Introduction
Lung cancer is not only the most commonly diagnosed
cancer, but also the leading cause of cancer-associated death
globally, with a predicted 2,28,150 novel cases and 1,47,510 deaths
annually (1,2). Non-small cell lung cancer (NSCLC) is
the most prevalent pathological subtype of lung cancer and accounts
for 80–85% of all lung cancer cases (3). NSCLC is classified into lung squamous
cell carcinoma, lung adenocarcinoma and large-cell lung cancer, and
all three pathological classifications manifest similar biological
behaviors and gene mutations (4).
Although vigorous attempts have been made to improve the quality of
NSCLC therapy, the long-term survival of patients remains low
(5). When diagnosed at an early
stage, NSCLC can be effectively treated by surgical resection,
chemotherapy and radiotherapy. However, most patients are diagnosed
at a later or advanced stage, and are typically treated with
first-line therapeutic strategies that have poor clinical
efficiency (6). Therefore,
advancements in diagnostic testing and therapy are necessary. An
improved understanding of the mechanisms underlying NSCLC
tumorigenesis may aid in the identification of promising diagnostic
biomarkers and therapeutic targets.
Long noncoding RNAs (lncRNAs) belong to a large and
diverse group of transcripts of >200 nucleotides that lack
protein-coding capacity (7). In
previous decades, lncRNAs have received increasing attention for
their roles in numerous biological activities and diseases,
particularly in human cancers (8).
Differentially expressed lncRNAs have been verified in almost all
human cancer types, including NSCLC (9,10). An
increasing number of studies have shown that the dysregulation of
lncRNAs plays a significant role in NSCLC tumorigenesis and
progression by exerting cancer-inhibiting or oncogenic effects
(11–13).
MicroRNAs (miRNAs) are a class of single-stranded
noncoding RNA that can directly bind to the 3′-untranslated region
of their target mRNAs, which triggers mRNA degradation and/or
translation depression (14).
Studies have shown that aberrant miRNA expression is closely
associated with the initiation and development of NSCLC, and is
implicated in the regulation of a wide range of tumorigenic
behaviors (15–17). Multiple studies have revealed that
lncRNAs are capable of interacting with miRNAs, thereby forming
competing endogenous RNA (ceRNA) regulatory networks to modulate
the expression and function of coding genes in NSCLC (18–20).
Hence, studying the detailed roles of lncRNAs and miRNAs in NSCLC,
as well as illustrating their cellular functions and interactions,
may aid the diagnosis, prognostic prediction and development of
therapies for NSCLC.
lncRNA CBR3 antisense RNA 1 (CBR3-AS1) has been
shown to promote the initiation and progression of osteosarcoma
(21). The present study aimed to
explore whether CBR3-AS1 is involved in the development of NSCLC.
CBR3-AS1 expression was detected in NSCLC tissues and cells lines
to reveal its prognostic significance in NSCLC, and to determine
its influence on aggressive phenotypes of NSCLC cells. The study
also aimed to elucidate the potential molecular mechanisms
underlying the oncogenic roles of CBR3-AS1, which may provide
potential targets for novel diagnostic and therapeutic
strategies.
Materials and methods
Patients and tissue samples
In total, 57 pairs of tumor and adjacent healthy
tissues were collected from patients (31 male and 26 female
patients; age range, 51–72 years) admitted to Weifang People's
Hospital (Weifang, China) between May 2014 and February 2015. The
study participants did not receive systemic or local anticancer
treatments prior to surgical resection. All surgical tissues were
immediately frozen and stored in liquid nitrogen for further use.
The present study was approved by the Human Ethics Committee of
Weifang People's Hospital and was performed in accordance with the
Declaration of Helsinki. Written informed consent was obtained from
all study participants.
Cell lines
A total of four NSCLC cell lines (SK-MES-1, H522,
H460 and A549) and the human BEAS-2B nontumorigenic bronchial
epithelial cell line were purchased from the American Type Culture
Collection. SK-MES-1 cells were cultured in Minimum Essential
Medium containing 10% fetal bovine serum (FBS) and 1%
penicillin-streptomycin (all Gibco; Thermo Fisher Scientific,
Inc.). H522, H460, and A549 cells were cultured in RPMI 1640
(Gibco; Thermo Fisher Scientific, Inc.), though the other
conditions were the same as those used for SK-MES-1 cells. BEAS-2B
cells were cultured in Bronchial Epithelial Cell Growth Medium
(Lonza Group Ltd.). All cells were maintained at 37°C in an
incubator supplied with 5% CO2.
Transfection
Specific small interfering RNAs (siRNAs) against
CBR3-AS1 (si-CBR3-AS1#1, #2 and #3) and a negative control siRNA
(si-NC) were acquired from Guangzhou RiboBio Co., Ltd. The
si-CBR3-AS1 sequences were as follows: si-CBR3-AS1#1,
5′-ATGCAATTTCTTTAAAAAGC-3′; si-CBR3-AS1#2,
5′-CAGTTTATTTTTATTTATTTTTT-3′; and si-CBR3-AS1#3,
5′-AGCTCAAATTTTTTATATATTTC-3′. The si-NC sequence was
5′-CACGATAAGACAATGTATTT-3′. Furthermore, the miR-509-3p agomir
(agomir-509-3p), negative control agomir (agomir-NC), miR-509-3p
antagomir (antagomir-509-3p) and negative control antagomir
(antagomir-NC) were synthesized by Shanghai GenePharma Co., Ltd.
The corresponding sequences were as follows: Agomir-509-3p,
5′-GAUGGGUGUCUGCAUGGUUAGU-3′; agomir-NC,
5′-UUGUACUACACAAAAGUACUG-3′; antagomir-509-3p,
5′-CUACCCACAGACGUACCAAUCA-3′; and antagomir-NC
5′-CAGUACUUUUGUGUAGUACAA-3′. The histone deacetylase 9 (HDAC9)
overexpression vector pcDNA3.1-HDAC9 and empty pcDNA3.1 plasmid
were purchased from Sangon Biotech Co., Ltd.
Cells were seeded into 6-well plates at a density of
6×105 cells per well, and then separately or
co-transfected with the aforementioned siRNAs (100 pmol), agomirs
(50 nM), antagomirs (100 nM) and plasmids (4 µg) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). All transfection procedures were conducted at
room temperature. After 48 h incubation at 37°C, reverse
transcription-quantitative (RT-q) PCR, flow cytometry and Transwell
migration and invasion assays were carried out. Cell Counting kit-8
(CCK-8) and western blot assays were performed at 24 and 72 h
post-transfection, respectively.
Nuclear-cytoplasmic fractionation, RNA
isolation and RT-qPCR
The nuclear and cytoplasmic fractions of NSCLC cells
were separated using the PARIS™ Kit (Invitrogen; Thermo Fisher
Scientific, Inc). Next, the total nuclear and cytoplasmic RNA was
extracted using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), after which the RNA was subjected to
RT-qPCR to determine the subcellular localization of CBR3-AS1. To
quantify CBR3-AS1 and HDAC9 mRNA expression, complementary DNA was
synthesized using the PrimeScript RT reagent Kit (Takara
Biotechnology Co., Ltd.) and qPCR was conducted using SYBR Premix
Ex Taq (Takara Biotechnology Co., Ltd.). The qPCR thermocycling
conditions were as follows: 5 min at 95°C, followed by 40 cycles of
95°C for 30 sec and 65°C for 45 sec. GAPDH was used for the
normalization of CBR3-AS1 and HDAC9 mRNA expression. To quantify
miR-509-3p expression, total RNA was reversed transcribed using the
miScript reverse transcription kit and qPCR was performed using the
miScript SYBR Green PCR kit (both Qiagen GmbH). The thermocycling
conditions were as follows: 95°C for 2 min, 95°C for 10 sec, 55°C
for 30 sec and 72°C for 30 sec, for 40 cycles. The expression of
miR-509-3p was normalized to that of U6 small nuclear RNA. All
expression levels were quantified using the 2−ΔΔCq
method (22). Each group contained
three replicates and RT-qPCR was repeated three times. The qPCR
primer sequences were as follows: CBR3-AS1 forward,
5′-CAATAGGGAAGCAGAGGGAGAA-3′ and reverse,
5′-TTAGAGATTCCTACAGACCCAGGTC-3′; HDAC9 forward,
5′-TCAGCTCAGTGGATGTGAAGTCA-3′ and reverse,
5′-GCTGTTTCTGAAACTCTGCTATCAG-3′; GAPDH forward,
5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse,
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′; miR-509-3p forward,
5′-TCGGCAGGUACUGCAGACGUG-3′ and reverse,
5′-CACTCAACTGGTGTCGTGGA-3′; and U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′.
CCK-8 assay
A total of 2×103/well transfected cells
resuspended in 100 µl culture medium were inoculated into 96-well
plates. The cells were incubated for 0, 24, 48 or 72 h, and 10 µl
of CCK-8 solution (Sigma-Aldrich, Merck KGaA) was added to each
well, after which the plates were incubated at 37°C (5%
CO2) for an additional 2 h. Absorbance was measured at
450 nm on a microplate reader (Multiscan MK3; Thermo Fisher
Scientific, Inc.). Each group contained five replicates and the
assay was repeated three times.
Flow cytometry
The Annexin V-Fluorescein Isothiocyanate (FITC)
Apoptosis Detection Kit (BioLegend, Inc.) was used to determine
apoptotic rate. Briefly, transfected cells were collected 48 h
post-transfection and washed twice with ice-cooled
phosphate-buffered solution (Gibco; Thermo Fisher Scientific,
Inc.). The cells were centrifugated at 800 × g for 5 min at room
temperature, and resuspended in 100 µl binding buffer. Then, 5 µl
Annexin V-FITC and 10 µl propidium iodide were added for double
staining. Following a 15-min incubation in the dark at room
temperature, the apoptotic rate was determined by flow cytometry
(FACScan; BD Biosciences). CellQuest software (version 2.9; BD
Biosciences) was used for data analysis. Each group contained three
replicates and the assay was repeated three times.
Transwell migration and invasion
assays
Transfected cells were harvested and resuspended in
serum-free culture medium. Next, 200 µl cell suspension
(5×104 cells) was added into the upper compartments of
Transwell inserts (pore size, 8 µm; BD Biosciences) and 600 µl
complete culture medium was added to the bottom chambers to induce
migration. After 24 h incubation at 37°C, non-migratory cells were
removed from the upper chamber using a cotton swab, and cells that
had migrated to the lower chamber were fixed with methanol at room
temperature for 30 min and stained with 0.1% crystal violet at room
temperature for 30 min. Following extensive rinsing, imaging of the
stained cells was performed using an inverted microscope (Olympus
Corporation; ×200 magnification), and five randomly selected fields
were counted. The same procedure was followed for the Transwell
invasion assay, but the membranes were precoated with Matrigel (BD
Biosciences) at 37°C for 2 h. The number of migratory and invasive
cells was counted, and the average values were used to determine
the migratory and invasive capacities, respectively. The assay was
repeated three times.
Tumor xenograft model
Plasmids carrying short hairpin RNA (shRNA) against
CBR3-AS1 (pLKO.1-sh-CBR3-AS1) or the negative control shRNA
(pLKO.1-sh-NC) were manufactured by Shanghai GenePharma Co., Ltd.
H460 cells were transfected with the aforementioned lentivirus
produced by Shanghai GenePharma Co., Ltd, and puromycin was used to
select a stable CBR3-AS1-knockdown cell line. Animal experimental
procedures were approved by the Institutional Animal Care and Use
Committee of Weifang People's Hospital. For the xenograft study,
H460 cells stably transfected with sh-CBR3-AS1 (sh-CBR3-AS1 group)
or sh-NC (sh-NC group) were collected and subcutaneously injected
into 4–6-week-old male BALB/c nude mice (20 g each; Beijing Vital
River Laboratory Animal Technology Co., Ltd). A total of six mice
were used in the assay (three mice per group). The animals were
maintained under specific pathogen-free conditions at 25°C and 50%
humidity, with a 10:14 light/dark cycle and ad libitum access to
food and water. The size of the subcutaneous tumors was monitored
and recorded weekly for 4 weeks. Tumor volume was calculated as
follows: Volume=0.5 × length × width2. All mice were
euthanized by cervical dislocation after the last measurement, and
the subcutaneous tumors were resected and weighed. Tumor xenografts
were collected and used for RT-qPCR and western blot analysis.
Bioinformatics analysis
The potential interaction between CBR3-AS1 and
miRNA(s) was predicted using the StarBase online tool (http://starbase.sysu.edu.cn/). For miRNA target
prediction, two online databases, miRDB (http://mirdb.org/) and TargetScan (http://www.targetscan.org/vert_72/), were used to
search for putative targets of miR-509-3p. lncLocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/) was
utilized to predict the localization of CBR3-AS1.
Luciferase reporter assay
Partial sequences of CBR3-AS1 carrying wild-type
(WT) miR-509-3p binding sequences were amplified and inserted into
the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega
Corporation) and termed WT-CBR3-AS1. The mutant (MUT) sequences of
CBR3-AS1 were also cloned into the same vector to generate the
MUT-CBR3-AS1 luciferase reporter vector. WT-HDAC9 and MUT-HDAC9
luciferase reporter vectors were designed and produced using the
same procedure. NSCLC cells were seeded into 24-well plates and 24
h later, were co-transfected with WT or MUT luciferase reporter
vectors along with agomir-509-3p or agomir-NC using
Lipofectamine® 2000. After 48 h, luciferase activity was
measured using the Dual-Glo® Luciferase Assay System
(Promega Corporation). The assay was repeated three times and
contained three replicates. Renilla luciferase activity was
used for the normalization of firefly luciferase activity.
RNA immunoprecipitation (RIP)
The Magna RIP™ RNA-Binding Protein
Immunoprecipitation kit (EMD Millipore) was used per the
manufacturer's protocol to determine potential interactions between
CBR3-AS1 with miR-509-3p in NSCLC. NSCLC cells were harvested and
lysed using the supplied RIP lysis buffer. Then, whole-cell
extracts were incubated with RIP buffer and human anti-Argonaute
(anti-Ago2) or anti-IgG antibodies (both cat. no. 03–110; EMD
Millipore). After overnight incubation, the magnetic beads were
collected, rinsed with washing buffer and treated with proteinase K
to digest the proteins. The immunoprecipitated RNA was analyzed by
RT-qPCR. The RIP assay was repeated three times and contained three
replicates.
Protein preparation and western
blotting
Cells were lysed in RIPA buffer (Beyotime Institute
of Biotechnology) supplemented with a protease inhibitor cocktail
(Roche Diagnostics), and the extracted protein was quantified using
a bicinchoninic acid protein assay kit (Nanjing KeyGen Biotech Co.,
Ltd.). Equal amounts of protein (30 µg per well) were separated by
electrophoresis on 10% SDS-PAGE gels, and transferred onto
polyvinylidene difluoride membranes (EMD Millipore). The membranes
were blocked for 2 h at room temperature with 5% nonfat dry milk in
Tris-buffered saline (0.1% Tween-20). After blocking, the membranes
were incubated with primary antibodies [anti-HDAC9 (cat. no.
ab109446; Abcam) and anti-GAPDH (cat. no. ab128915; Abcam), both
1:1,000] at 4°C overnight, and further incubated with a horseradish
peroxidase-conjugated secondary antibody (1:5,000; cat. no.
ab205718; Abcam). The blots were developed using the Immobilon
Western Chemilum HRP substrate (EMD Millipore) and the assay was
repeated three times. Quantity One software version 4.62 (Bio Rad
Laboratories, Inc.) was used for densitometric analysis.
Statistical analysis
The experimental results were analyzed using the
SPSS statistics software package (version 21.0; IBM Corp) and
expressed as the mean ± standard deviation. The χ2 test
was used to evaluate the association between CBR3-AS1 expression
and the clinicopathological characteristics of patients with NSCLC.
Differences in CBR3-AS1 expression between tissue samples were
assessed using paired Student's t-tests. One-way analysis of
variance followed by Tukey's post-hoc test was performed to compare
differences among multiple groups. Correlations between the
expression levels of CBR3-AS1 and miR-509-3p in NSCLC tissues were
analyzed using Spearman's correlation analysis. Patient overall
survival was analyzed using Kaplan-Meier survival analysis, and the
results were compared using the log-rank test. P<0.05 was
considered to indicate a statistically significant difference.
Results
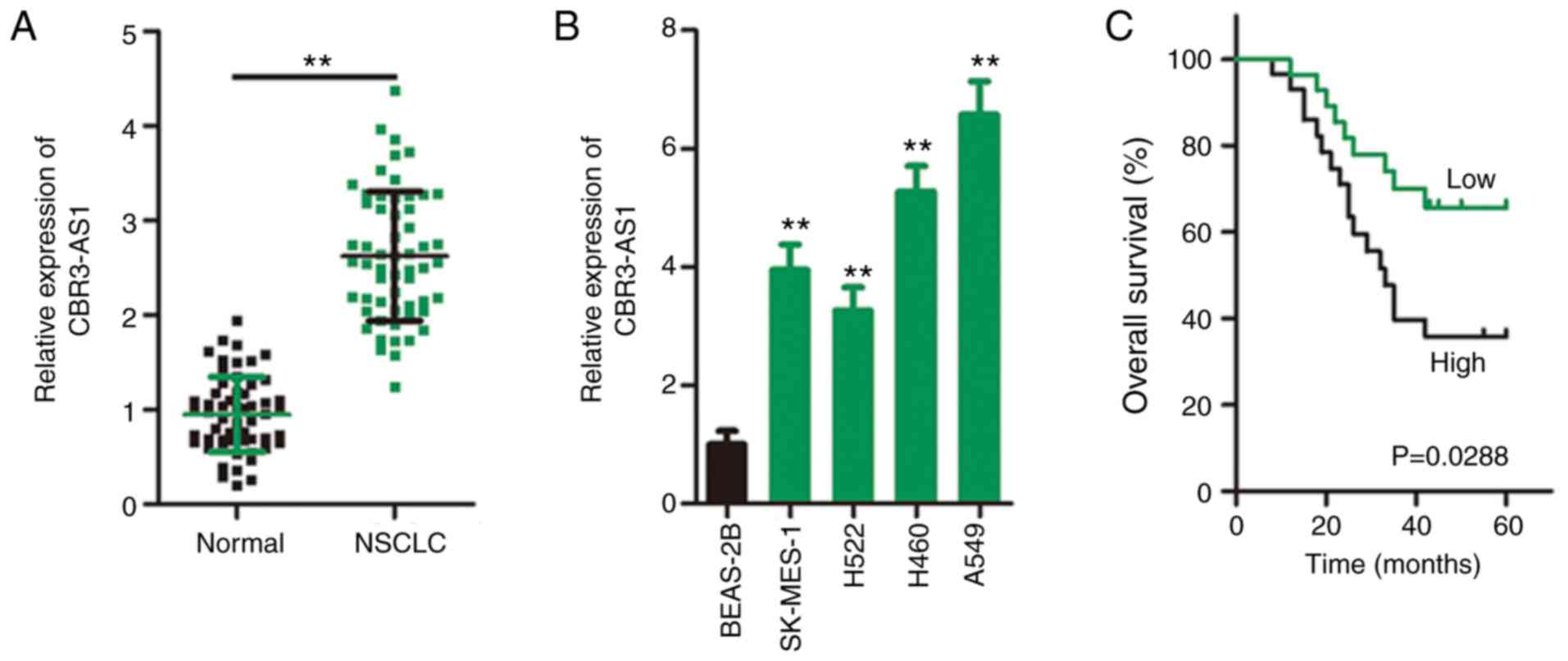
CBR3-AS1 is upregulated and closely
associated with poor prognosis in NSCLC
In total, 57 pairs of NSCLC tissues and adjacent
healthy tissues were collected during the present study, and
RT-qPCR was performed to determine CBR3-AS1 expression. CBR3-AS1
expression was prominently upregulated in NSCLC tissues compared
with that in adjacent healthy tissues (Fig. 1A). CBR3-AS1 expression was also
consistently higher in all four tested NSCLC cell lines (SK-MES-1,
H522, H460 and A549) than in the human BEAS-2B nontumorigenic
bronchial epithelium cell line (Fig.
1B).
To determine the clinical relevance of CBR3-AS1 in
NSCLC, the patients were divided into high (≥ median) or low (<
median) CBR3-AS1 expression groups based on the median CBR3-AS1
level of their NSCLC tissue samples. χ2 analysis
revealed that among the 57 patients, increased CBR3-AS1 expression
was closely correlated with larger tumor size (P=0.033), advanced
TNM stage (P=0.014), and increased incidence of lymph node
metastasis (P=0.024; Table I).
Furthermore, Kaplan-Meier survival analysis revealed a
significantly shorter overall survival time in NSCLC patients
exhibiting high CBR3-AS1 expression than in those with low CBR3-AS1
expression (Fig. 1C; P=0.0288).
Collectively, these findings indicate aberrant CBR3-AS1 expression
in NSCLC, which is strongly correlated with tumorigenesis and
progression.
 | Table I.Correlations between CBR3-AS1 and the
clinicopathological characteristics of 57 patients with NSCLC. |
Table I.
Correlations between CBR3-AS1 and the
clinicopathological characteristics of 57 patients with NSCLC.
|
| CBR3-AS1
expression |
|
|---|
|
|
|
|
|---|
| Clinicopathological
characteristic | High | Low | P-value |
|---|
| Sex |
|
| 0.599 |
|
Male | 17 | 14 |
|
|
Female | 12 | 14 |
|
| Age, years |
|
| 0.792 |
|
<60 | 14 | 12 |
|
|
≥60 | 15 | 16 |
|
| Tumor size, cm |
|
| 0.033a |
|
<3 | 12 | 20 |
|
| ≥3 | 17 | 8 |
|
|
Differentiation |
|
| 0.777 |
| High
and Moderate | 8 | 9 |
|
|
Poor | 21 | 19 |
|
| TNM stage |
|
| 0.014a |
|
I+II | 13 | 22 |
|
|
III+IV | 16 | 6 |
|
| Lymph node
metastasis |
|
| 0.024a |
|
Negative | 15 | 23 |
|
|
Positive | 14 | 5 |
|
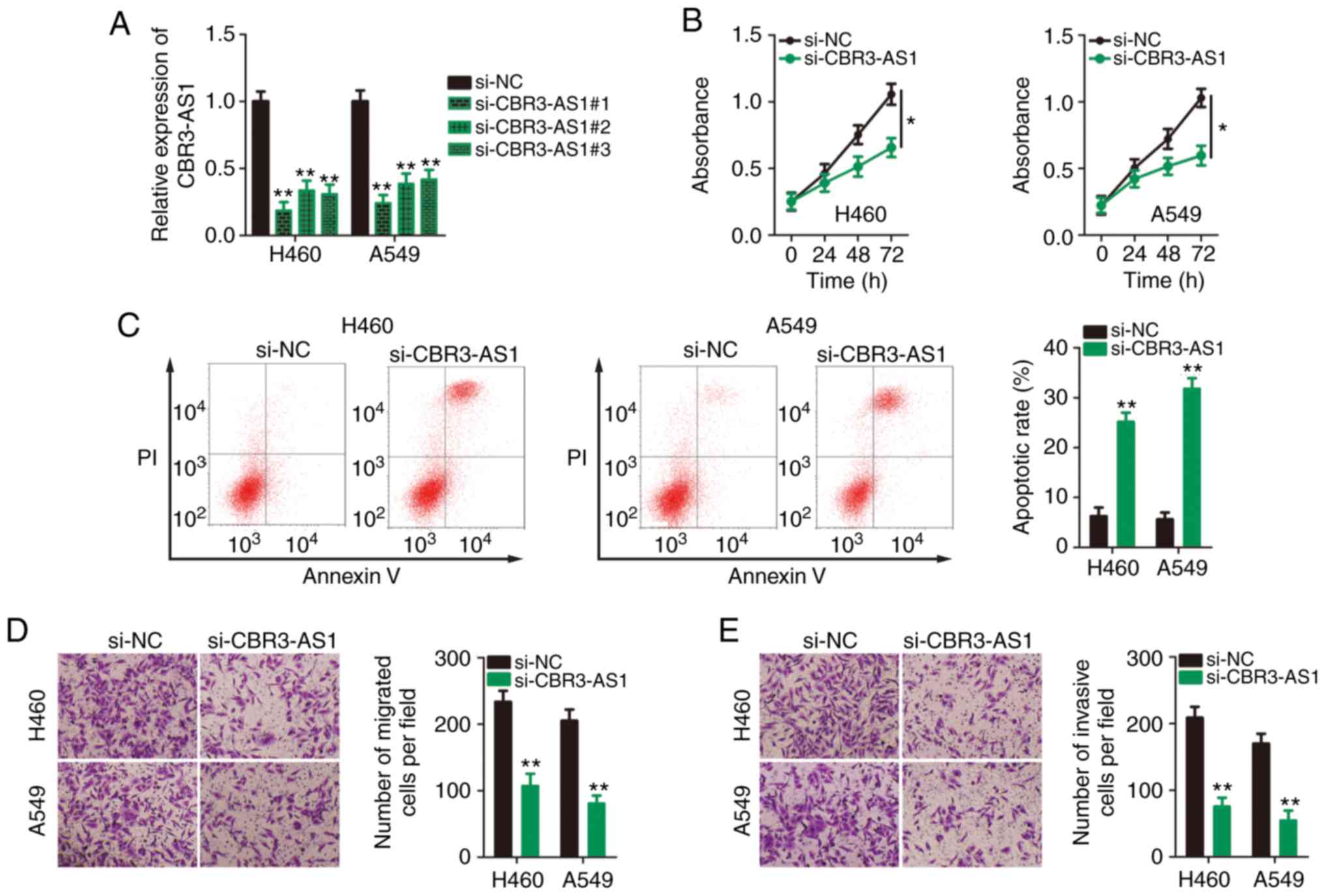
CBR3-AS1-knockdown inhibits the
aggressive phenotypes of NSCLC cells
Given the expression profile of CBR3-AS1 in NSCLC
cell lines and tissues, it was next determined whether CBR3-AS1 was
required for the malignant progression of NSCLC. To this end, three
siRNAs against CBR3-AS1 were transfected into H460 and A549 cells,
and knockdown efficiency was assessed by RT-qPCR. si-CBR3-AS1#1 was
shown to be the most effective at silencing CBR3-AS1 expression in
both H460 and A549 cells (Fig. 2A)
and was therefore used in the following functional assays, and is
henceforth referred to as si-CBR3-AS1. CCK-8 assays indicated that
CBR3-AS1-knockdown markedly inhibited proliferative ability
(Fig. 2B), and flow cytometry
revealed that CBR3-AS1-knockdown promoted apoptosis (Fig. 2C) in H460 and A549 cells.
Furthermore, the migration (Fig.
2D) and invasion (Fig. 2E)
capacities of CBR3-AS1-deficient H460 and A549 cells were hindered
compared with cells transfected with si-NC. Taken together, these
data indicate that the downregulation of CBR3-AS1 suppressed
proliferation, migration and invasiveness and promoted apoptosis in
NSCLC cells.
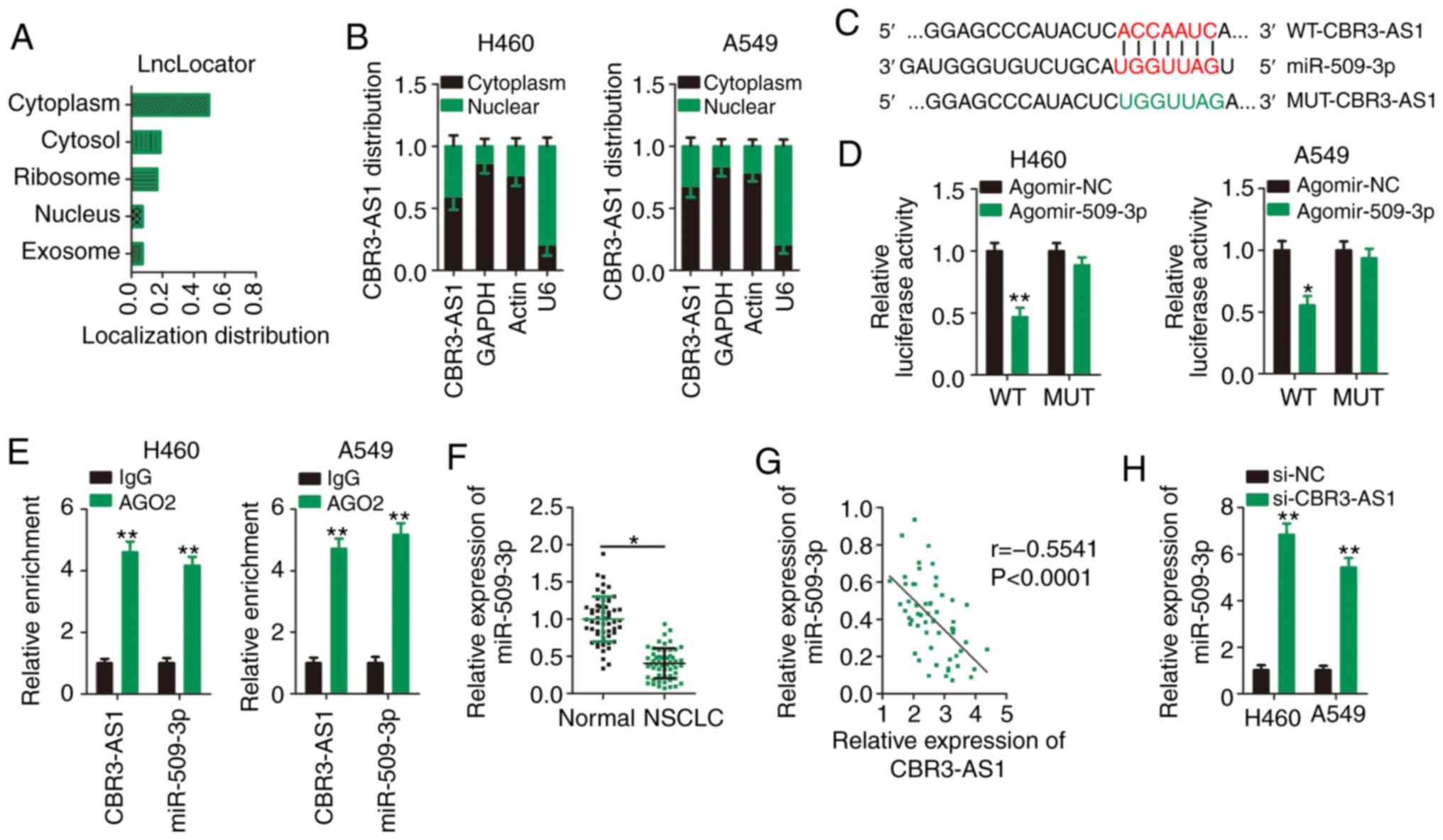
CBR3-AS1 acts as a ceRNA and sponges
miR-509-3p in NSCLC cells
To determine the molecular events implicated in the
CBR3-AS1-assocaited control of aggressive NSCLC cell phenotypes,
lncLocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/), an
lncRNA subcellular localization predictor, was used to predict the
cellular localization of CBR3-AS1. CBR3-AS1 was predicted to be
predominantly located in the cytoplasm (Fig. 3A), which was similar to the results
of nuclear-cytoplasmic fractionation (Fig. 3B). These results suggest that
CBR3-AS1 regulates the expression of target proteins at the
post-transcriptional level. An increasing number of studies has
illustrated that cytoplasmic lncRNAs function as ceRNAs or
molecular sponges for specific miRNAs, thereby liberating miRNAs
from their target RNA transcripts (23).
Bioinformatics analysis was performed to identify
miRNA(s) that could directly interact with CBR3-AS1. miR-509-3p was
predicted to contain putative complementary binding sequences for
CBR3-AS1 (Fig. 3C). Because
miR-509-3p plays a crucial role in tumorigenesis and cancer
progression (24–27), it was selected for further analysis.
As is evident from the luciferase reporter assay, miR-509-3p
overexpression successfully suppressed the luciferase activity of
the reporter vector (harboring WT miR-509-3p binding sites) in H460
and A549 cells (Fig. 3D), and this
suppression was abrogated when the miR-509-3p binding sequences
were mutated (MUT-CBR3-AS1). Next, a RIP assay was used to
determine whether CBR3-AS1 and miR-509-3p are present in the
RNA-induced silencing complex. The results indicated that compared
with the IgG control group, CBR3-AS1 and miR-509-3p were notably
enriched in the Ago2-containing magnetic beads in H460 and A549
cells (Fig. 3E).
To further elucidate the correlation between
CBR3-AS1 and miR-509-3p expression, RT-qPCR was performed to
determine the expression levels of miR-509-3p in 57 paired NSCLC
and adjacent healthy tissues. miR-509-3p expression was
downregulated in NSCLC tissues compared with adjacent healthy
tissues (Fig. 3F), which was
inversely correlated with that of CBR3-AS1 in NSCLC tissues
(Fig. 3G; r=−0.5541, P<0.0001).
Furthermore, miR-509-3p expression was increased following
CBR3-AS1-knockdown in H460 and A549 cells (Fig. 3H). Collectively, CBR3-AS1 was shown
to act as a molecular sponge that inhibits miR-509-3p expression in
NSCLC cells.
HDAC9 is a direct target gene of
miR-509-3p in NSCLC cells
To assess the role of CBR3-AS1 in NSCLC,
agomir-509-3p or agomir-NC were transfected into H460 and A549
cells. RT-qPCR analysis confirmed that miR-509-3p expression was
markedly increased in both cell lines following agomir-509-3p
transfection (Fig. 4A). CCK-8 assay
and flow cytometric assays revealed that exogenous miR-509-3p
expression significantly restrained proliferation (Fig. 4B) and promoted apoptosis (Fig. 4C) in H460 and A549 cells.
Furthermore, Transwell migration and invasion assays confirmed that
ectopic miR-509-3p expression reduced the migratory (Fig. 4D) and invasive (Fig. 4E) abilities of H460 and A549
cells.
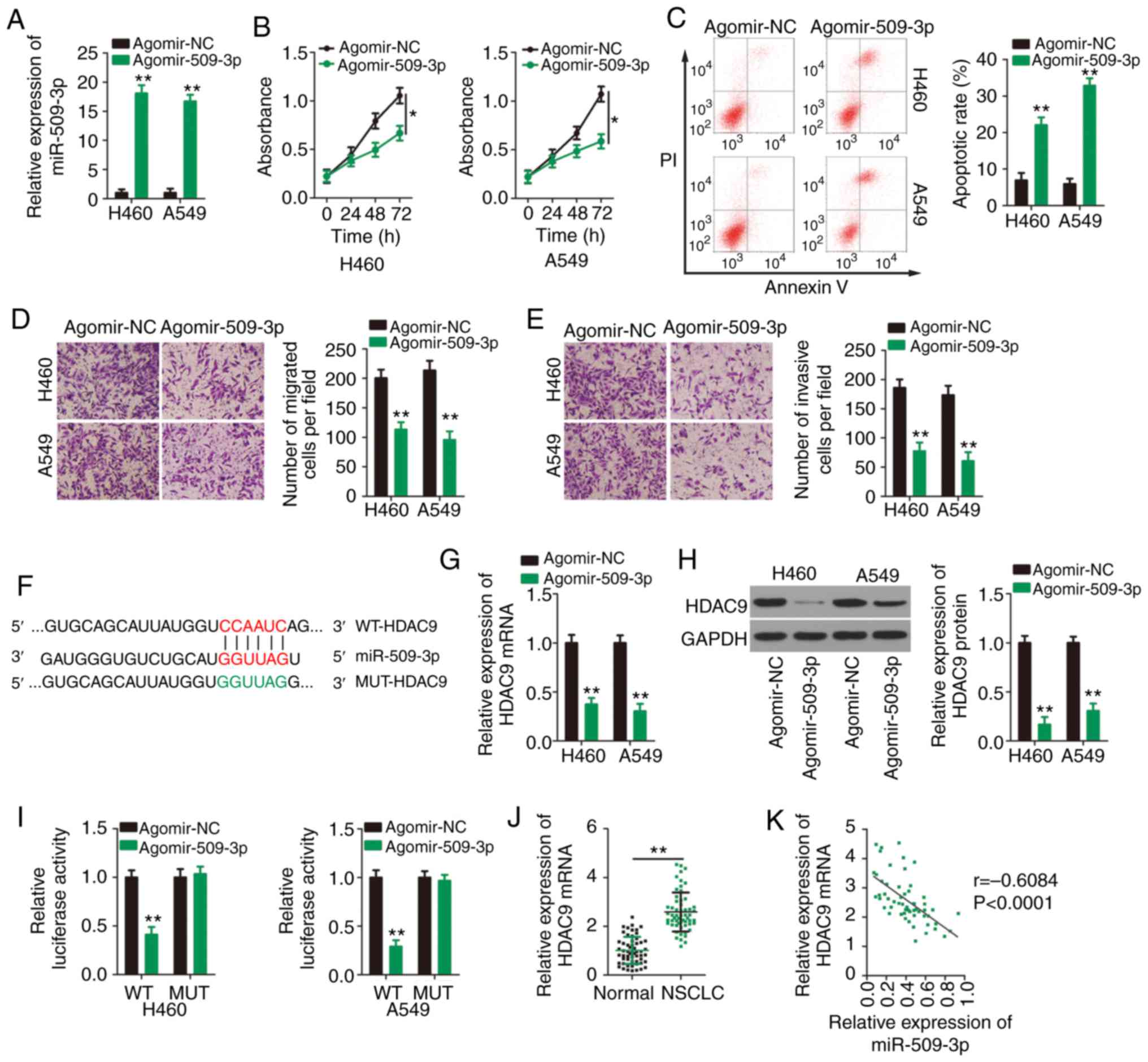 | Figure 4.miR-509-3p directly targets HDAC9 in
NSCLC cells. (A) Agomir-509 or agomir-NC was transfected into H460
and A549 cells and transfection efficiency was validated using
RT-qPCR. (B) Proliferation and (C) apoptosis of H460 and A549 cells
transfected with agomir-509 or agomir-NC were detected by Cell
Counting Kit 8 assays and flow cytometry, respectively. Transwell
(D) migration and (E) invasion assays were performed to assess the
effects of miR-509-3p overexpression on the migratory and invasive
abilities of H460 and A549 cells. Magnification, ×200. (F) WT and
MUT miR-509-3p target sites in the HDAC9 transcript. (G) RT-qPCR
and (H) western blotting were performed to detect HDAC9 mRNA and
protein expression, respectively, in H460 and A549 cells after
upregulation of miR-509-3p expression. (I) Luciferase reporter
assays were performed to determine the luciferase activities of
H460 and A549 cells after co-transfection with agomir-509 or
agomir-NC and WT-HDAC9 or MUT-HDAC9. (J) RT-qPCR for the expression
of HDAC9 mRNA in 57 pairs of NSCLC and adjacent healthy tissues.
(K) Spearman's correlation analysis was performed to analyze the
correlation between miR-509-3p and HDAC9 mRNA expression in 57
NSCLC tissues (r=−0.6084, P<0.0001). *P<0.05 and **P<0.01.
miR, microRNA; HDAC9, histone deacetylase 9; NSCLC, non-small cell
lung cancer; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; WT, wild-type; MUT, mutant. |
To investigate the mechanisms underlying the
tumor-suppressing roles of miR-509-3p in NSCLC cells, online
bioinformatics tools were used to predict the potential targets of
miR-509-3p. HDAC9 was predicted to contain a putative binding site
for miR-509-3p (Fig. 4F). RT-qPCR
and western blotting were then performed to analyze the impacts of
miR-509-3p upregulation on HDAC9 mRNA and protein expression,
respectively. Transfection with agomir-509-3p resulted in a
significant decrease in HDAC9 expression at the both mRNA (Fig. 4G) and protein (Fig. 4H) levels. Next, a luciferase
reporter assay was performed to address the direct binding between
miR-509-3p and HDAC9 in NSCLC cells. Overexpression of miR-509-3p
reduced the luciferase activity of WT-HDAC9 in H460 and A549 cells,
but not that of its mutated counterpart (Fig. 4I). Further analysis confirmed that
HDAC9 was highly expressed in NSCLC tissues compared with adjacent
healthy tissues (Fig. 4J), and a
negative expression correlation between HDAC9 mRNA and miR-509-3p
was identified in NSCLC tissues (Fig.
4K; r=−0.6084, P<0.0001). The aforementioned results
collectively confirm that HDAC9 is a direct target of miR-509-3p in
NSCLC cells.
CBR3-AS1 drives the tumorigenicity of
NSCLC cells through the miR-509-3p/HDAC9 axis
In the present study, CBR3-AS1 was found to function
as a ceRNA for miR-509-3p, and HDAC9 as a direct target of
miR-509-3p in NSCLC cells; accordingly, whether CBR3-AS1 positively
modulates HDAC9 expression in NSCLC via miR-509-3p sponging was
subsequently assessed. Silencing of CBR3-AS1 decreased HDAC9 mRNA
(Fig. 5A) and protein (Fig. 5B) expression in both H460 and A549
cells. Consistently, CBR3-AS1 expression was positively correlated
with HDAC9 mRNA expression in 57 NSCLC tissue samples (Fig. 5C; r=0.5546, P<0.0001). Rescue
experiments were then performed to determine whether miR-509-3p
sponging was required for the CBR3-AS1-mediated regulation of
HDAC9. Antagomir-509-3p was used to silence miR-509-3p expression
in H460 and A549 cells, and silencing efficiency was verified by
RT-qPCR (Fig. 5D). Antagomir-509-3p
or antagomir-NC (in combination with si-CBR3-AS1) were
co-transfected into H460 and A549 cells, and changes in HDAC9
expression were evaluated. In H460 and A549 cells,
CBR3-AS1-knockdown decreased HDAC9 mRNA (Fig. 5E) and protein (Fig. 5F) expression, which was mostly
restored after co-transfection with antagomir-509-3p.
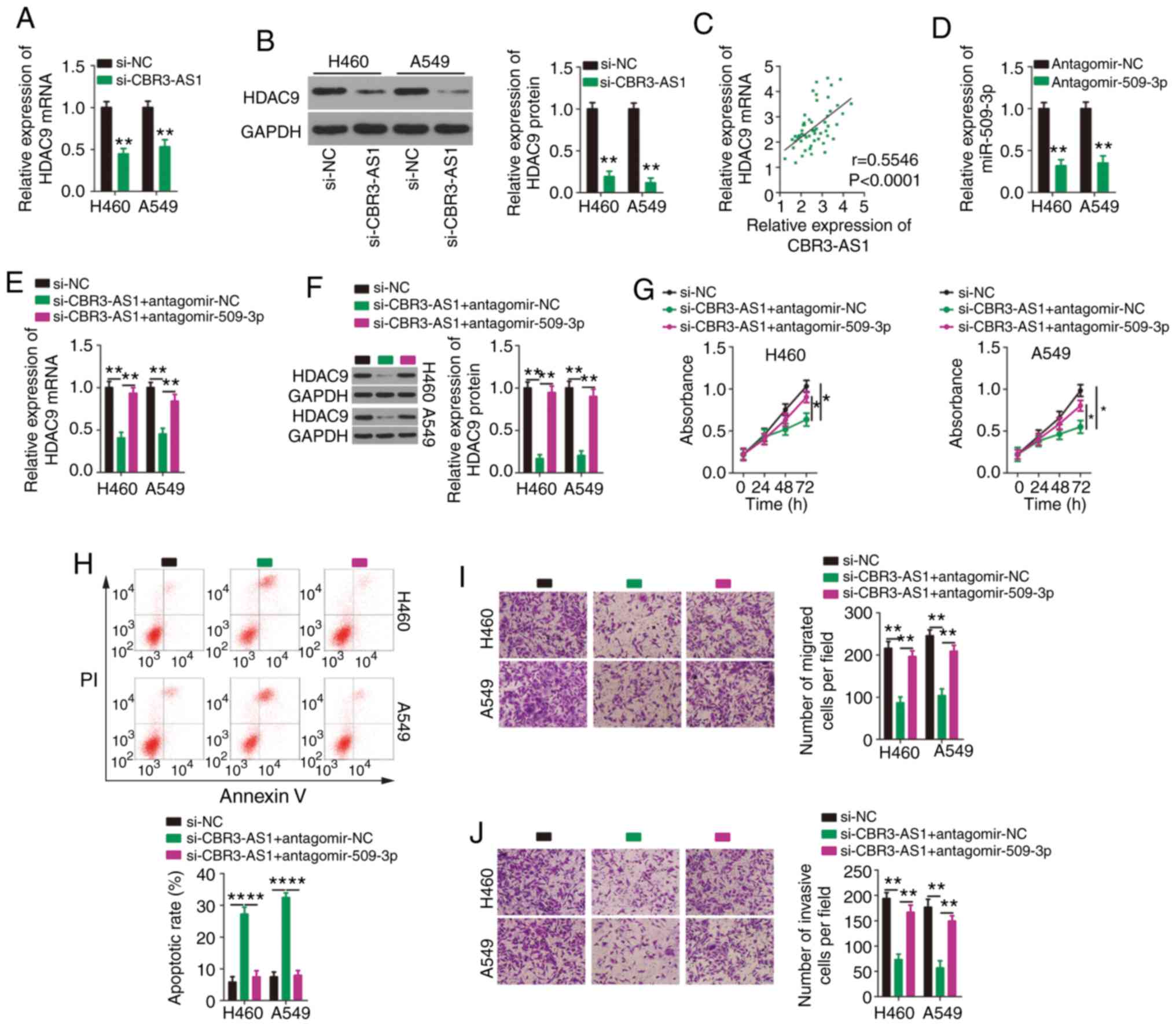 | Figure 5.Suppression of miR-509-3p expression
counteracts the effects of CBR3-AS1 knockdown on NSCLC cells. H460
and A549 cells were transfected with si-CBR3-AS1 or si-NC, and
HDAC9 mRNA and protein expression were measured by (A) RT-qPCR and
(B) western blotting, respectively. (C) Correlation between
CBR3-AS1 and HDAC9 mRNA expression in 57 paired NSCLC tissues was
examined using Spearman's correlation analysis (r=0.5546,
P<0.0001). (D) Silencing efficiency of antagomir-509-3p in H460
and A549 cells was evaluated by RT-qPCR. Antagomir-NC acted as the
control for antagomir-509-3p. CBR3-AS1-deficient H460 and A549
cells were transfected with antagomir-509-3p or antagomir-NC. (E)
RT-qPCR and (F) western blotting were used to assess changes in
HDAC4 mRNA and protein expression, respectively. (G) Proliferation
and (H) apoptosis were detected by Cell Counting Kit 8 and flow
cytometric assays, respectively. Transwell (I) migration and (J)
invasion assays were performed to determine the migratory and
invasive abilities of transfected H460 and A549 cells.
Magnification, ×200. *P<0.05, **P<0.01 and ****P<0.0001.
miR, microRNA; CBR3 antisense RNA 1; NSCLC, non-small cell lung
cancer; si(RNA), small interfering; NC, negative control; HDAC9,
histone deacetylase 9; RT-qPCR, reverse transcription-quantitative
PCR. |
CCK-8 and flow cytometric assays showed that
downregulating CBR3-AS1 expression restricted H460 and A549 cell
proliferation (Fig. 5G) and
promoted apoptosis (Fig. 5H), which
were abolished by co-transfecting cells with antagomir-509-3p.
Similarly, decreased CBR3-AS1 expression impaired the migratory
(Fig. 5I) and invasive (Fig. 5J) abilities of H460 and A549 cells,
and these effects were abrogated by synergistically knocking down
miR-509-3p expression.
Rescue experiments were also conducted to determine
whether upregulating HDAC9 reversed the malignant phenotypes of
NSCLC cells in conjunction with CBR3-AS1-knockdown. HDAC9
overexpression increased both HDAC9 mRNA (Fig. 6A) and protein (Fig. 6B) levels in H460 and A549 cells.
Next, pc-HDAC9 or pcDNA3.1 were introduced into H460 and A549 cells
in the presence of si-CBR3-AS1. Functionally, the impacts of
CBR3-AS1 inhibition on the proliferation (Fig. 6C), apoptosis (Fig. 6D), migration (Fig.6E) and invasiveness (Fig. 6E) of H460 and A549 cells were
counteracted by HDAC9 overexpression. Together, these data indicate
that the miR-509-3p/HDAC9 axis mediates the cancer-promoting
effects of CBR3-AS1 in NSCLC cells.
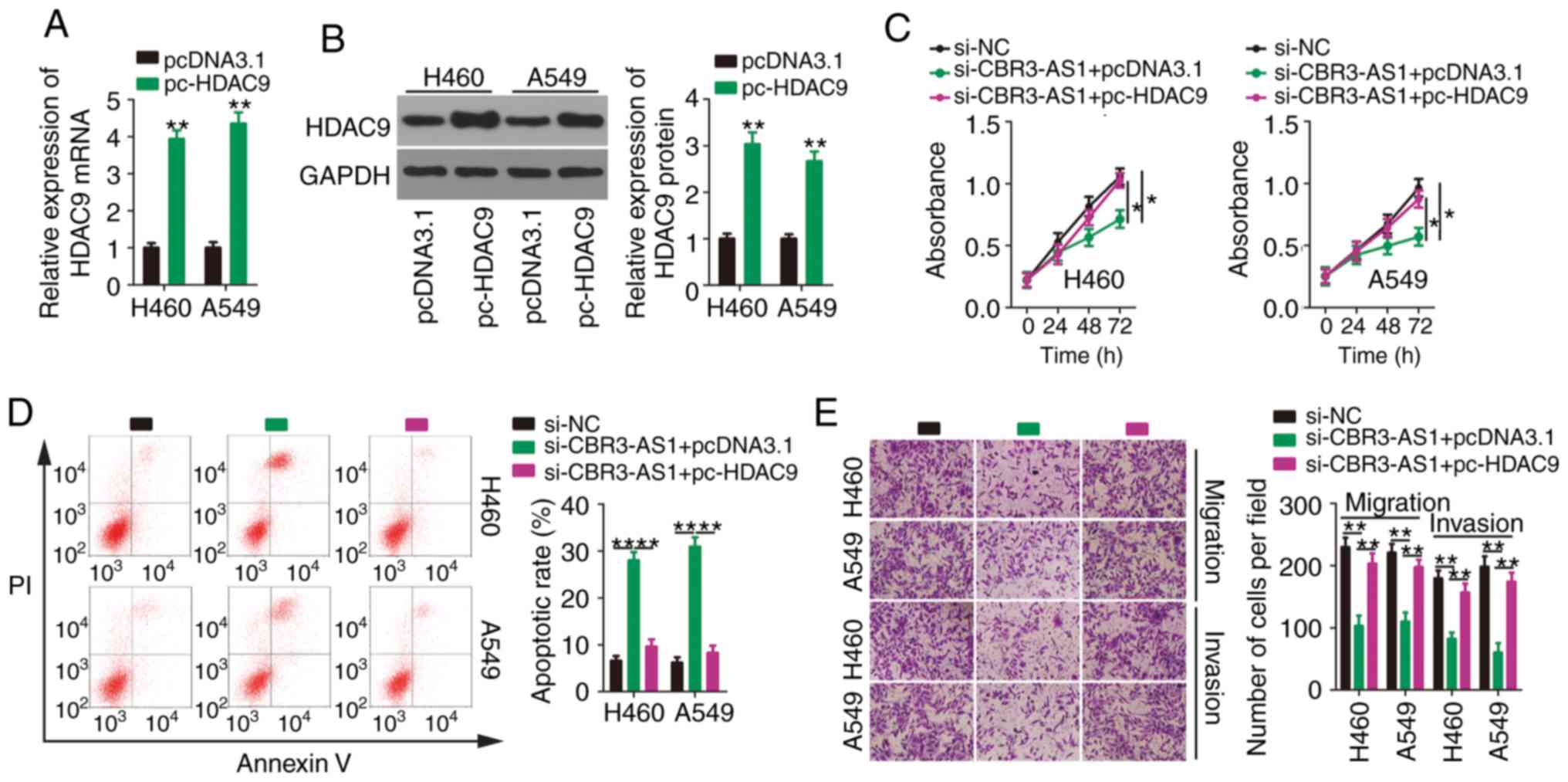 | Figure 6.Effects of CBR3-AS1-knockdown on
NSCLC cells are neutralized by HDAC9 reintroduction. (A) HDAC9 mRNA
and (B) protein expression were assessed by reverse
transcription-quantitative PCR and western blotting, respectively,
in H460 and A549 cells after pcDNA3.1 or pc-HDAC9 transfection.
si-CBR3-AS1 in parallel with pcDNA3.1 or pc-HDAC9 was
co-transfected into H460 and A549 cells. (C) Proliferation, (D)
apoptosis, (E) migration and invasiveness were evaluated by Cell
Counting Kit 8 assay, flow cytometry, and transwell migration and
invasion assays, respectively. Magnification, ×200. *P<0.05,
**P<0.01 and ****P<0.0001. CBR3 antisense RNA 1; NSCLC,
non-small cell lung cancer; HDAC9, histone deacetylase 9; si, small
interfering. |
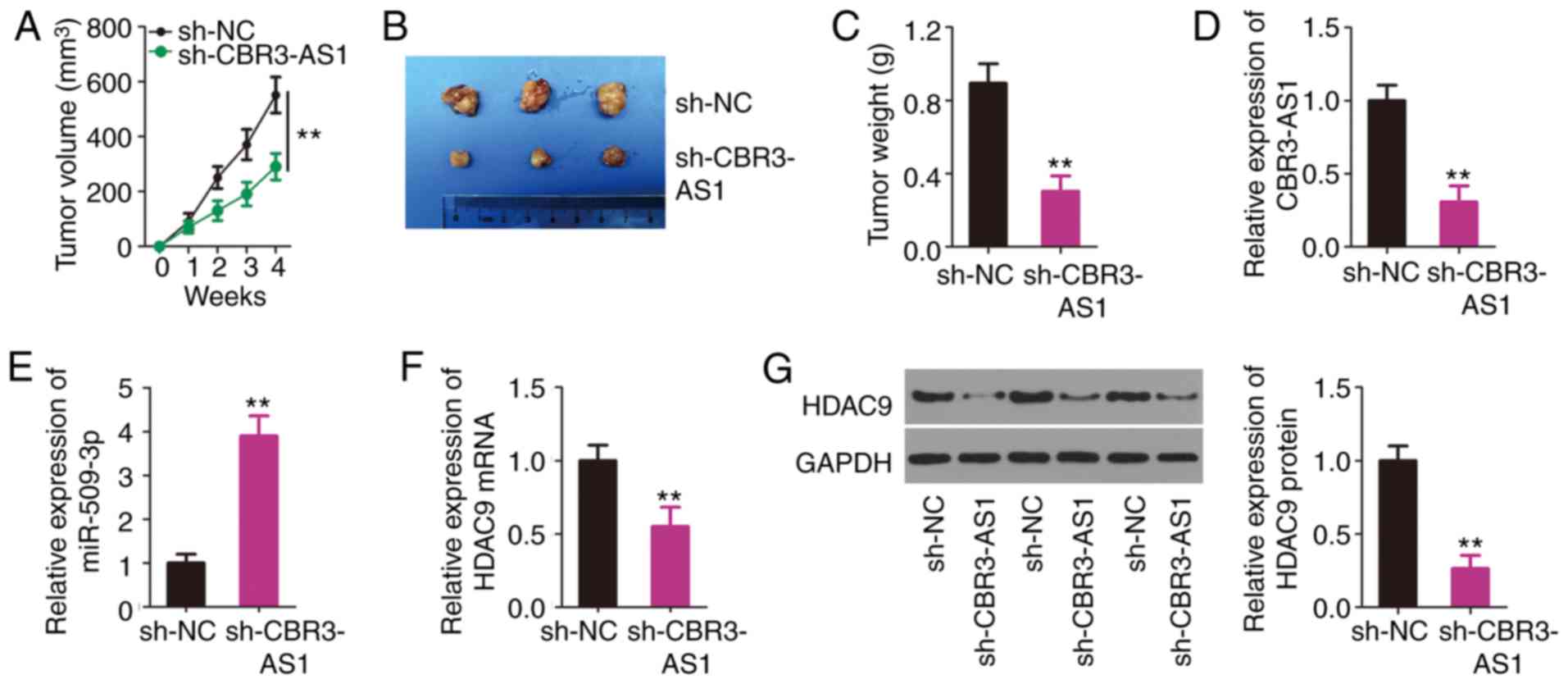
CBR3-AS1 interference impedes NSCLC
tumor growth in vivo
A tumor xenograft model was established to determine
tumor growth after a subcutaneous injection of H460 cells stably
transfected with sh-CBR3-AS1 or sh-NC. Tumor growth was inhibited
in the sh-CBR3-AS1 group compared with that in the sh-NC group
(Fig. 7A). After 4 weeks, all mice
were euthanized and the subcutaneous tumors were resected and
imaged (Fig. 7B). The average
weight of the subcutaneous tumors was found to be decreased in the
sh-CBR3-AS1 group compared with the sh-NC group (Fig. 7C). Furthermore, RT-qPCR was
performed to indicate the changes in CBR3-AS1 and miR-509-3p
expression in the sh-CBR3-AS1 and sh-NC group tumors. The results
indicated that CBR3-AS1 expression was downregulated (Fig. 7D), whereas miR-509-3p expression was
upregulated (Fig. 7E) in the
sh-CBR3-AS1 group. Furthermore, the mRNA (Fig. 7F) and protein (Fig. 7G) levels of HDAC9 were evidently
decreased in the sh-CBR3-AS1 group compared with those in the sh-NC
group. Thus, these results indicate that CBR3-AS1-knockdown
suppressed NSCLC cell tumor growth in in vivo by targeting
the miR-509-3p/HDAC9 axis.
Discussion
Over the last decade, numerous studies have reported
the aberrant expression of lncRNAs and their functions in NSCLC
tumorigenesis and progression (28–30);
thus, therapeutic strategies that target lncRNAs may hold promise
for the treatment of NSCLC. Although numerous lncRNAs have been
validated in the human genome (31), only a small number have been well
studied in relation to NSCLC. Therefore, further investigation into
the roles and relevant mechanisms of such lncRNAs is warranted. In
the present study, CBR3-AS1 expression was detected in NSCLC
tissues and its prognostic value was evaluated. The impacts of
CBR3-AS1 on the tumorigenicity of NSCLC cell lines, and the
mechanisms mediating the tumor-promoting activities of CBR3-AS1 in
NSCLC progression, were also assessed.
CBR3-AS1 expression is upregulated in osteosarcoma,
and this upregulation is closely associated with Enneking stage,
distant metastasis and histological grade (21). High CBR3-AS1 expression has also
been identified as an independent predictor of poor prognosis in
patients with osteosarcoma (21).
CBR3-AS1 is also highly expressed in breast cancer tissues and cell
lines (32); patients with breast
cancer and a high CBR3-AS1 expression level present with poorer
overall and disease-free survival outcomes than those with low
levels of CBR3-AS1 (32).
Nevertheless, the expression profile of CBR3-AS1 in NSCLC is yet to
be elucidated. In the present study, CBR3-AS1 was upregulated in
NSCLC tissues and cell lines, and high levels of CBR3-AS1 were
associated with a larger tumor size, advanced TNM stage and an
increased incidence of lymph node metastasis. The overall survival
times of NSCLC patients with high CBR3-AS1 expression were shorter
than those of patients with low CBR3-AS1 expression. These
observations suggest that the high CBR3-AS1 expression observed in
NSCLC is associated with poor postoperative prognosis in patients
with this malignancy. Thus, CBR3-AS1 may be developed as an
effective target for NSCLC diagnosis and prognosis.
Functionally, silencing CBR3-AS1 attenuates cellular
proliferation, migration and invasiveness, and promotes apoptosis
in osteosarcoma (21). In breast
cancer, CBR3-AS1 exerts an oncogenic role by regulating cell
proliferation, colony formation and apoptosis in vitro, and
tumor growth in vivo (32).
To better comprehend the detailed function(s) of CBR3-AS1 in NSCLC,
the impacts of CBR3-AS1-knockdown on NSCLC cells were determined
using a series of functional experiments in vitro and in
vivo. CBR3-AS1-knockdown resulted in an obvious decrease in
cellular proliferation, migration and invasiveness, as well as an
increase in apoptosis in vitro. Moreover, a tumor xenograft
model indicated that CBR3-AS1 inhibition hindered the
tumorigenicity of NSCLC cells in vivo. These results
collectively suggest that CBR3-AS1 is a potential target for NSCLC
anticancer therapy.
The functions of lncRNAs are largely determined by
their subcellular localization (33). To elucidate the functions of
CBR3-AS1 in NSCLC, an lncRNA subcellular localization predictor
(lncLocator) and nuclear-cytoplasmic fractionation were applied to
identify the localization of CBR3-AS1. The results confirmed
CBR3-AS1 as a cytoplasmic lncRNA and suggested that CBR3-AS1
functions as a ceRNA and sponges miRNA, thereby liberating miRNAs
from their binding sites on target mRNAs. Clarification of the
miRNA/mRNA axis will advance our understanding of the mechanisms by
which CBR3-AS1 promotes the oncogenic potential of NSCLC cells.
Bioinformatics analysis was performed to predict the
miRNAs that interact with CBR3-AS1. Among the candidates
identified, miR-509-3p was selected for further experimental
verification given its crucial roles in tumorigenesis and cancer
progression (24–27). Luciferase reporter and RIP assays
confirmed miR-509-3p as a target of CBR3-AS1 in NSCLC cells. In
addition, miR-509-3p was weakly expressed and inversely correlated
with CBR3-AS1 expression in NSCLC tissues. Furthermore, miR-509-3p
expression was increased in NSCLC cells after CBR3-AS1 silencing.
Mechanistic investigations identified HDAC9 as a direct target of
miR-509-3p in NSCLC cells; HDAC9 expression was also positively
regulated by CBR3-AS1, and these regulatory actions were exerted
through miR-509-3p sponging. These results validate that a ceRNA
regulatory pathway involving CBR3-AS1, miR-509-3p and HDAC9 exists
in NSCLC cells.
HDAC9, a member of the histone deacetylase family,
was discovered to play crucial roles in a number of malignant
characteristics of cancer progression. HDAC9 is highly expressed in
NSCLC and is associated with adverse clinicopathological
characteristics and shorter overall patient survival time (34). Functionally, HDAC9 exerts oncogenic
activities in NSCLC cells by promoting proliferation, colony
formation, migration and invasiveness, and by inducing apoptosis.
In the present study, rescue experiments demonstrated that
increasing the output of the miR-509-3p/HDAC9 axis counteracted
CBR3-AS1 depletion-induced inhibitory impacts on NSCLC cells.
Jointly, CBR3-AS1, miR-509-3p and HDAC9 constitute an interactive
regulatory network that exerts tumor-promoting effects in NSCLC
tumorigenesis and progression. Therefore, the
CBR3-AS1/miR-509-3p/HDAC9 pathway may be an effective target for
the improved control of NSCLC.
The present study has two limitations. Firstly,
CBR3-AS1 and miR-509-3p expression data in NSCLC from The Cancer
Genome Atlas (TCGA) database were not analyzed. Secondly, the
correlation between CBR3-AS1 and miR-509-3p expression in NSCLC was
not examined using data from TCGA database, limitations that will
be addressed in the near future. However, to the best of our
knowledge, the present study is the first to highlight the
cancer-promoting effects of CBR3-AS1 in NSCLC cells, both in
vitro and in vivo. Mechanistically, CBR3-AS1 was found
to function as a ceRNA that sponges miR-509-3p, thereby increasing
HDAC9 expression. These findings may positively impact the
development of novel targeted drugs and the enrichment of
therapeutic strategies for NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YG and LC provided substantial contributions to the
conception and design of the study. YC and JY performed flow
cytometry, Transwell migration and invasion assays, tumor xenograft
model construction, and RNA immunoprecipitation. All statistical
analysis was executed by LC. YG and LC drafted and critically
revised the manuscript for important intellectual content. All
authors read and approved the final draft.
Ethics approval and informed consent
The present study was approved by the Human Ethics
Committee of Weifang People's Hospital. The study was performed in
accordance with the Declaration of Helsinki, and written informed
consent was obtained from all participants. Animal experimental
procedures were approved by the Institutional Animal Care and Use
Committee of Weifang People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shin JY, Yoon JK and Marwaha G: Progress
in the treatment and outcomes for early-stage non-small cell lung
cancer. Lung. 196:351–358. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vargas AJ and Harris CC: Biomarker
development in the precision medicine era: Lung cancer as a case
study. Nat Rev Cancer. 16:525–537. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Inage T, Nakajima T, Yoshino I and
Yasufuku K: Early lung cancer detection. Clin Chest Med. 39:45–55.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen J, Wang R, Zhang K and Chen LB: Long
non-coding RNAs in non-small cell lung cancer as biomarkers and
therapeutic targets. J Cell Mol Med. 18:2425–2436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bian C, Yuan L and Gai H: A long
non-coding RNA LINC01288 facilitates non-small cell lung cancer
progression through stabilizing IL-6 mRNA. Biochem Biophys Res
Commun. 514:443–449. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Luo X, Liu Y, Han G and Sun D:
Long noncoding RNA RMRP promotes proliferation and invasion via
targeting miR-1-3p in non-small-cell lung cancer. J Cell Biochem.
120:15170–15181. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tang H, Han X, Li M, Li T and Hao Y:
Linc00221 modulates cisplatin resistance in non-small-cell lung
cancer via sponging miR-519a. Biochimie. 162:134–143. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang L, Hu J, Li J, Yang Q, Hao M and Bu
L: Long noncoding RNA LINC-PINT inhibits non-small cell lung cancer
progression through sponging miR-218-5p/PDCD4. Artif Cells Nanomed
Biotechnol. 47:1595–1602. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu X, Duan L, Liu H and Zhang L: Long
noncoding RNA LINC01296 induces non-small cell lung cancer growth
and progression through sponging miR-5095. Am J Transl Res.
11:895–903. 2019.PubMed/NCBI
|
|
14
|
Solé C and Lawrie CH: MicroRNAs and
metastasis. Cancers (Basel). 12:962019. View Article : Google Scholar
|
|
15
|
Deng H, Xie C, Ye Y and Du Z:
MicroRNA-1296 expression is associated with prognosis and inhibits
cell proliferation and invasion by Wnt signaling in non-small cell
lung cancer. Oncol Lett. 19:623–630. 2020.PubMed/NCBI
|
|
16
|
Zhou X, Liu S, Liu J, Zhang Z, Mao X and
Zhou H: MicroRNA-130a enhances the killing ability of natural
killer cells against non-small cell lung cancer cells by targeting
signal transducers and activators of transcription 3. Biochem
Biophys Res Commun. 523:481–486. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan L, Zhang Y, Li K, Wang M, Li J, Qi Z,
Wu J, Wang Z, Ling L, Liu H, et al: miR-593-5p inhibit cell
proliferation by targeting PLK1 in non small cell lung cancer
cells. Pathol Res Pract. 216:1527862020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qu CX, Shi XC, Zai LQ, Bi H and Yang Q:
LncRNA CASC19 promotes the proliferation, migration and invasion of
non-small cell lung carcinoma via regulating miRNA-130b-3p. Eur Rev
Med Pharmacol Sci. 23 (Suppl 3):S247–S255. 2019.
|
|
19
|
Wang X, Yin H, Zhang L, Zheng D, Yang Y,
Zhang J, Jiang H, Ling X, Xin Y, Liang H, et al: The construction
and analysis of the aberrant lncRNA-miRNA-mRNA network in non-small
cell lung cancer. J Thorac Dis. 11:1772–1778. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li H, Guo X, Li Q, Ran P, Xiang X, Yuan Y,
Dong T, Zhu B, Wang L, Li F, et al: Long non-coding RNA 1308
promotes cell invasion by regulating the miR-124/ADAM 15 axis in
non-small-cell lung cancer cells. Cancer Manag Res. 10:6599–6609.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Meng W and Cui H: LncRNA CBR3-AS1
predicts unfavorable prognosis and promotes tumorigenesis in
osteosarcoma. Biomed Pharmacother. 102:169–174. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang L, Cho KB, Li Y, Tao G, Xie Z and Guo
B: Long noncoding RNA (lncRNA)-mediated competing endogenous RNA
networks provide novel potential biomarkers and therapeutic targets
for colorectal cancer. Int J Mol Sci. 20:57582019. View Article : Google Scholar
|
|
24
|
Patil SL, Palat A, Pan Y, Rajapakshe K,
Mirchandani R, Bondesson M, Yustein JT, Coarfa C and Gunaratne PH:
MicroRNA-509-3p inhibits cellular migration, invasion, and
proliferation, and sensitizes osteosarcoma to cisplatin. Sci Rep.
9:190892019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Niu L, Ni H, Hou Y, Du Q and Li H:
miR-509-3p enhances platinum drug sensitivity in ovarian cancer.
Gene. 686:63–67. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen W, Du J, Li X, Su J, Huang Y, Ding N,
Zhang M and Jiang S: miR-509-3p promotes cisplatin-induced
apoptosis in ovarian cancer cells through the regulation of
anti-apoptotic genes. Pharmacogenomics. 18:1671–1682. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun J, Li J, Zhang W, Zhang J, Sun S, Li
G, Song H and Wan D: MicroRNA-509-3p inhibits cancer cell
proliferation and migration via upregulation of XIAP in gastric
cancer cells. Oncol Res. 25:455–461. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang L, Wang T, Zhang Y, Zhang J, Zhao H,
Wang H, Wu Y and Liu K: Long non-coding RNA AWPPH promotes
postoperative distant recurrence in resected non-small cell lung
cancer by upregulating transforming growth factor beta 1 (TGF-β1).
Med Sci Monit. 25:2535–2541. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bai Y, Zhang G, Chu H, Li P and Li J: The
positive feedback loop of lncRNA DANCR/miR-138/Sox4 facilitates
malignancy in non-small cell lung cancer. Am J Cancer Res.
9:270–284. 2019.PubMed/NCBI
|
|
30
|
Wang D and Hu Y: Long non-coding RNA PVT1
competitively binds microRNA-424-5p to regulate CARM1 in
radiosensitivity of non-small-cell lung cancer. Mol Ther Nucleic
Acids. 16:130–140. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu L, Zhu H, Gao F, Tang Y, Zhu Y, Sun Z
and Wang J: Upregulation of the long non-coding RNA CBR3-AS1
predicts tumor prognosis and contributes to breast cancer
progression. Gene X. 2:1000142019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ogunwobi OO and Kumar A: Chemoresistance
mediated by ceRNA networks associated with the PVT1 locus. Front
Oncol. 9:8342019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma Z, Liu D, Di S, Zhang Z, Li W, Zhang J,
Xu L, Guo K, Zhu Y, Li X, et al: Histone deacetylase 9
downregulation decreases tumor growth and promotes apoptosis in
non-small cell lung cancer after melatonin treatment. J Pineal Res.
67:e125872019. View Article : Google Scholar : PubMed/NCBI
|