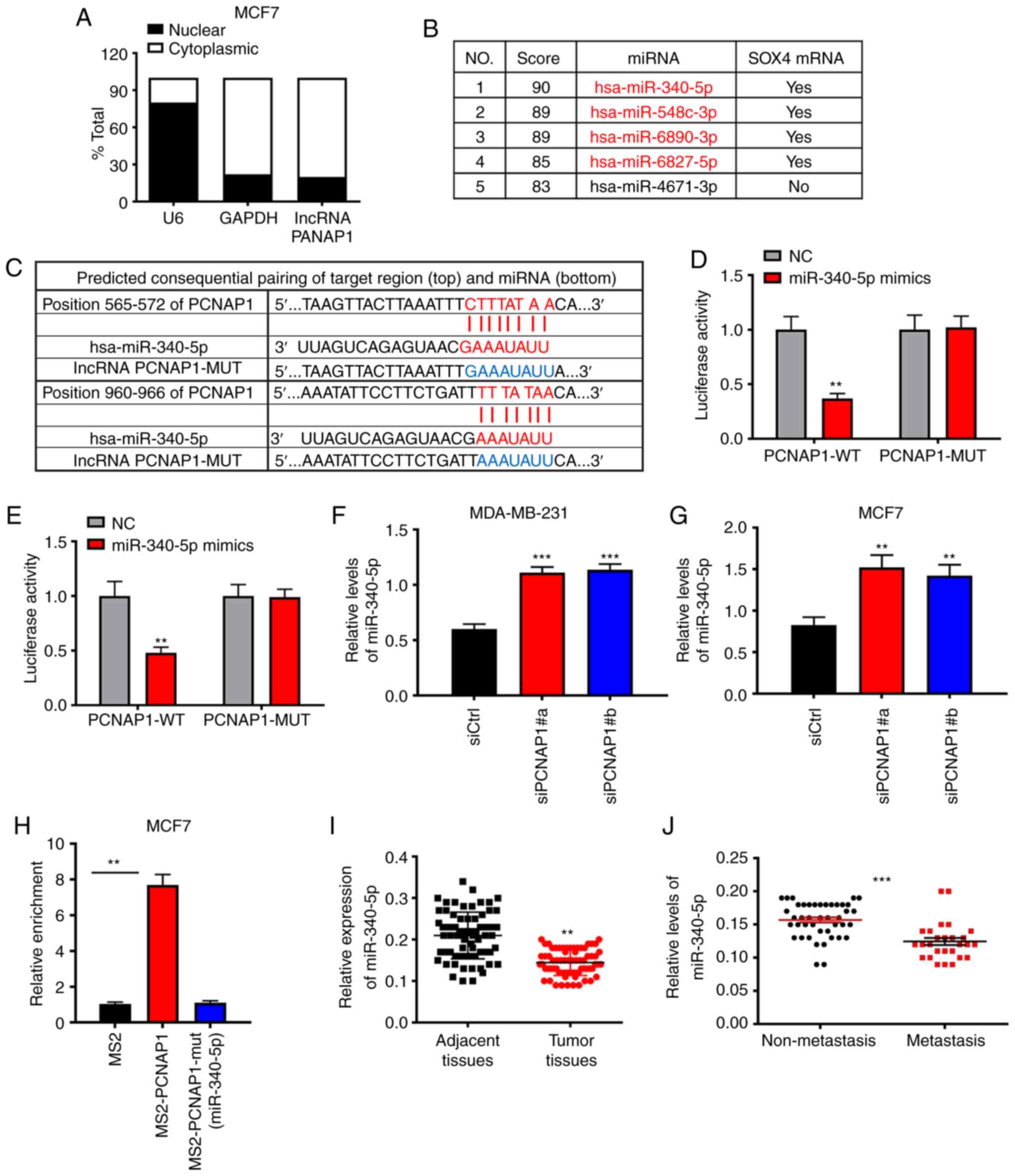
miR-340-5p has a target relationship
with lncRNA PCNAP1 and is downregulated in BC
It has been demonstrated that lncRNAs usually
function as miRNA sponges to regulate the binding of endogenous
miRNAs to their target mRNAs (9).
First, we separated MCF7 cells into nuclear and cytoplasmic
fractions and identified the cellular localization of lncRNA
PCNAP1. Both U6 and GAPDH were used as control groups. As shown in
Fig. 3A, 80.1% of lncRNA PCNAP1 was
detected in the cytoplasm fraction in the MCF7 cells, suggesting
that it may act at the post-transcriptional level. To further
explore the molecular mechanisms, we used bioinformatic tools to
predict miRNAs that could bind lncRNA PCNAP1. Surprisingly, the
first four miRNAs identified using the TargetScan software were all
predicted to target SOX4 directly (Fig.
3B). The association between SOX4 and BC migration has been
investigated previously (12,23).
Of these four miRNA candidates, we focused on miR-340-5p, which
could bind lncRNA PCNAP1 according to miRcode predictions (Fig. 3C). Furthermore, we cloned wild-type
(WT) lncRNA PCNAP1 luciferase plasmids containing potential
miR-340-5p binding sites or mutants (MUT) for each site. Luciferase
assays were performed to confirm the interaction between miR-340-5p
and lncRNA PCNAP1 after transfecting the plasmids with miR-340-5p
mimics into MBA-MD-231 and MCF7 cells. The miR-340-5p mimics
substantially inhibited the luciferase activity of WT lncRNA
PCNAP1, by ~60% (P<0.01); however, they did not affect the
luciferase activity of the lncRNA PCNAP1-MUT (Fig. 3D and E). To confirm these results,
lncRNA PCNAP1 was knocked down in both MBA-MD-231 and MCF7 cells;
the knockdown of lncRNA PCNAP1 increased the level of miR-340-5p
2-fold (P<0.01) as expected (Fig. 3F
and G). To validate the binding between these miRNA-340-5p and
lncRNA PCNAP1 at an endogenous level, MS2-RNA immunoprecipitation
(MS2-RIP) was used to pull down endogenous miRNAs associated with
lncRNA PCNAP1. For this purpose, an empty vector (MS2), a vector
containing the full sequence of lncRNA PCNAP1, a vector containing
lncRNA PCNAP1 with mutations in the miRNA-340-5p targeting binding
sites [designated PCNAP1 mut (miRNA-340-5p)], and a vector
containing lncRNA PCNAP1 with mutations in the miRNA-340-5p
targeting binding sites [PCNAP1-mut (miRNA-340-5p)] were
engineered. The RT-qPCR results showed that PCNAP1 RIP was
significantly enriched for miRNA-340-5p in MCF7 cells compared with
MS2 and the corresponding mutated vector (Fig. 3H). Taken together, these data
suggest that miR-340-5p directly binds to lncRNA PCNAP1. By
analyzing clinical samples, we found that miR-340-5p expression was
decreased in the BC tissues compared with that noted in the
adjacent non-tumor tissues (P<0.01), and in metastatic BC
tissues compared with non-metastatic tissues (P<0.01) (Fig. 3I and J), which further verified the
results of the above experiments.
lncRNA PCNAP1 promotes the migration
of BC cells by downregulating miR-340-5p
Next, we investigated whether lncRNA PCNAP1 promotes
breast cancer cell migration by interacting with miR-340-5p,
according to the bioinformatics predictions. To demonstrate this
hypothesis, we transfected miR-340-5p mimics and miR-340-5p
inhibitor in MDA-MB-231 and MCF7 cells (Fig. 4A and B). Overexpression of
miR-340-5p did not cause significant effects on BC cell viability;
this finding was confirmed by consistent results of the CCK-8 assay
(Fig. 4C and D). In wound healing
assays, the rate of scratch closure in cells overexpressing
miR-340-5p was significantly decreased compared to the negative
control: By ~66% (P<0.01) in MDA-MB-231 cells, and by ~72%
(P<0.01) in MCF7 cells (Fig.
4E-H). Transwell invasion assays showed that the degree of
invasion in the overexpressed group was significantly decreased
compared with the control group (Fig.
4I-K); this was highly consistent with the lncRNA
PCNAP1-knockdown results. We also found that miR-340-5p mimics and
inhibitor did not affect the expression of lncRNA PCNAP1 (Fig. 4L). These results indicate that
lncRNA PCNAP1 may promote the migration of BC cells by
downregulating miR-340-5p.
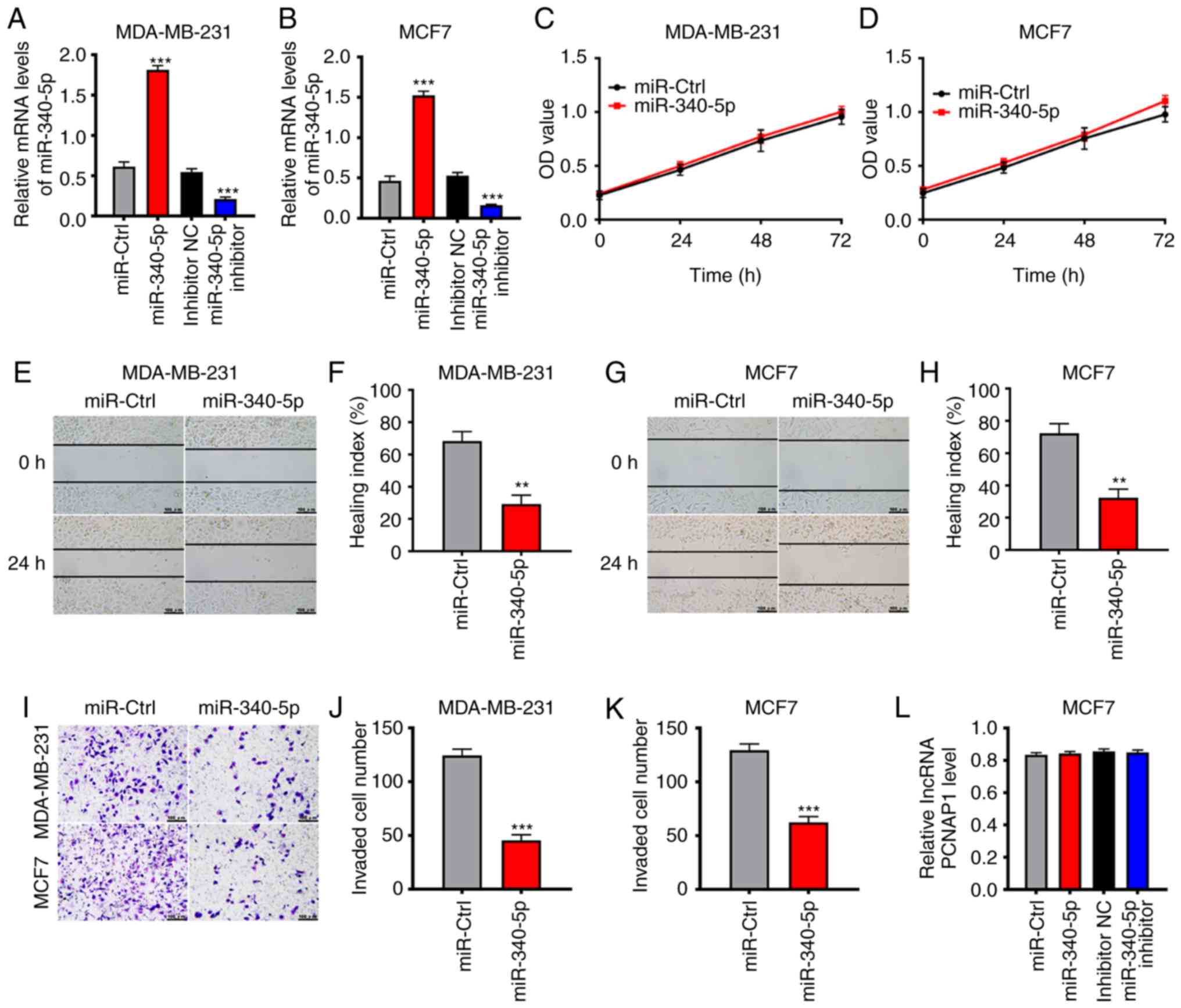 | Figure 4.lncRNA promotes the migration of BC
cells by downregulating miR-340-5p. (A and B) Transfection of
miR-340-5p mimics significantly upregulated the expression of
miR-340-5p in MDA-MB-231 and MCF7 cells compared with the miR-ctrl
group, ***P<0.001, compared with the miR-ctrl group.
Transfection of miR-340-5p inhibitor obviously downregulated the
expression of miR-340-5p compared with the inhibitor NC group,
indicating successful transfection. ***P<0.001, compared with
the inhibitor NC group. (C and D) Overexpression of miR-340-5p had
no significant effect on BC cell viability, as confirmed by CCK-8
assay results. (E-H) Overexpression of miR-340-5p mimics decreased
the speed of the wound closure by ~66% in MDA-MB-231 cells and by
~72% in MCF7 cells (both **P<0.01, compared with the miR-ctrl
group). (I-K) Transwell invasion assays showed that overexpression
of miR-340-5p mimics could inhibit cell invasion in MDA-MB-231 and
MCF7 cells compared with the negative control group. **P<0.01,
compared with the miR-ctrl group. (L) Overexpression or inhibition
of miR-340-5p had no influence on the expression of lncPCNAP1 in
MCF7 cells. Results are representative of three independent
experiments. Data are shown as mean ± SD. BC, breast cancer;
lncRNA, long noncoding RNA; PCNAP1, proliferating cell nuclear
antigen pseudogene 1. |
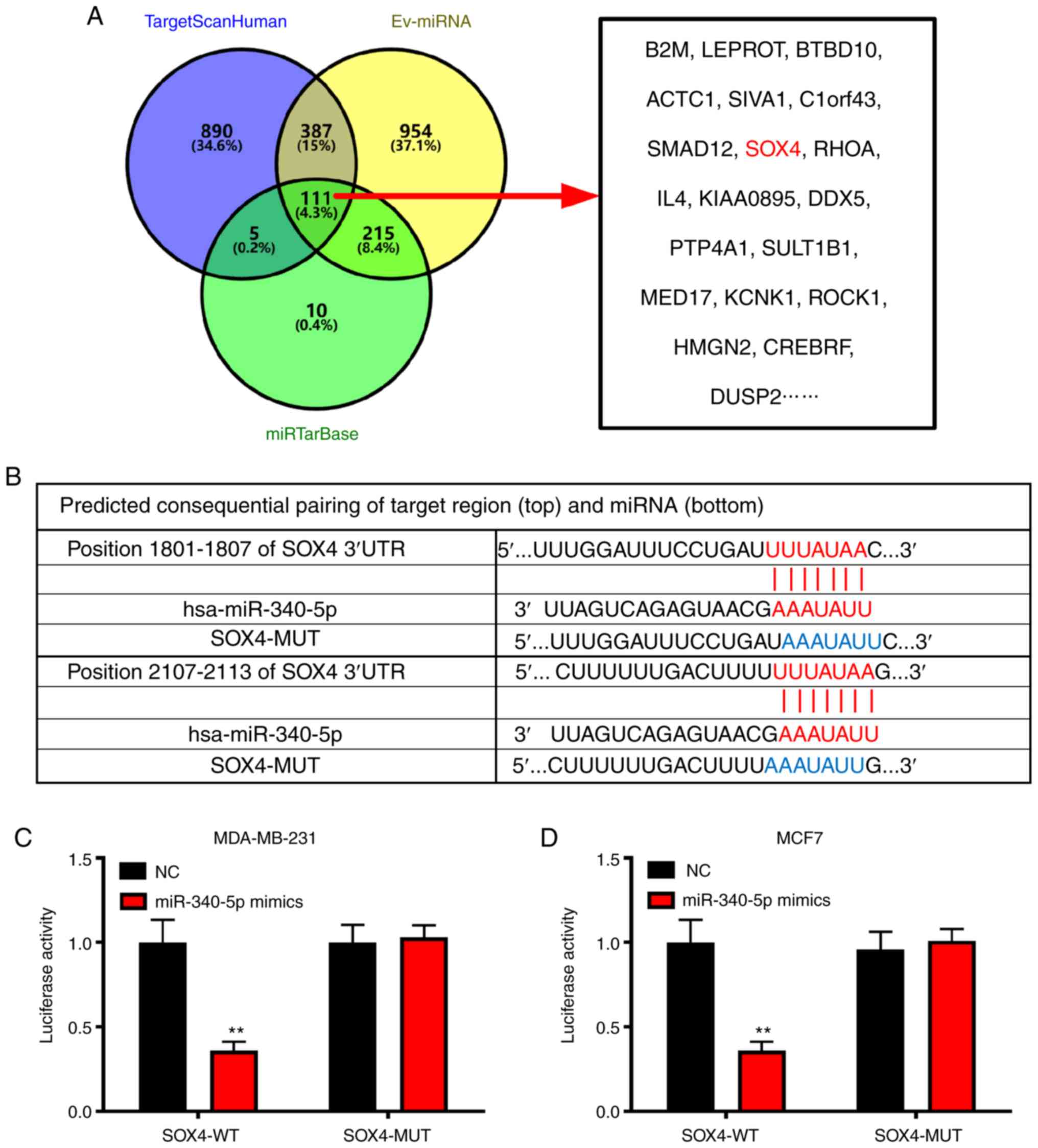
SOX4 has a target relationship with
miR-340-5p
Using TargetScan, we previously predicted that four
miRNAs binding to PCNAP1 may target SOX4 directly. In order to
verify whether the binding of lncRNA to miR-340-5p could regulate
SOX4, we performed in-depth bioinformatics predictions. TargetScan
Human, EV-miRNA, and miRTarBase jointly predicted 111 proteins that
miR-340-5p directly targeted, including SOX4, suggesting that it
was likely to be a miR-340-5p-targeted protein (Fig. 5A). According to miRcode predictions,
miR-340-5p can bind to the 3′ UTR of SOX4 (Fig. 5B). To verify this result, we
performed luciferase reporter assays in both MDA-MB-231 and MCF7
cells, and the reported luciferase contained SOX4-WT as well as
SOX4-MUT without potential binding sites. The vector was
co-transfected into MDA-MB-231 and MCF7 cells with miR-340-5p
mimics and miR-340-5p inhibitor. The miR-340-5p mimics inhibited
the luciferase activity of SOX4-WT by ~60 and ~64% in MDA-MB-231
and MCF7 cells, respectively, but did not inhibit the luciferase
activity of SOX4-MUT (P<0.01 in MDA-MB-231; P<0.01 in MCF7)
(Fig. 5C and D), indicating that
miRNA-340-5p can directly target SOX4.
lncRNA PCNAP1 promotes BC metastasis
by downregulating miR-340-5p, then upregulating SOX4
To test whether lncRNA PCNAP1 promotes the
metastasis of BC cells by regulating SOX4, we first analyzed the
clinical organization of BC. Immunohistochemistry results showed
that SOX4 expression was upregulated in BC tissues relative to
adjacent tissues (Fig. 6A), and
RT-qPCR results confirmed this finding (Fig. 6B). Furthermore, the expression of
SOX4 was significantly upregulated in tissues of metastatic
patients compared with those of non-metastatic patients (Fig. 6C). This result was consistent with
those obtained for lncRNA PCNAP1 in BC tissues. Therefore, we
performed a correlation analysis for lncRNA PCNAP1 and SOX4
expression in tumor tissues of metastatic patients. The results
showed a significant positive correlation between lncRNA PCNAP1 and
SOX4 expression in metastatic tissues (Fig. 6D). The expression of SOX4 was also
significantly downregulated after knockdown of lncRNA PCNAP in the
MDA-MB-231 and MCF7 cells, which confirmed the correlation between
the two (Fig. 6E-G). By contrast,
miR-340-5p expression was negatively correlated with SOX4
expression in metastatic tissues (Fig.
6H). Western blot analysis confirmed that overexpression of
miR-340-5p mimics in MDA-MB-231 and MCF7 cells resulted in a
significant downregulation of SOX4 expression; by contrast, the
miRNA inhibitor upregulated the expression of SOX4 (Fig. 6I-K). The Transwell assay results
also confirmed that invasion of MCF7 was significantly decreased by
the knockdown of lncRNA PCNAP1 and restored by the overexpression
of SOX4 (Fig. 6L-O). Thus, the
above results indicate that the lncRNA promotes the metastasis of
breast cancer by downregulating miR-340-5p and consequently
upregulating SOX4.
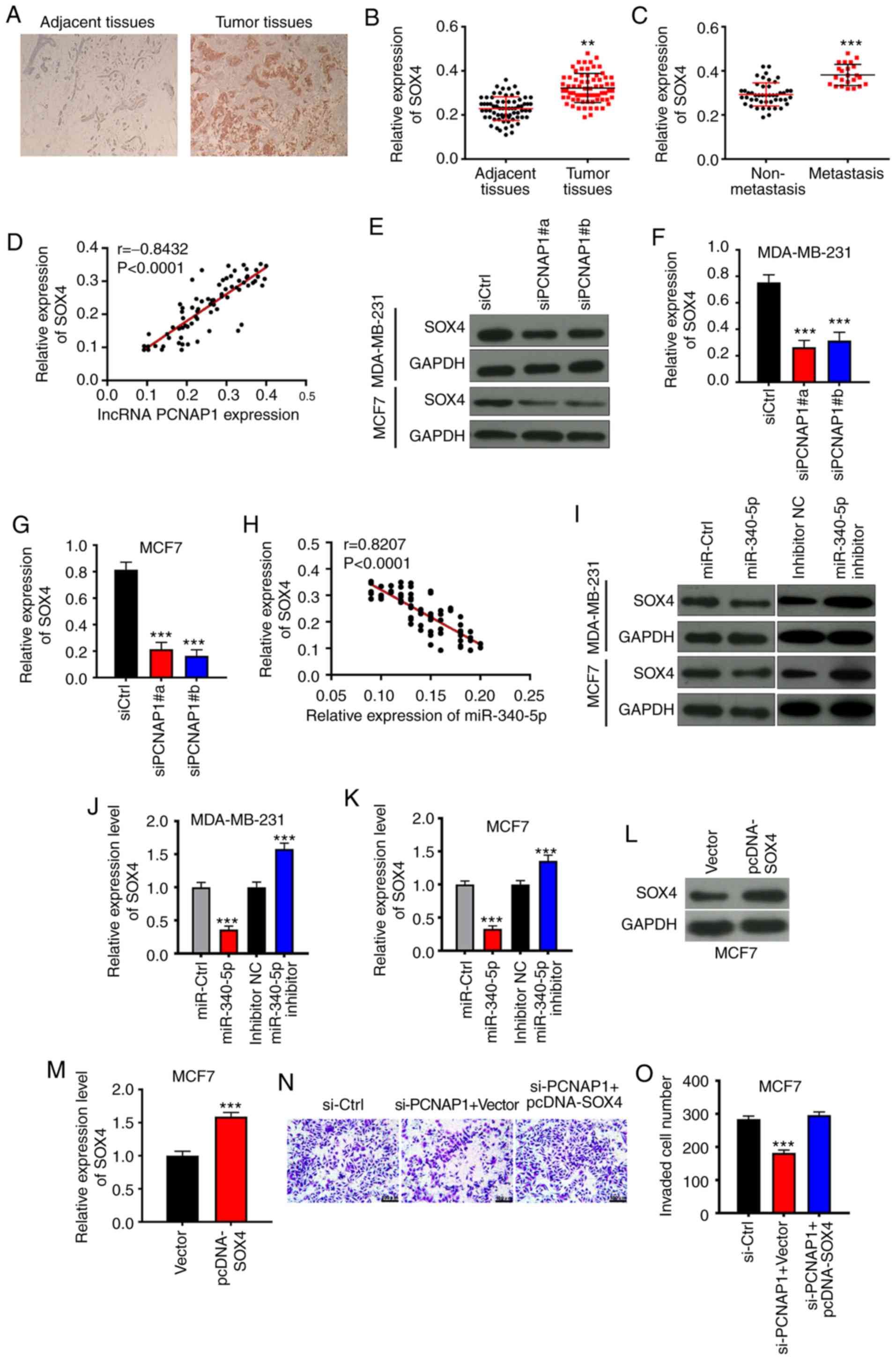 | Figure 6.lncRNA PCNAP1 promotes BC metastasis
by downregulating miR-340-5p, then upregulating SOX4. (A)
Immunohistochemistry results showed that SOX4 was significantly
increased in BC tissues compared with that in adjacent normal
tissues (magnification, ×100). (B) RT-qPCR validation of the
immunohistochemistry results. **P<0.01, compared with adjacent
tissues. (C) SOX4 expression was increased in metastatic BC tissues
compared with non-metastatic tissues. ***P<0.001, compared with
non-metastatic tissues. (D) There was a positive correlation
between the expression levels of lncRNA PCNAP1 and SOX4 in
metastatic tissues. (E-G) Knockdown of PCNAP1 (siPCNAP1#a and
siPCNAP1#b) significantly suppressed the protein expression of SOX4
in MDA-MB-231 and MCF7 cells compared with the negative control
group (siCtrl). ***P<0.001, compared with the siCtrl group. (H)
There was a negative correlation between miR-340-5p and SOX4
expression in metastatic tissues. (I-K) miR-340-5p mimics decreased
the protein expression of SOX4 compared with the miR-Ctrl group in
MDA-MB-231 and MCF7 cells. ***P<0.001, compared with the
miR-Ctrl group. In contrast, the miR-340-5p inhibitor increased the
expression compared with the inhibitor NC group. ***P<0.001,
compared with the inhibitor NC group. (L and M) Expression of SOX4
was upregulated by the overexpression plasmid. ***P<0.001,
compared with the vector group. (N and O) Knockdown of lncRNA
PCNAP1 decreased invasion which was restored by overexpression of
SOX4. ***P<0.001, compared with the siCtrl group. Results are
representative of three independent experiments. Data are shown as
mean ± SD. BC, breast cancer; lncRNA, long noncoding RNA; PCNAP1,
proliferating cell nuclear antigen pseudogene 1; SOX4, SRY-box
transcription factor 4. |
Discussion
It is well established that mammalian genomes encode
large numbers of long non-coding RNAs (lncRNAs) in addition to
protein-coding RNAs, and that the majority of these lncRNAs have
important functions, they are involved in chromatin remodeling, as
well as transcriptional and post-transcriptional regulation,
through a variety of chromatin-based mechanisms and via cross-talk
with other RNA species (24). With
ongoing research, newly identified lncRNAs have emerged as critical
factors in cellular development and human diseases including breast
cancer. For example, the ectopic expression of lncRNA Smad7 can
rescue apoptosis induced by a TGF-β receptor inhibitor in breast
cancer (25). Gupta et al
found that the expression of lncRNA HOTAIR was often high and could
be a powerful predictor of metastasis and survival in primary
breast cancer (9). Pseudogenes such
as proliferating cell nuclear antigen pseudogene 1 (PCNAP1) serve
as functional regulators of ancestral gene expression and have
important roles in regulating protein-coding transcripts (26), regulating genes via the piRNA
pathway for limiting transposable element damage to the genome
(27), or acting as a ceRNA
(28). PCNAP is exclusively
expressed in malignant tissues, including prostate cancer and
breast cancer, but not in normal cells, therefore can be used as a
cell marker for the classification of different tumors (13). Pseudogene-derived lncRNAs could
function as antisense RNAs or endo-siRNAs, or serve as sponges of
miRNAs and thus exert biological roles in cancer (29,30). A
recent study found that lncRNA PCNAP1 expression levels were
significantly increased in liver cancer tissues positive for
hepatitis B virus covalently closed circular DNA, suggesting an
oncogenic role in this cancer (29). However, the precise molecular
mechanisms by which lncRNA PCNAP1 modulates breast cancer (BC)
growth remain largely unknown.
Plants, animals, and some viruses also contain
miRNAs, which are small noncoding RNA molecules of about 22
nucleotides. The main role of miRNAs is RNA silencing and negative
transcriptional regulation of gene expression (31,32).
miRNAs are also key regulators of lncRNAs via complicated and
diverse mechanisms. Notably, lncRNAs have recently been found to
act as miRNA sponges or miRNA inhibitors (antagomirs), which
interact with miRNAs and modulate the expression of miRNA target
genes (33,34). Moreover, a lncRNA may act as a
competing endogenous RNA (ceRNA), effectively inhibiting the
expression of miRNAs (35). Liu
et al found that lncRNA HOTAIR could modulate the
de-repression of HER2, a target gene of miR-331-3p in gastric
cancer (36). There may be some
mechanisms by which lncRNAs can degrade miRNAs by binding to
miRNAs, similar to the function of miRNA sponges or antagomirs that
promote miRNA degradation (37).
However, the exact mechanism remains unclear.
In the present study, we found that the average
level of lncRNA PCNAP1 in BC was significantly higher than that
observed in corresponding non-tumor tissues. Interestingly,
patients with metastasis had increased lncRNA PCNAP1 levels
compared with patients without metastasis. In addition, we
investigated the correlation between lncRNA PCNAP1 levels and
prognosis in patients with BC. We found that high expression of
lncRNA PCNAP1 in BC tissues was associated with poor prognosis.
These findings suggest that lncRNA PCNAP1 plays a critical role in
the development and metastasis of BC. An RNA interference approach
was used to further analyze the role of lncRNA PCNAP1 in BC cells.
Although inhibition of lncRNA PCNAP1 did not affect the growth of
BC MDA-MB-231 or MCF7 cells in vitro, its knockdown
significantly inhibited the migration and invasion of BC cells,
which was further confirmed in mouse transplant models. These
results suggest that increased lncRNA PCNAP1 levels in samples with
metastasis may be the cause of BC metastasis, rather than a
consequence. Mechanistically, we showed that lncRNA PCNAP1
participates, at least partly, in these processes by regulating the
expression of SRY-box transcription factor 4 (SOX4) via
competitively binding miRNA-340-5p as a ceRNA. Collectively, the
present data demonstrated a key causal role of lncRNA PCNAP1 in
metastasis of BC, and suggest that it is a potential therapeutic
target for BC metastasis.
SOX4, a member of the Sry-related high mobility
group box family of transcription factors, is considered a master
regulator of tumorigenesis and cancer stemness (38,39).
Numerous studies have found increased SOX4 expression in various
cancers (including lung, colorectal, prostate, and esophageal
cancers) (40–43). In the present study, we demonstrated
that the knockdown of lncRNA PCNAP1 reduced mRNA and protein levels
of SOX4, suggesting that SOX4 may be a downstream target gene
regulated by lncRNA PCNAP1. More importantly, overexpression of
SOX4 rescued, at least in part, the impaired migration and invasion
caused by lncRNA PCNAP1 silencing. This finding indicates that SOX4
participates in the process of metastasis affected by lncRNA PCNAP1
in BC. These comprehensive data are consistent with the results of
previous studies. The main molecular functions of lncRNAs are as:
i) Decoys to locate transcription factors; ii) regulatory signals
for transcription; iii) scaffolds to aggregate different proteins;
iv) ‘sponges’ to interact with miRNAs; and v) guides for binding of
specific proteins to target genes (44). In the present study, it was
demonstrated that lncRNA PCNAP1 may act as a ‘sponge’ to interact
with miRNA-340-5p, leading to a reduction in miRNA-340-5p levels in
BC cells. This decrease in miRNA-340-5p levels abolishes the
inhibitory effect of lncRNA PCNAP1 on SOX4, which eventually causes
BC metastasis. This suggests that the lncRNA
PCNAP1/miRNA-340-5p/SOX4 axis is important in BC metastasis.
Whether other miRNAs function as the ‘bridge’ linking lncRNA PCNAP1
and SOX4 remains an open question, as 10 miRNAs bound by lncRNA
PCNAP1 were predicted to target SOX4 in a similar way. Furthermore,
there have been few reports on the role of miRNA-340-5p in cancer
development. Although the knockdown of miRNA-340-5p increased the
migration and invasion of BC cells, the detailed mechanism by which
this occurs requires further investigation in the future, as does
the question of whether miRNA-340-5p influences the development of
other cancers.
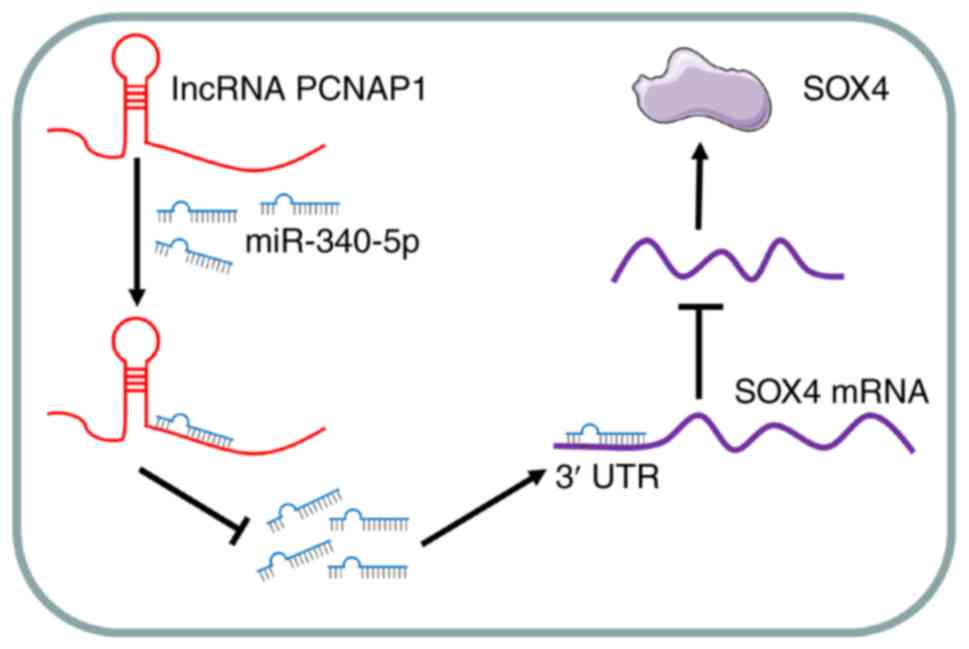
In summary, the results of the present study
demonstrated that high lncRNA PCNAP1 expression levels promote
malignant metastasis in BC cells in vitro and in vivo
(Fig. 7); thus, lncRNA PCNAP1 has
potential as a biomarker for the progression of BC. Moreover, our
results suggest that lncRNA PCNAP1 functions as a
metastasis-promoting gene in certain types of cancer, paving the
way for the development of new therapeutic modalities. Depletion of
lncRNA PCNAP1 and even use of an antagonist may be a promising
strategy for BC treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Henan Science
and Technology Project (grant no. 171022310005).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
Conception and design of the research study was
accomplished by YY and HL. Administrative support was provided by
YH. Collection and assembly of the data were carried out by YH and
YS. Data analysis and interpretation were conducted by YS and QC.
Manuscript writing was carried out by YY and HL. All authors read
and approved the manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
The clinical data collection and research procedures
were reviewed and approved by the Medical Ethics Committee of Henan
Provincial People's Hospital. In addition, BC donors participating
in the study provided written informed consent for their tissue
samples to be used for scientific research. All animal experiments
were performed at the Animal Laboratory Center of Henan Provincial
People's Hospital. All experimental procedures and protocols were
approved by the Institutional Animal Care and Use Committee at
Henan Provincial People's Hospital, in accordance with the Guide
for the Care and Use of Laboratory Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Playdon MC, Bracken MB, Sanft TB, Ligibel
JA, Maura H and Irwin ML: Weight gain after breast cancer diagnosis
and all-cause mortality: Systematic review and meta-analysis. J
Natl Cancer Inst. 107:djv2752015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Goding Sauer A, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen P, Wang R, Yue Q and Hao M: Long
non-coding RNA TTN-AS1 promotes cell growth and metastasis in
cervical cancer via miR-573/E2F3. Biochem Biophys Res Commun.
503:2956–2962. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Amaral PP, Dinger ME, Mercer TR and
Mattick JS: The eukaryotic genome as an RNA machine. Science.
319:1787–1789. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Amaral PP and Mattick JS: Noncoding RNA in
development. Mamm Genome. 19:454–492. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ginger MR, Shore AN, Contreras A, Rijnkels
M, Miller J, Gonzalez-Rimbau MF and Rosen JM: A noncoding RNA is a
potential marker of cell fate during mammary gland development.
Proc Natl Acad Sci USA. 103:5781–5786. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yuan SX, Yang F, Yang Y, Tao QF, Zhang J,
Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al: Long noncoding RNA
associated with microvascular invasion in hepatocellular carcinoma
promotes angiogenesis and serves as a predictor for hepatocellular
carcinoma patients' poor recurrence-free survival after
hepatectomy. Hepatology. 56:2231–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang EB, Kong R, Yin DD, You LH, Sun M,
Han L, Xu TP, Xia R, Yang JS, De W and Chen Jf: Long noncoding RNA
ANRIL indicates a poor prognosis of gastric cancer and promotes
tumor growth by epigenetically silencing of miR-99a/miR-449a.
Oncotarget. 5:2276–2292. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang F, Shen Y, Zhang W, Jin J, Huang D,
Fang H, Ji W, Shi Y, Tang L, Chen W, et al: An androgen receptor
negatively induced long non-coding RNA ARNILA binding to miR-204
promotes the invasion and metastasis of triple-negative breast
cancer. Cell Death Differ. 25:2209–2220. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Feng J, Yang G, Liu Y, Gao Y, Zhao M, Bu
Y, Yuan H, Yuan Y, Yun H, Sun M, et al: lncRNA PCNAP1 modulates
hepatitis B virus replication and enhances tumor growth of liver
cancer. Theranostics. 9:5227–5245. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Park SY, Jeong MS, Han CW, Yu HS and Jang
SB: Structural and functional insight into proliferating cell
nuclear antigen. J Microbiol Biotechnol. 26:637–647. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Albus U: Guide for the care and use of
laboratory animals (8th edn). Lab Anim. 46:267–268. 2012.
View Article : Google Scholar
|
|
16
|
Salmon HW, Guha A, Rojiani AM and Siemann
DW: Vascular development in mouse lung metastases. Am J Cancer Res.
2:581–588. 2012.PubMed/NCBI
|
|
17
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu T, Zhang Q, Zhang J, Li C, Miao YR,
Lei Q, Li Q and Guo AY: EVmiRNA: A database of miRNA profiling in
extracellular vesicles. Nucleic Acids Res. 47(D1): D89–D93. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC,
Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al: miRTarBase: A
database curates experimentally validated microRNA-target
interactions. Nucleic Acids Res. 39((Database Issue)): D163–D169.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jeggari A, Marks DS and Larsson E:
miRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao
X, Feng J, Zhang Y, Gao H, Liu DX, et al: SOX4 induces
epithelial-mesenchymal transition and contributes to breast cancer
progression. Cancer Res. 72:4597–4608. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fang Y and Fullwood MJ: Roles, functions,
and mechanisms of long non-coding RNAs in cancer. Genomics
Proteomics Bioinformatics. 14:42–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Arase M, Horiguchi K, Ehata S, Morikawa M,
Tsutsumi S, Aburatani H, Miyazono K and Koinuma D: Transforming
growth factor-β-induced lncRNA-Smad7 inhibits apoptosis of mouse
breast cancer JygMC(A) cells. Cancer Sci. 105:974–982. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chan WL and Chang JG: Pseudogene-derived
endogenous siRNAs and their function. Methods Mol Biol.
1167:227–239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Siomi MC, Sato K, Pezic D and Aravin AA:
PIWI-interacting small RNAs: The vanguard of genome defence. Nat
Rev Mol Cell Biol. 12:246–258. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Karreth FA, Reschke M, Ruocco A, Ng C,
Chapuy B, Léopold V, Sjoberg M, Keane TM, Verma A, Ala U, et al:
The BRAF pseudogene functions as a competitive endogenous RNA and
induces lymphoma in vivo. Cell. 161:319–332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dinger ME, Amaral PP, Mercer TR, Pang KC,
Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C,
et al: Long noncoding RNAs in mouse embryonic stem cell
pluripotency and differentiation. Genome Res. 18:1433–1445. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lou W, Ding B and Fu P: Pseudogene-derived
lncRNAs and their miRNA sponging mechanism in human cancer. Front
Cell Dev Biol. 8:852020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 lncRNA
antagonizes let-7 microRNAs. Mol Cell. 52:101–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang K, Long B, Zhou LY, Liu F, Zhou QY,
Liu CY, Fan YY and Li PF: CARL lncRNA inhibits anoxia-induced
mitochondrial fission and apoptosis in cardiomyocytes by impairing
miR-539-dependent PHB2 downregulation. Nat Commun. 5:35962014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang K, Liu F, Zhou LY, Long B, Yuan SM,
Wang Y, Liu CY, Sun T, Zhang XJ and Li PF: The long noncoding RNA
CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res.
114:1377–1388. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Parvani JG and Schiemann WP: Sox4, EMT
programs, and the metastatic progression of breast cancers:
Mastering the masters of EMT. Breast Cancer Res. 15:R722013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vervoort SJ, van Boxtel R and Coffer PJ:
The role of SRY-related HMG box transcription factor 4 (SOX4) in
tumorigenesis and metastasis: Friend or foe? Oncogene.
32:3397–3409. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bilir B, Osunkoya AO, Wiles WG IV,
Sannigrahi S, Lefebvre V, Metzger D, Spyropoulos DD, Martin WD and
Moreno CS: SOX4 is essential for prostate tumorigenesis initiated
by PTEN ablation. Cancer Res. 76:1112–1121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Koumangoye RB, Andl T, Taubenslag KJ,
Zilberman ST, Taylor CJ, Loomans HA and Andl CD: SOX4 interacts
with EZH2 and HDAC3 to suppress microRNA-31 in invasive esophageal
cancer cells. Mol Cancer. 14:242015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang B, Li Y, Tan F and Xiao Z: Increased
expression of SOX4 is associated with colorectal cancer
progression. Tumour Biol. 37:9131–9137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou Y, Wang X, Huang Y, Chen Y, Zhao G,
Yao Q, Jin C, Huang Y, Liu X and Li G: Down-regulated SOX4
expression suppresses cell proliferation, metastasis and induces
apoptosis in Xuanwei female lung cancer patients. J Cell Biochem.
116:1007–1018. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|