Introduction
Cancer is anticipated to be the leading cause of
death worldwide in the 21st century and is expected to be the main
barrier to increasing life expectancy (1). Esophageal cancer (EC) ranks seventh
according to incidence (572,000 new cases), and sixth in terms of
mortality (509,000 deaths) (1), while
it ranks third (477,900 new cases) and fourth (375,000 deaths) in
terms of incidence and mortality in China (2). EC primarily consists of two cell types,
esophageal squamous cell carcinoma (ESCC) in addition to esophageal
adenocarcinoma. ESCC is the most common (90%) histological subtype
of EC (3); ~70% of cases occur in men
(1). Over the past decade, the early
stage detection and overall survival rates have improved,
benefiting from the increased uptake of early referral schemes,
novel endoscopic therapies and perioperative treatment strategies
in some developed countries. However, the overall survival among
patients with EC remains low; the 5-year survival is 10–15% among
all patients, although it increases to 40% among patients who
undergo curative surgery (4,5). Therefore, the molecular mechanism of EC
underlying its development and progression is of great
significance.
It is commonly known that the microRNA (miRNA/miR)
family, which are composed of 18–25-nucleotide short non-coding
RNAs, can regulate the expression of target mRNAs by binding to the
3′-untranslated regions (3′-UTRs). These miRNAs can downregulate
the expression of target mRNA, which negatively modulates the gene
expression or mRNA degradation at the posttranscriptional level
(6–8).
For example, miR-1254 downregulates E3 ubiquitin-protein ligase
SMURF1 to reduce cell proliferation, migration and Matrigel
invasion in gastric cancer cell lines (9), while miRNA-146a downregulates VEGF to
reduce cancer metastasis in hepatocellular carcinoma (10). To date, miRNAs have been regarded as
vital factors in cancer progression, including tumour
proliferation, migration, invasion, metastasis and radiosensitivity
(11–15). It has been found that miR-485-5p,
which regulates different targets in various human cancers, has the
ability to function as a suppressor tumour gene. miR-485-5p
downregulates the expression of tumour protein D54 (16) and paired box 3 (17) to inhibit the proliferation and
invasion of glioma cells. However it can also reduce the
O-GlcNAcylation of polycomb complex protein BMI-1 (18) and inhibit proliferation by targeting
CD147 in colorectal cancer (19). Han
et al (20) reported that
O-linked N-acetylglucosamine transferase could be downregulated by
the tumour suppressor miR-485-5p to inhibit the progression of
ESCC, while in the present study another target was found,
flotillin-1 (FLOT-1).
Flotillins are a group of ubiquitously expressed,
evolutionarily conserved, membrane-associated scaffolding proteins
located on microdomain lipid rafts containing two homologous
isoforms, FLOT-1 and FLOT-2, which are involved in various
procedures, including cell proliferation, migration, cell adhesion,
survival, differentiation, endocytosis, signal transduction,
membrane trafficking and T-cell activation (21–27).
In the present study, it was verified that
miR-485-5p expression was reduced in ESCC. Cell proliferation,
locomotion, invasion and epithelial-mesenchymal metastasis (EMT)
were blocked by the overexpression of miR-485-5p. As FLOT-1 is a
direct target of miR-485-5p, and various reports have verified
FLOT-1 plays an important role in promoting cancer progression
(21–27), it was concluded that miR-485-5p
suppresses ESCC by targeting FLOT-1 and inhibiting the EMT.
Materials and methods
Subjects and tissue specimens
The Ethics Committee of The Fourth Hospital of Hebei
Medical University (Shijiazhuang, China) approved this study
(approval no. 2019053). Written informed consent was obtained from
all subjects or guardians. In this study, ESCC and adjacent tissues
(>1 cm away from the edge of the tumor) were surgically excised
from 80 patients with ESCC between August 2015 and August 2018.
None of the patients had previously received chemo- or
radiotherapy. After surgery, the samples were dipped in
RNAlater™ (Ambion; Thermo Fisher Scientific, Inc.) and
stored in a −80°C freezer immediately for further use. The
specimens were clinically and histologically diagnosed by the
Department of Thoracic Surgery and Pathology, The Fourth Hospital
of Hebei Medical University. The lymph node metastasis and staging
of patients was determined according to The AJCC Esophageal Cancer
Staging System, Eighth Edition (28).
Cell lines and culture
ESCC cell lines Eca 109, KYSE 170, KYSE 180 and TE-1
were obtained from the Shanghai Institutes for Biological Sciences.
KYSE 30, KYSE 510, TE-12 and YES-2 cell lines were provided by
Professor Masatoshi Tagawa (Department of Molecular Biology and
Cancer Biology, Chiba University, Chiba, Japan). All cells were
cultured in air containing 5% CO2 at 37°C in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% fetal bovine serum (Biological Industries), 1% penicillin and
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.).
Transfection
KYSE 30, Eca 109 and TE-1 cell lines (at 70%
density), were transfected with 100 nM miR-485-5p mimics (cat. no.
miR10002175-1-5; Guangzhou RiboBio Co., Ltd.), miRNA mimic negative
controls (mimics-NC; cat. no. miR1N0000001-1-5; Guangzhou RiboBio
Co., Ltd.), miR-485-5p inhibitor (cat. no. miR20002175-1-5;
Guangzhou RiboBio Co., Ltd.), miRNA inhibitor negative controls
(inhibitor-NC; cat. no. miR2N0000001-1-5; Guangzhou RiboBio Co.,
Ltd.), FLOT-1 plasmid (2 µg/well; YouBio) and pcDNA 3.1 (2 µg/well;
YouBio) using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions. The time interval between transfection and subsequent
experimentation was 48 h.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
According to the protocols reported previously
(29), RNA extraction and RT-qPCR
were carried out. Total RNA was extracted from cells or tissues
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
According to the manufacturers' instructions, the GoScript™ Reverse
Transcription System (Promega Corporation) was used to synthesize
cDNA from 2 µg total RNA It is worth noting that the reverse
transcription primer of U6 and miR-485-5p replacing random primer
were used to perform target-specific reverse transcription.
Meanwhile, cDNA, which was used to quantitate the expression of
mRNA, was reverse transcribed as described previously (29). qPCR was performed in triplicate with a
1:4 dilution of cDNA using the SYBR-Green PCR Kit (Promega
Corporation) with the 7500 Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling
conditions were as follows: Initial denaturation at 95°C for 2 min,
followed by 40 cycles of denaturation at 95°C for 15 sec, and
annealing and extension at 60°C for 1 min. Gene specific qPCR
primers were as follows: U6 snRNA reverse transcription primer,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; U6 snRNA forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; miR-485-5p reverse transcription
primer,
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGAATTCA-3′;
miR-485-5p forward, 5′-GGAGAGGCTGGCCGTGAT-3′ and reverse,
5′-CAGTGCGTGTCGTGGAGT-3′; N-cadherin forward,
5′-TGTTTGACTATGAAGGCAGTGG-3′ and reverse,
5′-TCAGTCATCACCTCCACCAT-3′; E-cadherin forward,
5′-CCGCCGGCGTCTGTAGGAA-3′ and reverse,
5′-AGGGCTCTTTGACCACCGCTCTC-3′; Vimentin forward,
5′-GAGAACTTTGCCGTTGAAGC-3′ and reverse, 5′-TCCAGCAGCTTCCTGTAGGT-3′;
zinc finger E-box-binding homeobox 1 (ZEB1) forward,
5′-TTGTAGCGACTGGATTTT-3′ and reverse, 5′-AGACGATAGTTGGGTCCCGGC-3′;
FLOT-1 forward, 5′-CCCATCTCAGTCACTGGCATT-3′ and reverse,
5′-CCGCCAACATCTCCTTGTTC-3′; and GAPDH forward,
5′-GGACCTGACCTGCCGTCTAG-3′ and reverse,
5′-GTAGCCCAGGATGCCCTTGA-3′.
All primers were purchased from Generay Biotech Co.,
Ltd. The relative expression levels of miRNA and mRNA were
normalized to U6 and GAPDH expression, respectively. The relative
mRNA expression levels were calculated using the 2−ΔΔCt
method (30).
Semi-quantitative PCR
After RNA extraction from ESCC cell lines and
target-specific reverse transcription to cDNA, as aforementioned,
cDNA was diluted with nuclear free water with a 1:4 dilution. qPCR
was performed with the diluted cDNA using the GoTaq® G2
Master Mixes (cat. no. M7822; Promega Corporation) with the
GeneAmp™ PCR System 9700 (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The thermocycling conditions were as follows:
Initial denaturation at 95°C for 5 min; followed by 35 cycles of
denaturation at 95°C for 15 sec, annealing at 60°C for 30 sec and
extension at 72°C for 30 sec; and a final extension at 72°C for 15
min. The agarose gels were 2% with 0.01% ethidium bromide. The gels
were visualized using GBOX EF2 gel doc system (Syngene). Sequences
of reverse transcription, forward and reverse primers were the same
as aforementioned for RT-qPCR.
Cell proliferation assay
After cell transfection with miR-485-5p mimics,
mimics-NC, miR-485-5p inhibitor and inhibitor-NC for 24 h, an MTS
assay (Promega Corporation) was performed to examine proliferation.
Harvesting and subculturing of KYSE 30, Eca 109 and TE-1 cells in
96-well plates (2,500 cells/well) were then performed. Cells were
incubated for 0, 24, 48, 72 and 96 h, and cell proliferation was
assessed using the MTS assay at each desired time point according
to the manufacturer's instructions. MTS reagent (20 µl/well) was
added into each well and cells were incubated at 37°C for 2 h in
the dark, and the absorbance was measured at 492 nm by a microplate
reader. The experiment was performed in triplicate.
Cell migration and invasion
assays
Cell migration and invasion were assessed by
migration and invasion assays in a Transwell chamber (Corning,
Inc.). Matrigel was used to coat the upper Transwell insert of the
chamber at 37°C overnight in the cell invasion assay. According to
the manufacturer's instructions, transiently transfected cells were
serum-starved for 2 h in RPMI-1640 without FBS, and
5×104 cells (Eca 109, KYSE 30 and TE-1) were suspended
in 200 µl serum-free RPMI-1640 and seeded onto a Transwell insert.
RPMI-1640 containing 20% FBS, as a chemoattractant, was added to
the Transwell insert below. The cells in the upper chamber migrated
to the lower chamber when cultured in humidified air containing 5%
CO2 at 37°C for 10 h. Then, cells were fixed with 4%
paraformaldehyde for 10–15 min and stained with 0.1% crystal violet
for 5–10 min at 25°C. A total of three medium magnification fields
(magnification, ×200; light microscope) were randomly selected, and
the number of migrated or invasive cells was counted. The
experiments above were performed in triplicate.
Wound healing assay
Wound healing assays were performed to assess the
migratory ability of cells. ESCC cell lines (5×105)
transfected with miR-485-5p mimics or vector were seeded in 6-well
plates (at 100% confluency). Linear wounds were scratched using a
200-µl sterile pipette tips when all wells were filled with cells.
To remove suspended cells, plates were washed several times with
PBS, and the cells were cultured in humidified air containing 5%
CO2 at 37°C with serum-free RPMI-1640. The wounds were
imaged using a light microscope at the same location at 0, 12 and
24 h, and the percentage of the wound area was calculated (wound
area/total area). The experiment was carried out three times.
Immunohistochemistry (IHC)
All specimens (5 µm) were fixed in 4% formalin at
25°C for 24 h, and then embedded in paraffin. Antigen retrieval was
achieved by boiling the specimen in Tris-EDTA buffer (cat. no.
C1038; Beijing Solarbio Science & Technology Co., Ltd.) at
120°C for 3 min, followed by blocking endogenous peroxide and
protein activity with endogenous peroxidase blocker (reagent I,
streptavidin-avidin-biotin detection system; cat. no. SP-9001,
ZSGB-BIO) for 20 min in the dark at 25°C. After blocking with
normal goat serum (reagent II, streptavidin-avidin-biotin detection
system; cat. no. SP-9001, ZSGB-BIO) at 25°C for 30 min, sections
were incubated with primary antibodies specific for FLOT-1 (1:100;
cat. no. 18634; Cell Signaling Technology, Inc.), E-cadherin
(1:100; cat. no. ab40772; Abcam), Ki-67 (1:100; cat. no.
27309-1-AP; ProteinTech Group, Inc.), N-cadherin (1:50; cat. no.
22018-1-AP; ProteinTech Group, Inc.) and Vimentin (1:100; cat. no.
10366-1-AP; ProteinTech Group, Inc.) overnight at 4°C, Next,
sections were incubated with the secondary antibody (reagent III,
streptavidin-avidin-biotin detection system; cat. no. SP-9001,
ZSGB-BIO) at 37°C for 30 min, followed by incubation with a
HRP-labelled streptavidin solution (reagent IV,
streptavidin-avidin-biotin detection system; cat. no. SP-9001,
ZSGB-BIO) at 25°C for 30 min. Sections were washed with PBS after
each step. After visualization of the positive antigen antibody
reaction by incubation with 3,3-diaminobenzidine-tetrachloride
(DAB, cat. no. ZLI-9018; ZSGB-BIO) at 25°C for 5 min, sections were
counterstained with haematoxylin at 25°C for 3 min and evaluated by
light microscopy at ×40 and 200 magnification.
The expression of FLOT-1 was evaluated using IHC
scores by choosing five views with high power lenses at random. The
total number of tumour cells and the number of positive cells were
calculated. The percentage of positive cells in the total number of
tumour cells was calculated. The corresponding scores of positive
cell percentages are shown as follows: i) 0, 0–25% positive cells;
ii) 1, 26–50% positive cells; iii) 2, 51~75% positive cells; and
iv) 3, 76~100%. In addition to the percentages of positive cells,
FLOT-1 expression was also scored according to the dyeing strength:
i) 0, no claybank; ii) 1, light claybank; iii) 2, medium claybank;
and iv) 3, dark claybank. FLOT-1 expression in tissues was ranked
according to the following scores: i) 0, -; ii) 1–2, +; iii) 3–4,
++; and iv) 5–6, +++. All the sections were interpreted by two
pathologists.
Dual-luciferase reporter assay
TargetScan (http://www.targetscan.org/vert_72/) was employed to
predict the targets of miR-485-5p. The FLOT-1 mRNA 3′-UTR target
sequence was cloned by Creative Biogene. The wild-type (WT) or
mutant (MUT) miR-485-5p binding sequence of FLOT-1 was inserted
into the pmirGLO plasmid (YouBio) to establish the recombinant
luciferase reporter plasmids (FLOT-1 WT and FLOT-1 MUT). 293T cells
(Procell Life Science & Technology Co., Ltd.) were
co-transfected with pmirGLO, FLOT-1 WT or FLOT-1 MUT and miR-485-5p
mimics or mimics-NC with Lipofectamine® 2000. Following
48 h of incubation, the firefly and Renilla luciferase
activities of 293T cells were examined using the
Dual-Luciferase® Reporter Assay System (cat. no. E1910;
Promega Corporation) following the manufacturer's protocols. The
relative luciferase activity was determined by normalizing the
firefly luciferase activity against the Renilla luciferase
activity.
Western blotting
Total protein was obtained from Eca 109 or KYSE 30
cells using RIPA buffer and PMSF (Beijing Solarbio Science &
Technology Co., Ltd.). Protein concentration was determined using a
BCA Protein Assay kit (cat. no. SI255483; Pierce; Thermo Fisher
Scientific, Inc.). Proteins (80 µg/lane) were separated via
SDS-PAGE on 10% gels, and then separated proteins were
electrophoretically transferred onto PVDF membranes (EMD
Millipore). Membranes were then blocked with 5% skimmed milk at
room temperature for 1 h. The membrane was incubated with
antibodies against FLOT-1 (1:1,000; cat. no. 18634; Cell Signaling
Technology, Inc.), E-cadherin (1:1,000; cat. no. ab40772; Abcam),
ZEB1 (1:500; cat. no. 21544-1-AP; ProteinTech Group, Inc.),
N-cadherin (1:500; cat. no. 22018-1-AP; ProteinTech Group, Inc.),
Vimentin (1:500; cat. no. 10366-1-AP; ProteinTech Group, Inc.) and
GAPDH (1:10,000; cat. no. AP0063; Bioworld Technology, Inc.) at 4°C
for 12 h. Next, the membranes were incubated at room temperature
for 1 h with a fluorochrome-labelled secondary anti-rabbit IgG
(1:10,000; cat. no. 926-32211; LI-COR Biosciences). The membranes
were visualized using the Odyssey® Infrared Imaging
System (LI-COR Biosciences). GAPDH served as a loading control.
Western blot analysis was carried out three times,
independently.
Lentiviral transfection
Eca 109 cells were cultured in 6-well plates.
HitransG P (40 µl/well; GeneChem, Inc.) together with
lentivirus-miR-485-5p (MOI 10) or lentivirus-vector (5 µl/well;
GeneChem, Inc.) labeled with enhanced green fluorescent protein
(EGFP) were added into the wells. HitransG P and
lentivirus-miR-485-5p or lentivirus-vector were removed 12–18 h
later. Then, cells were cultured in humidified air containing 5%
CO2 at 37°C for 72 h. Millesimal puromycin was used to
select cells for 24 h. The cells were observed under a fluorescence
microscope, if the EGFP-labeled cells were nearly at 100%, the
lentiviral transfection was successful.
Tumour xenograft in animals
A total of 12 male athymic Balb/c nude mice (4–6
weeks, 20–25 g) were purchased from Beijing Vital River Laboratory
Animal Technology Co., Ltd. All animal experiments were performed
with approval from the Animal Care Committee of The Fourth Hospital
of Hebei Medical University (approval. no. 110324200104285428). The
mice were housed at The Fourth Hospital of Hebei Medical University
Experiment Animal Centre (humidity, 50%; temperature, 25°C; light
cycle, 12 h light/12 h dark; ad libitum access to food and
water). The mice were randomly divided into two groups with six
mice per group. Eca 109 cells were stably transfected with
lentivirus-miR-485-5p or lentivirus-vector. Cells were suspended in
PBS (5×106 cells/200 µl) and injected into the right
flanks of mice subcutaneously. Tumour volumes were monitored with a
calliper every 5 days. Following which, the mice were sacrificed by
spinal dislocation on day 32. Finally, the tumours were removed and
weighed and fixed in formalin for IHC analysis, as described
above.
Statistical analysis
Statistics were calculated using GraphPad Prism
(v5.0) software (GraphPad Software, Inc.) and SPSS v22.0 (IBM
Corp.). The experimental data are presented as the mean ± standard
deviation from at least three separate experiments. For comparison
among the paired samples, paired t-tests were performed. For
comparison among the unpaired samples, an unpaired Student's t-test
was performed. One-way ANOVA followed by Dunnett's post hoc test
was used to examine the significant differences in the experimental
data between multiple groups. The relationship between
clinicopathological results and the expression of miR-485-5p was
examined using a Pearson's chi-squared test. The correlation
between miR-485-5p and FLOT-1 expression was analysed using
Spearman's correlation analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
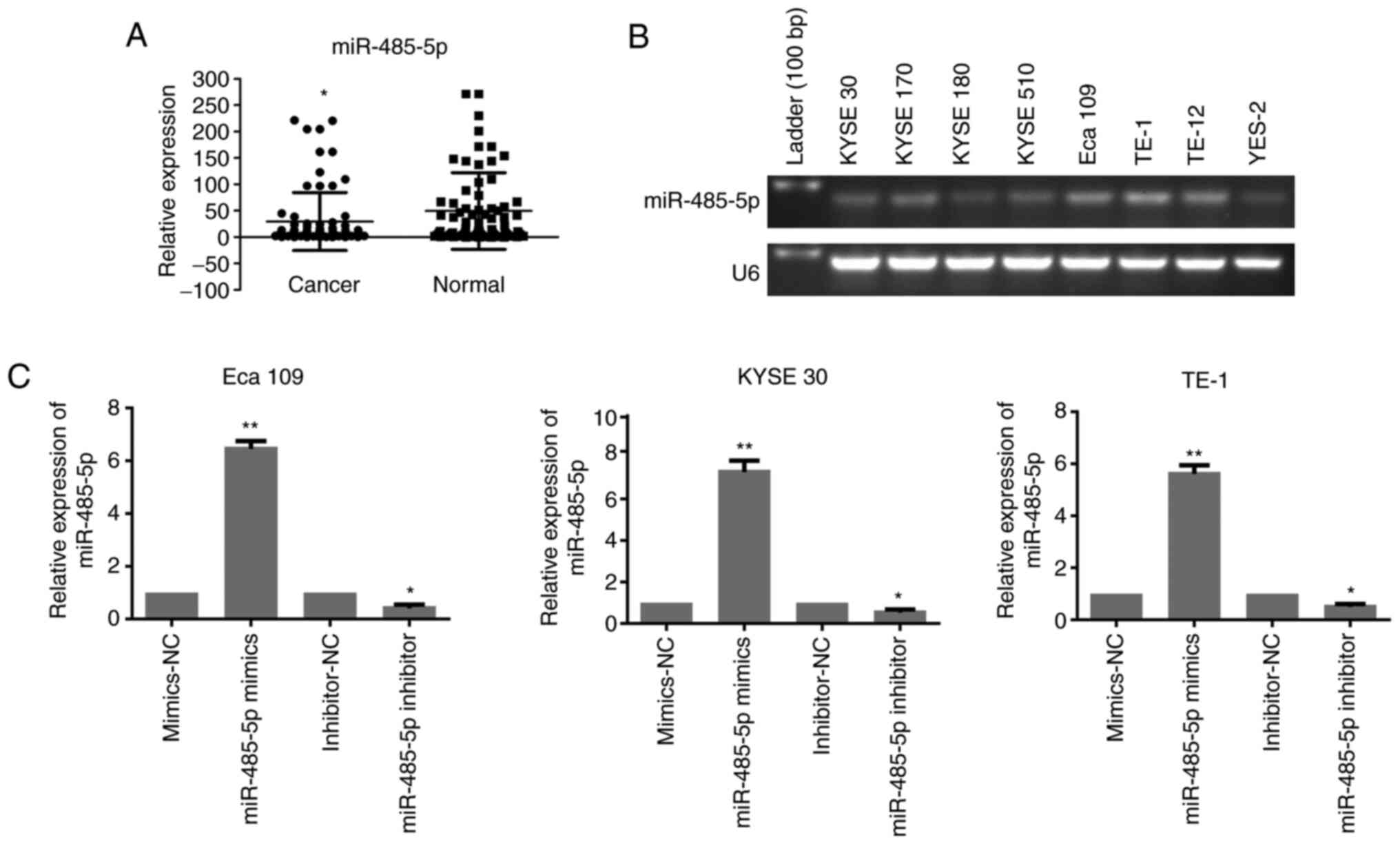
miR-385-5p is downregulated in ESCC
tissues
The relative expression of miR-485-5p in 80 pairs of
ESCC and paracarcinoma tissues was detected via RT-qPCR. As shown
in Fig. 1A, human ESCC tissues
exhibited lower expression of miR-485-5p than adjacent control
tissues.
It was also demonstrated that the expression of
miR-485-5p was strongly associated with the clinicopathological
features of ESCC. Based on the expression of miR-485-5p, 80 ESCC
samples were split into two groups. There were 44 cases in the high
expression group and 36 cases in the low expression group.
Decreased miR-485-5p expression was associated with a larger tumour
size and poor differentiation and stages III/IV. Moreover, a
decrease in miR-485-5p was more common in male patients (Table I). These data indicated that the
expression of miR-485-5p was downregulated in ESCC tissues.
 | Table I.Association between
clinicopathological characteristics of patients with esophageal
squamous cell carcinoma and miR-485-5p expression. |
Table I.
Association between
clinicopathological characteristics of patients with esophageal
squamous cell carcinoma and miR-485-5p expression.
|
| miR-485-5p
expression |
|
|---|
|
|
|
|
|---|
| Clinicopathological
characteristics | High | Low | P-value |
|---|
| Age, years |
|
| 0.369 |
|
≥65 | 24 | 16 |
|
|
<65 | 20 | 20 |
|
| Sex |
|
| 0.021a |
|
Male | 36 | 21 |
|
|
Female | 8 | 15 |
|
| Size,
cm3 |
|
| 0.014a |
|
≥10 | 16 | 23 |
|
|
<10 | 28 | 13 |
|
| Lymph node
metastasis |
|
| 0.805 |
| Present
(N1-N3) | 22 | 17 |
|
| Absent
(N0) | 22 | 19 |
|
| Histological
type |
|
| 0.001b |
|
Poor | 14 | 25 |
|
| Well to
moderate | 30 | 11 |
|
| Stage |
|
| 0.006b |
|
I/II | 27 | 11 |
|
|
III/IV | 17 | 25 |
|
Furthermore, the expression of miR-485-5p was
examined in ESCC cell lines (KYSE 30, KYSE 170, KYSE 180, KYSE 510,
Eca 109, TE-1, TE-12 and YES-2) by semi-quantitative PCR.
miR-485-5p expression was verified to be relatively high in Eca 109
and TE-1 cells, but relatively low in KYSE 30 cells, as shown in
Fig. 1B. Thus, these three cell lines
were used for the following experiments.
KYSE 30, Eca 109 and TE-1 cell lines were
transfected with miR-485-5p mimics and inhibitor to explore the
role of miR-485-5p in the biological functions of ESCC cells. The
ratio of cell line transfection was validated by RT-qPCR. As
indicated in Fig. 1C, the expression
of miR-485-5p was significantly upregulated in mimics group and
downregulated in the inhibitor group, compared with the control
groups.
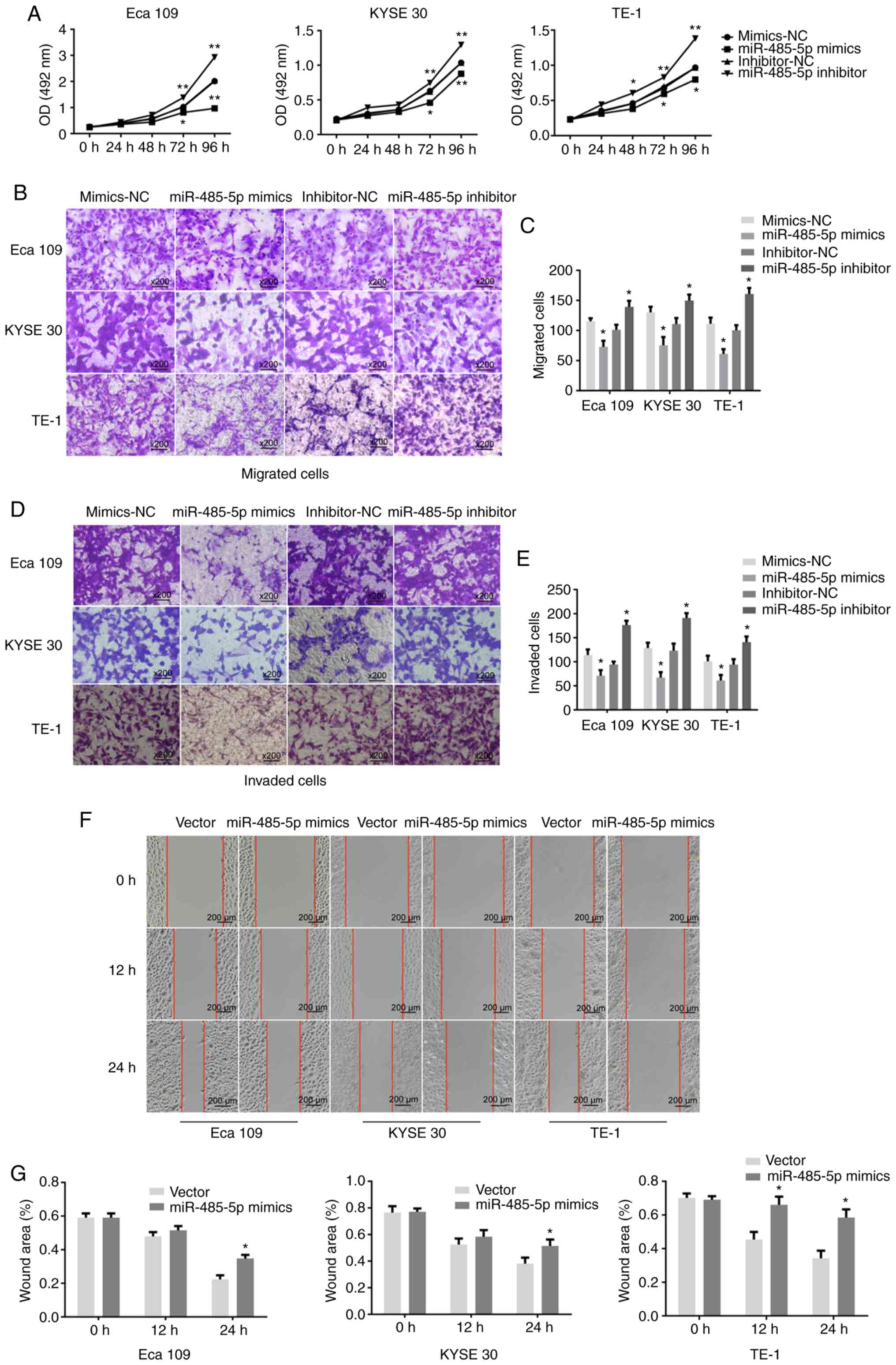
miR-485-5p inhibits the proliferation,
migration and invasion of ESCC cell lines
The influence of miR-485-5p on the proliferative
capacity of ESCC cells was verified using an MTS assay. Compared
with Eca 109, KYSE 30 and TE-1 cells transfected with the
inhibitor-NC, the proliferation rates of cells transfected with
miR-485-5p inhibitor were increased. Whereas, the inhibition rates
of Eca 109, KYSE 30 and TE-1 cells transfected with miR-485-5p
mimics were decreased in contrast to cells transfected with
mimics-NC. The proliferation rate was significantly reduced in Eca
109, KYSE 30 and TE-1 cells transfected with miR-485-5p mimics
compared with the mimics-NC group, while the opposite effect was
observed in the inhibitor group (Fig.
2A).
Further investigation demonstrated the inhibitory
role that miR-485-5p played in ESCC cells. The migration ratios of
Eca 109, KYSE 30 and TE-1 cells transfected with miR-485-5p mimics
were reduced, whereas the migration ratios of Eca 109, KYSE 30 and
TE-1 cells transfected with miR-485-5p inhibitor were significantly
increased (Fig. 2B and C). Moreover,
compared with the negative control, the invasion ratio was also
decreased in Eca 109, KYSE 30 and TE-1 cell lines overexpressing
miR-485-5p, but increased in the miR-485-5p knockdown group
(Fig. 2D and E).
Next, a wound healing assay was performed to confirm
these results. Compared with the negative control, the migratory
rates of Eca 109, KYSE 30 and TE-1 cells transfected with mimics at
24 h were decreased. The overexpression of miR-485-5p reduced the
migratory rate of ESCC cells (Fig.
2F). Additionally, the statistical results uncovered the
suppressive role of miR-485-5p in the migration of ESCC cells
(Fig. 2G).
FLOT-1 is a predicted target of
miR-485-5p
TargetScan (http://www.targetscan.org/vert_72/) was employed to
predict targets of miR-485-5p. According to TargetScan, FLOT-1,
which is an oncogene related to a number of malignancies (21–27,31), might
also be a target gene of miR-485-5p (Fig.
3A). As Guo et al (31)
verified that the upregulation of FLOT-1 enhanced the malignant
phenotype of lung adenocarcinoma cells in vitro, including
cell proliferation, migration and invasion, it is of significance
to explore whether the potential mechanism by which miR-485-5p
inhibits ESCC is related to inhibition of the expression of FLOT-1.
First, the FLOT-1 plasmid was constructed and transfection efficacy
was verified via western blotting (Fig.
3B). Compared with cells transfected with pcDNA3.1, the protein
expression of FLOT-1 was increased in cells transfected with the
FLOT-1 plasmid. To confirm the predicted result, FLOT-1 expression
was determined in ESCC cell lines transfected with miR-485-5p
mimics or inhibitor via RT-qPCR and western blotting. Both the mRNA
and protein levels were relatively high in cells transfected with
the inhibitor, whereas the cells transfected with miR-485-5p mimics
showed the opposite effect in comparison with the control group
(Fig. 3C and D).
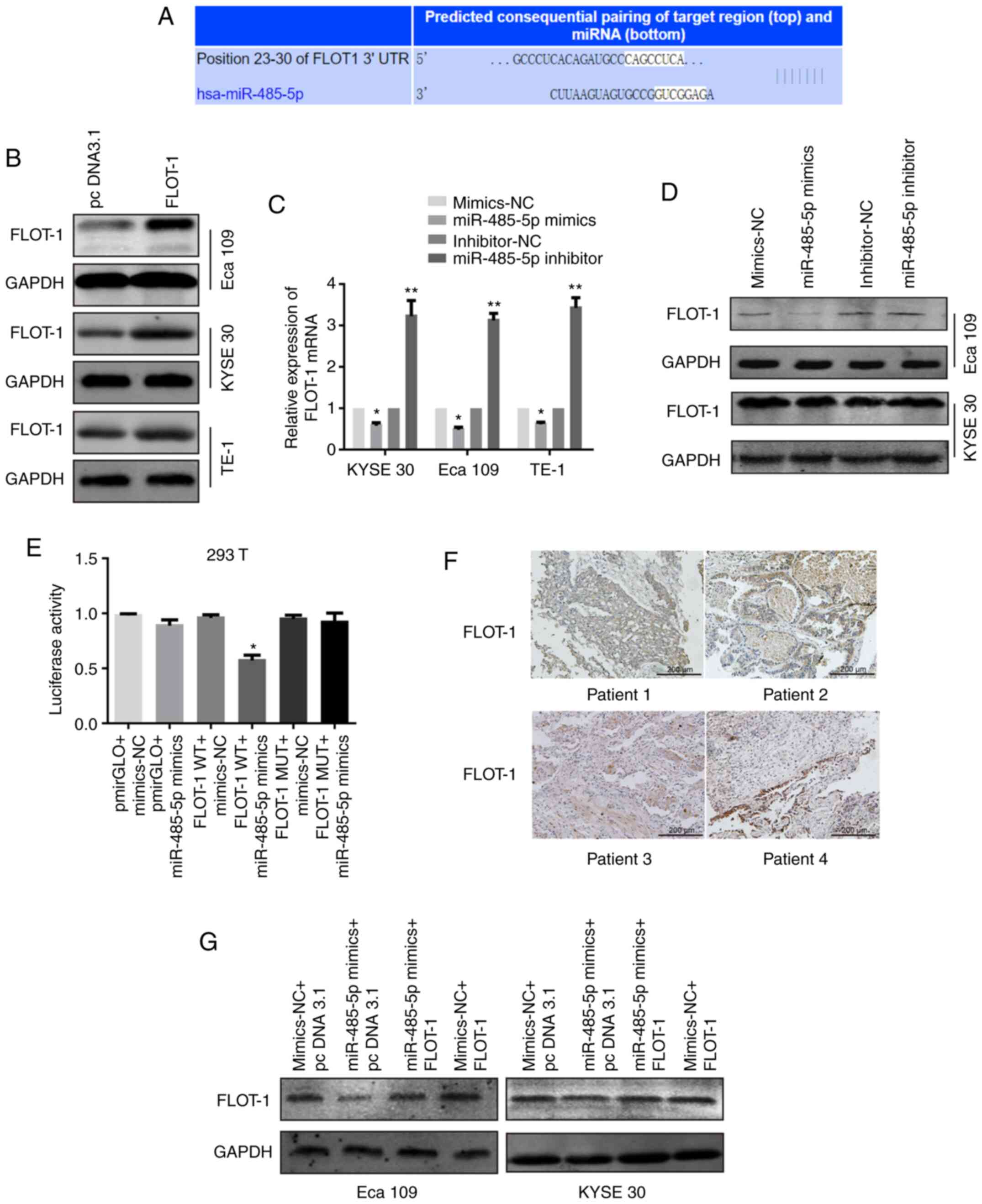 | Figure 3.FLOT-1 is a predicted target of
miR-485-5p. (A) Bioinformatics analysis predicted a
miR-485-5p-binding site in the 3′UTR of FLOT1 mRNA. (B) The FLOT-1
plasmid transfection efficacy was verified in Eca 109, KYSE 30 and
TE-1 cells transfected with FLOT-1 plasmid or pcDNA3.1 via western
blotting. (C) FLOT-1 mRNA levels in KYSE 30, Eca 109 and TE-1 cells
transfected with miR-485-5p mimics, miR-485-5p inhibitor or their
corresponding controls (miR-NC and inhibitor-NC). (D) FLOT-1
protein levels of Eca 109 and KYSE 30 cells transfected with
miR-485-5p mimics, miR-485-5p inhibitor or their corresponding
controls (miR-NC and inhibitor-NC). (E) Effects of miR-485-5p on
the translation of the reporter gene inserted downstream of the
3′UTR of FLOT-1 mRNA or the mutated 3′UTR of FLOT-1 mRNA in 293T
cells. 293T cells were co-transfected as shown in the figure, and
the luciferase activities were measured using a Dual-luciferase
reporter assay. The luciferase activity was normalized and
expressed as the ratio of firefly/Renilla luciferase
activities. (F) IHC staining of FLOT-1 expression in esophageal
squamous cell carcinoma tissue sections (scale bar, 200 µm). (G)
FLOT-1 protein levels in Eca 109 and KYSE 30 cells co-transfected
with mimics-NC and pcDNA3.1, miR-485-5p mimics and pcDNA3.1,
miR-485-5p mimics and FLOT-1, mimics-NC and FLOT-1. The data are
expressed as the mean ± SD. *P<0.05 and **P<0.01 vs. control
group. FLOT-1, flotillin-1; miR, microRNA; UTR, untranslated
region; NC, negative control; IHC, immunohistochemistry; WT,
wild-type; MUT, mutant. |
In addition, to further verify whether FLOT-1 is a
direct target of miR-485-5p, a dual-luciferase reporter assay
system was utilized. MUT and WT FLOT-1 3′UTR (site-directed
mutations were contained in the former) were cloned into pmirGLO
reporter plasmids. pmirGLO-WT-FLOT-1 3′UTR and miR-485-5p mimics
were co-transfected into 293T cells. As shown in Fig. 3E, a notably decreased luciferase
activity could be observed in the FLOT-1 WT + miR-485-5p mimics
group compared with the FLOT-1 WT + mimics-NC group. Moreover, the
overexpression of miR-485-5p did not have any influence on the
luciferase activity of cells transfected with pmirGLO-MUT-FLOT-1
3′UTR.
To determine the relationship between the expression
of miR-485-5p and FLOT-1 in ESCC, 40 ESCC samples were collected
for examination by RT-qPCR and IHC. These 40 samples were divided
into two different groups based on the median expression of
miR-485-5p: A high expression group and a low expression group. It
was found that there was a moderate negative correlation between
FLOT-1 expression and miR-485-5p expression (r=−0.560; Fig. 3F and Table
II).
 | Table II.Negative correlation between
miR-485-5p and FLOT-1 protein expression levels using Spearman's
correlation analysis. miR-485-5p expression was measured via
reverse transcription-quantitative PCR, and FLOT-1 protein levels
were measured by IHC. |
Table II.
Negative correlation between
miR-485-5p and FLOT-1 protein expression levels using Spearman's
correlation analysis. miR-485-5p expression was measured via
reverse transcription-quantitative PCR, and FLOT-1 protein levels
were measured by IHC.
|
| miR-485-5p
expression |
|
|
|---|
|
|
|
|
|
|---|
| FLOT-1
expression | High | Low | r-value | P-value |
|---|
| Negative (−/+) | 14 | 5 | −0.560 | P<0.01 |
| Positive
(++/+++) | 6 | 15 |
|
|
To further confirm that miR-485-5p inhibited tumour
progression through FLOT-1 inhibition in ESCC, functional recovery
experiments were carried out. miR-485-5p mimics and FLOT-1
plasmids, and their NCs, were co-transfected into the Eca 109 and
KYSE 30 cells. After transfection, western blotting was performed
for each group to determine FLOT-1 expression (Fig. 3G). The FLOT-1 expression levels in Eca
109 and KYSE 30 cells in the miR-485-5p mimics + pcDNA 3.1 group
were notably decreased compared with the cells in the mimics-NC +
pc DNA3.1 group, whereas the FLOT-1 expression of cells in the
miR-485-5p mimics + FLOT-1 group increased compared with cells in
the miR-485-5p mimics + pcDNA3.1 group. These results suggested
that the 3′UTR of FLOT-1 was a target of miR-485-5p, yet the
interaction was abolished in this sequence by point mutations.
Overexpression of miR-485-5p inhibits
the EMT
EMT is an important biological process that promotes
the invasion and metastasis of cancer cells. It has been reported
that miR-485-5p can reverse EMT in non-small cell lung cancer cells
(32). As the present study confirmed
that miR-485-5p overexpression could inhibit the migration and
invasion of ESCC cells, it was of significance to determine the
association between miR-485-5p and EMT in ESCC cells by evaluating
the mRNA and protein expression levels of EMT-associated factors.
The RT-qPCR and western blotting results revealed that E-cadherin
expression in Eca 109 and KYSE 30 cells transfected with miR-485-5p
mimics was increased, whereas Vimentin, N-cadherin and ZEB1
expression levels were decreased (Fig.
4A-C).
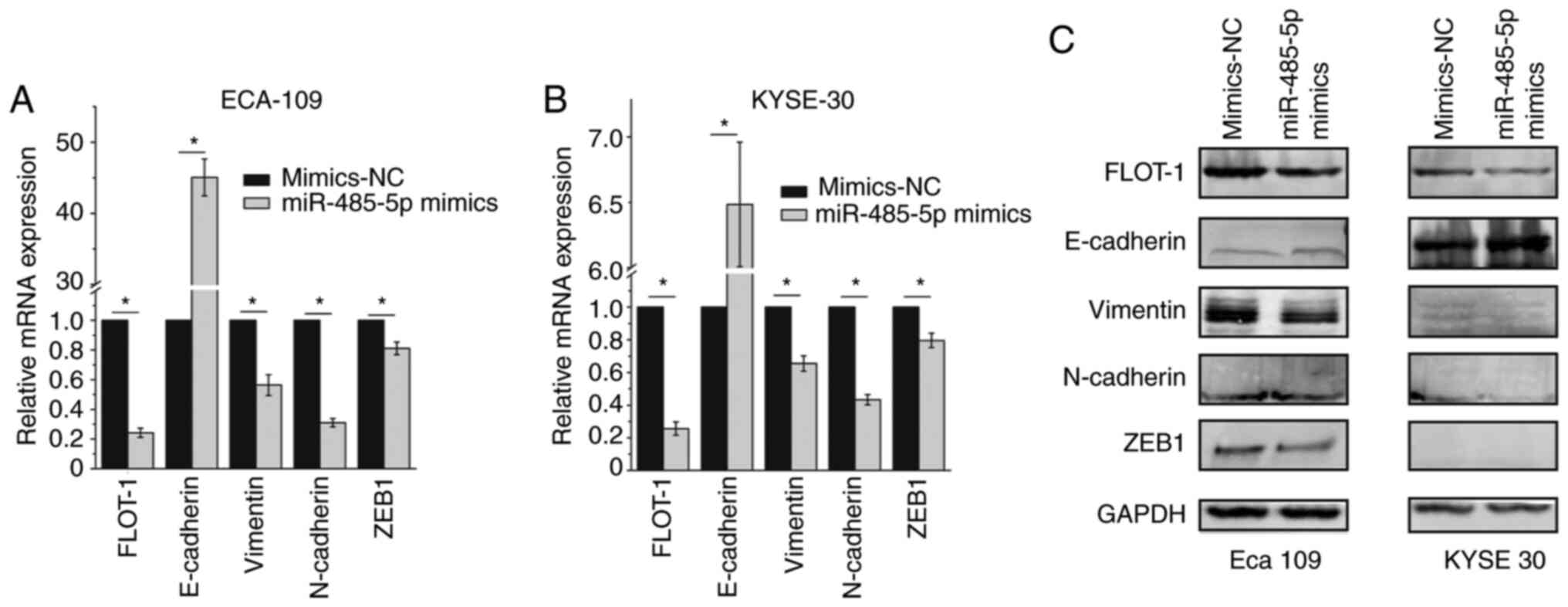 | Figure 4.Overexpression of miR-485-5p inhibits
the epithelial-mesenchymal transition. The mRNA levels of FLOT-1,
E-cadherin, Vimentin, N-cadherin and ZEB1 in (A) Eca 109 and (B)
KYSE 30 cells transfected with miR-485-5p or mimics-NC. (C) The
protein levels of FLOT-1, E-cadherin, Vimentin, N-cadherin and ZEB1
in Eca 109 (left) and KYSE 30 (right) cells transfected with
miR-485-5p or mimics-NC. The data are expressed as the mean ± SD.
*P<0.05. FLOT-1, flotillin-1; miR, microRNA; NC, negative
control; ZEB1, zinc finger E-box-binding homeobox 1. |
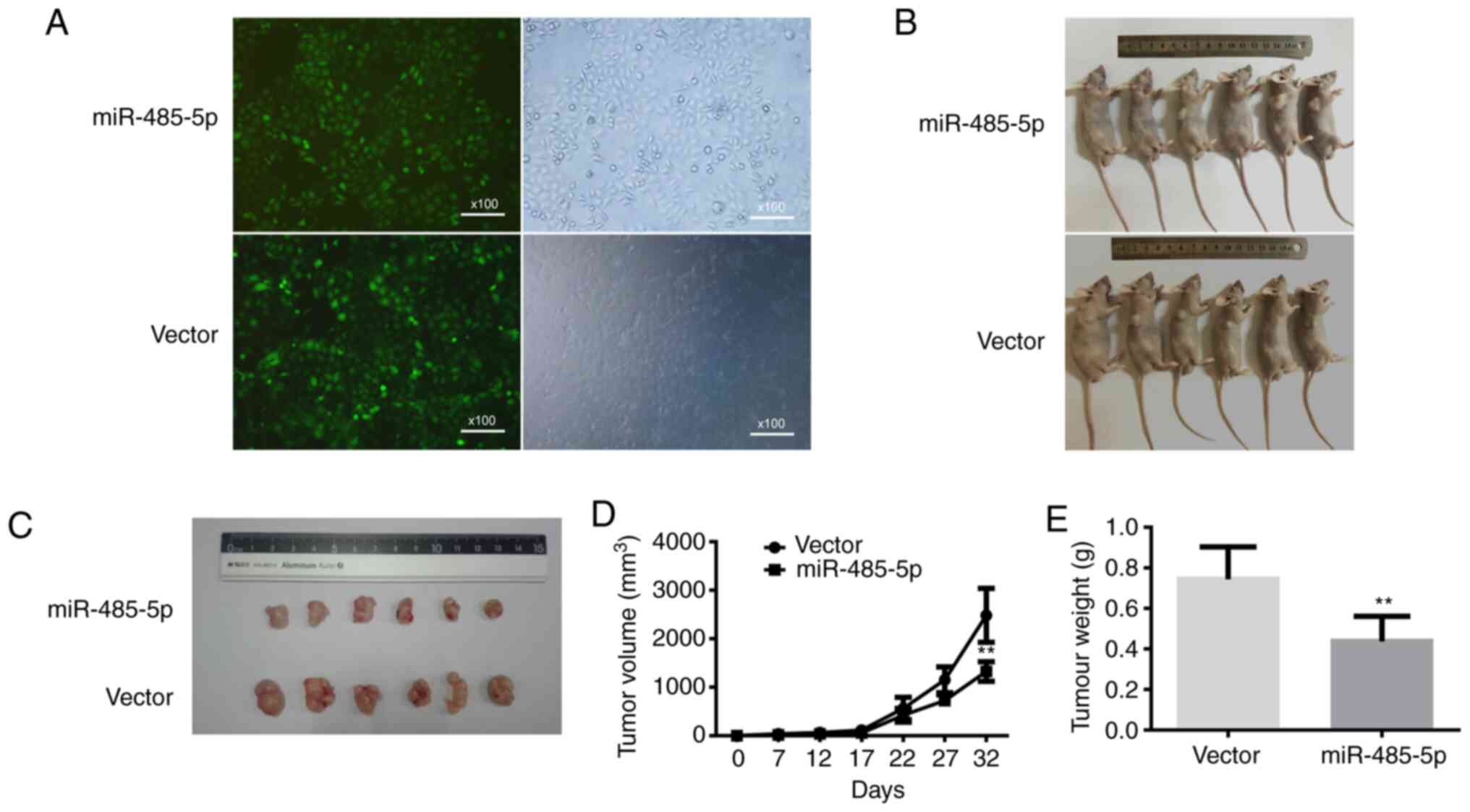
miR-485-5p suppresses the growth of
ESCC in vivo
To illustrate the influence of miR-485-5p on tumour
growth in vivo, Eca 109 cells transfected with
lentivirus-miR-485-5p were injected into the flanks of nude mice,
while cells transfected with lentivirus-NC were utilized as the
negative control (Fig. 5A and B). As
demonstrated in Fig. 5C-E, the
lentivirus-miR-485-5p group exhibited an observable decline in
tumour volume and weight compared with the control group. On day
32, the volume and weight of tumours transfected with
lentivirus-miR-485-5p were 1,324.5±201.39 mm3 and
0.45±0.11 g, respectively, while those of the negative control were
2,482.83±555.13 mm3 and 0.76±0.15 g, respectively.
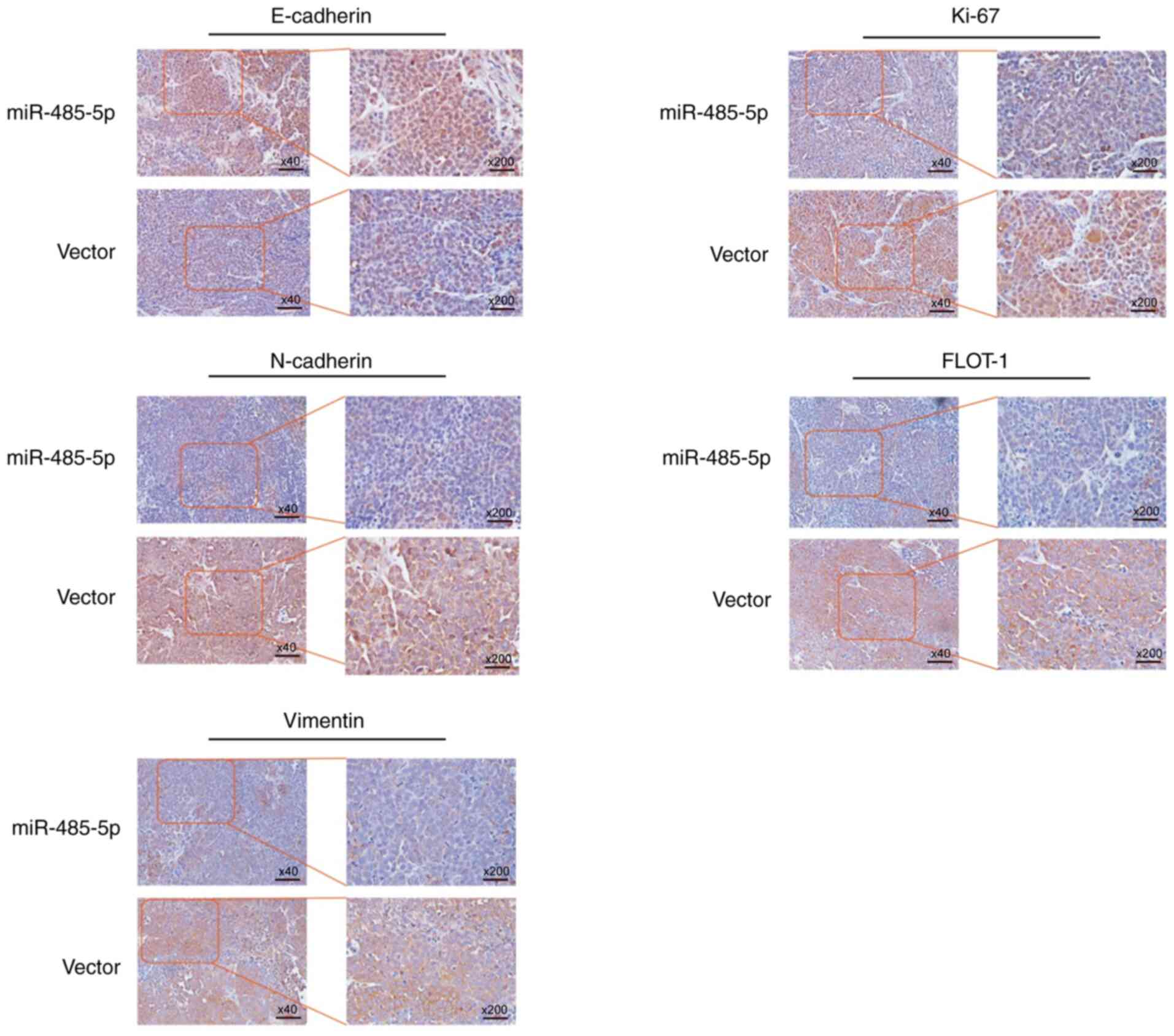
Moreover, the expression levels of FLOT-1, Ki-67 and
EMT-related proteins were investigated by IHC. The protein
expression levels of Ki-67, FLOT-1, N-cadherin and Vimentin,
compared with those in the control group, were all obviously
decreased in the lentivirus-miR-485-5p transfection group, while
E-cadherin was elevated (Fig. 6). In
summary, miR-485-5p played roles as a tumour suppressor and
apoptosis promotor of ESCC in vivo.
Discussion
The first miRNA was discovered in 1993 when the
heterochronic gene lin-4 from Caenorhabditis elegans was
identified as a small non-coding RNA (33). Over the past decades, as an increasing
number of miRNAs have been identified, investigators have gradually
realized that miRNAs are a cluster of short non-coding RNAs that
can inhibit the stability of mRNA structures, decrease the
translation ratio of proteins, disrupt the regulation of a number
of biological processes, and play a role in cancer development
(34). miRNAs have an impact on the
biological behaviour of cancer cells, including proliferation,
migration, invasion, cell cycle and apoptosis, and play a role in
tumour occurrence and development (34–38).
miR-485-5p has been demonstrated to be a functional
tumour suppressor. Duan et al (39) reported that miR-485-5p functions as a
tumour suppressor in gastric cancer by targeting
7,8-dihydro-8-oxoguanine triphosphatase. Gao et al (40) discovered that miR-485-5p blocks the WW
domain-binding protein 2/Wnt signalling pathway to inhibit the
progression of hepatocellular carcinoma. The inhibitory function of
miR-485-5p has also been reported in thyroid cancer (41,42),
cholangiocarcinoma (43), lung cancer
(32,44), osteosarcoma (45), breast cancer (46) and EC (20). Han et al (20) discovered that miR-485-5p, which can
downregulate the expression of O-linked N-acetylglucosamine
transferase, represses the proliferation and invasion of EC cells.
The present study showed that miR-485-5p was reduced in human ESCC,
and that overexpression of miR-485-5p could suppress the
proliferation, migration and invasion of ESCC cell lines. Through
bioinformatics prediction, FLOT-1 was identified as a potential
target of miR-485-5p.
Previous studies have revealed that the
dysregulation of FLOT is involved in various cancers. Several
reports have demonstrated that the overexpression of FLOT-1
increases the ratio of tumourigenicity as well as cell
proliferation by activating FOXO3a transcriptional activity in
breast cancer (47), whereas the
upregulation of FLOT-1 adjusted by NF-κB and Wnt/β-catenin promotes
cell invasion, motility and lymph metastasis of the pelvis in
cervical carcinoma (48).
Furthermore, Liu et al (49)
verified that highly expressed FLOT-2 facilitates proliferation,
migration and invasion in melanoma. Studies have found that FLOT-1
facilitates the signalling of tumour necrosis factor-α (TNF-α)
receptor and is relevant to ESCC progression (50). Guo et al (31) verified that FLOT-1 promotes the
malignant phenotype of lung adenocarcinoma in vitro,
including cell proliferation, migration and invasion. Moreover,
Jang et al (51) reported that
the sumoylation of FLOT-1 promotes EMT in metastatic prostate
cancer.
Previous studies have verified that FLOT-1
facilitates malignant phenotypes, such as cell proliferation,
migration, invasion and EMT in vitro, while in the present
study, the results of bioinformatics prediction showed that FLOT-1
could be a potential target of miR-485-5p. It was verified that the
expression of FLOT-1 could be directly reduced by the upregulation
of miR-485-5p in ESCC. As shown in the present study, miR-485-5p
overexpression inhibited cell proliferation, migration, invasion
and EMT in ESCC, thus it is reasonable to conclude that miR-485-5p
can inhibit these cell behaviours by repressing FLOT-1 expression.
There are some limitations in this article. It was verified that
miR-485-5p expression was higher in ESCC tissues than that in
paracancerous tissues, however miR-485-5p expression levels in ESCC
cell lines were not compared with a normal esophagus cell line.
Although other studies reported FLOT-1 facilitates malignant
phenotypes in various cancer types, the present study did not
verify its functions in ESCC directly. Overall, understanding the
underlying actions of miR-485-5p in the pathogenesis of ESCC will
increase the knowledge of the biological basis of tumour
progression, which will increase the possibility of developing a
novel diagnostic marker and original therapeutic strategy for
ESCC.
Acknowledgements
Not applicable.
Funding
This project was supported by the National Natural
Science Foundation of China (grant nos. 81973520 and 81673642).
Availability of data and materials
All data generated or analysed during this study are
included in this published article.
Authors' contributions
RiZ and YS conceived the hypothesis, designed and
performed the experiments. XZ and RuZ conducted the data
collection, analysis and interpretation, and wrote the manuscript.
CZ collected the specimens, performed the in vitro
experiments and edited the manuscript. LZ and BS performed the
animal experiments, treated the animals, supervised the findings of
this work, aided in interpreting the results, and provided the
funds and critical revision of the manuscript. YS and LZ confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
This project was approved by the Ethics Committee of
the Fourth Hospital of Hebei Medical University (Shijiazhuang,
China). Written informed consent was obtained from all subjects or
guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ESCC
|
esophageal squamous cell carcinoma
|
|
miRNA
|
microRNA
|
|
3′UTR
|
3′untranslated region
|
|
NC
|
negative control
|
|
FLOT-1
|
flotillin-1
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
EMT
|
epithelial-mesenchymal transition
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brock MV, Gou M, Akiyama Y, Muller A, Wu
TT, Montgomery E, Deasel M, Germonpré P, Rubinson L, Heitmiller RF,
et al: Prognostic importance of promoter hypermethylation of
multiple genes in esophageal adenocarcinoma. Clin Cancer Res.
9:2912–2919. 2003.PubMed/NCBI
|
|
4
|
Thrumurthy SG, Chaudry MA, Thrumurthy SSD
and Mughal M: Oesophageal cancer: Risks, prevention, and diagnosis.
BMJ. 366:143732019.
|
|
5
|
Mariette C, Markar SR, Dabakuyo-Yonli TS,
Meunier B, Pezet D, Collet D, D'Journo XB, Brigand C, Perniceni T,
Carrère N, et al: Hybrid minimally invasive esophagectomy for
esophageal cancer. N Engl J Med. 380:152–162. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang M, Shi L, Yang C, Ge Y, Lin L, Fan
H, He Y, Zhang D, Miao Y and Yang L: MiR-1254 inhibits cell
proliferation, migration, and invasion by down-regulating Smurf1 in
gastric cancer. Cell Death Dis. 10:322019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Z, Zhang Y, Sun XX, Ma X and Chen
ZN: MicroRNA-146a inhibits cancer metastasis by downregulating VEGF
through dual pathways in hepatocellular carcinoma. Mol Cancer.
14:11862015. View Article : Google Scholar
|
|
11
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Troschel FM, Böhly N, Borrmann K, Braun T,
Schwickert A, Kiesel L, Eich HT, Götte M and Greve B: MiR-142-3p
attenuates breast cancer stem cell characteristics and decreases
radioresistance in vitro. Tumour Biol. 40:10104283187918872018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu Y, Huang J, Xu H and Gong Z:
Over-Expression of miR-15a-3p enhances the radiosensitivity of
cervical cancer by targeting tumor protein D52. Biomed
Pharmacother. 105:1325–1334. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shao Y, Zhang D, Li X, Yang J, Chen L,
Ning Z, Xu Y, Deng G, Tao M, Zhu Y and Jiang J: MicroRNA-203
increases cell radiosensitivity via directly targeting Bmi-1 in
hepatocellular carcinoma. Mol Pharm. 15:3205–3215. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Peng X, Lu X, Wei Q, Chen M and
Liu L: Inhibition of hsa_circ_0001313 (circCCDC66) induction
enhances the radio-sensitivity of colon cancer cells via tumor
suppressor miR-338-3p: Effects of cicr_0001313 on colon cancer
radio-sensitivity. Pathol Res Pract. 215:689–696. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu J, Wu SW and Wu WP: A tumor-suppressive
microRNA, miRNA-485-5p, inhibits glioma cell proliferation and
invasion by down-regulating TPD52L2. Am J Transl Res. 9:3336–3344.
2017.PubMed/NCBI
|
|
17
|
Wang R, Zuo X, Wang K, Han Q, Zuo J, Ni H,
Liu W, Bao H, Tu Y and Xie P: MicroRNA-485-5p attenuates cell
proliferation in glioma by directly targeting paired box 3. Am J
Cancer Res. 8:2507–2517. 2018.PubMed/NCBI
|
|
18
|
Chai Y, Du Y, Zhang S, Xiao J, Luo Z, He F
and Huang K: MicroRNA-485-5p reduces O-GlcNAcylation of Bmi-1 and
inhibits colorectal cancer proliferation. Exp Cell Res.
368:111–118. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu XX, Xu XN, He BS, Sun HL, Xu T, Liu XX,
Chen XX, Zeng KX, Wang SK and Pan YQ: MicroRNA-485-5p functions as
a tumor suppressor in colorectal cancer cells by targeting CD147. J
Cancer. 9:2603–2611. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han DL, Wang LL, Zhang GF, Yang WF, Chai
J, Lin HM, Fu Z and Yu JM: MiRNA-485-5p, inhibits esophageal cancer
cells proliferation and invasion by down-regulating O-linked
N-acetylglucosamine transferase. Eur Rev Med Pharmacol Sci.
23:2809–2816. 2019.PubMed/NCBI
|
|
21
|
Dam DHM, Jelsma SA, Yu JM, Liu H, Kong B
and Paller AS: Flotillin and AP2A1/2 promote IGF-1 receptor
association with clathrin and internalization in primary human
keratinocytes. J Invest Dermatol. 140:1743–1752. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sonnino S and Prinetti A: Membrane domains
and the ‘lipid raft’ concept. Curr Med Chem X20. 4–21. 2013.
|
|
23
|
George KS and Wu S: Lipid raft: A floating
island of death or survival. Toxicol Appl Pharmacol. 259:311–319.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ficht X, Ruef N, Stolp B, Samson GPB,
Moalli F, Page N, Merkler D, Nichols BJ, Diz-Muñoz A, Legler DF, et
al: In vivo function of the lipid raft protein flotillin-1 during
CD8(+) T cell-mediated host surveillance. J Immunol. 203:2377–2387.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu SM and Fairn GD: Mesoscale organization
of domains in the plasma membrane-beyond the lipid raft. Crit Rev
Biochem Mol Biol. 53:192–207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Affentranger S, Martinelli S, Hahn J,
Rossy J and Niggli V: Dynamic reorganization of flotillins in
chemokinestimulated human T-lymphocytes. BMC Cell Biol. 12:282011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guillaume E, Comunale F, Do Khoa N,
Planchon D, Bodin S and Gauthier-Rouviere C: Flotillin microdomains
stabilize cadherins at cell-cell junctions. J Cell Sci.
126:5293–5304. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rice T, Ishwaran H, Hofstetter W, Kelsen
D, Apperson-Hansen C and Blackstone E; Worldwide Esophageal Cancer
Collaboration Investigators, : Recommendations for pathologic
staging (pTNM) of cancer of the esophagus and esophagogastric
junction for the 8th edition AJCC/UICC staging manuals. Dis
Esophagus. 29:897–905. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dai SL, Wei SS, Zhang C, Li XY, Liu YP, Ma
M, Lv HL, Zhang Z, Zhao LM and Shan BE: MTA2 promotes the
metastasis of esophageal squamous cell carcinoma via EIF4E-twist
feedback loop. Cancer Sci. 19:147782020.
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guo AY, Liang XJ, Liu RJ, Li XX, Bi W,
Zhou LY, Tang CE, Yan A, Chen ZC and Zhang PF: Flotilin-1 promotes
the tumorigenicity and progression of malignant phenotype in human
lung adenocarcinoma. Cancer Biol Ther. 18:715–722. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang RS, Zheng YL, Li C, Ding C, Xu C and
Zhao J: MicroRNA-485-5p suppresses growth and metastasis in
non-small cell lung cancer cells by targeting IGF2BP2. Life Sci.
199:104–111. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee RC, Feinbaum RL and Ambros V: The C.
Elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen H, Yao X, Di X, Zhang Y, Zhu H, Liu
S, Chen T, Yu D and Sun X: MiR-450a-5p inhibits autophagy and
enhances radiosensitivity by targeting dual-specificity phosphatase
10 in esophageal squamous cell carcinoma. Cancer Lett. 28:114–126.
2020. View Article : Google Scholar
|
|
36
|
Wang F, Li L, Piontek K, Sakaguchi M and
Selaru FM: Exosome miR-335 as a novel therapeutic strategy in
hepatocellular carcinoma. Hepatology. 67:940–954. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Qiu BQ, Lin XH, Ye XD, Huang W, Pei X,
Xiong D, Long X, Zhu SQ, Lu F, Lin K, et al: Long non-coding RNA
PSMA3-AS1 promotes malignant phenotypes of esophageal cancer by
modulating the miR-101/EZH2 axis as a ceRNA. Aging (Albany NY).
12:1843–1856. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Y, Sun L, Wang L, Liu Z, Li Q, Yao B,
Wang C, Chen T, Tu K and Liu Q: Long non-coding RNA DSCR8 acts as a
molecular sponge for miR-485-5p to activate wnt/β-catenin signal
pathway in hepatocellular carcinoma. Cell Death Dis. 9:8512018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Duan J, Zhang H, Li S, Wang X, Yang H,
Jiao S and Ba Y: The role of miR-485-5p/NUDT1 axis in gastric
cancer. Cancer Cell Int. 17:922017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gao J, Dai C, Yu X, Yin XB and Zhou F:
MicroRNA-485-5p inhibits the progression of hepatocellular
carcinoma through blocking the WBP2/wnt signaling pathway. Cell
Signal. 66:1094662020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y, Hu J, Zhou W and Gao H: LncRNA
FOXD2-AS1 accelerates the papillary thyroid cancer progression
through regulating the miR-485-5p/KLK7 axis. J Cell Biochem.
19:10022018.
|
|
42
|
Li G and Kong Q: LncRNA LINC00460 promotes
the papillary thyroid cancer progression by regulating the
LINC00460/miR-485-5p/raf1 axis. Biol Res. 52:612019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bao W, Cao F, Ni S, Yang J, Li H, Su Z and
Zhao B: lncRNA FLVCR1-AS1 regulates cell proliferation, migration
and invasion by sponging miR-485-5p in human cholangiocarcinoma.
Oncol Lett. 18:2240–2247. 2019.PubMed/NCBI
|
|
44
|
Gao F, Wu H, Wang R, Guo Y, Zhang Z, Wang
T, Zhang G, Liu C and Liu J: MicroRNA-485-5p suppresses the
proliferation, migration and invasion of small cell lung cancer
cells by targeting flotillin-2. Bioengineered. 10:1–12. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang FR, Xu SH, Wang BM and Wang F:
MiR-485-5p inhibits metastasis and proliferation of osteosarcoma by
targeting CX3CL1. Eur Rev Med Pharmacol Sci. 22:7197–7204.
2018.PubMed/NCBI
|
|
46
|
Wang M, Cai WR, Meng R, Chi JR, Li YR,
Chen AX, Yu Y and Cao XC: MiR-485-5p suppresses breast cancer
progression and chemosensitivity by targeting survivin. Biochem
Biophys Res Commun. 501:48–54. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lin C, Wu Z, Lin X, Yu C, Shi T, Zeng Y,
Wang X, Li J and Song L: Knockdown of FLOT1 impairs cell
proliferation and tumorigenicity in breast cancer through
upregulation of FOXO3a. Clin Cancer Res. 17:3089–3099. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li Z, Yang Y, Gao Y, Wu X, Yang X, Zhu Y,
Yang H, Wu L, Yang C and Song L: Elevated expression of flotillin-1
is associated with lymph node metastasis and poor prognosis in
early-stage cervical cancer. Am J Cancer Res. 6:38–50. 2016.
|
|
49
|
Liu R, Xie H, Luo C, Chen Z, Zhou X, Xia
K, Chen X, Zhou M, Cao P, Cao K and Zhou J: Identification of FLOT2
as a novel target for microRNA-34a in melanoma. J Cancer Res Clin
Oncol. 141:993–1006. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Song L, Gong H, Lin C, Wang C, Liu L, Wu
J, Li M and Li J: Flotillin-1 promotes tumor necrosis factor-alpha
receptor signaling and activation of NF-κB in esophageal squamous
cell carcinoma cells. Gastroenterology. 143:995–1005. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jang D, Kwon H, Choi M, Lee J and Pak Y:
Sumoylation of flotillin-1 promotes EMT in metastatic prostate
cancer by suppressing snail degradation. Oncogene. 38:3248–3260.
2019. View Article : Google Scholar : PubMed/NCBI
|