Introduction
Malignant pleural mesothelioma (MPM) is a rare
malignancy that originates from the pleural mesothelium and is
associated with an extremely poor patient prognosis. MPM is
strongly associated with asbestos exposure (1) and has a long latency period (20–40
years) following asbestos exposure (2). Asbestos has been banned in Japan since
2006, but there are countries where asbestos is still mined and
used. The incidence of MPM has increased over the last decade and
is predicted to peak sometime before 2030 (2). Moreover, the death toll from asbestos
exposure is expected to rise. The symptoms of MPM include chest
tightness, shortness of breath, and coughing; these symptoms are
often caused by pleural effusion. Unfortunately, MPM frequently
does not show early symptoms, and therefore, the disease is often
progressed by the time it is diagnosed. Therefore, it is extremely
important to diagnose MPM in its early stages.
Diagnosing MPM is often difficult. Comprehensive
diagnosis consists of biopsy of the suspected tumor lesion and
measuring pleural effusion. In the case of epithelioid MPM, a
cytological diagnosis can be conducted by examining pleural
effusion (3). In contrast,
sarcomatoid MPM is very difficult to diagnose via pleural effusion
examination. Secondary markers of MPM, such as mesothelin (4), osteopontin (5), and fibulin-3 (6) have been reported. However, they are
not necessarily reliable diagnostic markers due to their
variability in values. Therefore, there are still no useful markers
for MPM diagnosis.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs
(approximately 20–22 nucleotides) that function in diverse
biological processes and regulate the expression of target genes.
Many researchers have reported that miRNAs are associated with
cancer development, progression, and metastasis (7–9). In
MPM, miRNAs have recently gained attention as diagnostic and
therapeutic targets. For example, it has been reported that the
miR-200 family can be as potential candidates with which to
differentiate MPM from other cancers in a diagnostic setting
(10).
Extracellular vesicles (EVs) include exosomes,
microvesicles, and apoptotic bodies. EVs consist of a lipid bilayer
that envelops diverse proteins, mRNAs, and miRNAs. In terms of
diameter, exosomes range between 20 and 150 nm, microvesicles
between 100 nm and 1 µm, and apoptotic bodies are more than 1 µm.
While exosomes and microvesicles are produced differently, both are
secreted from intact cells and circulate throughout the body
(11). Therefore, EVs can be found
in any fluid (blood, saliva, urine, ascites fluid and pleural
effusion). Exosomes and microvesicles are taken up by other cells
via endocytosis, and it has been shown that miRNAs contained in EVs
function in cells that they are taken up by (12–15).
These findings have garnered a wave of attention towards
functionalized EVs as intercellular transmitters.
There is not yet a standard purification method to
individually isolate exosomes and microvesicles. For EV
purification, ultracentrifugation, exosome capturing using proteins
expressed on the exosome membrane, and size exclusion
chromatography are used. However, separating exosomes and
microvesicles may be difficult. In this study, we purified EVs
containing exosomes and microvesicles in a diameter-dependent
manner with membrane filters. This method is simple and easy. Using
this method, six miRNAs in EVs that were secreted by MPM cells but
not normal mesothelial cells were selected. We believe that these
miRNAs are promising targets for MPM diagnosis via blood and
pleural effusion measurements.
Materials and methods
Cell culture
We used six malignant pleural mesothelioma (MPM)
cell lines (ACC-MESO1, ACC-MESO4, MSTO-211H, L324, N407, and K921)
and a normal pleural mesothelial cell line (MeT-5A). ACC-MESO1 and
ACC-MESO4 were obtained from the Riken Cell Bank (Tsukuba, Japan).
MSTO-211H was purchased from the American Type Culture Collection
(ATCC, Manassas, VA, USA). L324, N407, and K921 cells were
generated in the laboratory and kindly donated by Dr Hidetaka
Uramoto, Kanazawa Medical University, Ishikawa (16). MeT-5A cells were purchased from
ATCC. Human prostate cancer PC3 cells were obtained as previously
described (17). Immortalized human
pulmonary fibroblast (IHPF) cells (T0490) were purchased from
Applied Biological Materials Inc. (Richmond, BC, Canada). MPM,
MeT-5A, PC3, and IHPF cells were maintained in RPMI-1640 medium,
Medium 199, DMEM (Thermo Fisher Scientific, Inc.), and Prigrow III
medium (Applied Biological Materials Inc.), respectively. All cells
were cultured with GlutaMAX supplement (Thermo Fisher Scientific,
Inc.), 10% heat-inactivated fetal bovine serum (FBS), and 1% (v/w)
penicillin/streptomycin, and were maintained at 37°C in 5%
CO2.
Preparation of GFP/Nluc-fused CD9,
CD63, CD81, hsa-miR-4728-5p, hsa-miR-193a-5p, and hsa-miR-551b-5p
expression plasmids
CD63-green fluorescent protein (GFP) (CYTO122-PA-1),
CD9-GFP (CYTO120-PA-1), and CD81-red fluorescent protein (RFP)
(CYTO125-PA-1) expression plasmids were purchased from System
Biosciences. The pNL1.1 vector containing nanoluciferase (Nluc) was
purchased from Promega Corp. CD63-Nluc, CD9-Nluc, CD81-Nluc, and
CD81-GFP expression plasmids were constructed with these plasmids.
GFP fusion proteins were used for qualitative analysis of
intracellular expression and intracellular uptake. Nluc fusion
proteins were used for quantitative analysis of extracellular
secretion and intracellular uptake. Double-stranded
oligonucleotides against hsa-miR-4728-5p, hsa-miR-193a-5p, and
hsa-miR-551b-5p were synthesized (Table SI) and ligated into multicloning
sites of pSIH1-H1-GFP-T2A-Puro, which was originally constructed
with pSIH1-H1-copGFP (SI501A-1, System Biosciences) and
pCDH-EF1-MCS-BGH-PGK-GFP-T2A-Puro (CD550A-1, System
Biosciences).
Expression by the lentivirus
system
Lentivirus-containing expression cassettes of
GFP/Nluc fusion genes and miRNAs were obtained using the System
Biosciences lentivirus packaging system. Lentiviral transduction
was performed according to the manufacturer's protocol. Briefly,
after infecting the MeT-5A cells, cells expressing CD63-GFP/Nluc,
CD9-GFP/Nluc, CD81-GFP/Nluc were selected using 10 µg/ml of
puromycin. In the same way, MeT-4728-5p, MeT-193a-5p and
MeT-551b-5p cells expressing hsa-miR-4728-5p, hsa-miR-193a-5p and
hsa-miR-551b-5p, respectively, were obtained by infection and
selection with MeT-5A cells. MeT-ctrl cells did not express miRNA.
Fused GFPs or independently expressed GFP of the Nluc fusion
protein and miRNAs were observed using an EVOS fluorescence
microscope (Thermo Fisher Scientific, Inc.). After confirming that
over 90% of the cells were expressing GFP using LUNA-FL™ (Logos
Biosystems, Korea), they were used in the assay.
Isolation and removal of EVs
Syringe filters with sizes of 220 nm (SFPES013022N),
50 nm (SF16008), and 20 nm (2.CF7103.0001) were purchased from
Membrane Solutions, TISCH Scientific, and ANPEL Laboratory
Technologies, respectively. In our laboratory, FBS was passed
through a 220-nm bottle-top vacuum filter (Thermo Fisher
Scientific, Inc.) (FBSpas220) to maintain normal cells. To remove
EVs from FBS, FBSpas220 was passed through 50-nm syringe filters
(FBSpas50). When cells grown in 10-cm dishes containing maintenance
medium were 80% confluent, they were collected and seeded into
three 10-cm dishes containing FBSpas50. After 48 h, the culture
medium was collected and passed sequentially through 220 nm and 50
nm syringe filters. EVs captured by the 50 nm syringe filter
(EVcap50/220) were partially collected by reverse-flow with
phosphate-buffered saline (PBS) (EVrev50/220). The EVrev50/220 and
EVs that had passed through a 50 nm syringe filter (EVpas50) were
analyzed by dynamic light scattering (DLS, Zetasizer, Malvern
Panalytical, UK) and transmission electron microscopy (TEM, JEOL
1200EX; JEOL, Japan). EVcap50/220 was used directly for RNA
extraction and protein extraction, as outlined in the procedures
below.
Dynamic light scattering (DLS) and
transmission electron microscopy (TEM) were performed
The diameter of EVs obtained by reverse flow was
measured by DLS (Zetasizer) according to the manufacturer's
instructions, and as previously described (18). Samples were prepared using an
Exosome TEM-easy kit (101 Bio, Mountain View, CA, USA) with EVs
obtained from reverse flow; observation was carried out on a TEM
(JEOL 1200EX).
Cell proliferation assay
PC3 cells (1,000 cells) were seeded into 12-well
plates. Cell number was measured the next day and was used as the
baseline at 0 h. PC3 cells were washed twice with PBS, and were
added to either medium without FBS, medium containing FBSpas220
(normal medium), or medium containing FBSpas50. Cells were
collected every 24 h, and the number of cells was calculated by
LUNA-FL™ (Logos Biosystems) according to manufacturer's
instructions. Experiments were performed in triplicates.
Protein extraction and western blot
analysis
When MeT-5A cells grown in 10-cm dishes containing
maintenance medium were 50% confluent, they were washed twice with
PBS, and added with FBSpas50-containing medium. After 48 h,
cultured medium or fresh medium containing FBSpas50 (for the
negative control) was collected and passed sequentially through 220
and 50 nm syringe filters. Proteins of EVcap50/220 were eluted with
2 ml CHAPS lysis buffer (Dojindo) containing 1 mM PMSF. Each eluted
solution was concentrated using Amicon Ultra 10 kDa (Merck
Millipore) until its volume was less than 30 µl. All samples were
subjected to sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and were transferred to polyvinylidene
difluoride (PVDF) membranes. The membrane was immunoblotted first
with anti-EpCAM antibody (EBA-1, dilution 1:1,000, Santa Cruz
Biotechnology, Inc.), CD63 antibody (TS63, dilution 1:1,000, Thermo
Fisher Scientific, Inc.), CD9 antibody (TS9, dilution 1:1,000,
Thermo Fisher Scientific, Inc.), CD81 antibody (M38, dilution
1:1,000, Thermo Fisher Scientific, Inc.). Secondary antibodies
conjugated with horseradish peroxidase were used for detection.
Signal intensity was obtained using LAS 4000 Mini and Multi Gauge
software version 3.0 (Fujifilm), as previously described (17).
Expression of GFP/Nluc fusion protein
and analysis of extracellular secretion
MeT-5A cells expressing and secreting CD9-GFP/Nluc,
CD63-GFP/Nluc, and CD81-GFP/Nluc were cultured in plates or dishes
with medium containing FBSpas50. These cells are called donor cells
because the fusion proteins secreted by these cells are taken up by
other cells. After the indicated hours, cultured medium or cells
were collected. Each medium was passed through a 220 nm or 50 nm
syringe filter and was used for Nluc assays. Cells were washed
twice with PBS and lysed with Reporter Lysis Buffer (Promega
Corp.). The cell lysate was vortexed, centrifuged (21,000 × g for 2
min), and the supernatant was collected for Nluc assays. Nluc
activity was measured using the Nano-Glo® Luciferase
Assay System (Promega Corp.) according to the manufacturer's
instructions. In order to observe secreted EVs with the GFP fusion
protein via an EVOS fluorescence microscope (Thermo Fisher
Scientific, Inc.), EVs were concentrated using a Total Exosome
Isolation kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions.
Efficiency of EV capture by membrane
filters
Culture medium of MeT-5A cells that expressed
CD81-Nluc was passed through a 220-nm syringe filter (EVpas220).
EVpas50/220 and EVpas20/50 were obtained by passing EVpas220
through a 50-nm syringe filter and by passing the EVpas50/220
through a 20-nm syringe filter, respectively. Nluc activities of
EVpas220, EVpas50/220, and EVpas20/50 were measured as described
above.
Uptake assay of the GFP/Nluc fusion
protein
To obtain the GFP fusion protein, MeT-5A cells
expressing CD9-GFP, CD63-GFP, and CD81-GFP (donor cells) were
cultured with 10 ml Opti-MEM medium without FBS for 48 h. Each
culture medium was passed through a 220-nm syringe filter and
concentrated to 30 µl or less by Amicon Ultra-4 (Merck Millipore).
They were added to the culture medium of recipient cells that were
able to take up the GFP fusion protein. After 8 h, cells were
washed twice with PBS, and were observed under an EVOS fluorescence
microscope (Thermo Fisher Scientific, Inc.). For the Nluc fusion
protein, MeT-5A cells expressing CD81-Nluc were cultured in medium
containing FBSpas50. After 48 h, each culture medium was passed
through 220 and 50-nm syringe filters, and was independently
replaced with media of recipient cells. After the allocated hours,
Nluc activities in cell lysates were measured as described above,
and were standardized to cell protein concentration. Uptake of EVs
with diameters between 50 and 220 nm was determined by
subtraction.
RNA extraction
The medium that passed through the 220-nm syringe
filter was passed through a 50-nm syringe filter to obtain
EVcap50/220. Total RNA of EVcap50/220 was eluted with 1 ml ISOGEN
(Nippongene). The eluted solution was mixed with 200 µl chloroform,
and was centrifuged at 12,000 × g for 15 min. Total RNA was
purified with 500 µl supernatant using a NucleoSpin®
miRNA Plasma Kit (Macherey-Nagel), which was used according to the
manufacturer's instructions. Total cell RNA was eluted in 100 µl
H2O, and was precipitated with 10 µl of 3.0 M potassium
acetate, 100 µl isopropanol, and 1 µl Pellet Paint. After washing
and drying the pellet, RNA was dissolved in 2 µl or 10 µl
H2O for microarray or quantitative real-time polymerase
chain reaction (qPCR), respectively. Verification of microRNA
purification was performed with Eukaryote Total RNA Nano Series II
using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). All
RNAs, including the miRNAs from cells, were purified using the
miRNeasy Mini Kit (Qiagen), according to the manufacturer's
instructions.
Microarray
Microarray analyses with the Human miRNA Oligo
chip-4 plex (TORAY, Tokyo, Japan) containing 2,565 probe sets and
the Human Oligo chip 25k (TORAY) containing 24,460 probe sets were
carried out using a 3D-Gene array system, according to the
manufacturer's protocol. For the miRNA array, 2 µl RNA solution
obtained from 30 ml medium or 250 ng total cell RNA were hybridized
with the Human miRNA Oligo chip. For the mRNA array, 1 µg total
cell RNA was used to hybridize the Human Oligo Chip 25k.
Quantitative real-time polymerase
chain reaction (qPCR)
For RNA obtained from EVcap50/220, the dry pellet
was dissolved with 10 µl of H2O, and 1 µl RNA was used
for each probe. For RNA obtained from cell lysates, RNA was
prepared according to the manufacturer's instructions. cDNA for the
miRNA was synthesized using the TaqMan MicroRNA Reverse
Transcription Kit (Thermo Fisher Scientific, Inc.) in a StepOne
Plus real-time PCR system (Thermo Fisher Scientific, Inc.). qPCR
was performed using a specifically designed TaqMan probe. Primer
sequences used to analyze human miRNAs and gene expression via qPCR
were provided by the following sources: miR-4728-5p, Thermo Fisher
Scientific, Inc., 461811_mat; miR-193a-5p, Thermo Fisher
Scientific, Inc., 002281; miR-551b-5p, Thermo Fisher Scientific,
Inc., 002346; U6, Thermo Fisher Scientific, Inc., 001973. The
cycling conditions were as follows: Denaturing, hold at 95°C for 20
sec, 40 cycles of amplification (denaturation at 95°C for 30 sec,
annealing and extension at 60°C for 30 sec). U6 was used as an
internal control. The Ct values for each miRNA were normalized to
U6, and relative gene expression was calculated using the ∆∆Ct
method. All samples were run in duplicates in each experiment.
Statistics
Statistical analysis was performed using GraphPad
Prism7 (GraphPad Software, Inc.). Results were compared using a
two-tailed Student's t-test and one-way analysis of variance
(ANOVA) followed by Turkey's test and Dunnett's test. Data are
expressed as mean ± SD. P<0.05 was considered to indicate a
statistically significant difference.
Results
Structural analysis of extracellular
vesicles (EVs)
In general, the cell culture medium was made by
adding fetal bovine serum (FBS) to the basic culture medium. Since
FBS contains EVs derived from bovine, it is necessary to remove
them in order to analyze cultured cell-derived EVs. We usually use
FBS passed through 220 nm (FBSpas220) for maintenance culture
medium. In this experiment, to remove EVs that were under 50 nm in
diameter, FBSpas220 were further passed through 50-nm filters
(FBSpas50). The efficiency of EV removal was analyzed via dynamic
light scattering (DLS) with EVs captured by 50-nm filters
(EVcap50/220), as well as EVs that passed through those filters
(EVpas50). As shown in Fig. 1A, the
diameter of EVcap50/220 was approximately 170 nm; however, this
peak disappeared as they flowed through the 50-nm filters (Fig. 1B). To evaluate the shape of
EVcap50/220, the EVs were collected by backflow (EVrev50/220). As
shown in Fig. 1C, EVrev50/220 was
spherical when analyzed using transmission electron microscopy
(TEM). These results suggest that EVs from 50 to 220 nm diameter
can be captured with 50 nm filters by passing the liquid
sequentially through 220 and 50 nm filters (Fig. S1). Next, we investigated the
effects of using FBSpas50 medium on cell growth. For this analysis,
we used hormone-insensitive PC3 cells (19) with markedly rapid cell growth. As
shown in Fig. 1D, cell
proliferation was increased with a culture medium containing
FBSpas50 when compared to FBSpas220-containing medium. This result
suggested that FBSpas50 can be used for analysis of cultured cells,
and that bovine-derived EVs inhibited cell growth. In this study,
when isolating cell-derived EVs, cells were cultured in medium
containing FBSpas50, and EVs secreted from the cells were captured
with filters of various diameters. By this method, FBS-derived EVs
are not contaminated during analysis with cell culture medium when
filters greater than 50 nm are used.
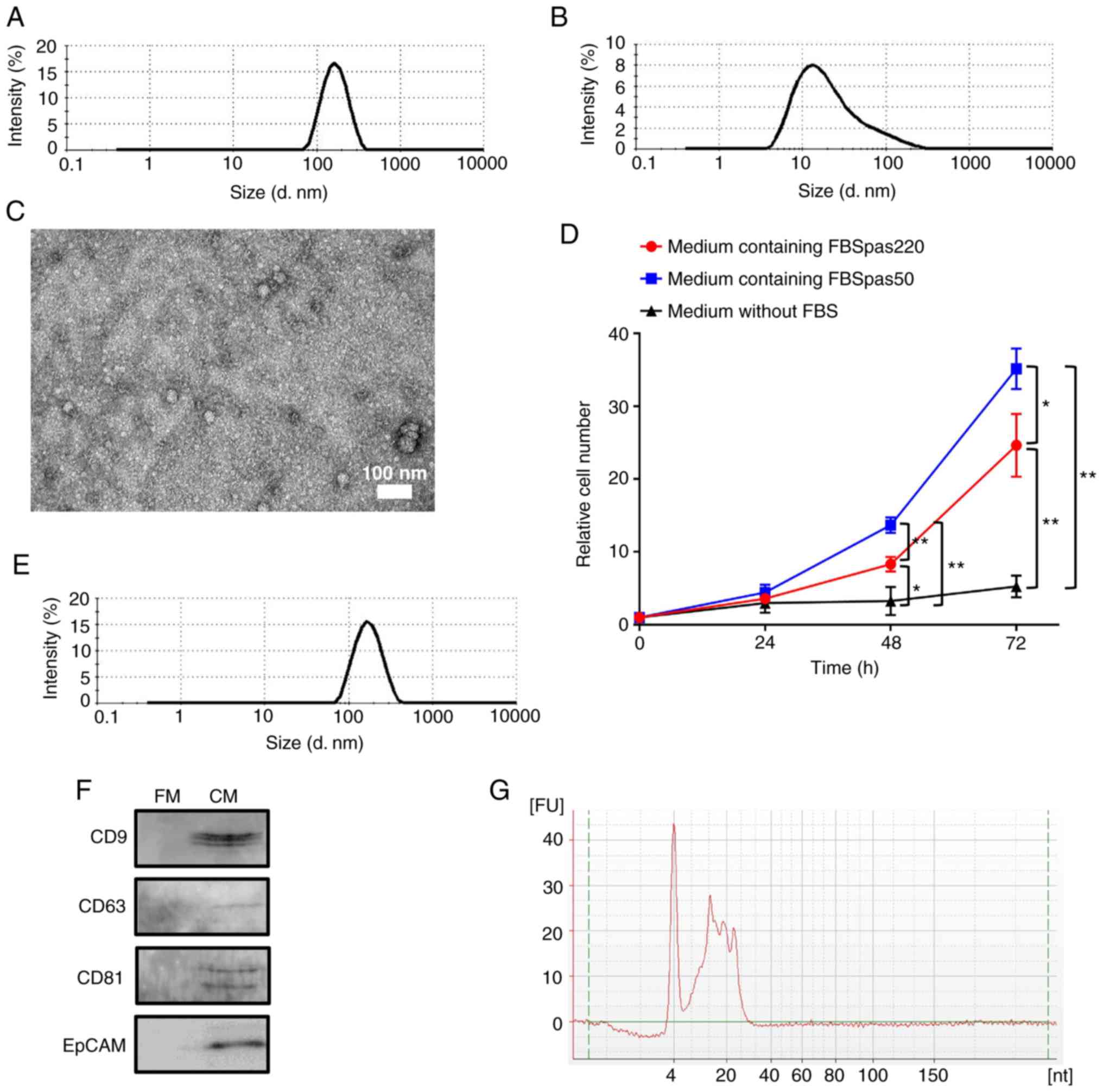 | Figure 1.Characterization of EVs derived from
fetal bovine serum (FBS). (A) EVs captured with a 50-nm filter
(EVcap50/220) were collected by reverse flow (EVrev50/220), and
their size was analyzed by dynamic light scattering (DLS). (B) A
flow-through 50-nm filter (EVpas50/220) was also analyzed by DLS.
(C) EVrev50/220 were observed using a transmission electron
microscope (TEM). (D) PC3 cells were cultured in indicated media
and cell numbers were counted after the indicated number of hours.
The results were normalized to cell numbers at 0 h. Points, mean of
at least three independent experiments; bars, SD. *P<0.05.
**P<0.01. The experiment was performed three times
independently, and cell number at 0 h was set to 1. (E) EVrev50/220
of cell culture medium were collected and their size was analyzed
by DLS. (F) Eluted proteins obtained from EVcap50/220 of each fresh
medium (FM) and medium cultured for 48 h (CM) were subjected to
SDS-PAGE, and western blotting was performed with anti-EpCAM,
anti-CD9, anti-CD63, and anti-CD81 antibodies. (G) Total RNA
purified from EVcap50/220 was analyzed by Bioanalyzer 2100. FU and
nt indicate fluorescence units and nucleotides, respectively. EVs,
extracellular vesicles. |
MeT-5A cells were cultured in medium with FBSpas50
for 48 h and EVs in the medium were captured by 50-nm filters. The
diameter of EVcap50/220 was approximately 170 nm when measured by
DLS (Fig. 1E). Proteins and RNAs of
EVcap50/220 were analyzed. As shown in Fig. 1F, EVcap50/220 contained CD9, CD63,
CD81, and EpCAM proteins, which are markers of enriched exosomes.
EVcap50/220 also contained microRNAs (miRNAs) less than 30 bp in
length (Fig. 1G). Particle
diameter, shape, specific protein expression, and miRNA inclusion
obtained from FBS and cell culture medium suggested that
EVcap50/220 consisted of exosomes and microvesicles.
Functional analysis of the EVs
As EVs function as intercellular mediators, it is
very important that EVs derived from cells (donor cells) are taken
up by other cells (recipient cells). We generated MeT-5A cells that
expressed CD9-green fluorescent protein (GFP), CD63-GFP, and
CD81-GFP proteins, which are representative marker proteins of
exosomes. Exosomes are membrane vesicles released by the fusion of
organelles in the endocytic pathway, forming multivesicular bodies
(20). Localization similar to
multivesicular bodies was observed in cells expressing CD81-GFP
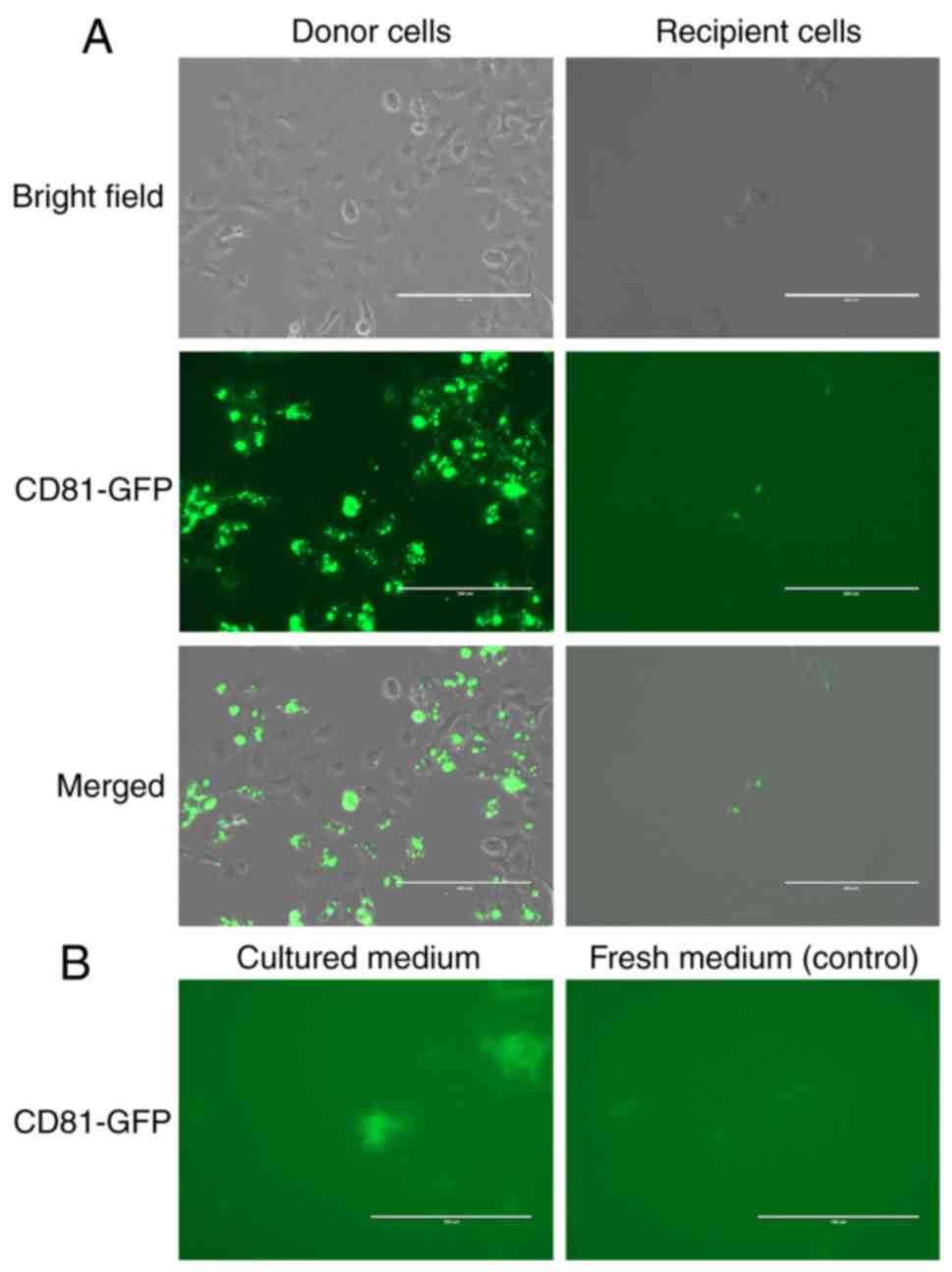
(Fig. 2A, donor cells), CD9-GFP,
and CD63-GFP (Figs. S2A and
S3A). Next, we examined secretion
of EVs containing CD81-GFP, CD9-GFP, and CD63-GFP. EVpas220 of
culture medium was captured using the Total Exosome Isolation Kit
as the EVpas220 was too small to be directly observed via a
microscope. As shown in Fig. 2B,
the GFP signal was observed in medium in which the cells expressed
CD81-GFP, but not in the control medium. Results with CD9-GFP and
CD63-GFP are shown in Figs. S2B
and S3B. These results suggested
that the EVs contained CD63-GFP, CD9-GFP, and CD81-GFP. Since we
observed that EVs contained GFP fusion protein in the culture
medium, we investigated whether MeT-5A cells could take up EVs in
the medium. As shown in Fig. 2A
(recipient cells) and Figs. S2A
and S3A, multivesicular bodies
were observed 8 h after addition of the medium. These results
suggested that EVs in the medium could be taken up by other cells.
To quantify extracellular secretion and endocytosis of EVs,
nanoluciferase (Nluc) fusion proteins, CD63-Nluc, CD9-Nluc, and
CD81-Nluc were expressed in cells and analyzed. The amount of
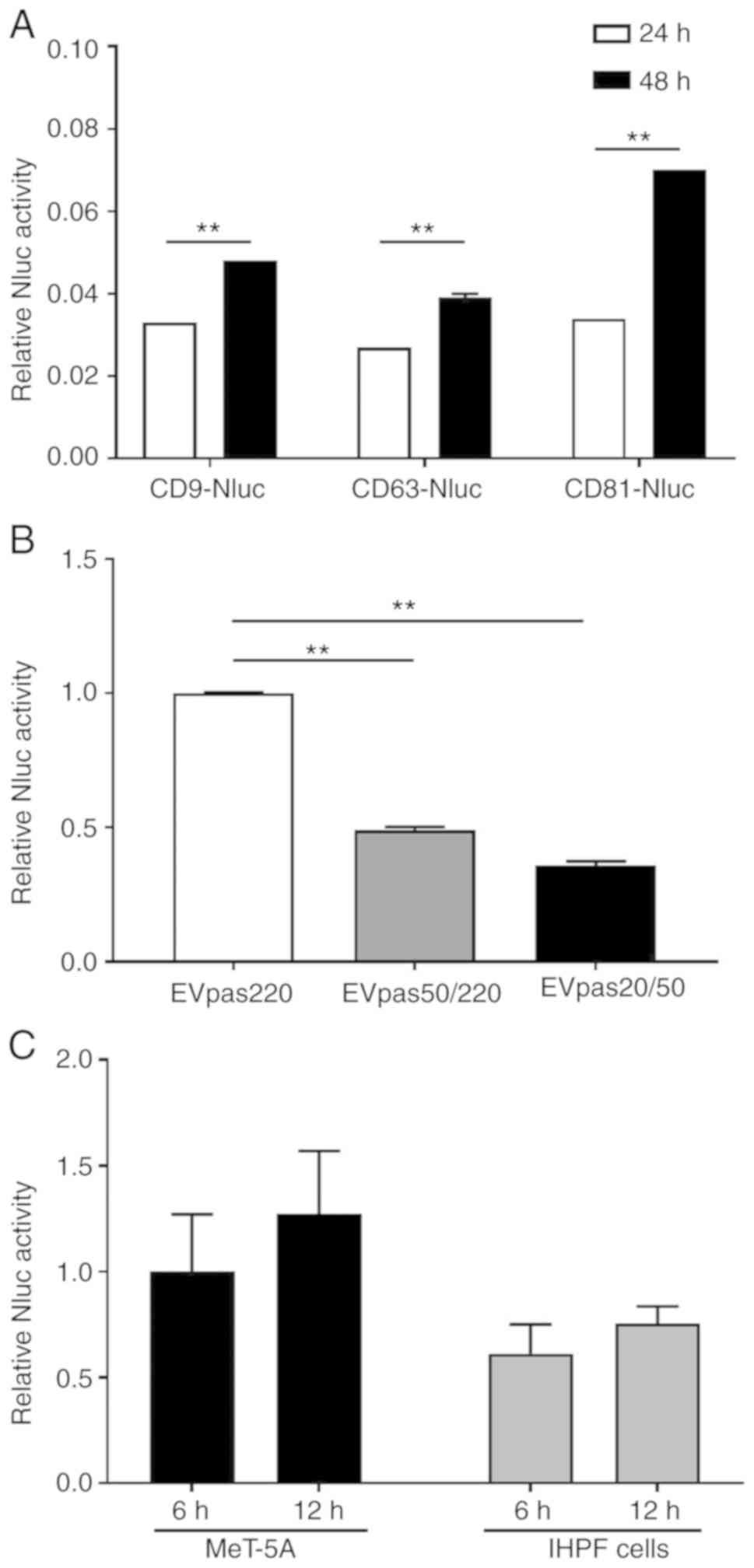
extracellular secretion of each Nluc fusion protein was found to be
increased in a time-dependent manner (Fig. 3A). Since the luciferase activity of
CD81-Nluc was the highest among the three, the capture rate of the
syringe filter was calculated using cells that expressed CD81-Nluc.
As shown in Fig. 3B, 64% of
EVpas220 were captured at 20 nm, of which 80% of the EVs were
captured at 50 nm. Next, we investigated whether MeT-5A and IHPF
cells were able to take up EVs with diameters between 50 and 220
nm. As shown in Fig. 3C, EV uptake
increased in a time-dependent manner, but the amount varied between
cells.
Microarray analysis with
EV50-miRNA
As shown in Fig. 3B,
EVcap50/220 captured 80% of EVs with diameters between 20 and 220
nm, which included exosomes and microvesicles, but not apoptotic
bodies. Considering the collection efficiency and filter handling
(pressure power, flow volume), miRNAs in EVs were purified from
EV50/220 (EV50-miRNAs). Expression profiles of the EV50-miRNAs were
obtained by a 3D-Gene microarray system, and were compared between
6 types of malignant pleural mesothelioma (MPM) and MeT-5A cells.
This chip contained a set of 2,565 probes derived from human cells,
and signals detected for each miRNA were normalized by a global
normalization method that adjusted the median of the detected
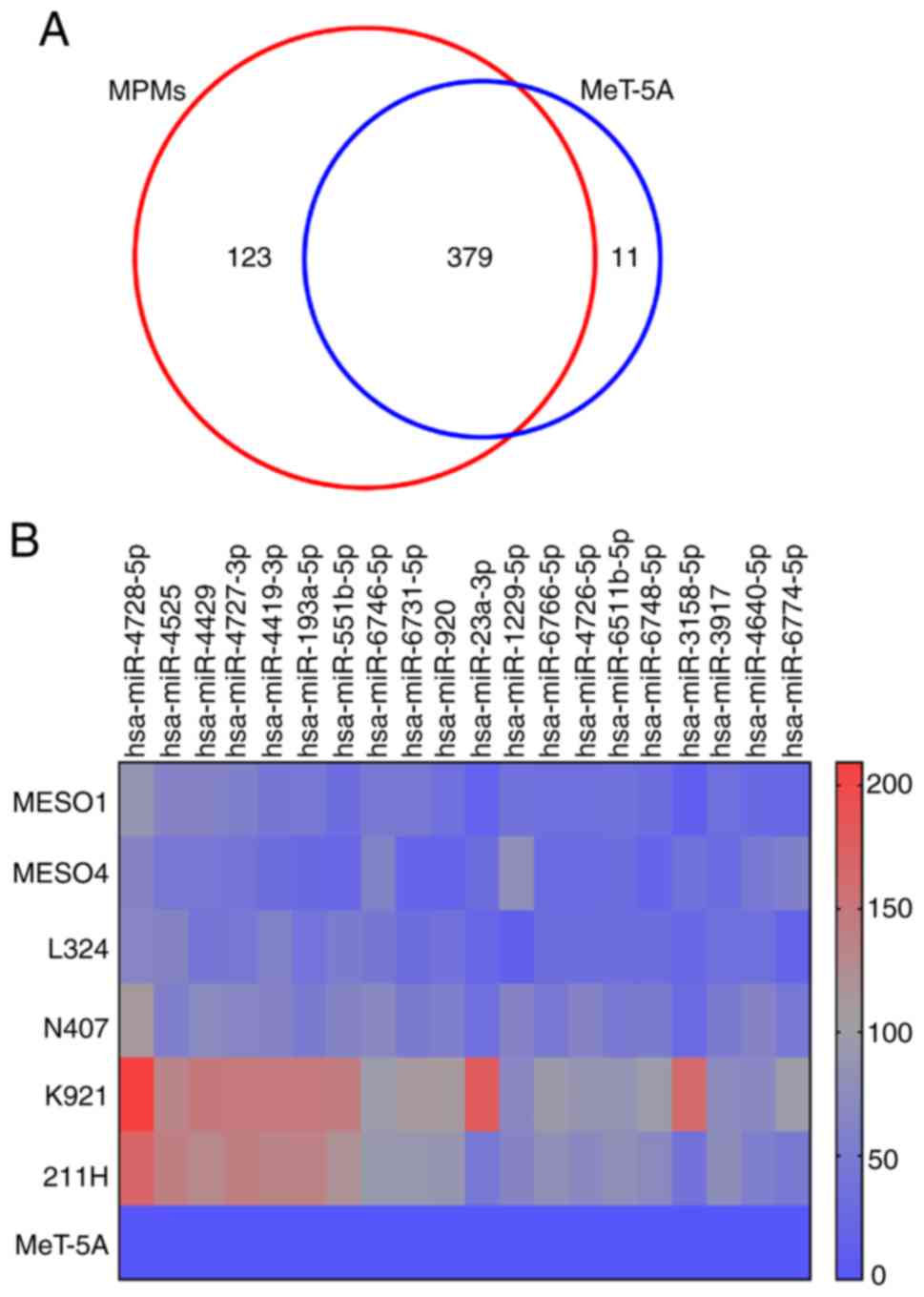
signal intensity to 25. From the results of the microarray, 390 and
502 EV50-miRNAs were detected in MeT-5A and all six MPM cell lines,
respectively. As shown in Fig. 4A,
11 EV50-miRNAs were detected exclusively in MeT-5A cells, but not
in MPM cells. On the other hand, 123 EV50-miRNAs were detected in
MPM cells but not in MeT-5A cells. These EV50-miRNAs may be markers
of MPM. Among the 123 EV50-miRNAs, the 20 most common variants are
shown in Table I in descending
order of expression. The ratio was calculated using the mean values
of MPM cells. Expression of these EV50-miRNAs are also shown on a
heat map (Fig. 4B). Since the
expression value of hsa-miR-4728-5p was the highest among all
EV50-miRNAs, we focused on this miRNA and searched for its target
genes.
 | Table I.EV50-microRNAs expressed in 6 MPM
cell lines but not expressed in EV50-microRNAs of MeT-5A cells (the
20 most common variants). |
Table I.
EV50-microRNAs expressed in 6 MPM
cell lines but not expressed in EV50-microRNAs of MeT-5A cells (the
20 most common variants).
| MicroRNA | Average expression
in six MPM cell lines |
|---|
|
hsa-miR-4728-5p | 117.93 |
| hsa-miR-4525 | 83.58 |
| hsa-miR-4429 | 83.18 |
|
hsa-miR-4727-3p | 82.95 |
| hsa-miR-4419b | 79.99 |
|
hsa-miR-193a-5p | 73.78 |
|
hsa-miR-551b-5p | 71.64 |
|
hsa-miR-6746-5p | 69.19 |
|
hsa-miR-6731-5p | 58.20 |
| hsa-miR-920 | 56.70 |
| hsa-miR-23a-3p | 54.11 |
|
hsa-miR-1229-5p | 54.08 |
|
hsa-miR-6766-5p | 52.48 |
|
hsa-miR-4726-5p | 52.37 |
|
hsa-miR-6511b-5p | 51.69 |
|
hsa-miR-6748-5p | 50.54 |
|
hsa-miR-3158-5p | 49.87 |
| hsa-miR-3917 | 49.03 |
|
hsa-miR-4640-5p | 48.77 |
|
hsa-miR-6774-5p | 48.22 |
Search for targets of hsa-miR-4728-5p
in MeT-5A cells
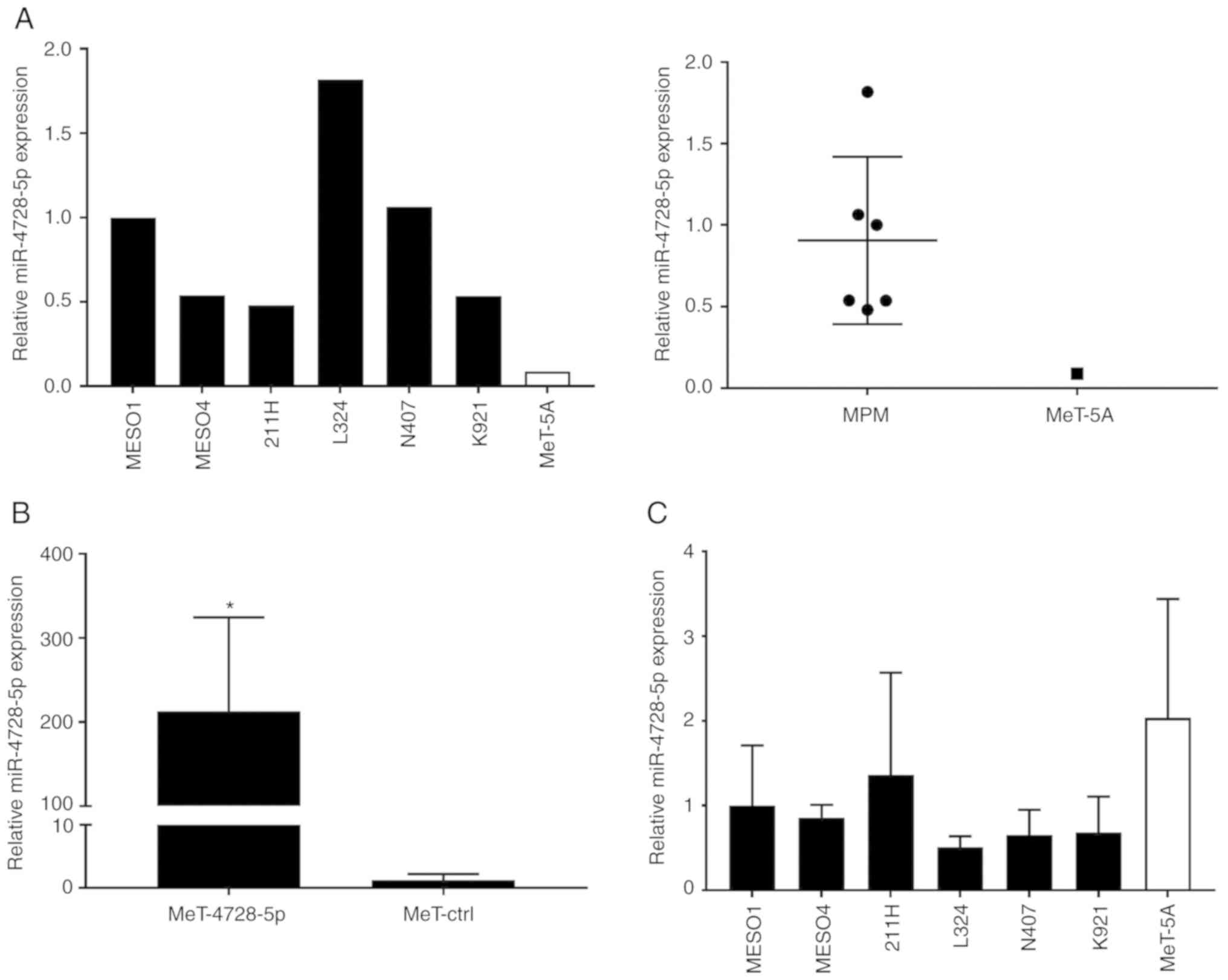
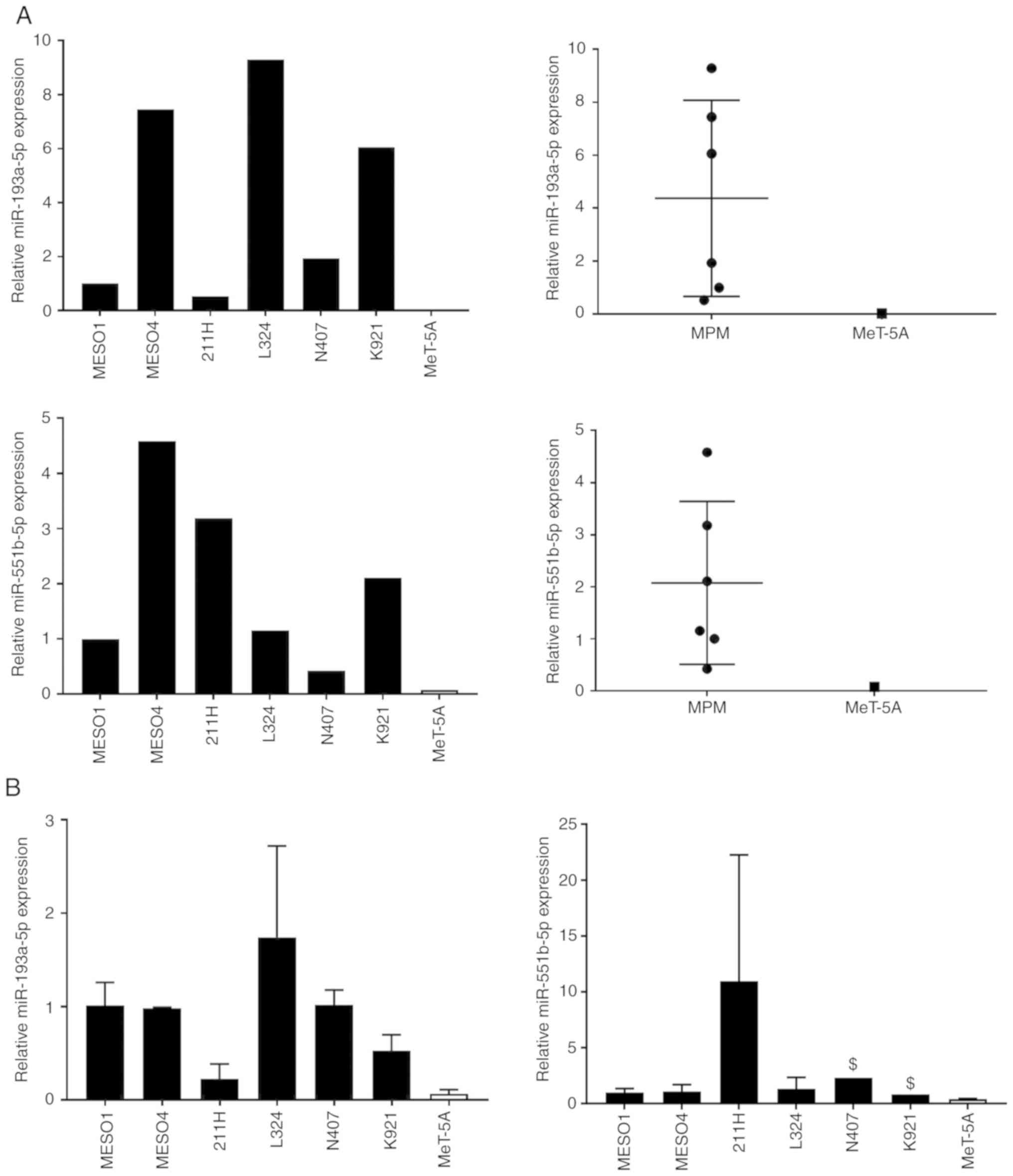
First, we confirmed the expression of
hsa-miR-4728-5p in EVcap50/220 of MPM and MeT-5A cells by qPCR. As
shown in Fig. 5A, expression levels
of hsa-miR-4728-5p in MPM cells were 5–17 times higher as compared
with those of MeT-5A cells. To investigate the target genes of
hsa-miR-4728-5p in normal cells, we established MeT-5A cells that
overexpressed this miRNA (MeT-4728 cells) as well as control
miRNA-free cells (MeT-ctrl). Since it was confirmed that
hsa-miR-4728-5p was overexpressed in the MeT-4728-5p cells
(Fig. 5B), microarray analysis was
performed using mRNA obtained from MeT-4728-5p and MeT-ctrl cells.
Based on the result, only two mRNAs, which came from EIF4H
and TMEM87A, satisfied the following two conditions: i)
Expression level of genes after global normalization was higher
than 100 in MeT-ctrl cells; ii) expression levels of genes in
MeT-4728 cells decreased to less than 0.5 when compared to MeT-ctrl
cells. Since there are few mRNAs whose expression is suppressed by
hsa-miR-4728-5p, the intracellular expression of hsa-miR-4728-5p in
each cell was analyzed by qPCR. Contrary to our expectations, its
expression was found to be highest in MeT-5A cells (Fig. 5C). These data suggest that MeT-5A
cells express hsa-miR-4728-5p, but do not secrete it
extracellularly, and function only in the cell.
Search for targets of hsa-miR-193a-5p
and hsa-miR-551b-5p in MeT-5A cells
As described above, hsa-miR-4728-5p, which showed
the highest expression in EV50-miRNAs of MPM cells, was highly
expressed in MeT-5A cells. Therefore, we selected EV50-miRNAs that
were not found inside MeT-5A cells from 123 EV50-miRNAs via
microarray analysis. As a result, 6 miRNAs were listed as
candidates (Table II). Based on
these results, we selected hsa-miR-193a-5p and hsa-miR-551b-5p,
which exhibited the highest expression. It was confirmed that these
miRNAs were contained in EV50-miRNAs of many MPM cells, and were
not often present in the MeT-5A cells (Fig. 6A). We also examined the expression
of hsa-miR-193a-5p and hsa-miR-551b-5p in cells, but found no
correlation in miRNA expression between MPM and MeT-5A cells
(Fig. 6A and B). Next, we
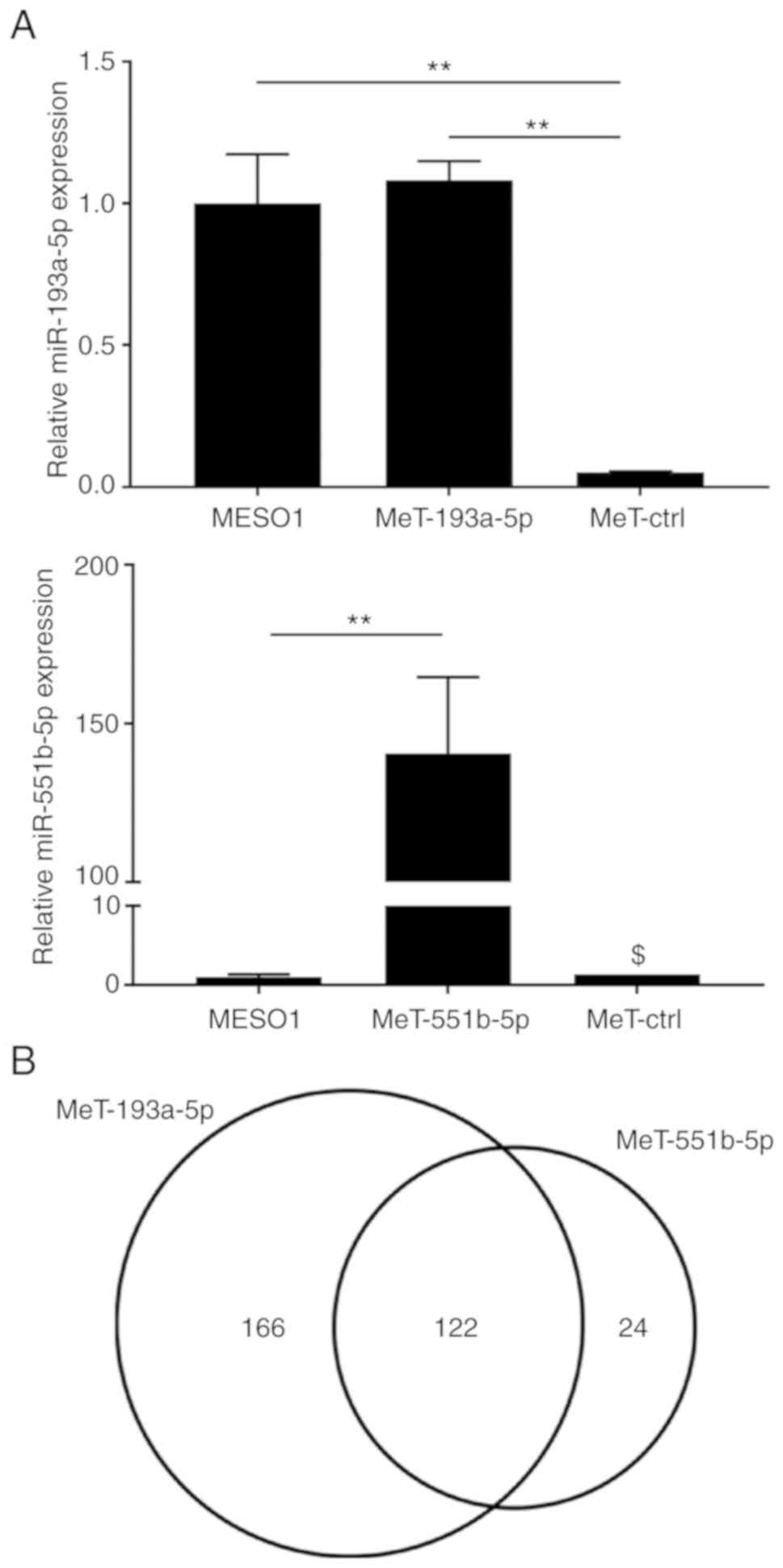
established MeT-5A cells that overexpressed hsa-miR-193a-5p
(MeT-193a cells) or hsa-miR-551b-5p (MeT-551b cells) to investigate
their influence on gene expression (Fig. 7A). There were 288 mRNAs in the
MeT-193a-5p cells and 146 mRNAs in the MeT-551b-5p cells that
satisfied the following two conditions: i) Expression levels of
genes after global normalization were higher than 100 in MeT-ctrl
cells; ii) expression levels of genes in MeT-193a-5p cells or
MeT-551b-5p cells decreased to less than 0.2 when compared to
MeT-ctrl cells. Interestingly, the expression of 122 mRNAs was
found to be suppressed (Fig. 7B).
The function of these genes was analyzed with the GeneCoDis3
software (http://genecodis.cnb.csic.es/), and it was found that
these genes are involved in several pathways (Table III). Among these functions, we
focused on genes involved in the binding between cells as well as
between the cell and the extracellular matrix, as MPM cells may
reduce the adhesion ability of normal mesothelial cells. Invading
the space between these cells is important for malignant
transformation. A summary of each gene is shown in Table IV.
 | Table II.EV50-microRNAs expressed in 6 MPM
cell lines but not expressed in both EV50-microRNAs and cellular
microRNAs of MeT-5A cells. |
Table II.
EV50-microRNAs expressed in 6 MPM
cell lines but not expressed in both EV50-microRNAs and cellular
microRNAs of MeT-5A cells.
| MicroRNA | Average expression
in six MPM cell lines |
|---|
|
hsa-miR-193a-5p | 73.78 |
|
hsa-miR-551b-5p | 71.64 |
| hsa-miR-3122 | 39.34 |
| hsa-miR-4771 | 24.62 |
| hsa-miR-195-3p | 17.62 |
|
hsa-miR-3180-5p | 15.75 |
 | Table III.Pathway analysis of hsa-miR-193a-5p
and hsa-miR-551b-5p target genes (GeneCodis analysis, KEGG
pathways). |
Table III.
Pathway analysis of hsa-miR-193a-5p
and hsa-miR-551b-5p target genes (GeneCodis analysis, KEGG
pathways).
| No. of genes | NGR | Hyp | Hyp* | Annotations |
|---|
| 8 | 197 | 3.20E-07 | 2.82E-05 | Focal adhesion |
| 4 | 71 | 9.25E-05 | 2.71E-03 | Adherens
junction |
| 5 | 149 | 1.41E-04 | 3.10E-03 | Wnt signaling
pathway |
| 4 | 67 | 7.37E-05 | 3.24E-03 | p53 signaling
pathway |
| 4 | 102 | 3.74E-04 | 6.58E-03 | Amoebiasis |
| 4 | 113 | 5.51E-04 | 8.08E-03 | Leukocyte
transendothelial migration |
| 4 | 130 | 9.31E-04 | 1.02E-02 | Tight junction |
| 4 | 128 | 8.79E-04 | 1.10E-02 | Axon guidance |
| 3 | 68 | 1.50E-03 | 1.47E-02 | Long-term
potentiation |
| 3 | 82 | 2.57E-03 | 2.26E-02 | TGF-β signaling
pathway |
| 4 | 186 | 3.44E-03 | 2.75E-02 | Chemokine signaling
pathway |
| 3 | 98 | 4.25E-03 | 2.88E-02 | GnRH signaling
pathway |
| 3 | 98 | 4.25E-03 | 2.88E-02 | Melanogenesis |
| 2 | 33 | 5.34E-03 | 3.36E-02 | African
trypanosomiasis |
| 3 | 113 | 6.31E-03 | 3.70E-02 | Vascular smooth
muscle contraction |
| 2 | 42 | 8.54E-03 | 4.70E-02 |
Aldosterone-regulated sodium
reabsorption |
 | Table IV.Suppression of genes associated with
cell-cell and cell-matrix adhesion by hsa-miR-913a-5p and
hsa-miR-551b-5p. |
Table IV.
Suppression of genes associated with
cell-cell and cell-matrix adhesion by hsa-miR-913a-5p and
hsa-miR-551b-5p.
| Gene | Genes suppressed by
miR-193a-5p | Genes suppressed by
miR-551b-5p | Genes associated
with focal adhesion | Genes associated
with tight junction | Genes associated
with adherens junction |
|---|
| ACTN1 | ○ | ○ | ○ | ○ | ○ |
| PRKCB | ○ | ○ | ○ | ○ |
|
| CCND1 | ○ |
| ○ |
|
|
| THBS1 | ○ | ○ | ○ |
|
|
| CAPN2 | ○ | ○ | ○ |
|
|
| CAV1 | ○ | ○ | ○ |
|
|
| LAMB3 | ○ | ○ | ○ |
|
|
| JUN | ○ | ○ | ○ |
|
|
| ROCK1 | ○ | ○ | ○ |
|
|
| FLNB |
| ○ | ○ |
|
|
| PARD3 | ○ | ○ |
| ○ | ○ |
| MLLT4 | ○ | ○ |
| ○ | ○ |
| PPP2R1B | ○ |
|
| ○ |
|
| CREBBP | ○ | ○ |
|
| ○ |
Discussion
Malignant pleural mesothelioma (MPM) is a disease
associated with a poor patient prognosis, and the development of
diagnostic markers for early detection is urgently required. Since
initial studies have shown that extracellular vesicles (EVs) can
mediate intercellular transfer of RNA and proteins (12–15),
many studies have focused on the contents of EVs, and have tried to
deduce their involvement in intercellular communications (21,22).
The purpose of this study was to compare the profiles of EV-derived
miRNAs between MPM cell lines and a non-malignant cell line, and to
propose microRNAs (miRNAs) that would be useful in diagnosing
MPM.
Recently, EVs have attracted great attention in both
therapeutic and diagnostic applications (23–26).
To isolate EVs such as exosomes, there are various methods,
including ultracentrifugation, commercial kits that use antibodies
against exosome membrane surface proteins, and size-exclusion
chromatography (27–29). These methods have advantages and
disadvantages in terms of steps and time. We should keep in mind
the possibility that EVs could influence analyses because it is
difficult to exclude the captured substances and eluates using
antibodies against exosome membrane surface proteins. When using
these to isolate EVs from liquids, a choice must be made based on
the advantages and disadvantages of the method. In the present
study, we employed filter membranes to isolate EVs based on their
diameters. This method is simple and logical because EVs such as
exosomes, microvesicles, and apoptotic bodies are classified by
diameter. A 50-nm membrane filter could capture 80% of EVs that
passes through a 220-nm membrane filter. We believe that this
capture rate is sufficient for analyzing RNAs contained in exosomes
and microvesicles. The EVs captured by a 50-nm filter membrane were
analyzed using dynamic light scattering (DLS), transmission
electron microscopy (TEM), western blot analysis, and BioAnalyzer
2100. Based on our analysis on diameter, form, specific proteins,
and inclusion of miRNAs, the isolated EVcap50/220 and EVrev50/220
were considered structurally to be EVs. Furthermore, endocytosis
analysis of EVs with diameters between 50 and 220 nm found them to
be functionally EVs. Therefore, we consider this filter method to
be useful for EV analysis because it can not only capture EVs in
liquid, but also easily remove EVs from liquid. For example, we
removed EVs derived from fetal bovine serum (FBS) using this
method. This means that EVs can be easily removed from the cell
maintenance medium used in the laboratory.
MPM originates in pleura mesothelial cells, and many
MPM patients have pleural effusion. We speculated that EVs secreted
from MPM cells exist in both the blood and pleural effusion.
Therefore, we used a culture medium that resembles pleural
effusion. We performed miRNA array analyses using EV50-miRNAs of
six MPM cell lines and normal pleura mesothelial cells (MeT-5A) to
select specific EV50-miRNAs for each cell. Since it is more
convenient to test for EV50-miRNAs that proliferate in MPM cells
for diagnosis, we focused on EV50-miRNAs that could be detected
only in MPM cells. There were 123 EV50-miRNAs that were present
exclusively in MPM-derived EVs when they underwent microarray
analysis. It was speculated that EVs secreted by MPM cells are
taken up by normal mesothelial cells or fibroblasts, and
MPM-derived miRNAs regulate the gene expression of these cells. For
this reason, we selected the most highly expressed miRNA,
hsa-miR-4728-5p, to be overexpressed in MeT-5A cells. Contrary to
expectations, most mRNA expression levels did not vary, and
overexpression of hsa-miR-4728-5p reduced only two genes to less
than 0.5 compared to the control cells. We hypothesized that
hsa-miR-4728-5p is heavily expressed inside MeT-5A cells but has
little effect on the suppression of genes.
Even if a cell synthesizes a particular miRNA and
does not encapsulate it in EVs under normal conditions, it is
possible that the miRNA may be encapsulated in EVs under other
environmental conditions (inflammation, etc.) and secreted.
Therefore, we thought that it was necessary to select miRNAs that
were not synthesized in normal cells. Excluding the miRNAs that are
synthesized in MeT-5A cells from the original 123 EV50-miRNAs
derived from MPM cells, six miRNAs remained.
Cancer cells secrete EVs containing exosomes,
incorporate them into normal cells, and use internal mRNAs and
miRNAs to alter the gene expression of normal cells. This suggested
that cancer cells changed the microenvironment to be more favorable
for them. It has been reported that when exosomes containing MMP1
(matrix metalloproteinase 1) mRNA are secreted from ovarian cancer
cells and taken up by normal peritoneal mesothelial cells, MMP1
mRNA induces apoptosis in these healthy cells and contributes to
ovarian cancer metastasis (30). In
order to investigate the effect of EV50-miRNA uptake on gene
expression of cells, MeT-5A cells overexpressing hsa-miR-193a-5p or
hsa-miR-551b-5p were established. These two EV50-miRNAs were
selected as they were highly expressed in MPM cells but not in
normal pleural mesothelial cells (MeT-5A cells). Since these two
EV50-miRNAs reduced thousands of genes to less than 0.5 when
compared to control cells, genes were selected by narrowing down
the selection margin to less than 0.2. A few hundreds of genes were
selected, and despite the fact that they came from reads of two
different miRNAs, they suppressed the expression of many common
genes. Suppression of mRNA expression by miRNAs is permanent rather
than transient, so both primary (direct) and secondary (indirect)
suppression are included in our study. Genes whose expression was
commonly suppressed by hsa-miR-193a-5p and hsa-miR-551b-5p were
analyzed using pathways from the Kyoto Encyclopedia of Genes and
Genomes (KEGG). Many genes were found to be involved in cell-cell
and cell-matrix adhesion. For cancer cells to invade normal
tissues, it is necessary to destroy cell–cell and cell–matrix
adhesion, and there have been many studies that suggest that
secreted or membrane-bound types of MMPs are capable of digesting
matrix proteins (31). Therefore,
decreasing the expression of proteins associated with tight
junctions, adherens junctions, and focal adhesion may contribute to
successful invasion of MPM cells. Zhou et al (32) reported that PARD3 contributes to the
epithelial-mesenchymal transition (EMT) and invasion of
non-small-cell lung cancer (NSCLC). PARD3 is associated with tight
junctions and adherens junctions, and our results showed that PARD3
mRNA could be repressed by both hsa-miR-193a-5p and
hsa-miR-551b-5p.
Tight junctions are one of the intercellular
adhesion structures that control traffic of substances between
normal cells (33). Tight junctions
also play an important role in cancer cells. Downregulation or loss
of tight junctions can both contribute to cancer progression by
altering cell migration, proliferation, and differentiation
(34–37). In current studies, reduction of
tight junction-associated ZO-1 in breast tumors was associated with
metastasis in breast cancers (38).
In addition, Zhou et al (9)
showed that cancer-secreted miR-105 was capable of suppressing ZO-1
and promoting metastasis in breast cancers. In our study,
hsa-miR-193a-5p and hsa-miR-551b-5p suppressed genes associated
with tight junctions, which may contribute to metastasis in MPM
cells.
Many studies have suggested that EVs may be
attractive targets for both therapeutic and diagnostic applications
(23–26). Previously, proteomic analysis of EVs
isolated from human malignant pleural effusions has been reported
(39), but there are no reports on
miRNA-based analyses of EVs. Our results indicated that six
EV50-miRNAs in blood and pleural effusions may serve as novel
diagnostic markers for MPM. In particular, since the expression of
hsa-miR-193a-5p and hsa-miR-551b-5p are high in MPM-derived EVs,
these are very promising diagnostic markers. To date, there have
been no reports of using EV-derived hsa-miR-193a-5p and
hsa-miR-551b-5p for MPM diagnoses. Plasma-derived and serum-derived
hsa-miR-193a-5p have been reported as promising indicators for
diagnosing amyotrophic lateral sclerosis (ALS) (40), ovarian cancer (41), chronic myeloid leukemia (42), human T-cell leukemia virus type 1
(HTLV-1) infection (43),
colorectal cancer metastasis (44),
and aortic aneurysms (45). On the
other hand, plasma-derived or serum-derived hsa-miR-551b-5p has
been reported as a promising indicator of acute pancreatitis
(46,47) and gastric cancer (48). As far as we are aware, there are no
reports of using EV-derived miRNAs to diagnose MPM, and our report
will be the first to do so.
In this experiment, miRNAs were selected from the
microarray results and verified by qPCR, but there was no
correlation in the expression ratio between cells in each analysis.
It has been reported that miRNA expression does not necessarily
correlate between qPCR and microarrays (49). The major difference is that qPCR
amplifies miRNA samples, but microarray analysis does not. What is
important is that the results between tumor and normal cells were
trend-matched and not reversed between qPCR and the microarray in
this study.
In summary, we showed that many EVs could be easily
isolated and removed from liquids using 50 and 220-nm membrane
filters. Using this method, we identified six miRNAs that could be
specifically secreted from MPM cells and used to diagnose MPM.
Among them, hsa-miR-193a-5p and hsa-miR-551b-5p strongly inhibited
expression of genes related to cell-cell interactions and
cell-matrix interactions in normal cells. These results suggest
that hsa-miR-193a-5p and hsa-miR-551b-5b may contribute to invasion
of MPM cells, and may help elucidate the mechanism of malignant
acquisition of MPM. Since this study is an in vitro
analysis, verification analysis using clinical specimens is
necessary in the future.
Supplementary Material
Supporting Data
Acknowledgements
We would like to thank Yukiko Yoshiura (Center for
Stress-related Disease Control and Prevention, University of
Occupational and Environmental Health, Japan.) for her technical
assistance.
Funding
This research received no external funding.
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request. Supplementary materials have been provided by the authors
and are published online. They are linked to our website and can be
accessed by the reader.
Authors' contributions
Conceptualization of the study was accomplished by
HI. Formal analysis was carried out by TJ. Methodology was designed
by HI. Project administration of the experiments was carried out by
YM and KY. Supervision of the research was the responsibility of YM
and KY. Writing of the original draft was carried out by TJ and
writing and review and edited was conducted by HI. All authors read
and approved the manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Robinson BM and Lake RA: Advances in
malignant mesothelioma. N Engl J Med. 353:1591–1603. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Robinson BM: Malignant pleural
mesothelioma: An epidemiological perspective. Ann Cardiothorac
Surg. 1:491–496. 2012.PubMed/NCBI
|
|
3
|
Jamrozik E, de Klerk N and Musk AW:
Asbestos-related disease. Intern Med J. 41:372–380. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Creaney J and Robinson BW: Serum and
pleural fluid biomarkers for mesothelioma. Curr Opin Pulm Med.
15:366–370. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pass HI, Lott D, Lonardo F, Harbut M, Liu
Z, Tang N, Carbone M, Webb C and Wali A: Asbestos exposure, pleural
mesothelioma, and serum osteopontin levels. N Engl J Med.
353:1564–1573. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ren R, Yin P, Zhang Y, Zhou J, Zhou Y, Xu
R, Lin H and Huang C: Diagnostic value of fubulin-3 for malignant
leural mesothelioma: A systematic review and meta-analysis.
Oncotarget. 7:84851–84859. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang L, Zhang S, Yao J, Lowery FJ, Zhang
Q, Huang WC, Li P, Li M, Wang X, Zhang C, et al:
Microenvironment-induced PTEN loss by exosomal microRNA primes
brain metastasis outgrowth. Nature. 527:100–104. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Harazono Y, Muramatsu T, Endo H, Uzawa N,
Kawano T, Harada K, Inazawa J and Kozaki K: miR-655 Is an
EMT-suppressive microRNA targeting ZEB1 and TGFBR2. PLoS One.
8:e627572013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou W, Fong MY, Min Y, Somlo G, Liu L,
Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR, et al:
Cancer-secreted miR-105 destroys vascular endothelial barriers to
promote metastasis. Cancer Cell. 25:501–515. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gee GV, Koestlr DC, Christensen BC,
Sugarbaker DJ, Ugolini D, Ivaldi GP, Resnick MB, Houseman EA,
Kelsey KT and Marsit CJ: Downregulated microRNAs in the
differential diagnosis of malignant pleural mesothelioma. Int J
Cancer. 127:2859–2869. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Juri A, Zanoaga O, Braicu C, Tomuleasa C,
Irimie A and Berindan-Neagoe I: A comprehensive picture of
extracellular vesicles and their contents. Molecular transfer to
cancer cells. Cancers (Basel). 12:2982020. View Article : Google Scholar
|
|
12
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pegtel DM, Cosmopoulos K, Thorley-Lawson
DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD,
Würdinger T and Middeldorp JM: Functional delivery of viral miRNAs
via exosomes. Proc Natl Acad Sci USA. 107:6328–6333. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Liu D, Chen X, Li J, Li L, Bian
Z, Sun F, Lu J, Yin Y, Cai X, et al: Secreted monocytic miR-150
enhances targeted endothelial cell migration. Mol Cell. 39:133–144.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kosaka N, Iguchi H, Yoshioka Y, Takeshita
Y, Matsuki Y and Ochiya T: Secretory mechanisms and intercellular
transfer of microRNAs in living cells. J Biol Chem.
285:17442–17452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yasuda M, Hanagiri T, Shigematsu Y,
Onitsuka T, Kuroda K, Baba T, Mizukami M, Ichiki Y, Uramoto H,
Takenoyama M and Yasumoto K: Identification of a tumor associated
antigen in lung cancer patients with asbestos exposure. Anticancer
Res. 30:2631–2639. 2010.PubMed/NCBI
|
|
17
|
Koi C, Izumi H, Kurita T, Nguyen TT,
Murakami M, Yoshiura Y, Hachisuga T and Morimoto Y: Lovastatin
induced Kruppel like factor 2 (KLF2), Kruppel like factor 6 (KLF6)
and Ras homolog family member B (RHOB) genes and preferentially led
to viability reduction of Cisplatin-resistant cells. Oncotarget.
8:106429–106442. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Morimoto Y, Izumi H, Yoshiura Y, Tomonaga
T, Oyabu T, Myojo T, Kawai K, Yatera K, Shimada M, Kubo M, et al:
Pulmonary toxicity of well-dispersed cerium oxide nanoparticles
following intratracheal instillation and inhalation. J Nanopart
Res. 17:4422015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Samadder P, Byun HS, Bittman R and Arthur
G: A fluorescent alkyllysophospholipid analog exhibits selective
cytotoxicity against the hormone-insensitive prostate cancer cell
line PC3. Anticancer Agents Med Chem. 14:528–538. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jabbari N, Akbariazar E, Feqhhi M,
Rahbarghazi R and Rezaie J: Breast cancer-derived exosomes: Tumor
progression and therapeutic agents. J Cell Physiol. 235:6345–6356.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo Y, Ji X, Liu J, Fan D, Zhou Q, Chen C,
Wang W, Wang G, Wang H, Yuan W, et al: Effects of exosomes on
pre-metastatic formation in tumors. Mol Cancer. 18:392019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kling CM: Non-coding RNAs in breast
cancer: Intracellular and intercellular communication. Noncording
RNA. 4:402018.
|
|
23
|
Jayaseelan VP: Emerging role of exosomes
as promising diagnostic tool for cancer. Cancer Gene Ther.
27:395–398. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu Z, Yang Z, Dai Y, Zhu Q and Chen LA:
Update on liquid biopsy in clinical management of non-small cell
lung cancer. Onco Targets Ther. 12:5097–5109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fortunato O, Gasparini P, Boeri M and
Sozzi G: Exo-miRNAs as a new tool for liquid biopsy in lung cancer.
Cancers (Basel). 11:8882019. View Article : Google Scholar
|
|
26
|
Jiang L, Gu Y, Du Y and Liu J: Exosomes:
Diagnostic biomarkers and therapeutic delivery vehicles for cancer.
Mol Pharm. 16:3333–3349. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Purushothaman A: Exosomes from cell
culture-conditioned medium: Isolation by ultracentrifugation and
characterization. Methods Mol Biol. 1952:233–244. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Skottvoll FS, Berg HE, Bjørseth K, Lund K,
Roos N, Bekhradnia S, Thiede B, Sandberg C, Vik-Mo EO,
Roberg-Larsen H, et al: Ultracentrifugation versus kit exosome
isolation: nanoLC-MS and other tools reveal similar performance
biomarkers, but also contaminations. Future Sci OA. 5:FSO3592018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baranyai T, Herczerg K, Onódi Z, Voszka I,
Módos K, Marton N, Nagy G, Mäger I, Wood MJ, EI Andaloussi S, et
al: Isolation of exosomes from blood plasma: Qualitative and
quantitative comparison of ultracentrifugation and size exclusion
chromatography methods. PLoS One. 10:e01456862015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yokoi A, Yoshioka Y, Yamamoto Y, Ishikawa
M, Ikeda SI, Kato T, Kiyono T, Takeshita F, Kajiyama H, Kikkawa F
and Ochiya T: Malignant extracellular vesicles carrying MMP1 mRNA
facilitate peritoneal dissemination in ovarian cancer. Nat Commun.
8:144702017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang H: Matrix metalloproteinase-9
(MMP-9) as a cancer biomarker and MMP-9 biosensors: Recent
advances. Sensors (Basel). 18:32492018. View Article : Google Scholar
|
|
32
|
Zhou Q, Dai J, Chen T, Dada LA, Zhang X,
Zhang W, DeCamp MM, Winn RA, Sznajder JI and Zhou G: Downregulation
of PKCζ/Pard3/Pard6b is responsible for lung adenocarcinoma cell
EMT and invasion. Cell Signal. 38:49–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tsukita S, Furuse M and Itoh M:
Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol.
2:285–293. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Brennan K, Offiah G, McSherry EA and
Hopkins AM: Tight junctions: A barrier to the initiation and
progression of breast cancer? J Biomed Biotechnol. 2010:4606072010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Georgiadis A, Tshernutter M, Bainbridge
JW, Balaggan KS, Mowat F, West EL, Munro PM, Thrasher AJ, Matter K,
Balda MS and Ali RR: The tight junction associated signalling
proteins ZO-1 and ZONAB regulate retinal pigment epithelium
homeostasis in mice. PLoS One. 5:e157302010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Itoh M and Bissell MJ: The organization of
tight junctions in epithelia: Implications for mammary gland
biology and breast tumorigenesis. J Mammary Gland Biol Neoplasia.
8:449–462. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Martin TA and Jiang WG: Loss of tight
junction barrier function and its role in cancer metastasis.
Biochim Biophys Acta. 1788:872–891. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Polette M, Gilles C, Nawrocki-Raby B, Lohi
J, Hunziker W, Foidart JM and Birembaut P: Membrane-type 1 matrix
metalloproteinase expression is regulated by zonula occludens-1 in
human breast cancer cells. Cancer Res. 65:7691–7698. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bard MP, Hegmans JP, Hemmes A, Luider TM,
Willemsen R, Severijnen LA, van Meerbeeck JP, Burgers SA,
Hoogsteden HC and Lambrecht BN: Proteomic analysis of exosomes
isolated from human malignant pleural effusions. Am J Respir Cell
Mol Biol. 31:114–121. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Saucier D, Wajnberg G, Roy J, Beauregard
AP, Chacko S, Crapoulet N, Fournier S, Ghosh A, Lewis SM, Marrero
A, et al: Identification of a circulating miRNA signature in
extracellular vesicles collected from amyotrophic lateral sclerosis
patients. Brain Res. 1708:100–108. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ren X, Zhang H, Cong H, Wang X, Ni H, Shen
X and Ju S: Diagnostic model of serum miR-193a-5p, HE4 and CA125
improves the diagnostic efficacy of epithelium ovarian cancer.
Pathol Oncol Res. 24:739–744. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Prinsloo A, Pool R and Van Niekerk C:
Preliminary data on microRNA expression profiles in a group of
South African patients diagnosed with chronic myeloid leukaemia.
Mol Clin Oncol. 7:386–390. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fayyad-Kazan M, EIDirani R, Hamade E, EI
Majzoub R, Akl H, Bitar N, Fayyad-Kazan H and Badran B: Circulating
miR-29c, miR-30c, miR-193a-5p and miR-885-5p: Novel potential
biomarkers for HTLV-1 infection diagnosis. Infect Genet Evol.
74:1039382019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qu A, Yang Y, Zhang X, Wang W, Liu Y,
Zheng G, Du L and Wang C: Development of a preoperative prediction
nomogram for lymph node metastasis in colorectal cancer based on a
novel serum miRNA signature and CT scans. EBioMedicine. 37:125–133.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Moushi A, Michalidou K, Soteriou M,
Cariolou M and Bashiardes E: MicroRNAs as possible biomarkers for
screening of aortic aneurysms: A systematic review and validation
study. Biomarkers. 23:253–264. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang Y, Yan L and Han W: Elevated level
of miR-551b-5p is associated with inflammation and disease
progression in patients with severe acute pancreatitis. Ther Apher
Dial. 22:649–655. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu P, Xia L, Zhang WL, Ke HJ, Su T, Deng
LB, Chen YX and Lv NH: Identification of serum microRNAs as
diagnostic and prognostic biomarkers for acute pancreatitis.
Pancreatology. 14:159–166. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jiang X, Jiang M, Xu M, Xu J and Li Y:
Identification of diagnostic utility and molecular mechanisms of
circulating miR-551b-5p in gastric cancer. Pathol Res Pract.
215:900–904. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen Y, Gelfond JA, McManus LM and
Shireman PK: Reproducibility of quantitative RT-PCR array in miRNA
expression profiling and comparison with microarray analysis. BMC
Genomics. 10:4072009. View Article : Google Scholar : PubMed/NCBI
|