Introduction
Renal cell carcinoma (RCC) is one of the most common
malignant cancers in the adult urinary system worldwide (1). There are approximately 270,000 newly
diagnosed and approximately 116,000 deaths annually, worldwide
(2), making it the second most
malignant cancer of the urinary system, with a 40% mortality rate
(3). Surgical resection is the best
treatment for RCC. However, RCC is insidious, and approximately 30%
of RCC patients have metastasis at the time of diagnosis, losing
the chance of surgery (4). In
addition, up to 40% of patients have local recurrence and/or
distant metastasis (5). Unlike
other urinary system tumors, RCC is resistant to radiotherapy and
chemotherapy, leading to difficulties in the treatment of
metastatic RCC (6). In recent
years, molecular targeted drugs, as the first-line treatment of
metastatic RCC, have prolonged patient survival. However, this is
still merely effective for 6–15 months (7). Therefore, a more efficient therapeutic
strategy for the metastasis of RCC and the underlying molecular
mechanism of the initiation and development of RCC requires further
exploration.
MicroRNAs (miRNAs/miRs) are a group of endogenous,
short single-stranded non-coding RNA molecules containing
approximately 18–25 nucleotides (8). miRNAs regulate the expression of
targeted genes at the transcriptional or translational level mainly
through binding to the 3′ untranslated regions (3′UTR) of targeted
mRNAs, in order to regulate post-transcriptional expression of
target genes (9). Furthermore, they
are involved in cell growth, cell cycle control, apoptosis, tumor
tissue infiltration and metastasis, angiogenesis and infinite
proliferative potential (6). Tumor
angiogenesis is a critical process during cancer progression, which
modulates tumor growth and metastasis. Recent studies have reported
that certain miRNAs may become involved in this procedure. Liang
et al indicated that miR-153 suppresses the tube formation
and the migration of endothelial cells by targeting angiopoietin 1
(ANG1) directly in breast cancer cells (10). miR-205 was found to significantly
suppress angiogenesis and epithelial-mesenchymal transition (EMT)
through the simultaneous targeting of vascular endothelial growth
factor A (VEGFA), zinc finger e-box binding homeobox 1 (ZEB1), and
downstream products in anaplastic thyroid carcinoma (11).
Studies have shown that microRNA-218 (miR-218) can
inhibit the proliferation, migration, invasion and metastasis of
cancer cells, which is a tumor suppressor in oral squamous cell
carcinoma, gastric cancer, head and neck squamous cell carcinoma
and other tumors (12–14). Small and Olson reported that the
generation of the vascular system involves the regulation of
miRNAs, in which miR-218 is an important regulator (15). Guan et al also confirmed that
miR-218 regulates the angiogenesis of prostate cancer through the
RPTOR-independent companion of MTOR complex 2 (RICTOR)/VEGFA axis,
thereby inhibiting prostate cancer malignant progression (16). However, the role of miR-218 in RCC
angiogenesis remains unclear.
In the present study, it was found that miR-218
could specifically bind to the target site of the 3′UTR of
GRB2-associated binding protein 2 (GAB2), and negatively regulate
its expression. By inhibiting the PI3K/AKT pathway, the expression
of angiogenic factor VEGFA was suppressed. In addition, the
interaction between renal carcinoma cells and vascular endothelial
cells was disrupted, suppressing the proliferation, invasion,
migration and angiogenesis of RCC. This result provides new insight
into the mechanism of renal cell carcinogenesis and progression,
suggesting that miRNA-218 may serve as a target for the treatment
of RCC.
Materials and methods
Cell lines and cell culture
The human RCC cell lines ACHN, 769P and 786O were
obtained from the American Type Culture Collection (ATCC). Human
umbilical vein endothelial cells (HUVECs), human kidney 2 (HK-2)
and 293T cells were kindly provided by Dr Jer-Tsong Hsieh
(University of Texas Southwestern Medical Center). The 293T cells
and HUVECs were cultured in DMEM (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher
Scientific, Inc.). ACHN, 786O and 769P cells were cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing
10% FBS. All cells were cultured at 37°C, under 5% CO2
culture conditions.
Bioinformatics
Data for a total of 591 samples were downloaded from
the TCGA website (https://portal.gdc.cancer.gov/). The downloaded data
were the isoform expression quantification data of miRNA-Seq in the
The Cancer Genome Atlas Kidney Renal Clear Cell Carcinoma
(TCGA-KIRC) data collection. The mature miRNAs were calculated
using the mature human miRNA annotation information (V21 version)
from miRBase (http://www.mirbase.org/). The
pathological parameters were statistically analyzed by Chi-square
test, and the P-value of survival analysis was tested by log rank
method. The Kaplan-Meier (KM) survival curve graph made by R
package, and the expression level was distinguished by the median.
The TargetScan (http://www.targetscan.org/) database was used to
predict the targeted gene and binding site of miR-218, and GAB2 was
selected.
Construction of stably
miR-218-overexpressing and GAB2-knockdown cell lines
The lentivirus LV-miR-218 vector and the scrambled
lentiviral vector LV-NC were constructed by GenePharma (Shanghai,
China), which contained the green fluorescent protein (GFP) and
anti-puromycin sequence. Lentiviral vectors that encoded short
hairpin RNA (shRNA) targeting human GAB2 were also constructed by
GenePharma. After these were verified through the DNA sequence, the
lentivirus vectors were used to infect the ACHN, 786O and 769P cell
lines. Then, 2×106 RCC cells were seeded in 6-cm dishes.
Afterwards, the medium was replaced after the cells were adhered to
the wall, and 20-µl lentivirus vectors (1×108 TU/ml)
were added to the medium according to the optimal multiplicity of
infection (MOI) value of 10, along with 8 µg/ml of polybrene. The
medium was changed after 48 h, and 2 µg/ml of puromycin was added
to select and maintain the stably miR-218-overexpressing and
GAB2-knockdown RCC cell lines.
Total RNA and microRNA isolation
RCC cells were harvested when 50% confluence was
reached. The isolation of cellular total RNA and microRNA was
separately performed using a total RNA isolation kit (Feijie;
RNAfast200) and a miRcute miRNA isolation kit (Tiangen; DP501)
according to manufacturer's protocol.
RNA reverse transcription
The reverse transcription of total RNA and microRNA
was separately preformed using a PrimeScript™ RT reagent kit
(Takara; RR037A) and miRcute Plus miRNA First-Strand cDNA Synthesis
Kit (Tiangen; KR211), according to the manufacturer's
instructions.
Analysis of gene expression
The transcriptional expression of the target genes
was detected using SYBR Premix Ex Taq II (Takara; RR820A) using the
CFX96 Real-time PCR system (Bio-Rad Laboratories, Inc.). The PCR
reaction mixtures were incubated at 95°C for 30 sec for initial
denaturation; followed by 40 cycles at 95°C for 5 sec, 60°C for 30
sec. The relative gene expression was calculated using the
2−ΔΔCq method (17). The
primers used for PCR are provided in Table SI.
The protein expression of target genes was analyzed
by western blot analysis. The RCC cells were gently washed three
times using pre-cooled PBS until 80% confluence was reached, and
lysed by RIPA containing 2% protease inhibitor PMSF. Approximately
30 µg of each protein sample was added to 10–12% SDS-PAGE. Then,
these proteins were separated and blotted to polyvinylidene
fluoride membranes. Afterwards, the membranes were blocked with 5%
skim milk for 1 h at room temperature, and incubated with primary
antibodies against GAB2 (1:500, product code ab32365; Abcam),
protein kinase B (AKT) (1:500; product code ab8805; Abcam),
phosphorylated (p)Akt (1:2,500; product code ab81283; Abcam), mTOR
(1:1,000, product code ab2732; Abcam), pmTOR (1:1,000; product code
ab109268; Abcam), hypoxia inducible factor 1 subunit α (HIF1α)
(Abcam; [H1alpha67] (ab1), VEGFA (1:200; product code ab1316;
Abcam) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
(1:1,000; cat. no. SS12002; Kangcheng) at 4°C overnight.
Afterwards, these membranes were washed with TBST before incubation
with horseradish peroxidase-conjugated secondary antibodies
(1:2,000, cw0102 and cw0103; CWBIO) for 1 h at room temperature.
Finally, the protein bands were subjected to the ChemiDoc XRS
Visualize System (Bio-Rad Laboratories) for visualization and image
collection.
Luciferase reporter assay
In order to determine whether miR-218 directly
targets the 3′UTR of GAB2, two types of reporter plasmids were
constructed using the GP-miRGLO vector (GenePharma): Wild-type GAB2
3′UTR reporter plasmid (WT) and mutated GAB2 3′UTR reporter plasmid
(MUT). A total of 8×104 293T cells were seeded in a
24-well plate, and co-transfected with 50 nM of miR-218 or NC
mimic, and the luciferase reporter plasmid WT or MUT. At 48 h after
transfection, the luciferase activity was measured using a Dual
Luciferase Assay kit (Promega Corp.), according to the
manufacturer's instructions.
Conditioned medium collection
A total of 8×105 RCC cells were seeded in
a 6-cm culture dish. After adhesion, these cells were gently washed
three times with serum-free medium (SFM), fed 4 ml of SFM, and
cultured for 24 h. Then, the supernatant was centrifuged and
collected as conditioned medium (CM).
In vitro HUVEC migration assays
A total of 4×104 HUVECs in SFM were
placed into the upper chamber of a 24-well plate, while the lower
chamber was filled with 1 ml of different CMs, or seeded with
1×104 RCC cells in SFM. After 16 h of incubation, the
migrated cells were fixed with 4% paraformaldehyde and stained with
0.1% crystal violet at room temperature for 10 min. The number of
migrated cells was counted by upright microscopic imaging system
(Olympus, Japan) in six random fields per well (×200
magnification).
HUVEC tube formation assays
A total of 1×105 HUVECs mixed with CM or
SFM were seeded in a Matrigel-coated 24-well plate per well. After
incubation for 4 h, tube-like vascular structures were observed by
inverted microscope (Olympus, Japan). Merely perfectly continuous
tubes between two branching points were considered as a tube.
ELISA assay
The secreted VEGFA in CMs was assessed using a
RayBio® Human VEGF ELISA kit (RayBiotech Inc.),
according to the manufacturer's instructions.
CCK-8 assay
CCK-8 assay was performed to mesure the growth rate
of cells using Cell Counting Kit-8 (Selleck Chemicals), according
to the manufacturer's instructions.
In vivo studies
A total of 8 male New Zealand white rabbits, three
months old, weighing approximately 2 kg, and 10 male mice, six
weeks old, weighing about 20 g, approximately used in the present
study and obtained from Xi'an Jiaotong University Animal
Eperimental Center. Housing condition consisted of 23°C, 15 Pa
higher than room air pressure and 12-h light/dark cycle. All
procedures for animal experiments were performed in accordance to
the Guidelines of the Institutional Animal Care and Use Committee
of Xi'an Jiaotong University (18).
In order to develop the xenograft tumor models, 2×106
cells were subcutaneously injected into both sides of the plank
region of nude mice, with five mice in each group. The tumor size
was measured once a week, and the tumor volume (V) was calculated
by the formula V=(length × width2)/2. After four weeks,
the mice were euthanized by cervical dislocation, and the tumors
were harvested and weighed, and then fixed with 4%
paraformaldehyde. For the rabbit cornea assays, six male New
Zealand white rabbits were randomly divided into two groups.
Pentobarbital sodium (30 mg/kg) was injected into the auricular
vein of rabbits, and then tetracaine eye drops were used for local
anesthesia. After that, 1×105 cells were injected into
the surgically produced micro pocket (1.5×3.0 mm) in the 6 and 12
o'clock points of the right eye. After four weeks, the cornea
angiogenic responses were observed. When the observation was
completed, rabbits were euthanized by injection of pentobarbital
sodium (100 mg/kg). The animal experiments conducted in the present
study were approved by the Ethics Committee of The First Affiliated
Hospital of Xi'an Jiaotong University (2013065).
Immunohistochemistry
The nude mouse xenograft specimens were analyzed by
immunohistochemistry assay (IHC) using the EnVision™ System (Dako),
according to the manufacturer's instructions. The primary
antibodies were GAB2 (Proteintech, 22549-1-AP, dilution 1:50),
VEGFA (Abcam, ab1316, dilution 1:50), proliferating cell nuclear
antigen (PCNA) (Proteintech, 10205-2-AP, dilution 1:200), and CD31
(Abcam, ab28364, dilution 1:50). According to the intensity of the
nuclear or cytoplasmic staining, the average intensity score of the
positive cells was divided into four levels: None, 0; weak, 1;
intermediate, 2 and strong, 3. According to the percentage of
positively stained cells, the score was divided into four grades:
0-25=1; >25-50=2; >50-75=3, and >75-100%=4. These two
scores were multiplied to obtain the total staining score, which
ranged from 0 to 12. Three fields were randomly selected for the
examination.
Statistical analysis
For the differences between two groups (Student's
t-test) and multiple groups (one-way ANOVA followed by Dunnett's
multiple comparison tests), Pearson's correlation and linear
regression were analyzed using the GraphPad Prism version 6.0
software (GraphPad Software, Inc.). A P-value of <0.05 was
considered as indicative of statistical significance.
Results
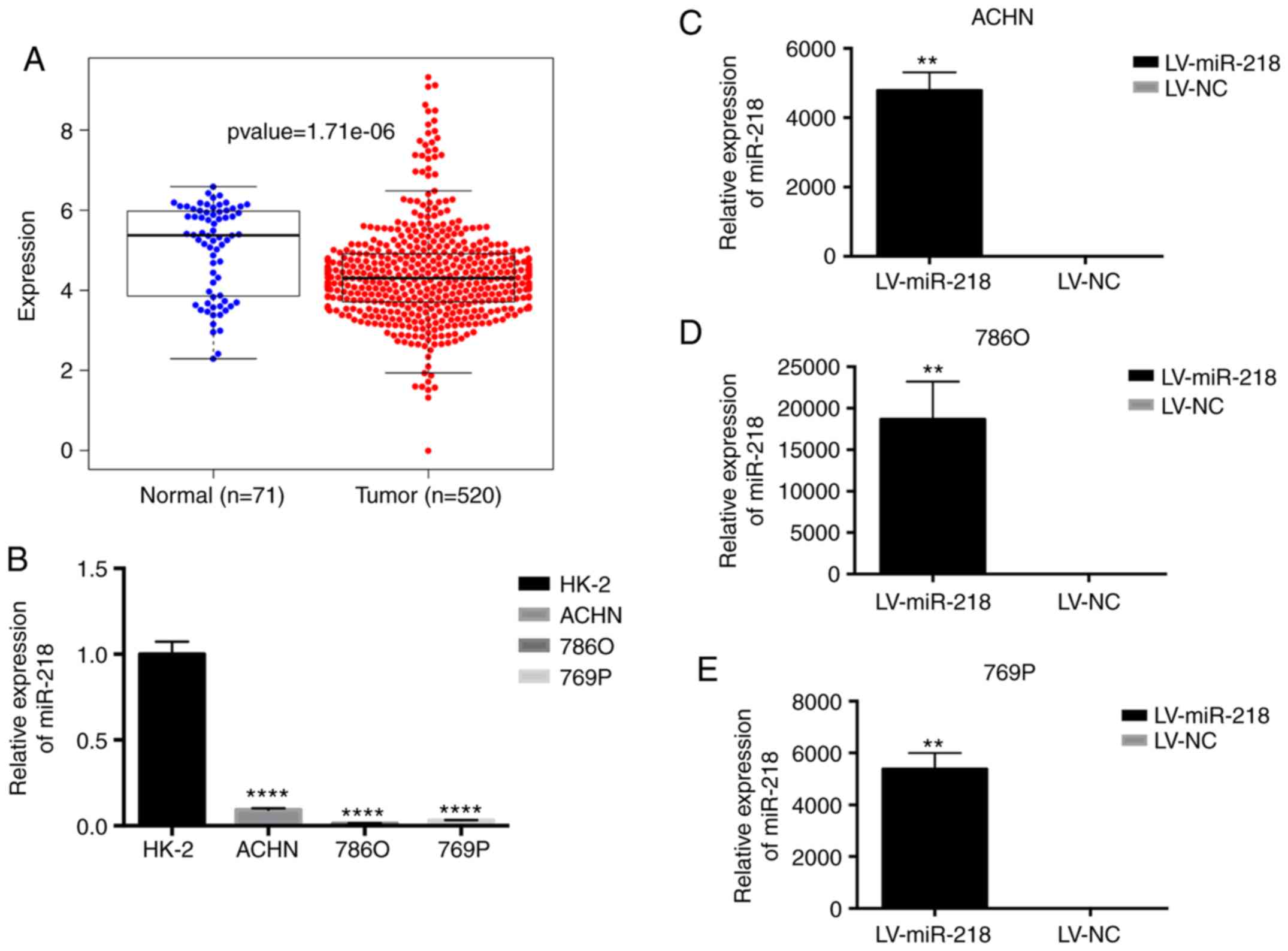
Expression of miR-218 in renal cell
carcinoma
The expression of miR-218 in human RCC tissue
samples and adjacent normal tissues was analyzed using the TCGA
database. The results found that miR-218 expression was
significantly lower in the tissue samples when compared to that in
the adjacent normal tissues (Fig.
1A). In addition, real-time PCR results also revealed a
significantly higher miR-218 expression in human renal cortical
proximal tubular epithelial HK-2 cells, when compared to the RCC
ACHN, 786O and 769P cell lines (Fig.
1B). Intriguingly, the overall survival rate of RCC samples
with high expression of miR-218 was lower than the overall survival
rate of the RCC samples with low expression (Fig. S1), and the expression of miR-218
was higher in the more malignant stages and grades (Table SII).
miR-218 inhibits RCC angiogenesis in
vitro
Three different RCC cell lines stably overexpressing
miR-218 (LV-miR-218) were established (Fig. 1C-E). Endothelial cell recruitment
assay was performed to explore the effect of miR-218 on the
migration of HUVECs. Under the condition of conditioned medium (CM)
or HUVEC co-culture, the migration ability of the HUVECs were
significantly restrained by overexpression of miR-218 (Fig. 2A and B). Furthermore, CCK-8 assay
was conducted to evaluate the effect of miR-218 on HUVEC
proliferation. Compared to the LV-NC group, the CM of
miR-218-overexpressing ACHN cells inhibited the proliferation of
HUVECs (Fig. 2C-E). The tube
formation assay also revealed that HUVECs formed less tube-like
structures in miR-218-overexpressing cells compared to the LV-NC
group, indicating that miR-218 could inhibit the angiogenesis of
RCC (Fig. 2F). In addition, the
expression of VEGFA, which is the most important angiogenic factor
in tumor angiogenesis, was decreased in RCC cells with miR-218
overexpression, when compared to the LV-NC group (Fig. 2G and H). The ELISA assay results
revealed that the concentration of VEGFA protein secreted by
miR-218-overexpressing RCC cells was also decreased (Fig. 2I), illustrating that miR-218
inhibited the secretion of VEGFA in RCC cells. In addition, the
mRNA expression of EMT-related markers E-cadherin and vimentin was
investigated, and the expression of vimentin was significantly
decreased while E-cadherin expression was significantly increased
in the LV-miR-218 group. The results indicated that the
overexpression of miR-218 induced a slight migration and invasion
change (Fig. S2, left panel).
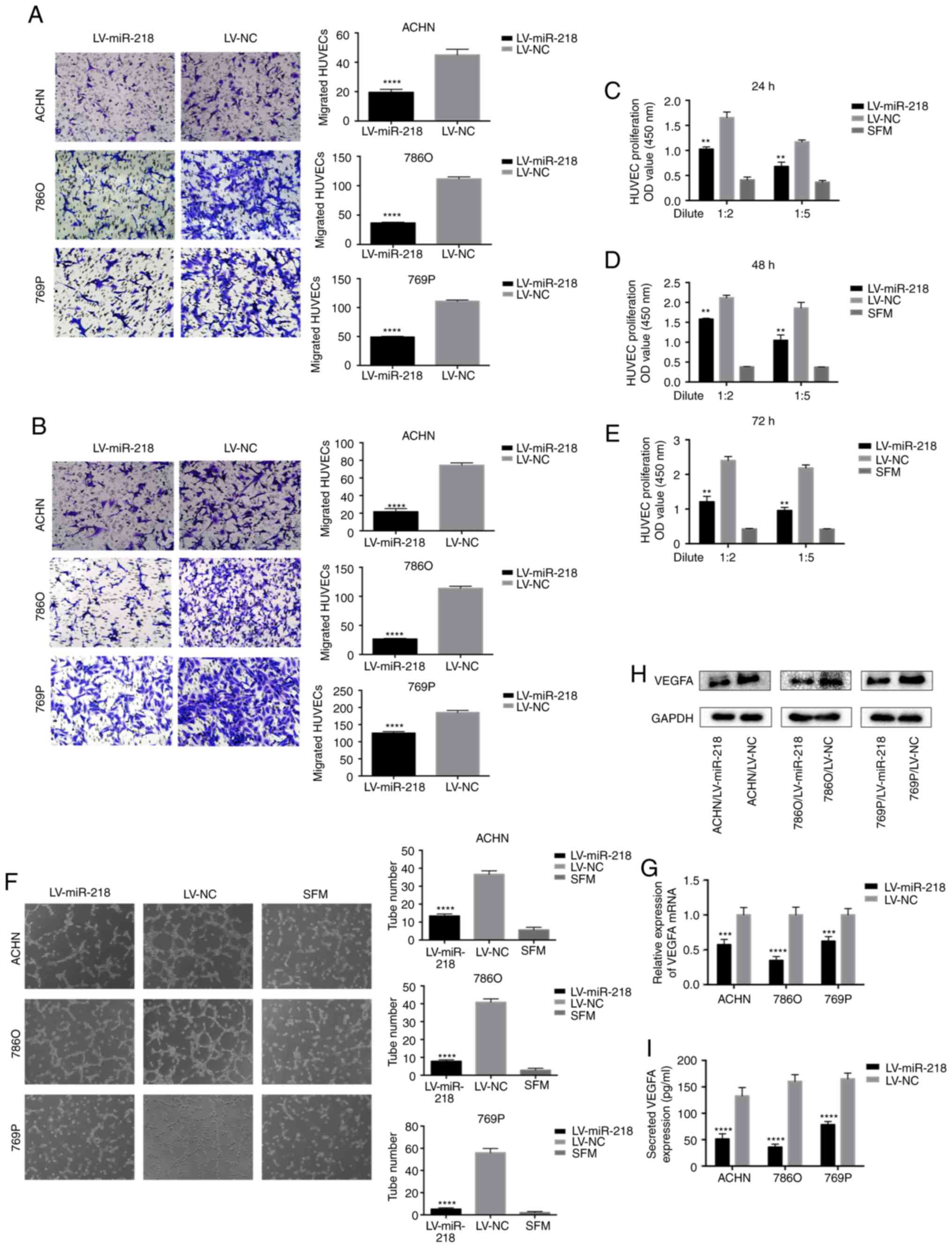 | Figure 2.miR-218 inhibits HUVEC migration,
proliferation and tube formation in vitro, and inhibits
VEGFA expression. (A) Overexpression of miR-218 suppresses the
recruitment of HUVECs in a co-cultured system. Cancer cells
cultured in the bottom of a 24-well plate were used to recruit
HUVEC seeding on the upper chamber of Transwell insets.
****P<0.0001 compared with the LV-NC group. (B) Overexpression
of miR-218 decreased the recruitment of HUVECs through the
conditioned medium (CM), which was collected from the ACHN, 786O
and 769P/LV-miR-218 or LV-NC cells. HUVECs were seeded on the upper
chamber of the Transwell insets within 16 h. The migrated cells in
six random fields per well were counted (magnification, ×200).
****P<0.0001 compared with the LV-NC group. (C-E) miR-218
overexpression reduced the proliferation of HUVECs. HUVECs were
treated with serum-free medium (SFM) or diluted CMs for 24, 48 and
72 h before the CCK-8 assay. **P<0.01 compared with the LV-NC
group. (F) miR-218 overexpression suppressed the tube formation of
HUVECs. HUVECs diluted in SFM or CMs were added into
Matrigel-coated wells and incubated for 4 h. The representative
images of tube-like structures were captured (left), and the tube
number in the whole field was counted (magnification, ×100)
(right). ****P<0.0001 compared with the LV-NC group. (G)
Real-time PCR was used to analyze the expression level of VEGFA in
ACHN, 786O and 769P cells transfected with LV-NC or LV-miR-218.
***P<0.001 and ****P<0.0001 compared to the LV-NC group. (H)
The VEGFA protein level was analyzed by western blot analysis. (I)
The concentration of secreted VEGFA protein in the CMs was
determined by ELISA. ****P<0.0001 compared to the LV-NC group.
These data are representative of three independent experiments.
HUVECs, human umbilical vein endothelial cells; VEGFA, vascular
endothelial growth factor A. |
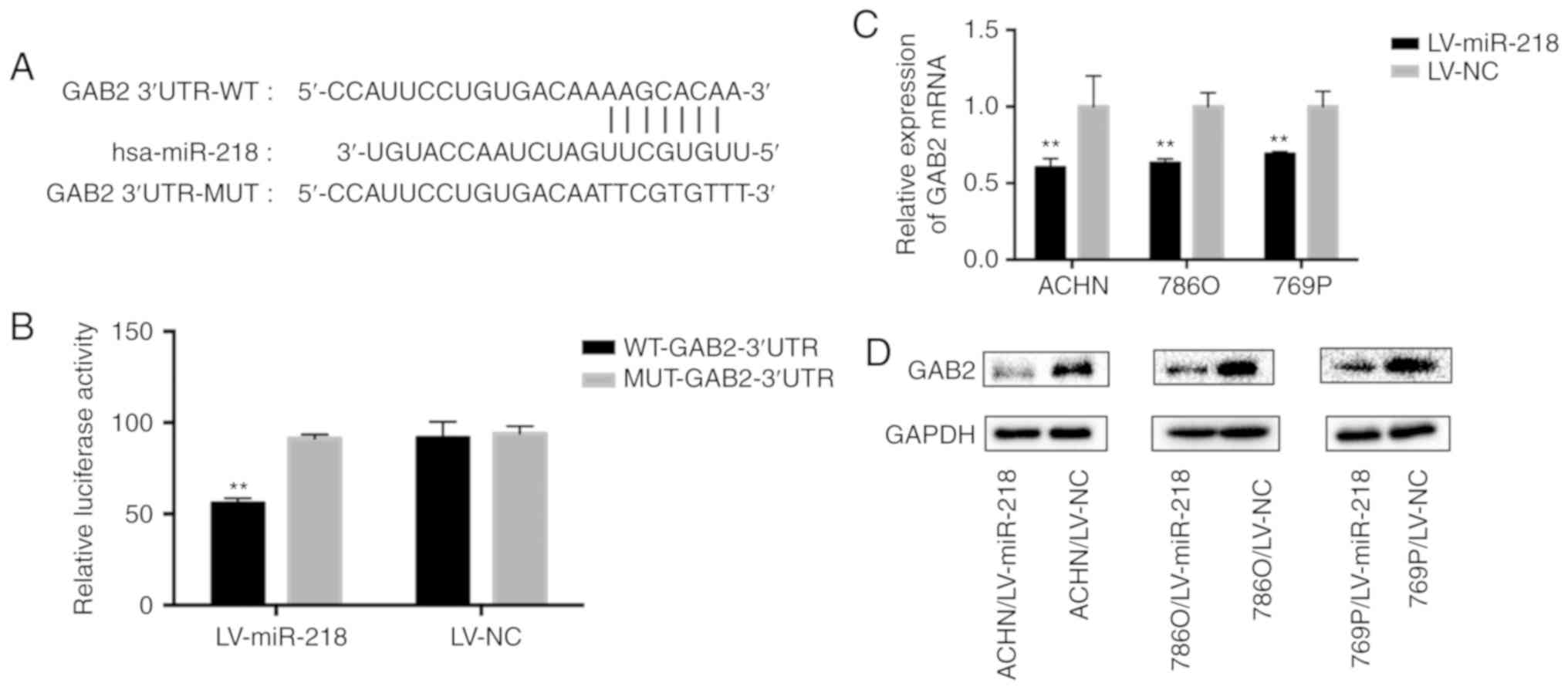
miR-218 targets GAB2 by binding to
3′-UTR
In order to find the binding site of miR-218, we
searched the TargetScan database and selected GAB2 (Fig. 3A). Then, a dual luciferase reporter
gene assay was performed. The wild-type (WT) and mutant (MUT)
sequences of the GAB2 3′UTR were cloned into the reporter plasmids
(Fig. 3A). Overexpression of
miR-218 was able to repress the luciferase activity (55%) of the
wild-type (WT) reporter, but not the mutant (MUT) reporter
(Fig. 3B), indicating that miR-218
is capable of specifically targeting the 3′UTR of GAB2.
Furthermore, the expression of GAB2 in RCC cell lines at both the
mRNA and protein level was indeed significantly suppressed by the
overexpression of miR-218 (Fig. 3C and
D), when compared to the LV-NC group.
GAB2 plays a crucial role in RCC
angiogenesis
In order to further investigate the mechanism of
miR-218 in regulating RCC angiogenesis by targeting GAB2,
GAB2-knockdown RCC cell lines were constructed using ACHN and 786O
cells, and the knockdown efficiency was confirmed at both the mRNA
and protein levels (Fig. 4A and B).
Consistently, after downregulating GAB2, the migration (Fig. 4C and D) and tube formation (Fig. 4E) ability of HUVECs were
significantly inhibited, and the EMT of RCC cells was also slightly
suppressed (Fig. S2, right panel).
In addition, western blot analysis (Fig. 4B) and ELISA (Fig. 4F) assays revealed a significant
decrease in VEGFA protein expression and secretion in the shGAB2
group when compared with the shNC group.
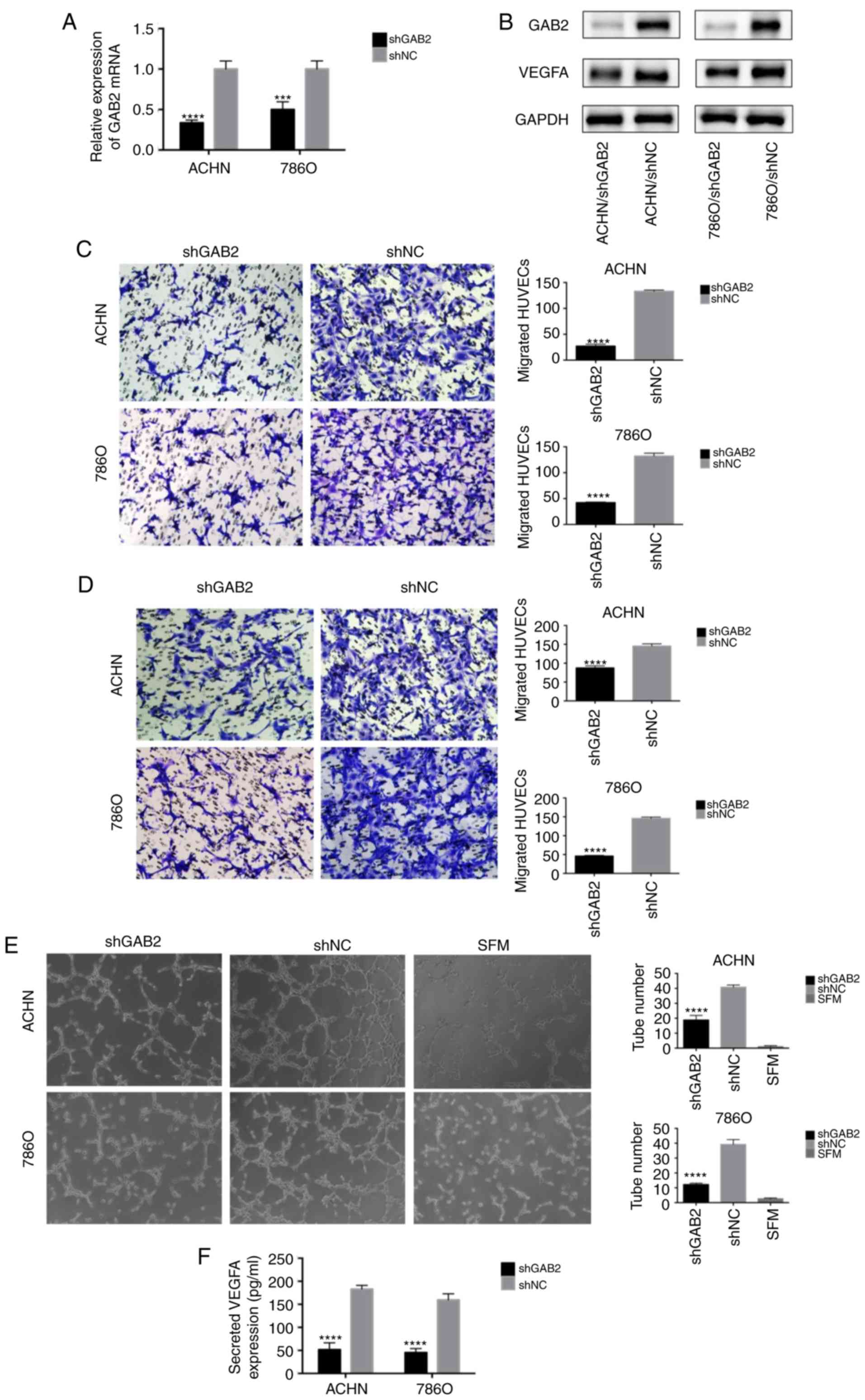 | Figure 4.GAB2 plays a crucial role in RCC
angiogenesis. (A and B) Real-time PCR and western blot analysis
were used to confirm the knockdown of GAB2 in ACHN and 786O cells
transfected with LV-shGAB2 both at the mRNA and protein level. GAB2
knockdown decreased VEGFA at the protein level. ***P<0.001 and
****P<0.0001 compared to the shNC group. (C) GAB2 knockdown
decreased the recruitment of HUVECs in a co-cultured system. Cancer
cells that were cultured in the bottom of a 24-well plate were used
to recruit HUVECs. ****P<0.0001 compared to the shNC group. (D)
GAB2 knockdown decreased the recruitment of HUVECs through the
conditioned medium (CM) collected from the ACHN, 786O/LV-shGAB2,
and ACHN, 786O/LV-shNC cells. The migrated cells in six random
fields per well were counted (magnification, ×200). ****P<0.0001
compared to the shNC group. (E) GAB2 knockdown reduced the tube
formation of HUVECs diluted in SFM or CMs. The representative
images of tube-like structures are shown, and the tube numbers in
the whole field were counted (magnification, ×100). ****P<0.0001
compared to the shNC group. (F) The concentration of secreted VEGFA
protein in the CMs was determined by ELISA. The values were
presented as the mean ± standard deviation (SD). GAB2,
GRB2-associated binding protein 2; RCC, renal cell carcinoma;
HUVECs, human umbilical vein endothelial cells; SFM, serum-free
medium; CM, conditioned medium; VEGFA, vascular endothelial growth
factor A. |
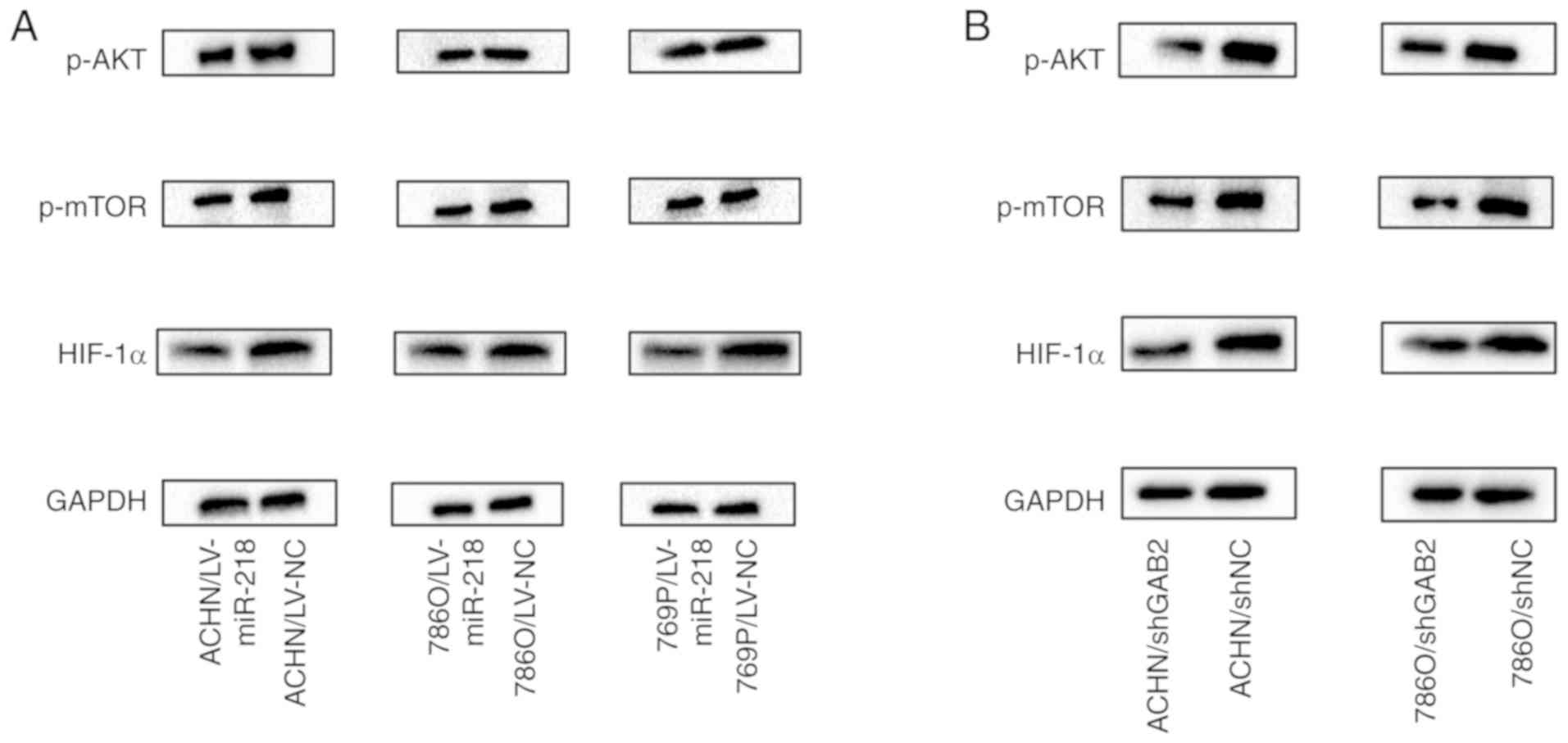
miR-218 regulates the PI3K/AKT/mTOR
axis by targeting GAB2
Overexpression of miR-218 in RCC cells not only
reduced GAB2 expression, but also inhibited the phosphorylation of
AKT (p-AKT) at position 473, the phosphorylation of mTOR (p-mTOR),
and the protein expression of HIF-1α (Figs. 5A and S3). Consistently, knockdown of GAB2 in
RCC cells exhibited similar results (Fig. 5B), indicating that miR-218 exerts
its role on inhibition of RCC angiogenesis through the
GAB2/PI3K/AKT/mTOR/HIF-1α/VEGFA axis.
miR-218 inhibits RCC tumor
angiogenesis in vivo
In order to investigate the effect of miR-218 on the
tumorigenicity of RCC cells in vivo, miR-218-overexpressing
(LV-miR-218)/control (LV-NC) 786O cells were subcutaneously
injected into the flanks of nude mice. The tumor growth was slower
in the 786O/LV-miR-218 group, showing significantly smaller tumor
size and lower tumor weight, when compared to the control group
(Figs. 6A and B and S4). In addition, the immunohistochemistry
(IHC) staining exhibited fewer CD31 (endothelial cell
marker)-positive tissues in the 786O/LV-miR-218 tumors, when
compared to the LV-NC group. Similar results were also observed in
the PCNA (proliferating cell nuclear antigen), GAB2 and VEGFA
staining in these tumor tissues (Fig.
6C-F).
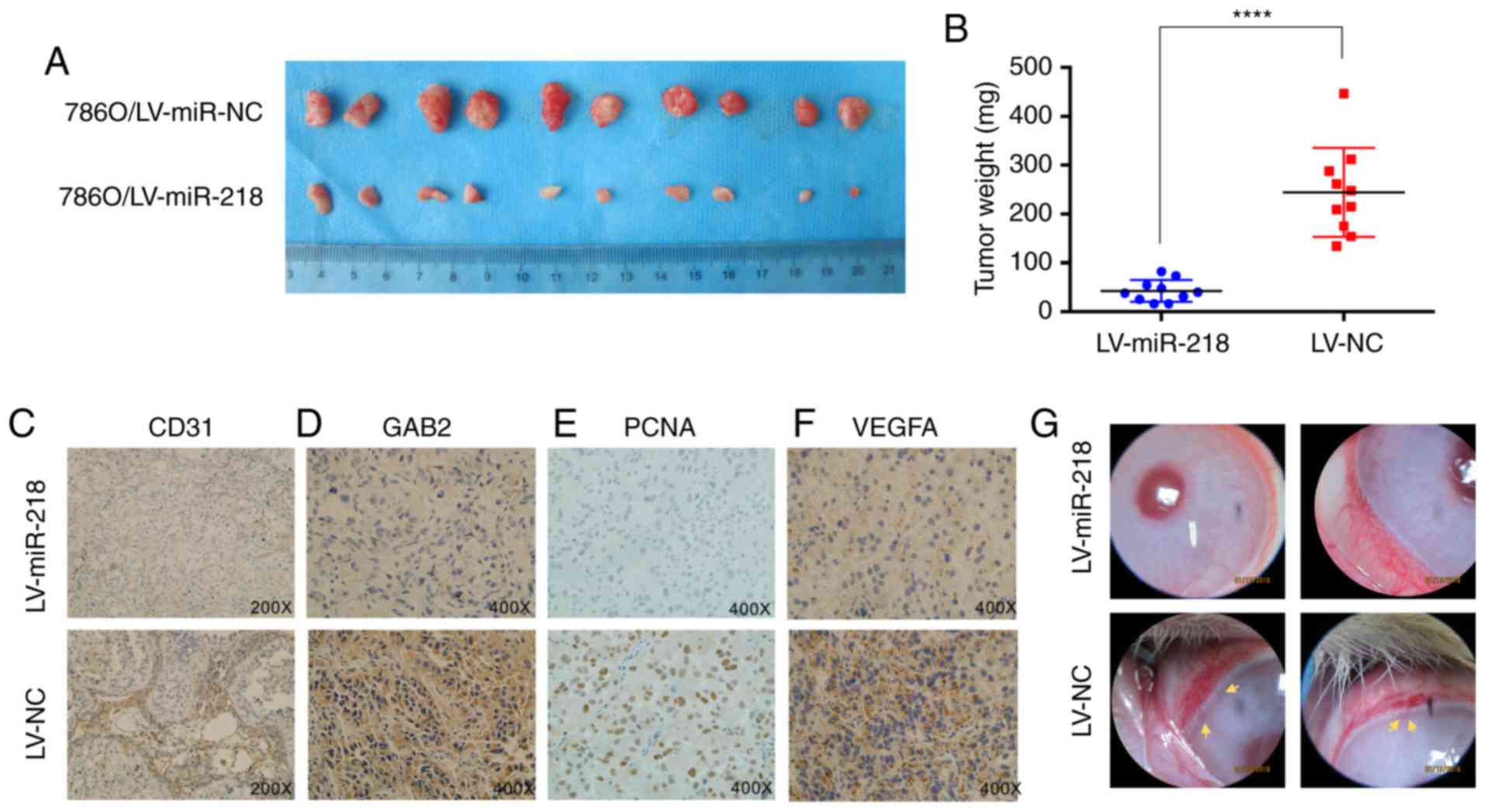 | Figure 6.miR-218 inhibits tumor growth and
angiogenesis in vivo. (A) Subcutaneous xenografts of
786O/LV-miR-218 and 786O/LV-NC subclones were harvested at four
weeks after inoculation. (B) The tumor weights between the two
groups. The values are presented as mean ± standard deviation (SD).
****P<0.0001. (C-F) Expression of CD31, GAB2, PCNA and VEGFA
were analyzed in paraffin-fixed tumor sections from 786O/LV-miR-218
and 786O/LV-NC xenografts by immunohistochemistry. Representative
images are shown at magnification ×200 (CD31) or ×400 (GAB2, PCNA
and VEGFA). (G) The 786O/LV-NC cells, but not the 786O /LV-miR-218
cells, induced the angiogenesis in rabbit cornea (n=3). Neonatal
vessels invaded the cornea, connecting the tumor (yellow arrow) and
the cornea limbal vascular plexus. GAB2, GRB2-associated binding
protein 2; RCC, renal cell carcinoma; PCNA, proliferating cell
nuclear antigen; VEGFA, vascular endothelial growth factor A. |
The rabbit cornea angiogenesis assay is a more
specific animal model to evaluate tumor angiogenesis. Therefore, a
tumorigenic test in the cornea of rabbit eyes was performed using
786O/LV-miR-218 and 786O/LV-NC cells. At four weeks after tumor
transplantation, 786O/LV-NC cells induced a neovascular response
and visible tumors in the cornea. However, 786O/LV3-miR-218 cells
lost this ability (Fig. 6G).
Overall, this demonstrated that miR-218 not only regulates RCC
tumorigenicity, but also plays an important role in suppressing RCC
angiogenesis.
Discussion
With the deepening investigation on microRNAs
(miRNAs/miRs), the basic characteristics and biological functions
of miRNAs in regards to development have been recognized. As for
miR-218, studies have confirmed that it can inhibit cancer cell
proliferation, migration, invasion and metastasis, and that it is a
tumor suppressor in prostate cancer, oral squamous cell carcinoma,
lung cancer, gastric cancer, head and neck squamous cell carcinoma,
and several other tumors (12–14,16).
There is an obvious inhibition or mutation of miR-218 expression in
tumors, which may be correlated to the
hypermethylation/demethylation of CpG islands around the miRNA
promoter and histone H3 acetylation (19). Leite et al (20) reported that metastatic prostate
cancer presents with sharp decreases in miR-218, when compared to
high grade prostate intraepithelial neoplasia (HGPIN) and localized
prostate cancer, indicating the expression change of miR-218
involved in the progression of metastatic prostate cancer, and that
angiogenesis is one of the key steps in this process. Researchers
have used HITS-CLIP technology to demonstrate that miR-218 targets
multiple functional genes, including nuclear factor-κB subunit 1
(NF-κB), roundabout guidance receptor 1 (ROBO1), LIM and SH3
protein 1 (LASP1), paxillin (PXN), cyclin-dependent kinase 6 (CDK6)
and cathepsin B (CTSB), and regulates multiple signaling pathways,
including cell proliferation, circulation, metabolism and cell
viability (21,22). In the present study, it was
demonstrated that miR-218 is lowly expressed in RCC tissues and
cells, and that RCC samples with high expression of miR-218
presented with extended overall survival, suggesting that the loss
of miR-218 may play a key regulatory role in the development of
RCC. In addition to the high miR-218 expression at lower ages,
intriguingly, in the TNM staging and grading of RCC, miR-218
presented a reverse tendency: Higher in the more malignant phases.
Moreover, the overall survival rate of RCC samples also shown an
opposite trend: Lower miR-218 expression in higher overall survival
rate, which probably means that the miR-218-related regulation is
complex, and that its clinical role needs to be explored from
multiple aspects (Table SII).
It is known that one main mechanism of RCC
metastasis is tumor angiogenesis, which is an essential
prerequisite for tumor growth and metastasis. As early as 1971,
Folkman first proposed that tumor growth depends on angiogenesis
(23). RCC has been widely accepted
as a vascular-rich tumor (24). In
the early stage of tumorigenesis, the tumor grows slowly due to the
lack of a neovascular system. However, as tumor-secreted factors
increase, the neovasculature begins to form. Thus, the tumor
rapidly grows. Without the support of neovascularization, these
tumors would not exceed 2–3 mm. On the other hand, with
neovascularization, some tumor cells can invade into the vascular
system, resulting in metastasis. Therefore, attenuating this
process can reduce or even prevent tumor growth and metastasis
(25,26). Vascular endothelial growth factor
(VEGF) family is one of the most important factors in all
angiogenic molecules. In general, VEGF refers to VEGFA, which is a
highly specific factor that promotes vascular permeability,
extracellular matrix degeneration, vascular endothelial cell
proliferation, migration and angiogenesis (27). In most RCC patients, there is an
excessive expression of VEGFA (28), Therefore, inhibition of VEGF
expression is one of the sources that can inhibit RCC angiogenesis.
In the present study, miR-218 was found to be overexpressed in RCC
cells. This inhibited HUVEC migration, proliferation and tube
formation in vitro, and repressed tumor growth and
angiogenesis in vivo. Therefore, it was hypothesized that
miR-218 may affect the function of HUVECs by regulating the
secretion of VEGFA in RCC cells. Indeed, the overexpression of
miR-218 in RCC cells reduced the expression and secretion of VEGFA,
thereby abrogating the angiogenic ability of the HUVECs. Since the
effect of miR-218 on angiogenesis in RCC has not been reported to
date, the present study demonstrated for the first time that
miR-218 is of great significance in regulating angiogenesis in
RCC.
In recent years, it has been found that GAB2 protein
is highly expressed in many malignant tumors, and its abnormal
expression can mediate the interaction between proteins, affecting
various signaling pathways, and regulating tumor cell
proliferation, apoptosis and migration (29–32).
It has been confirmed that a specific tyrosine residue in the GAB2
protein binds to the P85 subunit in PI3K, activating PI3K and
producing PIPs that bind to the PH domain in the GAB2 structure.
The enhancement of the membrane receptor recruitment of GAB2
results in a positive feedback loop that ultimately leads to
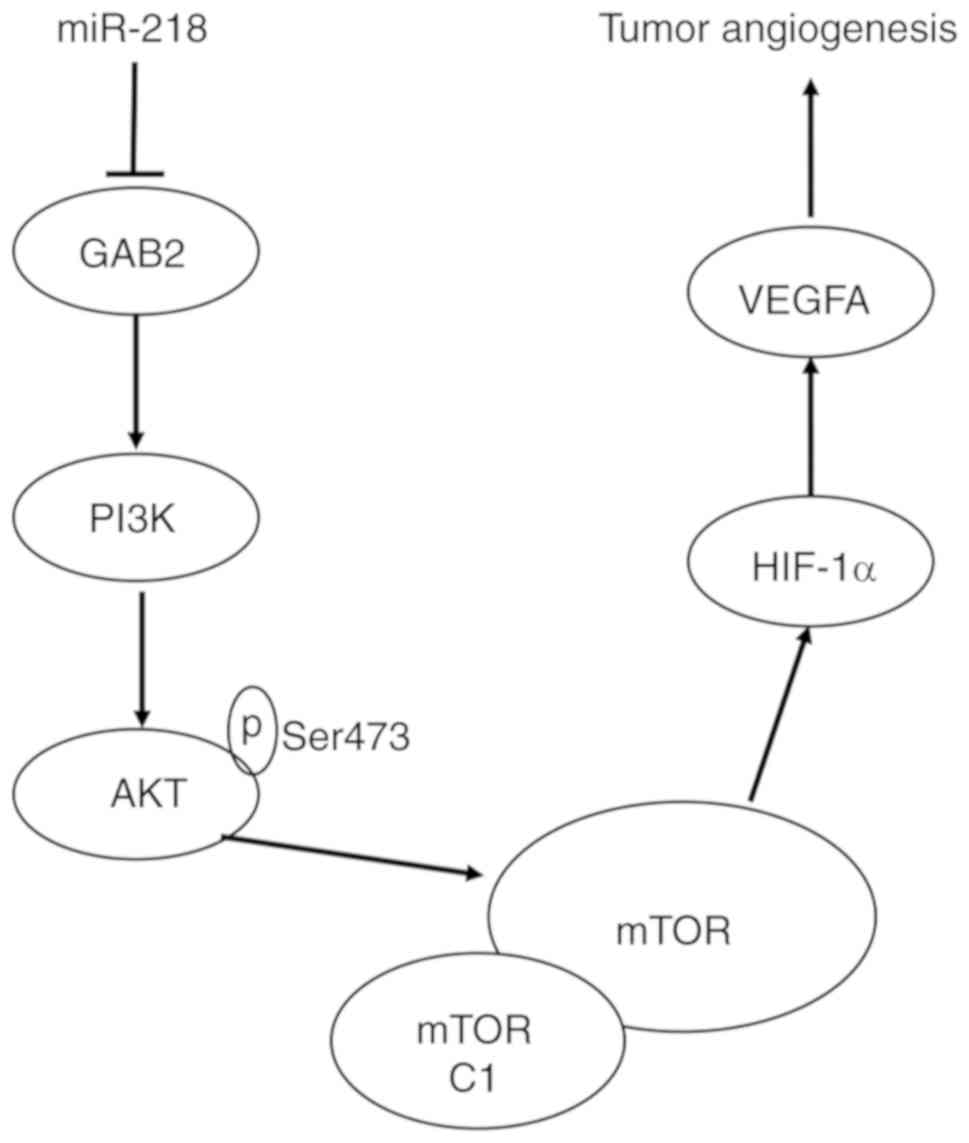
GAB2-mediated PI3K/AKT signaling pathway amplification (33). Downstream molecules AKT, mTOR,
HIF-1α, and VEGFA are important components of the angiogenic
pathway. In the present study, it was demonstrated that miR-218
negatively regulates GAB2 by binding to the specific target sites
within the 3′UTR of GAB2, repressing the expression of GAB2
protein. In addition, the phosphorylation status of AKT at Ser473,
and the expression of mTOR and HIF-1α are decreased after
overexpression of miR-218. Based on these results, it can be
concluded that miR-218 exerts its role in inhibiting RCC
angiogenesis by inhibiting the GAB2/PI3K/AKT/mTOR/HIF-1α/VEGFA
pathway (Fig. 7). In addition,
research has revealed that the upregulation of HIF-1α and VEGF
mediated by the overexpression of GAB2 in mouse xenograft models
promotes tumor angiogenesis (34).
This is consistent with the present results, and further proves the
validity of the present conclusions. However, this research still
has limitations. We did not detect the expression of miR-218 in
clinical patient samples. Yet, we will be fully prepared and
incorporate clinical patient sample information in future research
to further confirm our results.
As endogenous non-coding small RNAs, miRNAs play
important roles in a variety of pathophysiological processes. With
the advancement of science technology, it is hoped that miRNAs may
provide new biomarkers for the diagnosis and prognosis of RCC, and
therapeutic targets for the drug development for RCC treatment.
Many miRNAs associated with RCC have not been reported to date, and
the known miRNAs, such as miR-218, are still not well-researched in
regards to RCC development and metastasis. The present study
demonstrated that miR-218 significantly repressed RCC
tumorigenicity and angiogenesis, providing new insights into the
mechanism of RCC carcinogenesis and progression, and suggesting
that miRNA-218 may be a target for the treatment of RCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National
Natural Science Foundation of China grants (no. 81372736 to
YD).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available in the TCGA database.
Authors' contributions
DW and YD were involved in the design and
conceptualization of the study. LM and BG performed the experiments
and data generation. JT and XL were involved in the database
analysis. QL, MW and WW contributed to the methodology and data
curation. LM, BG, JS, XL and YD were involved in the writing,
reviewing and editing of the manuscript. JS and XL critically
revised the manuscript. YD supervised the study. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Xi'an Jiaotong
University (2013065).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lebacle C, Pooli A, Bessede T, Irani J,
Pantuck AJ and Drakaki A: Epidemiology, biology and treatment of
sarcomatoid RCC: Current state of the art. World J Urol.
37:115–123. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Posadas EM and Figlin RA: Systemic therapy
in renal cell carcinoma: Advancing paradigms. Oncology (Williston
Park). 26:290–301. 2012.PubMed/NCBI
|
|
6
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gregory RI and Shiekhattar R: MicroRNA
biogenesis and cancer. Cancer Res. 65:3509–3512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liang H, Ge F, Xu Y, Xiao J, Zhou Z, Liu R
and Chen C: miR-153 inhibits the migration and the tube formation
of endothelial cells by blocking the paracrine of angiopoietin 1 in
breast cancer cells. Angiogenesis. 21:849–860. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vosgha H, Ariana A, Smith RA and Lam AK:
miR-205 targets angiogenesis and EMT concurrently in anaplastic
thyroid carcinoma. Endocr Relat Cancer. 25:323–337. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kinoshita T, Hanazawa T, Nohata N, Kikkawa
N, Enokida H, Yoshino H, Yamasaki T, Hidaka H, Nakagawa M, Okamoto
Y and Seki N: Tumor suppressive microRNA-218 inhibits cancer cell
migration and invasion through targeting laminin-332 in head and
neck squamous cell carcinoma. Oncotarget. 3:1386–1400. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Uesugi A, Kozaki K, Tsuruta T, Furuta M,
Morita K, Imoto I, Omura K and Inazawa J: The tumor suppressive
microRNA miR-218 targets the mTOR component Rictor and inhibits AKT
phosphorylation in oral cancer. Cancer Res. 71:5765–5778. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S,
Guo X, Wang B, Gang Y, Zhang Y, et al: MiR-218 inhibits invasion
and metastasis of gastric cancer by targeting the Robo1 receptor.
PLoS Genet. 6:e10008792010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Small EM and Olson EN: Pervasive roles of
microRNAs in cardiovascular biology. Nature. 469:336–342. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guan B, Wu K, Zeng J, Xu S, Mu L, Gao Y,
Wang K, Ma Z, Tian J, Shi Q, et al: Tumor-suppressive microRNA-218
inhibits tumor angiogenesis via targeting the mTOR component RICTOR
in prostate cancer. Oncotarget. 8:8162–8172. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao Y, Wu K, Chen Y, Zhou J, Du C, Shi Q,
Xu S, Jia J, Tang X, Li F, et al: Beyond proliferation: KLF5
promotes angiogenesis of bladder cancer through directly regulating
VEGFA transcription. Oncotarget. 6:43791–43805. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Incoronato M, Urso L, Portela A, Laukkanen
MO, Soini Y, Quintavalle C, Keller S, Esteller M and Condorelli G:
Epigenetic regulation of miR-212 expression in lung cancer. PLoS
One. 6:e277222011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leite KR, Tomiyama A, Reis ST,
Sousa-Canavez JM, Sañudo A, Camara-Lopes LH and Srougi M: MicroRNA
expression profiles in the progression of prostate cancer-from
high-grade prostate intraepithelial neoplasia to metastasis. Urol
Oncol. 31:796–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grammatikakis I, Gorospe M and Abdelmohsen
K: Modulation of cancer traits by tumor suppressor microRNAs. Int J
Mol Sci. 14:1822–1842. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Venkataraman S, Birks DK, Balakrishnan I,
Alimova I, Harris PS, Patel PR, Handler MH, Dubuc A, Taylor MD,
Foreman NK and Vibhakar R: MicroRNA 218 acts as a tumor suppressor
by targeting multiple cancer phenotype-associated genes in
medulloblastoma. J Biol Chem. 288:1918–1928. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Folkman J: What is the evidence that
tumors are angiogenesis dependent? J Natl Cancer Inst. 82:4–6.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Karumanchi SA, Merchan J and Sukhatme VP:
Renal cancer: Molecular mechanisms and newer therapeutic options.
Curr Opin Nephrol Hypertens. 11:37–42. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Folkman J: Angiogenesis in cancer,
vascular, rheumatoid and other disease. Nat Med. 1:27–31. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Aishima S, Taguchi K, Sugimachi K, Asayama
Y, Nishi H, Shimada M, Sugimachi K and Tsuneyoshi M: The role of
thymidine phosphorylase and thrombospondin-1 in angiogenesis and
progression of intrahepatic cholangiocarcinoma. Int J Surg Pathol.
10:47–56. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ferrara N: VEGF and the quest for tumour
angiogenesis factors. Nat Rev Cancer. 2:795–803. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jacobsen J, Grankvist K, Rasmuson T, Bergh
A, Landberg G and Ljungberg B: Expression of vascular endothelial
growth factor protein in human renal cell carcinoma. BJU Int.
93:297–302. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wöhrle FU, Daly RJ and Brummer T:
Function, regulation and pathological roles of the Gab/DOS docking
proteins. Cell Commun Signal. 7:222009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sattler M, Mohi MG, Pride YB, Quinnan LR,
Malouf NA, Podar K, Gesbert F, Iwasaki H, Li S, Van Etten RA, et
al: Critical role for Gab2 in transformation by BCR/ABL. Cancer
Cell. 1:479–492. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y, Sheng Q, Spillman MA, Behbakht K
and Gu H: Gab2 regulates the migratory behaviors and E-cadherin
expression via activation of the PI3K pathway in ovarian cancer
cells. Oncogene. 31:2512–2520. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bentires-Alj M, Gil SG, Chan R, Wang ZC,
Wang Y, Imanaka N, Harris LN, Richardson A, Neel BG and Gu H: A
role for the scaffolding adapter GAB2 in breast cancer. Nat Med.
12:114–121. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gu H, Pratt JC, Burakoff SJ and Neel BG:
Cloning of p97/Gab2, the major SHP2-binding protein in
hematopoietic cells, reveals a novel pathway for cytokine-induced
gene activation. Mol Cell. 2:729–740. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang Y, Wu J, Demir A, Castillo-Martin M,
Melamed RD, Zhang G, Fukunaga-Kanabis M, Perez-Lorenzo R, Zheng B,
Silvers DN, et al: GAB2 induces tumor angiogenesis in NRAS-driven
melanoma. Oncogene. 32:3627–3637. 2013. View Article : Google Scholar : PubMed/NCBI
|