Introduction
Pulmonary malignancies, including lung and bronchus
cancer, rank first and second among different cancer types in terms
of mortality and morbidity, respectively, in both men and women
(1,2). Furthermore, ~85% of lung cancer cases
are categorized as non-small cell lung cancer (NSCLC), while the
remaining 15% are classified as SCLC (3). Although diagnostic methods and
therapeutic strategies based on traditional surgical excision,
chemotherapy and chest radiotherapy have continuously improved, the
prognosis of lung carcinoma remains at 15% for an overall 5-year
survival (4). Therefore, an
increased understanding of the malignant progression and studies on
novel therapeutic targets for the improved management of this
disease are essential.
Long non-coding RNAs (lncRNAs) are >200
nucleotides in length and have little or no protein coding capacity
(5). The mechanisms via which
lncRNAs regulate gene expression are diverse and include regulating
the transcription of target genes, functioning as transcriptional
precursors of small RNAs, generating different splice variants via
regulating mRNA splicing patterns, modulating protein activity and
subcellular localization and scaffolding for the assembly of
multiple component complexes (6,7).
In recent years, previous studies have reported that
various human cancer types exhibit lncRNAs dysfunction, and these
lncRNAs are involved in different aspects of pathogenesis such as
the proliferation, metastasis and apoptosis of tumor cells
(8,9). In lung cancer, lncRNA
metastasis-associated lung adenocarcinoma transcript 1 is found to
be upregulated in patients with advanced lung adenocarcinoma, and
may serve as a prognostic marker to predict the survival outcome of
patients with cancer (10,11). lncRNA HOX transcript antisense RNA
is also highly expressed in lung cancer (12), and it enhances the aggressiveness of
lymph node metastasis and indicates a short disease-free survival
in patients with NSCLC (13,14).
Furthermore, studies have shown that the expression of lncRNA
Urothelial carcinoma-associated 1 is significantly upregulated in
NSCLC, and may induce resistance to treatment of EGFR-tyrosine
kinase inhibitors by activating the AKT/mTOR pathway (15,16).
lncRNA RP11-284F21.9 was primarily discovered in a Pan-cancer
transcriptomic analysis (17).
lncRNA RP11-284F21.9 exists as a cluster of three annotated lncRNAs
(RP11-284F21.9/.10/.7) antisense to brevican, which is a
proteoglycan linked to invasiveness in glioma but lacks expression
in squamous cell lung carcinomas (18). However, the specific function and
the underlying mechanism of RP11-284F21.9 in lung carcinoma remain
unknown.
To the best of our knowledge, the present study
demonstrated for the first time that lncRNA RP11-284F21.9 was
significantly upregulated in lung carcinoma tissues and cell lines,
and was involved in the carcinogenesis of lung cancer. Together
with microRNA (miRNA/miR)-627-3p and cell division cycle and
apoptosis regulator 1 (CCAR1), the regulatory axis of
RP11-284F21.9/miR-627-3p/CCAR1 exists both in the lung carcinoma
cells in vitro and in the tumor growth model in vivo.
The present study aimed to investigate RP11-284F21.9 function in
lung carcinoma and demonstrate the molecular mechanism underlying
the regulation process via the RP11-284F21.9/miR-627-3p/CCAR1
axis.
Materials and methods
Tissue samples and cell lines
Between May 2017 and Jan 2019, paired tumor and
adjacent healthy tissues were isolated from 13 patients with lung
carcinoma (age range, 35–57 years; nine male patients; four female
patients) who were diagnosed and treated in First Affiliated
Hospital of Xi'an Jiaotong University. The samples were dissected
during the surgery and immediately flash-frozen in liquid nitrogen
and transferred to −80°C storage for further extraction of both RNA
and protein. All the tissue samples were obtained with written
informed consent from the patients. The protocol was approved by
The First Affiliated Hospital of Xi'an Jiaotong University
(approval no. 201810053).
A normal lung epithelial cell line (BEAS-2B) and
lung carcinoma cell lines NCI-H460, NCI-H1299, and A549 were
purchased from American Type Culture Collection (ATCC) and cultured
according to the ATCC guidelines. 293T cells were purchased from
Procell Life Science&Technology Co., Ltd., and cultured in DMEM
supplemented with 10% FBS (cat. no. 30-2020; ATCC) and 1X
Penicillin-streptomycin (Thermo Fisher Scientific, Inc.). BEAS-2B
cells were cultured in bronchial epithelial growth medium (BEGM;
cat. no. CC-3170; Clonetics Corporation), according to the
manufacturer's instructions. NCI-H460 and NCI-H1299 cells were
cultured in RPMI-1640 medium (cat. no. 30-2001; ATCC), and A549
cells in F12K medium (cat. no. 30-2004; ATCC) supplemented with 10%
FBS (cat. no. 30-2020, ATCC) and 1X Penicillin-streptomycin (Thermo
Fisher Scientific, Inc.). All cells were culture at 37°C with 5%
CO2.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA from both tissue samples and cell lines
were extracted using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). For each sample, 500 ng total RNA was
reverse transcribed to synthesize the first-strand cDNA using the
PrimeScript RT reagent kit (Takara Bio, Inc.).
cDNA samples were diluted 40 times to perform the
RT-qPCR using SYBR Premix Ex Taq (Takara Bio, Inc.) on a CFX96
real-time PCR detection system (Bio-Rad Laboratories, Inc.).
Expression levels of mRNAs, lncRNAs and miRNAs were normalized to
GAPDH. The primers used for RT-qPCR analyses were as follows: GAPDH
forward, 5′-AACGACCCCTTCATTGACC-3′ and reverse,
5′-TCCACGACATACTCAGCACC-3′; RP11-284F21.9 forward,
5′-AGGATTGGCACTCACTTCGG-3′ and reverse, 5′-TCTCTCACCACGTCTGGTCT-3′;
and CCAR1 forward, 5′-CTGATGGCTAGCCCTAGTATGGA-3′ and reverse,
5′-TGCCTTTCATGCCCACTAAAA−3′. The temperature protocol used to
perform RT was 42°C for 1 h followed by 70°C for 10 min. Thermal
conditions of PCR reactions were: Initial denaturation at 95°C for
1 min, followed by 40 cycles for 20 sec at 95°C and 60 sec at 60°C.
The mRNA expression levels were determined using the
2−ΔΔCq method (19).
Oligonucleotides and cell
transfection
The small interfering RNA (siRNA) synthetic negative
control (si-NC), RP11-284F21.9 siRNAs (si-RP11-284F21.9), miR-NC,
miR-627-3p mimics and miR-627-3p inhibitor were purchased from
Shanghai GenePharma Co., Ltd. All primer sequence information is
presented in Table I. At a density
of 2×105 cells/well, the cells were plated in 6-well
plates 24 h before transfection and were transfected at 60%
confluency. All of the oligonucleotides were transfected at a final
concentration of 50 nM using Lipofectamine® 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instruction. Cells were collected at 24 h
post-transfection for subsequent experiments.
 | Table I.Sequence of siRNAs and miRNA mimics
and inhibitors. |
Table I.
Sequence of siRNAs and miRNA mimics
and inhibitors.
|
Oligonucleotides | Sequence
(5′→3′) |
|---|
| si-NC |
UUCUCCGAACGUGUCACGUTT |
|
si-RP11-284F21.9 |
UAUUGGCACCAAGGAUAGC |
| miR-NC |
UCGUUAAUCGGCUAUAAUACGC |
| miR-627-3p
mimics |
UCUUUUCUUUGAGACUCACU |
| miR-627-3p
inhibitor |
UCUUUUCUUUGAGACUCACU |
Cell Counting Kit (CCK)-8 assay and
EdU labeling of proliferating cells
A CCK-8 was used for cell proliferation assay, the
cells were seeded into 96-well plates (2×103 cells/well)
and observed for 1, 2, 3 and 4 days, or indicated time points,
following the manufacturer's instructions (Dojindo Molecular
Technologies, Inc.). The optical density was measured at 450 nm
using a spectrophotometer (Thermo Fisher Scientific, Inc.).
For the EdU assay, cells were incubated with 10 µM
EdU (cat. no. ab219801; Abcam) for 2 h at 37°C and fixed with 4%
formaldehyde at room temperature for 10 min. After a brief washing
with PBS, click reagent was added into each well and incubated in
the dark for 30 min at room temperature. Followed by PBS washing,
the cells were stained with 1 µg/ml DAPI at room temperature for 10
min. Images were captured using a fluorescence microscope (Nikon
Corporation) and measured using Adobe Photoshop 6.0 software (Adobe
Systems, Inc.). The EdU labeled cells were analyzed with MoFlo
Astrios (Beckman-Coulter, Inc.; Magnification, ×200).
Transwell assay and flow cytometry
measurement of cell apoptosis
Transwell assays were performed with a coating of
Matrigel (BD Biosciences) mixed with culture medium mixed at 1:1
ratio at 37°C for 1 h. A total of 1×105 cells in 200 µl
serum-free medium were added to the upper layer of the Transwell
chambers (8 µm pore size; Corning, Inc.) and cultured for 24 h. The
lower chamber contained the culture medium with 10% FBS. The
migrated cells were fixed with 4% paraformaldehyde for 30 min at
room temperature, stained with 0.1% crystal violet for 20 min at
room temperature and images of six randomly selected fields in each
well were captured under a light microscope (Magnification,
×200).
Cellular apoptosis was detected using the Apoptosis
Detection kit (cat. no. KGF001; Nanjing KeyGen Biotech Co., Ltd.)
according to the manufacturer's instructions. Cells were stained
with fluorescein isothiocyanate-conjugated annexin V and PI. After
incubated for 15 min at 37°C in the dark, 400 µl 1X Binding Buffer
was added to each tube and stained cells were analyzed using BD
FACS Canto II flow cytometry (FACS Calibur; BD Biosciences). Data
were analyzed using FlowJo software version 8.8.6 (Tree Star,
Inc.).
Luciferase reporter assay
The RP11-284F21.9 wild-type (wt) or mutant (mut)
3′-untranslated region (3′-UTR), and CCAR1 wt or mut 3′-UTR
sequences were cloned into the pmirGLO plasmid (Youbio; http://www.youbio.cn/; cat. no. VT1439). The vectors
(2 µg/ml) were co-transfected with miR-NC or miR-627-3p mimic (50
nM) and Renilla plasmids (5 ng/well), used as an internal
control, into cells seeded in a 48-well plate
(1×104/well) using Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Cell lysates
were collected at 48 h after transfection and the luciferase
activities were detected with the Dual-Luciferase Reporter Assay
system (Promega Corporation) according to the manufacturer's
instructions.
Western blotting
Cell were lysed using RIPA lysis buffer
(Sigma-Aldrich; Merck KGaA) and protein concentrations were
assessed with the BCA Protein Assay kit according to the
manufacturer's instructions (Beyotime Institute of Biotechnology,
Shanghai, China). Equal amounts (25 µg) of cell protein lysates
were loaded and separated by 10% SDS-PAGE, transferred to a PVDF
membrane and blocked with 5% non-fat milk at room temperature for 2
h. The membranes were then incubated with CCAR1 primary antibody
(1:1,000; cat. no. ab70243; Abcam) overnight at 4°C, followed by
incubation with goat anti-mouse or goat anti-rabbit IgG-horseradish
peroxidase conjugate secondary antibodies (1:5,000; cat. no.
ab205718; Abcam) at room temperature for 2 h. GAPDH (1:2,000; cat.
no. ab181602; Abcam) was used as loading control. The signals were
detected using the ECL system (Protein Simple) according to the
manufacturer's instructions.
In vivo tumorigenicity analysis in mice. Male
BALB/c nude mice (age, 8 weeks; weight, 21–25 g) were obtained from
Beijing Vital River Laboratory Animal Technology Co., Ltd., and
housed at a room temperature of 25°C with a 12 h light/dark cycle.
The mice were maintained in an individually ventilated cage system
under specific pathogen-free conditions (temperature; 25°C;
humidity: 55%), and fed with sterile food and water (free access).
To evaluate the effect of RP11-284F21.9 knockdown on the growth of
lung carcinoma in vivo, 5×106 si-NC or
si-RP11-284F21.9 treated NCI-H1299 cells in 200 µl serum-free
medium were subcutaneously injected into each mouse (n=5 per group)
under anesthesia, which was induced by 5% isoflurane and maintained
by 2% isoflurane (flow rate, 1l/min). The animals were monitored
daily and the following criteria for humane endpoint was used:
Severe tumor burden (>20 mm in diameter), difficulty breathing,
significant body-weight loss and clinical signs such as
prostration, hypothermia and significant abdominal distension.
Tumors were measured on days 1, 5, 9 and 13, and the volumes were
calculated using the formula: a × b2/2 [the largest
diameter (a) and the smallest diameter (b)]. Then, 2 weeks after
inoculation, the mice were euthanized by CO2 inhalation
(CO2 flow rate, 10% of cage volume) and the death of
animals were confirmed by cessation of heartbeat. The xenografts
were imaged and weighed.
The total RNA was then extracted from the xenografts
as aforementioned. Animal care and study were approved by the
Institutional Animal Care and Use Committee of The First Affiliated
Hospital of Xi'an Jiaotong University (approval no. 201902013).
Target prediction
Potential target miRNAs of RP11-284F21.9 were
predicted using LncBase V2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index).
The target genes of miR-627-3p were predicted using three
bioinformatics algorithms: TargetScanV7.2 (http://www.targetscan.org/vert_72/) and miRDB
(http://www.mirdb.org/mining.html).
Statistics analysis
Data were analyzed using the GraphPad Prism 5.0
software (GraphPad Software, Inc.), and presented as the mean ± SD
from ≥3 independent experiments. A two-tailed unpaired Student's
t-test or one-way ANOVA with Tukey's post-hoc analysis were
performed to evaluate the statistical significance. P<0.05 was
considered to indicate a statistically significant difference.
Results
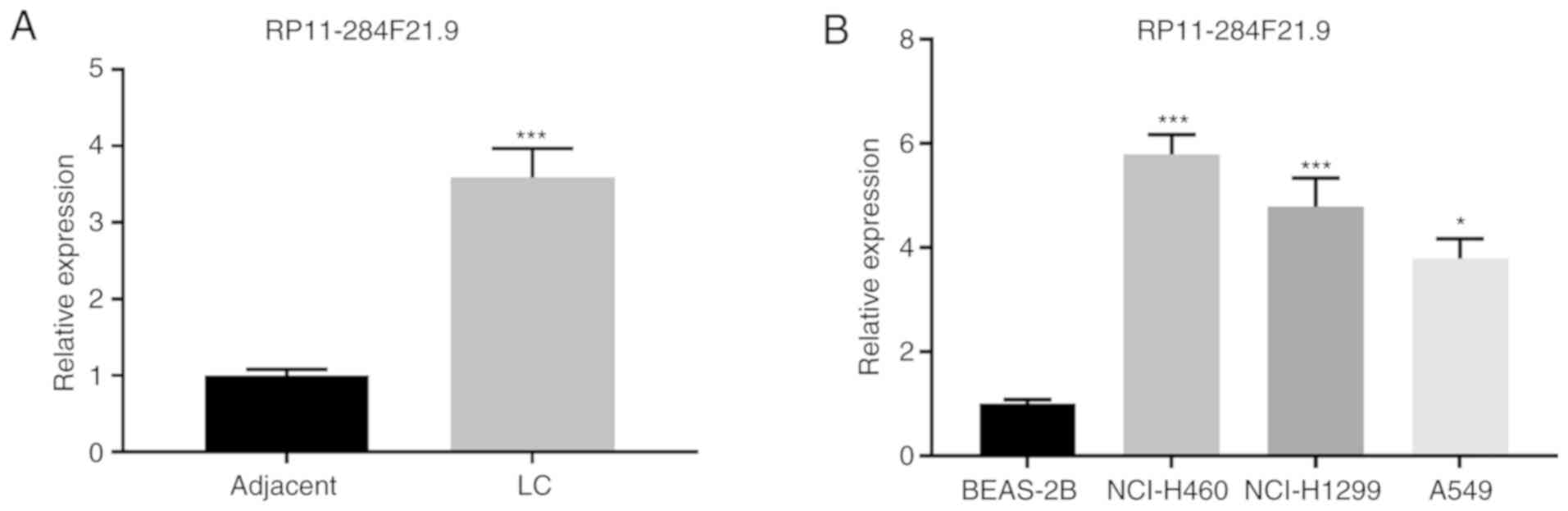
Expression of RP11-284F21.9 is
upregulated in lung carcinoma
To investigate the potential role of RP11-284F21.9
in lung carcinoma, its expression was analyzed in tissue samples
and matched adjacent healthy tissues from 13 patients with lung
carcinoma. The results demonstrated that the expression of
RP11-284F21.9 was significantly upregulated in tumor tissues
compared with healthy tissues (Fig.
1A). The expression of RP11-284F21.9 was also analyzed in human
lung carcinoma cell lines (NCI-H460, NCI-H1299 and A549) and normal
human lung epithelial cell line (BEAS-2B). Consistent with the
findings in the tissue samples, the expression of RP11-284F21.9 was
significantly increased in carcinoma cell lines compared with the
normal epithelial cell line (Fig.
1B). These results indicated that RP11-284F21.9 may serve an
oncogenic role in lung carcinoma.
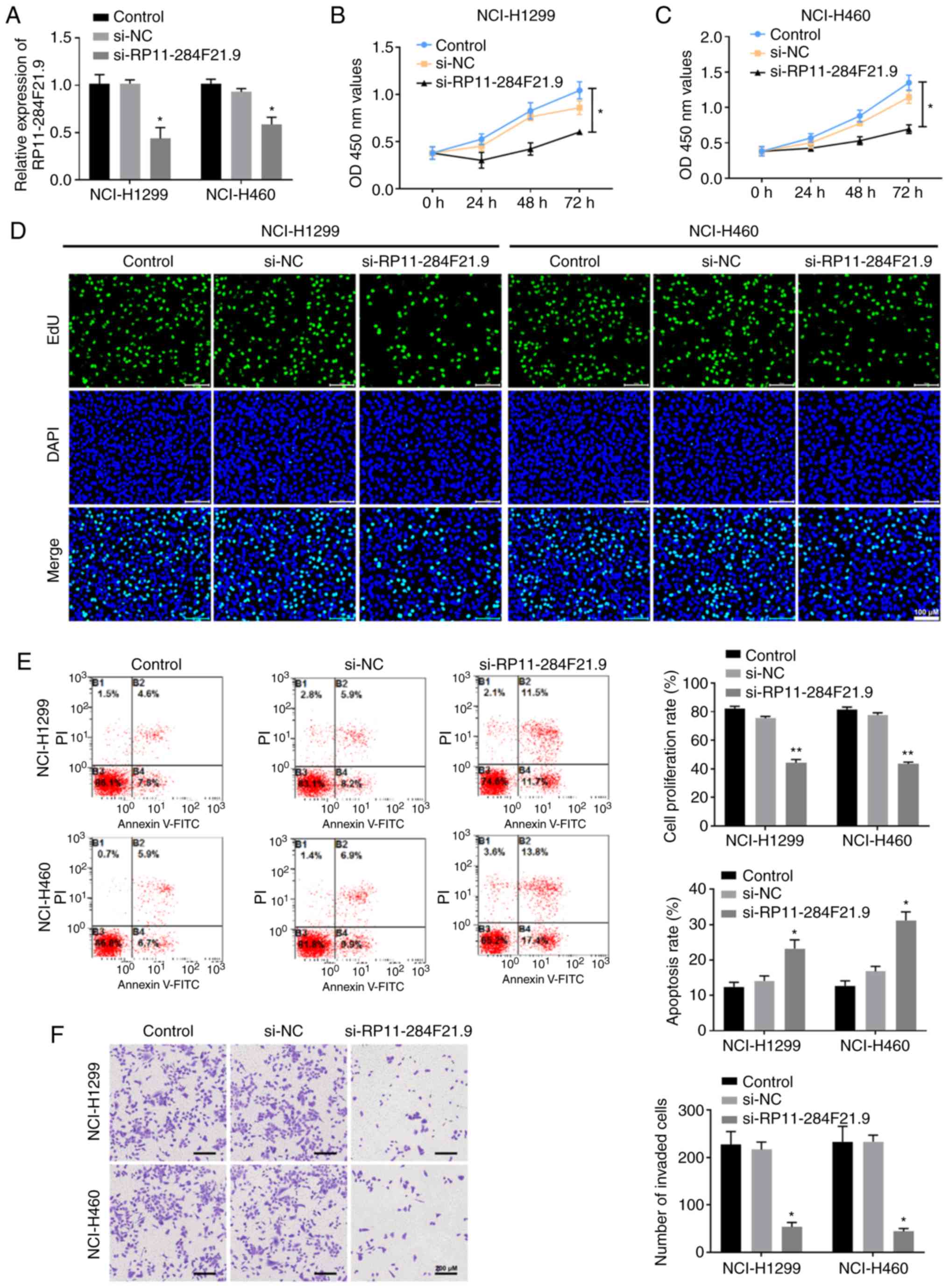
Knockdown of RP11-284F21.9 exerts
anti-oncogenic effects in lung carcinoma cells
To study the specific role of RP11-284F21.9 in lung
carcinoma cells, RP11-284F21.9 siRNA was transfected into NCI-H1299
and NCI-H460 cells (Fig. 2A). After
transfection, the proliferation of these cells was measured using
CCK-8 and EdU assays (Fig. 2B-D).
The results suggested that knocking down RP11-284F21.9
significantly reduced the proliferation of lung carcinoma cells
compared with the NC group (Fig.
2B-D). The invasiveness of si-RP11-284F21.9 transfected cells
also significantly decreased, as indicated by the data from the
Transwell assay (Fig. 2F). To
further validate the invasive capability, a RT-qPCR assay was
performed to detect the expression levels of invasion-related
genes, and the results identified that both MMP2 and MMP9 were
significantly decreased when RP11-284F21.9 was downregulated
(Fig. S1).
The results of flow cytometry measurement based
apoptosis assay suggested that cells transfected with
si-RP11-284F21.9 had a higher apoptotic rate compared with the
si-NC transfected group (Fig. 2E).
These data demonstrated the anti-tumor effects of RP11-284F21.9
knockdown in lung carcinoma cells, indicating an oncogenic role of
RP11-284F21.9.
RP11-284F21.9 directly interacts with
miR-627-3p
Based on the prediction of the online tool lncBase
v.2 from DIANA (Prediction module; http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index),
which was used to identify the downstream miRNAs of RP11-284F21.9,
the first five miRNAs in the output list were tested. Among the
predicted potential targets, it was found that miR-627-3p had the
most significant upregulation in NCI-H1299 cells transfected with
si-RP11-284F21.9 (Fig. S2).
Using sequence alignment, it was identified that
miR-627-3p was partially complementary with the 3′-UTR of
RP11-284F21.9 (Fig. 3A).
Subsequently, 293T cells were transfected with the
pmirGLO-RP11-284F21.9-wt or mut vector, containing the wt or mut
sequence of RP11-284F21.9 3′-UTR, with or without miR-627-3p
mimics. Results from the luciferase reporter assay suggested that
miR-627-3p mimics significantly decrease the signal of
RP11-284F21.9-wt transfected cells but not the RP11-284F21.9-mut
transfected cells, indicating a direct interaction between the two
non-coding RNAs (Fig. 3A).
Furthermore, transfection of si-RP11-284F21.9 into NCI-H1299 and
NCI-H460 cells resulted in the suppression of endogenous
RP11-284F21.9, leading to a significant increase in miR-627-3p
expression (Fig. 3B). Thus, these
findings suggested an inhibitory effect of RP11-284F21.9 on the
expression of miR-627-3p in lung carcinoma cells.
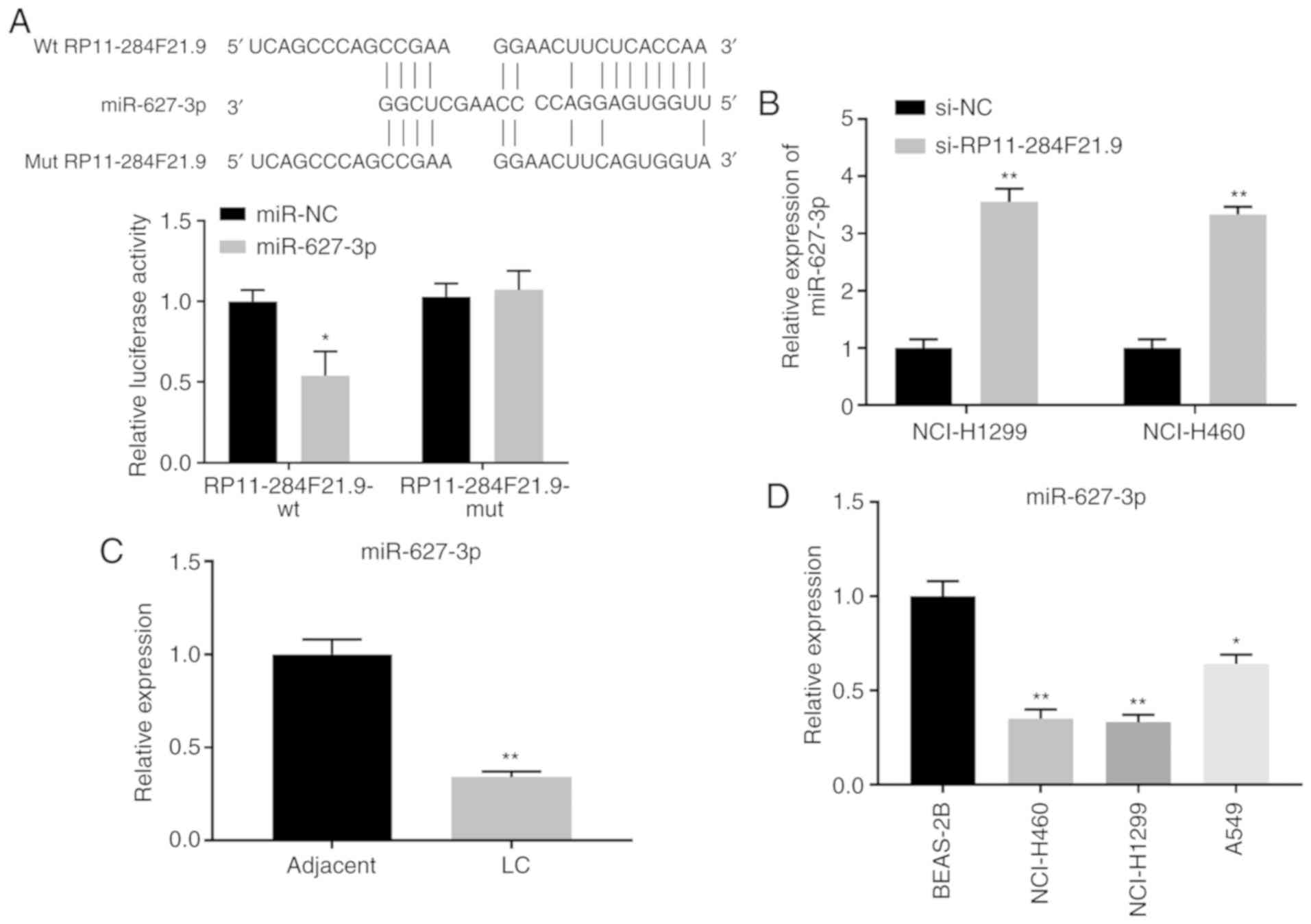 | Figure 3.RP11-284F21.9 directly interacts with
miR-627-3p. (A) Binding site between RP11-284F21.9 and miR-627-3p
that was identified using the DIANA tools, and a luciferase
reporter assay was conducted in pmirGLO-RP11-284F21.9-wt or mut
treated 293 cells in the presence of miR-627-3p mimics or miR-NC
(n=3). *P<0.05 vs. miR-NC. (B) Expression of miR-627-3p in
NCI-H1299 and NCI-H460 cells transfected with si-RP11-284F21.9 was
analyzed using RT-qPCR. **P<0.01 vs. si-NC (n=3). miR-627-3p
expression in (C) LC tissues and (D) NCI-H460, NCI-H1299 and A549
cells, compared with adjacent healthy tissues and normal lung
epithelial cells, was analyzed using RT-qPCR (n=3). *P<0.05,
**P<0.01 vs. adjacent tissue or BEAS-2B cells. NC, negative
control; siRNA, small interfering RNA; wt, wild-type; mut, mutant;
miR, microRNA; LC, lung carcinoma. |
The expression of miR-627-3p was detected in both
lung carcinoma tissues and cell lines. It was demonstrated that
miR-627-3p was significantly downregulated in carcinoma tissues
(Fig. 3C) and NCI-H460, NCI-H1299,
and A549 cells (Fig. 3D) compared
with healthy tissues and cells. Collectively, these data suggested
a direct interaction between RP11-284F21.9 and miR-627-3p, in which
RP11-284F21.9 suppresses the expression of miR-627-3p.
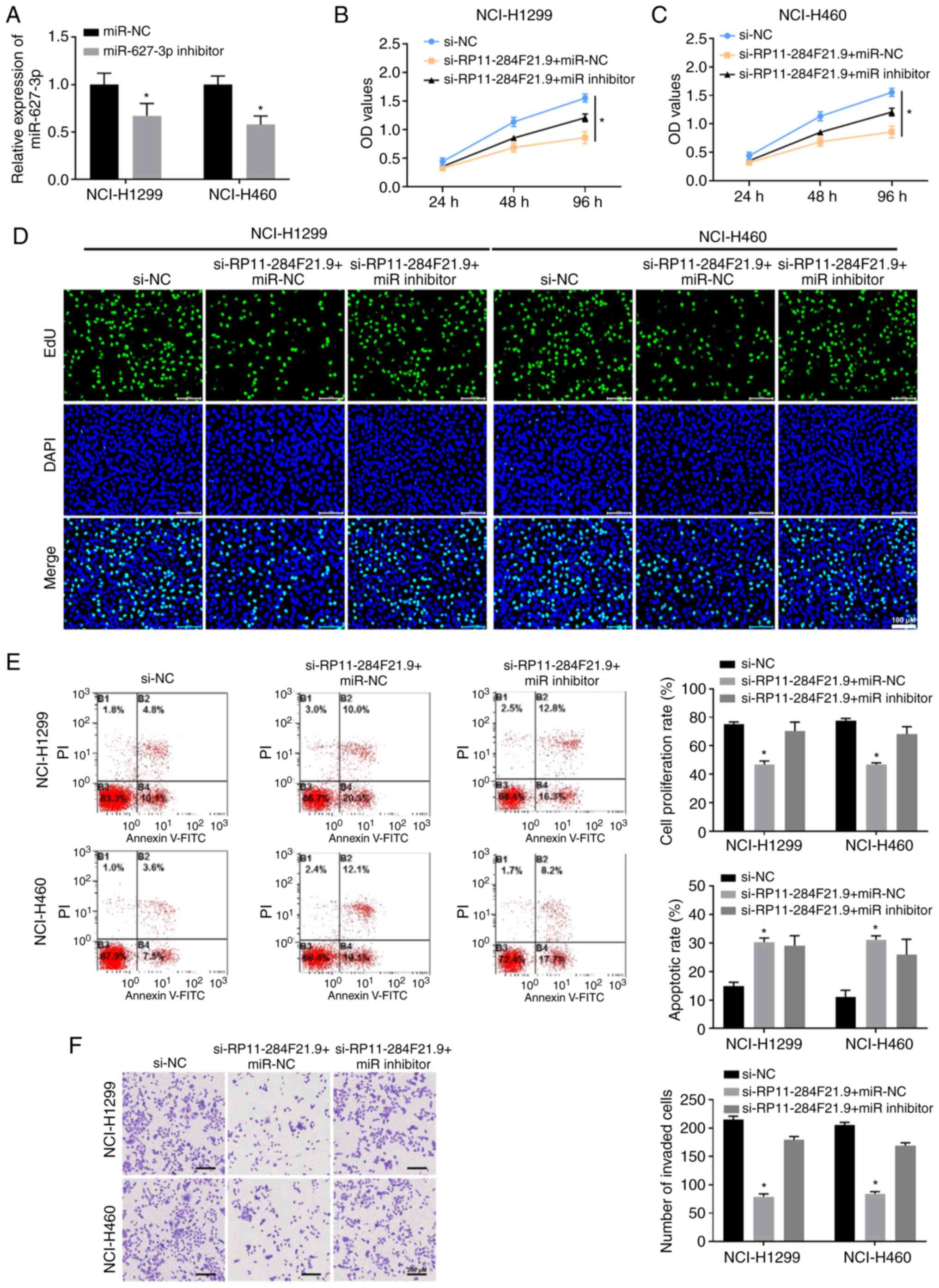
RP11-284F21.9 regulates the
proliferation and invasiveness of lung carcinoma cells via
miR-627-3p
To rescue the anti-tumor effects of si-RP11-284F21.9
in lung carcinoma cells, the miR-627-3p inhibitor, which
specifically downregulates the expression of miR-627-3p, was
transfected into NCI-H1299 and NCI-H460 cells (Fig. 4A). The results from the CCK-8 and
EdU assays demonstrated that treatment with si-RP11-284F21.9 and
miR-NC significantly decrease the proliferation of both NCI-H1299
and NCI-H460 cells (Fig. 4B-D).
However, the administration of miR-627-3p inhibitor partially
reversed the anti-proliferative effect of si-RP11-284F21.9,
indicating that RP11-284F21.9 regulates the proliferation of lung
carcinoma cells partially via miR-627-3p (Fig. 4B-D). In addition, the miR-627-3p
inhibitor restored the reduction in the number of NCI-H1299 and
NCI-H460 cells that migrated through the Transwell membrane induced
by si-RP11-284F21.9 treatment (Fig.
4F). These data indicated the participation of miR-627-3p in
the RP11-284F21.9-mediated invasive effect.
The qPCR assay results identified that both MMP2 and
MMP9 expression levels were restored in RP11-284F21.9-downregulated
cells when miR-627-3p was inhibited, compared with the miR-NC group
(Fig. S3). In addition,
transfection with miR-627-3p inhibitor also diminished the
pro-apoptosis effect of si-RP11-284F21.9 in both NCI-H1299 and
NCI-H460 cells (Fig. 4E).
Therefore, it was suggested that RP11-284F21.9 promoted the
proliferation and invasion, as well as suppressed the apoptosis of
lung carcinoma cells by inhibiting the expression of
miR-627-3p.
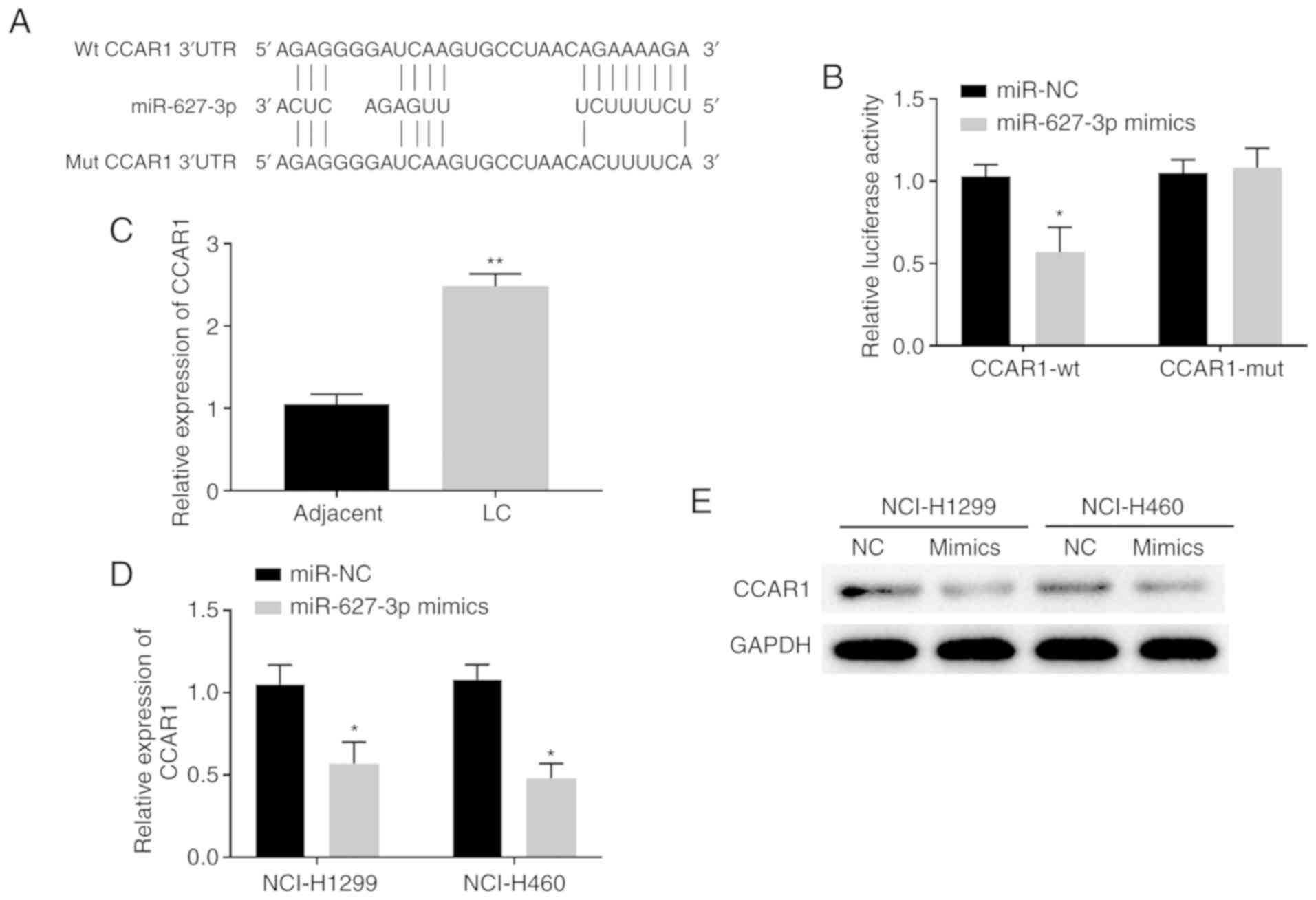
RP11-284F21.9 regulates CCAR1 via
targeting miR-627-3p
To further evaluate how RP11-284F21.9 exerts an
oncogenic role via miR-627-3p, the publicly available algorithms of
TargetScan (http://www.targetscan.org/) and miRDB were used, which
identified CCAR1 as a potential target for miR-627-3p (Fig. 5A). In order to validate this
prediction, miR-627-3p mimic was transfected into 293 cells and the
transfection efficiency was assessed. The results demonstrated that
transfection of miR-627-3p mimic increased the expression of
miR-627-3p by >70 times compared with cells transfected with
miR-NC (Fig. S4).
After validating the upregulation of miR-627-3p
mimic, a CCAR1-wt vector was constructed, which contained the wt
binding site between miR-627-3p and the CCAR1 3′-UTR, and CCAR1-mut
vector containing the mut sequence (Fig. 5A). The results from luciferase
reporter assays indicated that, compared with the miR-NC group, the
miR-627-3p mimic significantly decreased the luciferase activity of
CCAR1-wt treated cells but not the CCAR1-mut treated cells,
suggesting a direct binding of miR-627-3p to the 3′-UTR of CCAR1
(Fig. 5B). Increased expression
levels of CCAR1 were present in the lung carcinoma tissues compared
with the adjacent healthy tissues (Fig.
5C). Moreover, a significant decrease in both mRNA and protein
expression levels of CCAR1 was detected upon transfecting NCI-H1299
and NCI-H460 cells with miR-627-3p mimics (Fig. 5D and E). Thus, CCAR1 may be a direct
target of miR-627-3p in lung carcinoma cells and tissues.
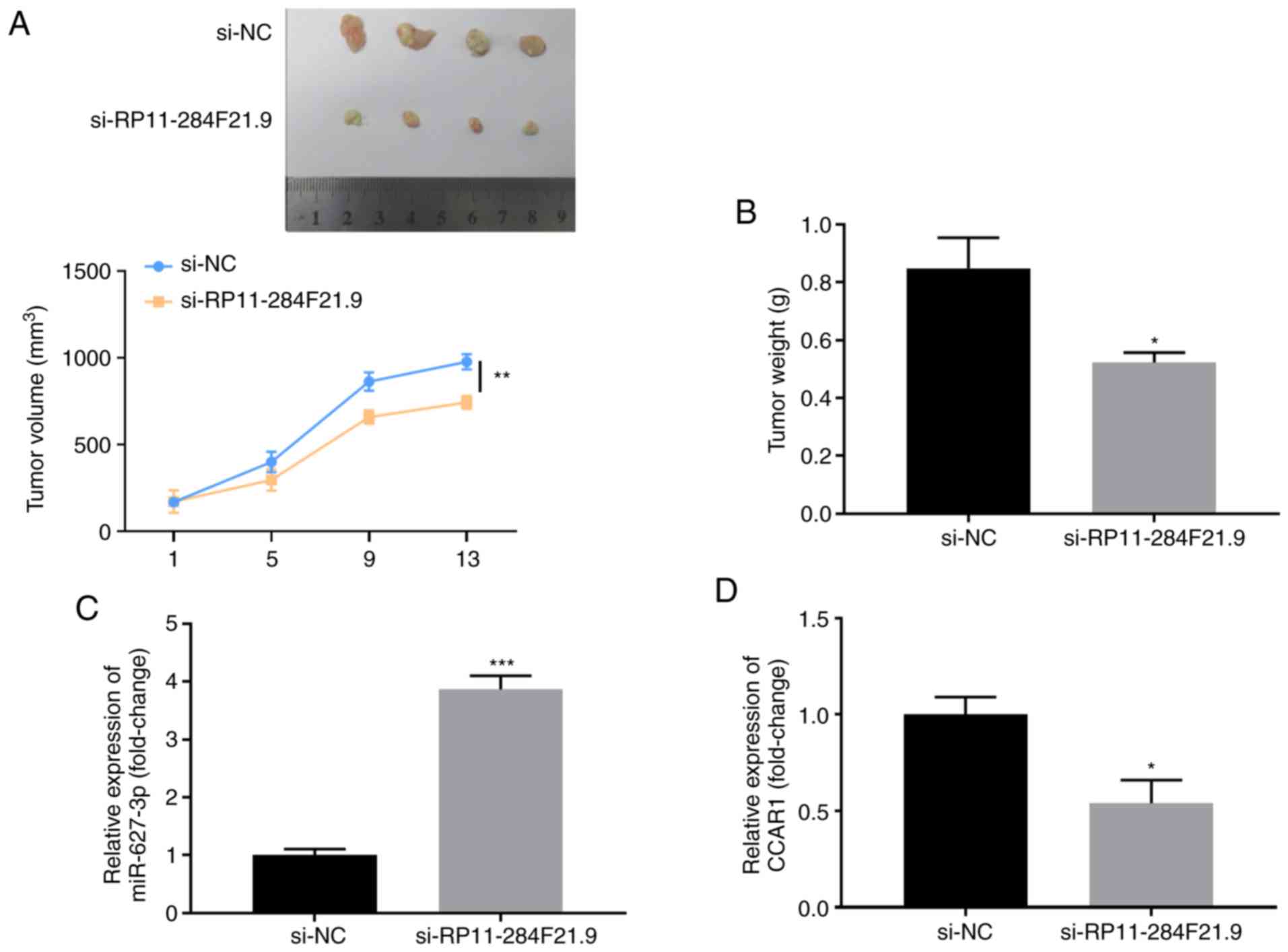
RP11-284F21.9 knockdown inhibits tumor
growth and the expression of CCAR1 in vivo
In order to investigate the effect of RP11-284F21.9
on in vivo tumorigenicity, NCI-H1299 cells were transfected
with si-NC or si-RP11-284F21.9 and injected into the nude mice.
After 2 weeks, a significantly slower proliferative rate of the
tumors was observed in the si-RP11-284F21.9 group compared with the
si-NC group (Fig. 6A and B).
Furthermore, the tumor volume and weight were significantly
decreased in the si-RP11-284F21.9 group compared with the control
group (Fig. 6A and B). RT-qPCR
analysis also demonstrated that, compared with the si-NC group, the
tumors in the si-RP11-284F21.9 group expressed higher levels of
miR-627-3p (Fig. 6C) and lower
levels of CCAR1 (Fig. 6D),
providing further evidence to the existence of the
RP11-284F21.9/miR-627-3p/CCAR1 regulatory axis in lung carcinoma
tumor tissues.
Discussion
The present study investigated the function of
RP11-284F21.9 in lung carcinoma. It was initially found that
RP11-284F21.9 was significantly upregulated in both lung cancer
tissues and cell lines. Following the deduction of a potential
oncogenic role of this lncRNA, si-RP11-284F21.9 was transfected
into NCI-H460 and NCI-H1299 cells, and it was demonstrated that
knockdown of RP11-284F21.9 inhibited the proliferation and
invasion, while promoting apoptosis of lung carcinoma cells. In the
mechanistic studies, using online prediction tools and in
vitro assays, the results indicated that miR-627-3p directly
interacts with RP11-284F21.9 by binding to its 3′-UTR.
The function of miR-627 was initially reported in
colorectal cancer (CRC). Padi et al (20) found that when upregulated by
calcitriol, miR-627 targets the histone demethylase Jumonji domain
containing 1A to increase methylation of histone H3K9 and
suppresses the proliferative factors of CRC cells, thus inhibiting
the proliferation of CRC both in vitro and in vivo.
Moreover, in CRC, Sun et al (21) discovered the role of miR-627 in
vitamin D-enhanced efficacy of irinotecan via inhibition of the
cytochrome P450 enzyme-mediated intratumoral drug metabolism.
miR-627 is also reported to be a potential non-invasive diagnostic
marker in gastric and breast cancer types (22,23).
In pulmonary diseases, miR-627 is downregulated in patients with
chronic obstructive pulmonary disease and targets the high-mobility
group box protein 1 to inhibit its expression, thus improving
transforming growth factor-β1-induced pulmonary fibrosis (24,25).
The present results demonstrated the inhibitory effect of
RP11-284F21.9 on the expression of miR-627-3p. In addition, it was
identified that the miR-627-3p inhibitor can neutralize the
anti-tumor effects of RP11-284F21.9 knockdown, indicating that
RP11-284F21.9 promotes the proliferation and invasiveness of lung
carcinoma cells partially by regulating miR-627-3p. This anti-tumor
role of miR-627-3p under the regulation of RP11-284F21.9 in lung
carcinoma tissues and cells is in accordance with the previous
aforementioned findings on human CRC, gastric and breast cancer
types.
Using the publicly available RNA interaction
prediction algorithms, the current study identified that CCAR1,
which was initially shown as the target gene of miR-627-3p, is also
regulated by RP11-284F21.9. Furthermore, the regulatory axis of
RP11-284F21.9/miR-627-3p/CCAR1 exists in the lung carcinoma cells
both in vitro and in vivo in the tumor growth model.
The interaction between RP11-284F21.9 and miR-627-3p, and the
interaction between miR-627-3p and CCAR1 were demonstrated by the
dual-luciferase assay. Although this method has been used to
validate RNA-RNA interactions in previous studies (26–28),
other assays, such as RNA pull-down and RNA binding protein
immunoprecipitation that would provide more direct evidence for the
RNA-RNA and RNA-protein interactions, should be performed.
CCAR1 was initially reported as a protein essential
for cancer cell apoptosis induced by retinoids or
chemotherapeutics, such as Adriamycin and etoposide (29). Subsequently, Kim et al
(30) revealed that this protein
functions as a transcriptional co-activator of nuclear receptors.
In human breast cancer cells, as CCAR1 interacts and cooperates
with the co-activators of estrogen receptor signaling, it promotes
the estrogen-dependent proliferation of cancer cells. In CRC cells,
Ou et al (31) reported that
CCAR1 can be recruited by β-catenin to act as a co-activator for
the transcriptional activation of lymphoid enhancer binding factor
1. CCAR1 is essential for the expression of Wnt target genes, as
well as the neoplastic transformation of CRC cells (31,32).
In gastric cancer cells, researchers have revealed the cooperation
between CCAR1 and β-catenin, which leads to the promotion of the
proliferation and migration of cancer cells (33). In lung cancer, CCAR1 was reported to
be an effector of Doxorubicin-induced apoptosis (34). Moreover, Muthu et al
(35) demonstrated that certain
chemical compounds that bind with CCAR1 can increase the expression
of CCAR1 and induce apoptosis. However, a contradictory conclusion
was reported in a recent study, which observed that CCAR1 was
promoted by serine and arginine rich splicing factor 5, which is
activated by glucose intake, and further enhanced tumorigenesis by
increasing the glucose consumption rate (36). Corroborating this finding, in the
current study, via the targeting of miR-627-3p, the expression of
CCAR1 was positively associated with that of RP11-284F21.9,
suggesting a carcinogenic role in lung carcinoma tissues and cells.
However, a limitation and future direction of the present study is
to investigate the function of CCAR1 in lung cancer, including the
identification of its interactome and the potential pathways that
trigger the tumorigenesis of lung cancer.
In conclusion, the present results demonstrated the
promoting effect of RP11-284F21.9 on the proliferation and invasion
of lung carcinoma cells. It was found that the oncogenic function
of RP11-284F21.9 was partially mediated via the
RP11-284F21.9/miR-627-3p/CCAR1 axis in both lung cancer cells and
tissues. By investigating the role and mechanism of a newly
identified lncRNA, the current study revealed a novel treatment
target and a potential therapeutic strategy for patients with lung
carcinoma.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the Natural Science
Foundation of Shaanxi Province (grant no. 2018JM7026), Xi'an
Science and Technology Project [grant no. 201805102YX10SF36(3)] and
the Fundamental Research Funds for the Central Universities in
Xi'an Jiaotong University (grant no. xjj2018272).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DL and YW conceived and designed the experiments.
DL, LW and JF performed the experiments. DL, LL and YS analyzed and
interpreted the data. DL wrote the manuscript. LL and YW revised
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All the tissue samples were obtained with written
informed consent from the patients. The protocol was approved by
The First Affiliated Hospital of Xi'an Jiaotong University
(approval no. 201810053). Animal care and study were approved by
the Institutional Animal Care and Use Committee of The First
Affiliated Hospital of Xi'an Jiaotong University (approval no.
201902013).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCAR1
|
cell division cycle and apoptosis
regulator 1
|
|
NSCLC
|
non-small cell lung cancer
|
|
SCLC
|
small cell lung cancer
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chheang S and Brown K: Lung cancer
staging: Clinical and radiologic perspectives. Semin Intervent
Radiol. 30:99–113. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pao W and Girard N: New driver mutations
in non-small-cell lung cancer. Lancet Oncol. 12:175–180. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alizadeh A, Moztarzadeh F, Ostad SN, Azami
M, Geramizadeh B, Hatam G, Bizari D, Tavangar SM, Vasei M and Ai J:
Synthesis of calcium phosphate-zirconia scaffold and human
endometrial adult stem cells for bone tissue engineering. Artif
Cells Nanomed Biotechnol. 44:66–73. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peng W, Wang J, Shan B, Peng Z, Dong Y,
Shi W, He D, Cheng Y, Zhao W, Zhang C, et al: Diagnostic and
prognostic potential of circulating long non-coding RNAs in non
small cell lung cancer. Cell Physiol Biochem. 49:816–827. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Du Z, Fei T, Verhaak RG, Su Z, Zhang Y,
Brown M, Chen Y and Liu XS: Integrative genomic analyses reveal
clinically relevant long noncoding RNAs in human cancer. Nat Struct
Mol Biol. 20:908–913. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tian X and Xu G: Clinical value of lncRNA
MALAT1 as a prognostic marker in human cancer: Systematic review
and meta-analysis. BMJ Open. 5:e0086532015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang JZ, Xiang JJ, Wu LG, Bai YS, Chen ZW,
Yin XQ, Wang Q, Guo WH, Peng Y, Guo H and Xu P: A genetic variant
in long non-coding RNA MALAT1 associated with survival outcome
among patients with advanced lung adenocarcinoma: A survival cohort
analysis. BMC Cancer. 17:1672017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Loewen G, Jayawickramarajah J, Zhuo Y and
Shan B: Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol.
7:902014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu XH, Liu ZL, Sun M, Liu J, Wang ZX and
De W: The long non-coding RNA HOTAIR indicates a poor prognosis and
promotes metastasis in non-small cell lung cancer. BMC Cancer.
13:4642013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nakagawa T, Endo H, Yokoyama M, Abe J,
Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T and Satoh K: Large
noncoding RNA HOTAIR enhances aggressive biological behavior and is
associated with short disease-free survival in human non-small cell
lung cancer. Biochem Biophys Res Commun. 436:319–324. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao
X, Chen WS and Li B: LncRNA-UCA1 exerts oncogenic functions in
non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett.
371:99–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng N, Cai W, Ren S, Li X, Wang Q, Pan
H, Zhao M, Li J, Zhang Y, Zhao C, et al: Long non-coding RNA UCA1
induces non-T790M acquired resistance to EGFR-TKIs by activating
the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer.
Oncotarget. 6:23582–23593. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ashouri A, Sayin VI, Van den Eynden J,
Singh SX, Papagiannakopoulos T and Larsson E: Pan-cancer
transcriptomic analysis associates long non-coding RNAs with key
mutational driver events. Nat Commun. 7:131972016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dwyer CA, Bi WL, Viapiano MS and Matthews
RT: Brevican knockdown reduces late-stage glioma tumor
aggressiveness. J Neurooncol. 120:63–72. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Padi SK, Zhang Q, Rustum YM, Morrison C
and Guo B: MicroRNA-627 mediates the epigenetic mechanisms of
vitamin D to suppress proliferation of human colorectal cancer
cells and growth of xenograft tumors in mice. Gastroenterology.
145:437–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun M, Zhang Q, Yang X, Qian SY and Guo B:
Vitamin D enhances the efficacy of irinotecan through
miR-627-mediated inhibition of intratumoral drug metabolism. Mol
Cancer Ther. 15:2086–2095. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shin VY, Ng EK, Chan VW, Kwong A and Chu
KM: A three-miRNA signature as promising non-invasive diagnostic
marker for gastric cancer. Mol Cancer. 14:2022015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao J, Luo C, Yan R, Peng R, Wang K, Wang
P, Ye H and Song C: rs15869 at miRNA binding site in BRCA2 is
associated with breast cancer susceptibility. Med Oncol.
33:1352016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li J, Kong X, Jiang S, Liao W, Zhang Z,
Song J, Liang Y and Zhang W: miR-627/HMGB1/NF-κB regulatory loop
modulates TGF-β1-induced pulmonary fibrosis. J Cell Biochem.
120:2983–2993. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Musri MM, Coll-Bonfill N, Maron BA,
Peinado VI, Wang RS, Altirriba J, Blanco I, Oldham WM, Tura-Ceide
O, García-Lucio J, et al: MicroRNA dysregulation in pulmonary
arteries from chronic obstructive pulmonary disease. relationships
with vascular remodeling. Am J Respir Cell Mol Biol. 59:490–499.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu Z, Yu Y, Ding X, Jin D, Wang G, Zhou Y,
Zhu Y, Na L, He Y and Wang Q: LncRNA FLJ33360 accelerates the
metastasis in hepatocellular carcinoma by targeting miRNA-140/MMP9
axis. Am J Transl Res. 12:583–591. 2020.PubMed/NCBI
|
|
27
|
Huang W, Huang F, Lei Z and Luo H: LncRNA
SNHG11 promotes proliferation, migration, apoptosis, and autophagy
by regulating hsa-miR-184/AGO2 in HCC. Onco Targets Ther.
13:413–421. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sui G, Zhang B, Fei D, Wang H, Guo F and
Luo Q: The lncRNA SNHG3 accelerates papillary thyroid carcinoma
progression via the miR-214-3p/PSMD10 axis. J Cell Physiol. Feb
11–2020.(Epub ahead of print). View Article : Google Scholar
|
|
29
|
Rishi AK, Zhang L, Boyanapalli M, Wali A,
Mohammad RM, Yu Y, Fontana JA, Hatfield JS, Dawson MI, Majumdar AP
and Reichert U: Identification and characterization of a cell cycle
and apoptosis regulatory protein-1 as a novel mediator of apoptosis
signaling by retinoid CD437. J Biol Chem. 278:33422–33435. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim JH, Yang CK, Heo K, Roeder RG, An W
and Stallcup MR: CCAR1, a key regulator of mediator complex
recruitment to nuclear receptor transcription complexes. Mol Cell.
31:510–519. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ou CY, Kim JH, Yang CK and Stallcup MR:
Requirement of cell cycle and apoptosis regulator 1 for target gene
activation by Wnt and beta-catenin and for anchorage-independent
growth of human colon carcinoma cells. J Biol Chem.
284:20629–20637. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yan L, You WQ, Sheng NQ, Gong JF, Hu LD,
Tan GW, Chen HQ and Wang ZG: A CREB1/miR-433 reciprocal feedback
loop modulates proliferation and metastasis in colorectal cancer.
Aging (Albany NY). 10:3774–3793. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chang TS, Wei KL, Lu CK, Chen YH, Cheng
YT, Tung SY, Wu CS and Chiang MK: Inhibition of CCAR1, a
coactivator of β-catenin, suppresses the proliferation and
migration of gastric cancer cells. Int J Mol Sci. 18:4602017.
View Article : Google Scholar
|
|
34
|
Cheriyan VT, Alsaab H, Sekhar S, Venkatesh
J, Mondal A, Vhora I, Sau S, Muthu M, Polin LA, Levi E, et al: A
CARP-1 functional mimetic compound is synergistic with
BRAF-targeting in non-small cell lung cancers. Oncotarget.
9:29680–29697. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Muthu M, Somagoni J, Cheriyan VT, Munie S,
Levi E, Ashour AE, Yassin AE, Alafeefy AM, Sochacki P, Polin LA, et
al: Identification and testing of novel CARP-1 functional mimetic
compounds as inhibitors of non-small cell lung and triple negative
breast cancers. J Biomed Nanotechnol. 11:1608–1627. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen Y, Huang Q, Liu W, Zhu Q, Cui CP, Xu
L, Guo X, Wang P, Liu J, Dong G, et al: Mutually exclusive
acetylation and ubiquitylation of the splicing factor SRSF5 control
tumor growth. Nat Commun. 9:24642018. View Article : Google Scholar : PubMed/NCBI
|