Introduction
Breast cancer is the most commonly diagnosed type of
cancer in women worldwide. Triple negative breast cancer (TNBC) is
defined as breast cancer lacking expression of specific hormone
receptors, and is associated with a high degree of malignancy, a
poor prognosis and drug resistance, and accounts for 15–20% of all
breast cancer cases (1). Significant
progress has been made in identifying oncogenes and tumor
suppressor genes involved in the pathogenesis of breast cancer
(2); however, the molecular
mechanisms underlying this process remain poorly understood in
TNBC. Therefore, novel and treatments are required to improve
patient prognosis and survival.
Previous studies have revealed that centrosomes are
associated with cancer development, as oncoproteins and tumor
suppressor proteins are located in this organelle and drive
aberrations in the centrosome (3–6). The
centrosome is essential for cellular motility and intracellular
transport, and centrosome defects can cause abnormal microtubule
nucleation, formation of disorganized mitotic spindles, abnormal
segregation of chromosomes and aneuploidy (7,8). Spindle
assembly abnormal protein 6 homolog (SASS6) is an important
centrosomal protein required for centrosome duplication (9). SASS6 serves an important role in the
initiation of centriole formation and stabilization of centriole
intermediates (10), and acts to
ensure that each centriole seeds the formation of a single
procentriole in each cell cycle. However, upregulated expression of
SASS6 results in the seeding of more than one procentriole per
centriole (11). It has recently been
revealed that SASS6 expression is upregulated in several different
types of cancer, often only modestly increased, including in
invasive breast carcinomas, and SASS6 overexpression has been
revealed to cause excess foci-bearing centriolar markers in
colorectal cancer (12). However, it
is unclear whether SASS6 serves a role in TNBC.
To explore the role of SASS6 in human TNBC,
expression analysis of SASS6 in TNBC tissues was performed by
immunohistochemical staining, and then proliferation of human TNBC
cells after SASS6 knocked out was analyzed by MTT method. The cell
cycle distribution and apoptosis were detected by flow cytometry.
Furthermore, the possible underlying mechanisms were assessed using
PathScan intracellular signaling arrays and western blotting.
Materials and methods
Cell culture
Human breast cancer cell lines MDA-MB-231,
MDA-MB-468 and HCC-1937 cells (The Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences) were maintained in
RPMI-1640 or DMEM (Gibco; Thermo Fisher Scientific, Inc.),
supplemented with 10% (v/v) FBS, L-glutamine, a 2-fold vitamin
solution (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin and 0.1 mg/ml streptomycin (Sangon Biotech, Co., Ltd.).
Primary normal breast epithelial cells (PNBEC) were isolated from
the unaffected contralateral breast of patients undergoing surgical
procedures as previously described (13) and were cultured in DMEM/F12
supplemented with 5% FBS with antibiotics. All cells were
maintained with 5% CO2 at 37°C in a humidified
incubator.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted from cells using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. RNA was
reverse-transcribed to cDNA by GoScript™ Reverse Transcription
System (Promega Corporation) according to the manufacturer's
procedure. Quantitative real-time PCR was performed on a Roche
LightCycler 480 II Real-Time PCR platform using SYBRGreen MasterMix
Kit (Takara Biotechnology Co., Ltd.) according to the
manufacturer's instructions. The sequences of the primers used for
amplification were: SASS6 forward, 5′-GCGGCTAATAAAGACTTAACCGA-3′
and reverse, 5′-CTTCTTGCTTAGTCCGCTGTAG-3′; and GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′. The thermocycling conditions for qPCR
were: Denaturation at 95°C for 15 sec; followed by 45 cycles at
95°C for 5 sec and 60°C for 30 sec. All samples were assessed in
triplicate. The 2−ΔΔCq method (14) was used to calculate the relative
expression of the gene of interest.
Tissue samples and immunohistochemical
staining
TNBC tissue (n=18) and the matching normal breast
tissue (n=18) were obtained as 4-µm paraffin-embedded tissue block
slides from female patients diagnosed with TNBC who underwent
surgery at Shanxi Cancer Hospital (Shanxi, China) between October
2003 and August 2007 (age range, 38–57 years). The use of tissues
from patients was retrospective in the present study. TNBC patients
did not receive any chemotherapy or endocrine therapy prior to
tumour removal. Ethics approval was obtained from the Ethics
Committee of Shanxi Cancer Hospital and written informed consent
was obtained from each patient prior to sample collection. The
slides were deparaffinized, dehydrated, immersed in Tris-EDTA
buffer (pH 9.0) and incubated at 4°C overnight with anti-SASS6
(dilution 1:400; cat. no. 21377-1-AP; ProteinTech Group, Inc.), and
anti-cyclin dependent kinase 1 (CDK1) (dilution 1:400; cat. no.
19532-1-AP; ProteinTech Group, Inc.) separately, and subsequently
incubated for 15 min at 37°C with a poly-horseradish peroxidase
anti-rabbit antibody (dilution 1:500; cat. no. SA00004-2;
ProteinTech Group, Inc.). After visualization of the reaction using
DAB as a chromogen (Shanghai Gene Technology Co., Ltd.), the slides
were counterstained with hematoxylin for 30 sec at room temperature
and mounted using glycerin gel. The expression of SASS6 and CDK1 in
tissues was observed using a light microscope (magnification, ×40;
B41; Olympus Corporation). The results were evaluated and scored by
two blinded pathologists. Expression in tissues was stratified as
follows: <10%, -; 10–30%, +; 31–80%, ++; and >80%, +++.
Recombinant lentiviral vector
production and infection
The complementary DNA sequence
(5′-TGGCACTTTAGGAGCATTA-3′) of SASS6 and the control short hairpin
(sh)RNA (shCtrl) sequence (5′-TTCTCCGAACGTGTCACGT-3′) were designed
and inserted into a pGCSIL-GFP lentiviral vector (Shanghai GeneChem
Co., Ltd.), and the 3rd recombinant lentivirus was generated. To
generate the 3rd recombinant lentivirus, the reconstructed SASS6
silencing plasmid/control plasmid (20 µg) and packaging vector (15
µg) and envelope vector (10 µg) were co-transfected into 293T cells
(The Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences) using Lipofectamine 2000 according to the manufacturer's
procedure. Approximately 72 h after transfection, recombinant
lentivirus was harvested and purified by ultracentrifugation. Then,
cells were cultured in 6-well plates for lentivirus infection, and
the shSASS6-lentivirus or negative control lentivirus, was added at
multiplicity of infection (MOI) of 10 for 4 h. Cells were observed
under a fluorescence microscope (MicroPublisher 1X71; Olympus
Corporation) after 72 h of infection, and cells were harvested
after 120 h of infection to determine knockdown efficiency using
western blotting and RT-qPCR.
Western blotting
The cultured cells were lysed in a buffer (100 mM
Tris-HCl, pH 6.8, 2% mercaptoethanol, 4% SDS, 20% glycerin)
containing 1 mM protein inhibitor, 1 mM phenylmethylsulfonyl
fluoride and complete protease inhibitors cocktail (MedChemExpress)
for 10–15 min on ice. The lysates were centrifuged at 12,000 × g at
4°C for 15 min and the supernatants were collected. The protein
content was measured by bicinchoninic acid (BCA) method. Aliquots
(20 µg total protein/lane for each sample) were loaded on a 12.5%
SDS-gel, resolved using SDS-PAGE and transferred to PVDF membranes.
After blocking the membranes with 5% non-fat milk for 1 h at room
temperature, they were incubated with anti-GAPDH (cat. no.
sc-32233; Santa Cruz Biotechnology, Technology, Inc.), anti-SASS6
and anti-CDK1 (cat. nos. 21377-1-AP and 19532-1-AP; ProteinTech,
Group, Inc.), anti-S6 ribosomal protein (rpS6) and anti-BAD
(product codes ab225676 and ab32445; Abcam, Inc.), anti-PCNA
(product no. 2586), anti-STAT3 (product no. 9139), phosphorylated
(p)-STAT3 (Thr705) (product no. 9145), p-BAD (Ser116) (product no.
5284) and p-rpS6 (Ser235/236) (product no. 4858; all from Cell
Signaling Technology, Inc.) at 4°C overnight followed by incubation
with goat anti-rabbit/mouse IgG HRP-linked antibodies (cat. nos.
7074 and 7076; Cell Signaling Technology, Inc.) for 1 h at room
temperature. The antibodies were used in the following dilutions:
Anti-SASS6, anti-CDK1, anti-rpS6), anti-BAD, anti-p-BAD,
anti-p-STAT3, anti-STAT3 (all 1:1,000); anti-PCNA, anti-GAPDH,
anti-p-rpS6 (all 1:2,000). Anti-GAPDH protein was used as an
internal control. The signals were visualized using enhanced
chemiluminescence reagent (Beijing BioChange Co., Ltd.). The
protein expression level was analyzed by ImageJ software 1.8.0
(National Institutes of Health).
Cell growth assay
Cells in the logarithmic phase of growth infected
with either the shCtrl lentivirus or shSASS6 lentivirus were plated
in 96-well plates (2,000 cells/well). The cells were maintained in
RPMI-1640 medium supplemented with 10% FBS for 5 days. The number
of cells were counted using Celigo® Image Cytometer
(Nexcelom Bioscience LLC). In each well, ≥800 cells were counted,
and the cell growth assay was performed in triplicate.
MTT assay
Cell proliferation was assessed using an MTT assay
according to the manufacturer's protocol. Briefly, the MDA-MB-231
cells at a density of 3×103 cells/well were seeded in
96-well plates and incubated at 37°C overnight. Then the cells were
infected with shSASS6 lentivirus or shCtrl lentivirus. Cell
proliferation was measured every 24 h using 0.5 mg/ml MTT solution
at 37°C for 4 h. Subsequently, 100 µl of dimethyl sulfoxide
(Sigma-Aldrich; Merck KGaA) was added to each well after discarding
the supernatants. The plates were read at 490 nm using an ELISA
reader (Tecan Infinite; Tecan Group, Ltd.). Each experiment was
performed in triplicate.
Cell cycle distribution analysis
Flow cytometry following propidium iodide staining
was used to analyze cell cycle distribution. After 4 days of
lentiviral infection, MDA-MB-231 cells were plated in 60-mm dishes
(2×105 cells/well). The cells were collected and fixed
in 70% ice-cold ethanol for 12 h at 4°C. Subsequently, cells were
stained with 0.05 mg/ml propidium iodide containing 100 U/ml RNase
A (Fermentas; Thermo Fisher Scientific, Inc.) in the dark at room
temperature for 30 min. DNA content of samples was measured on a
FACSAria flow cytometer (BD Biosciences). The data were analyzed by
FlowJo 7.6.3 software (FlowJo LLC).
Intracellular signaling array
To investigate the intracellular signaling pathways
following SASS6 knockdown, a PathScan intracellular signal array
(product no. 7323; Cell Signaling Technology, Inc.) was used to
detect the expression of intracellular signaling proteins according
to the manufacturer's protocol.
Caspase activity assay
A caspase assay system (Promega Corporation) was
used to detect Caspase3/7 activity. Cells were maintained at 37°C
in a humidified incubator with 5% CO2 in 96-well plates
and then the activities of Caspase-3/7 were measured according to
the manufacturer's protocol. The reaction products were detected
using an M2009PR microplate reader (Tecan infinite; Tecan Group,
Ltd.) for the measurement of luminescence. Experiments were
performed at least three times in duplicate.
Apoptosis analysis
An FITC Annexin V Apoptosis Detection kit I (BD
Biosciences) was used to measure the proportion of viable cells
according to the manufacturer's protocol and analyzed using a
FACScan flow cytometer. After 4 days of lentiviral infection,
MDA-MB-231 cells (5×105 cells/sample) were harvested,
washed using cold PBS, resuspended in Annexin V-binding buffer and
stained using propidium iodide and FITC Annexin V in the dark at
room temperature for 15 min. After incubating, all the samples were
analyzed by FACScan flow cytometry (Beckton Dickinson; BD
Biosciences).
Statistical analysis
Fisher's exact test was used compare differences in
SASS6 expression between cancer tissues and paired paracarcinoma
tissues. The SASS6 mRNA expression was analyzed with ANOVA followed
by Dunnett's post hoc test. The statistical significance between
two groups (shContrl vs. shSASS6) was determined using a Student's
t-test for raw data. Data are presented as the mean ± standard
deviation. Spearman's rank correlation coefficient analysis was
used to analyze the correlation between SASS6 and CDK1 expression.
P<0.05 was considered to indicate a statistically significant
difference.
Results
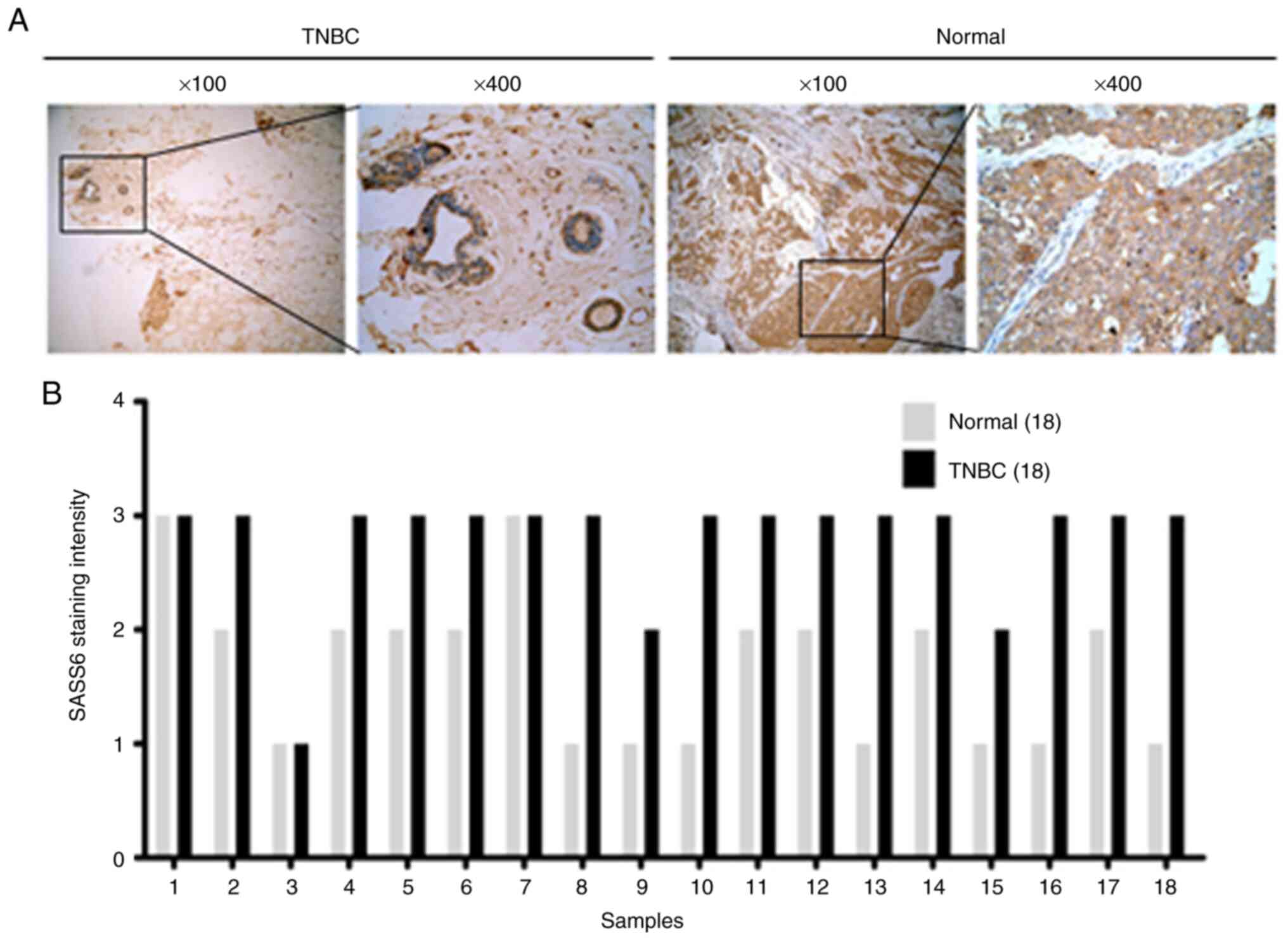
SASS6 expression in TNBC tissues
It has previously been reported that SASS6 mRNA
expression levels are upregulated in invasive breast carcinoma
compared with normal tissues based on data obtained from The Cancer
Genome Atlas (10). To analyze the
status of SASS6 expression in TNBC tissues, the expression levels
of SASS6 in clinical TNBC tissues and paired normal tissues were
determined using immunohistochemical staining. SASS6 expression
levels in TNBC tissues were significantly higher compared with the
normal tissues (Table I, P<0.001;
Fig. 1B). Representative images of
SASS6 staining are presented in Fig.
1A.
 | Table I.Immunohistochemical staining of SASS6
expression in TNBC and paired normal tissues. |
Table I.
Immunohistochemical staining of SASS6
expression in TNBC and paired normal tissues.
|
|
| Positive level |
|
|---|
|
|
|
|
|
|---|
| Tissue | No. of
specimens | + | ++ | +++ |
P-valuea |
|---|
| TNBC | 18 | 1 | 2 | 15 |
|
| Normal | 18 | 8 | 8 | 2 | <0.001 |
Lentivirus-mediated knockdown of SASS6
in MDA-MB-231 TNBC cells
To compare the expression levels of SASS6 in
different TNBC cell lines, the levels of SASS6 mRNA were detected
in three TNBC cell lines. The results revealed that SASS6 mRNA
expression was significantly increased in MDA-MB-231 and MDA-MB-468
cells compared with the PNBEC at the mRNA level (P<0.01;
Fig. 2A). The MDA-MB-231 cells
expressed higher levels of SASS6 expression compared with the
HCC-1937 and MDA-MB-468 cells (Fig.
2A), and were thus used for the knockdown experiments, to
explore the functional significance of SASS6. Lentiviral vectors
are an efficient gene delivery method due to their unique ability
to deliver target molecules into host cell DNA and replicate in
non-dividing cells (15). Therefore,
a control (shCtrl) and shSASS6 lentiviral vector was constructed.
SASS6 knockdown was confirmed using RT-qPCR and western blotting in
MDA-MB-231 cells. As revealed in Fig. 2B
and C, SASS6 mRNA and protein expression levels were
significantly reduced following transfection with SASS6-specific
shRNA in MDA-MB-231 cells.
SASS6 knockdown reduces the
proliferation of breast cancer cells
To observe the effect of SASS6 on cell growth,
MDA-MB-231 cells infected with shSASS6 or shCtrl lentivirus were
seeded into 96-well plates and subjected to 5 days of Celigo or MTT
analysis. As shown in Fig. 2E, the
knockdown of SASS6 significantly inhibited the growth of MDA-MB-231
cells. The MTT assay on MDA-MB-231 cells further revealed that
during the 5 days, the control-transfected cells grew notably
faster than the shSASS6-transfected cells (Fig. 2E). These data revealed that SASS6
knockdown potently suppressed cell proliferation of breast cancer
cells.
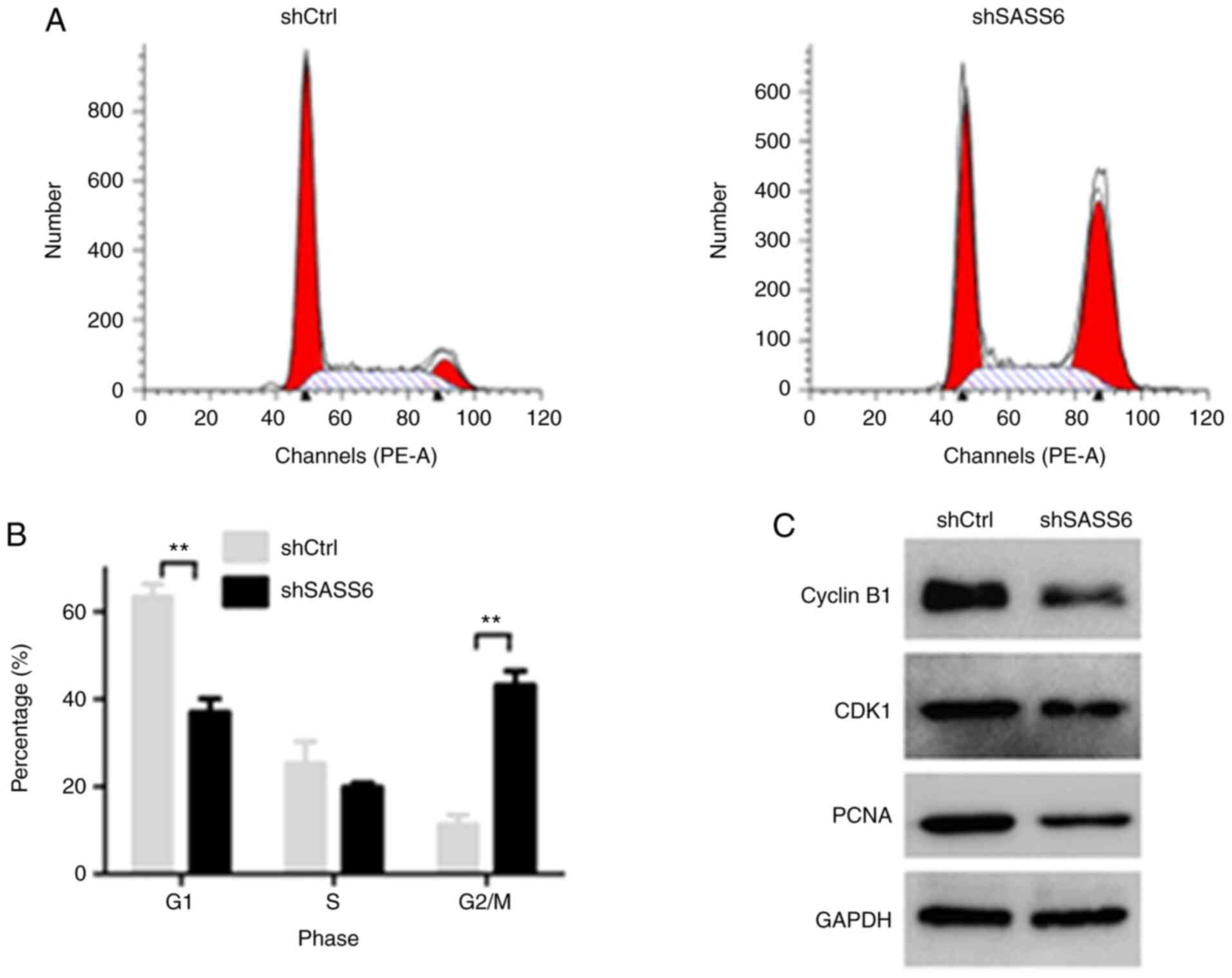
SASS6 knockdown arrests cell cycle
progression of MDA-MB-231 cells at the G2/M phase and downregulates
the expression of cell cycle-related proteins
To study the mechanism by which SASS6 knockdown
reduced growth, cell cycle distribution of MDA-MB-231 cells was
detected using a flow cytometer. As revealed in Fig. 3A and B, the proportion of cells in the
G0/G1 phase of the shSASS6-treated cells was lower than the
shCtrl-treated cells (P<0.01), whereas the proportion of cells
in the G2/M phase was increased (P<0.01). The percentage of
cells in the G2/M phase was increased from 11.34±2.19% in the
shCtrl-treated cells to 43.19±3.28% in the shSASS6-treated cells,
indicating an arrest of cell cycle progression at the G2/M phase
(Fig. 3A and B). These data indicated
that cell cycle arrest served an important role in growth
inhibition of breast cancer cells, and highlighted the involvement
of SASS6 in this process.
In order to further determine the potential
mechanism of G2/M-phase arrest, western blot analysis was used to
detect the changes in the expression of cell cycle regulators in
MDA-MB-231 cells following SASS6 knockdown. As revealed in Fig. 3C, in the shSASS6 treatment group, the
expression levels of CDK1 and cyclin B1, which are associated with
the G2-M transition were reduced, and PCNA expression was also
reduced.
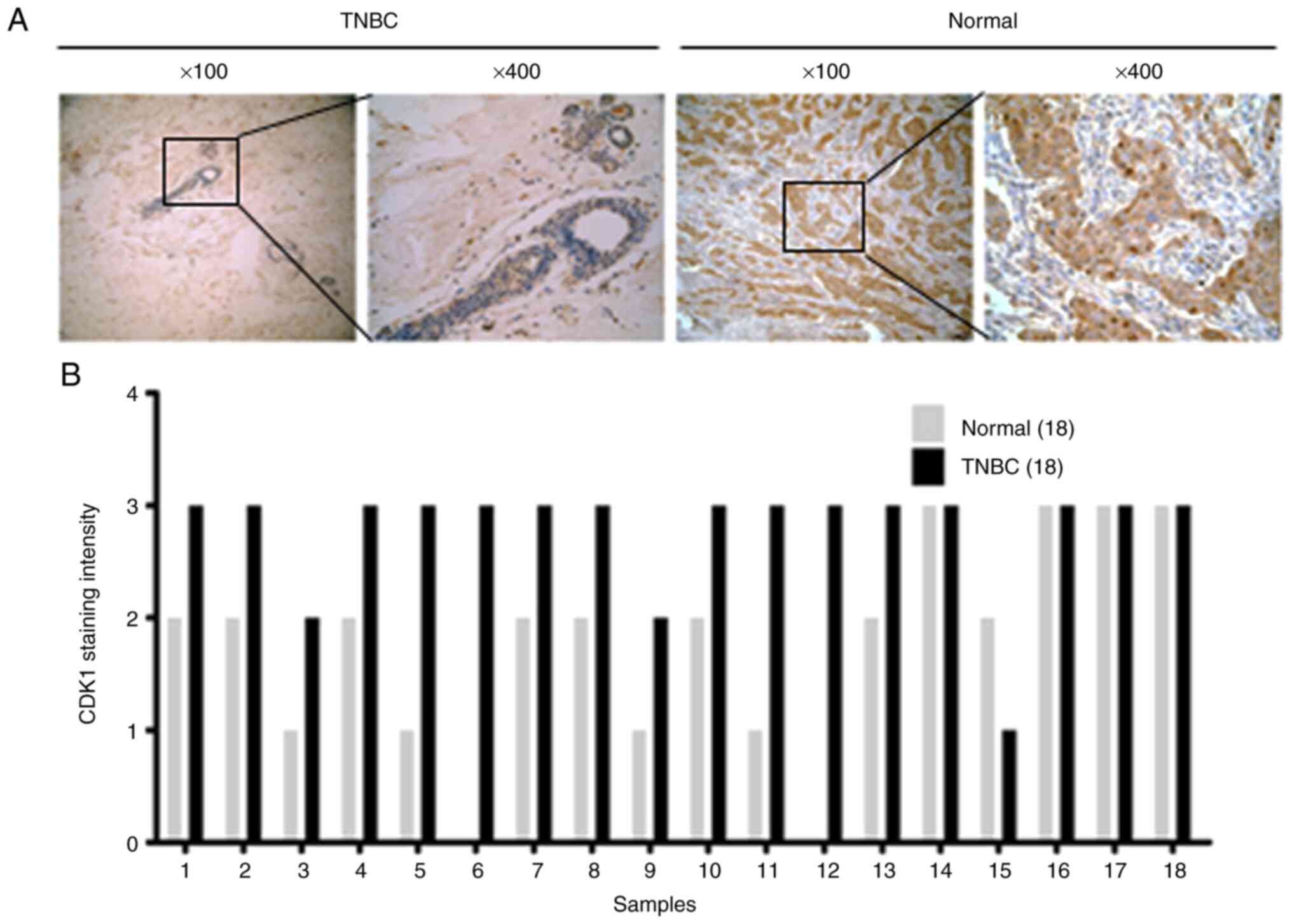
CDK1 is overexpressed in TNBC tissues
and is positively correlated with SASS6 expression
The expression of CDK1, a potential downstream
signaling molecule of SASS6 was investigated, and the relationship
between CDK1 and SASS6 was determined. CDK1 expression levels in
TNBC tissues were significantly higher compared with the normal
tissues (Table II, P=0.002; Fig. 4B). Representative images of SASS6
staining are presented in Fig. 4A. In
addition, as revealed in Table III,
SASS6 expression was positively correlated with CDK1 expression
(R=0.989; P<0.001), suggesting a strong correlation between
SASS6 and CDK1 expression in TNBC, providing evidence at an in
vivo level that SASS6 may regulate cell cycle progression in
TNBC.
 | Table II.Immunohistochemical staining of CDK1
expression in TNBC and paired normal tissues. |
Table II.
Immunohistochemical staining of CDK1
expression in TNBC and paired normal tissues.
|
|
| Positive level |
|
|---|
|
|
|
|
|
|---|
| Tissue | No. of
specimens | +/- | + | ++ | +++ |
P-valuea |
|---|
| TNBC | 18 | 0 | 1 | 2 | 15 |
|
| Normal | 18 | 2 | 4 | 8 | 2 | 0.002 |
 | Table III.SASS6 expression is positively
correlated with CDK1 expression in TNBC tissues (n=18). |
Table III.
SASS6 expression is positively
correlated with CDK1 expression in TNBC tissues (n=18).
|
| Expression of
SASS6 |
|
|
|---|
|
|
|
|
|
|---|
| Expression of
CDK1 | + | ++ | +++ | R | P-value |
|---|
| + | 0 | 1 | 0 |
|
|
| ++ | 1 | 1 | 0 |
|
|
| +++ | 0 | 0 | 15 | 0.989 | <0.001 |
Effects of SASS6 knockdown on STAT3,
BAD and rpS6 protein phosphorylation
To further elucidate the molecular mechanism by
which SASS6 reduced breast cancer cell growth, intracellular
signaling arrays were used to analyze changes in the levels of
signaling molecules in MDA-MB-231 cells following SASS6 knockdown.
As revealed in Fig. 5A and B, the
phosphorylation of STAT3 (Thr705), BAD (Ser116) and rpS6
(Ser235/236), was downregulated in shSASS6-treated cells
(P<0.01). Subsequently p-STAT3 (Thr705), STAT3, p-rpS6, rpS6,
p-BAD and BAD protein expression levels were determined using
western blotting (Fig. 5C). The
results revealed that SASS6 knockdown reduced the phosphorylation
of STAT3, BAD and rpS6. These results supported the hypothesis that
SASS6 serves a crucial role in TNBC cell growth by blocking the
activation of STAT3 and rpS6 protein.
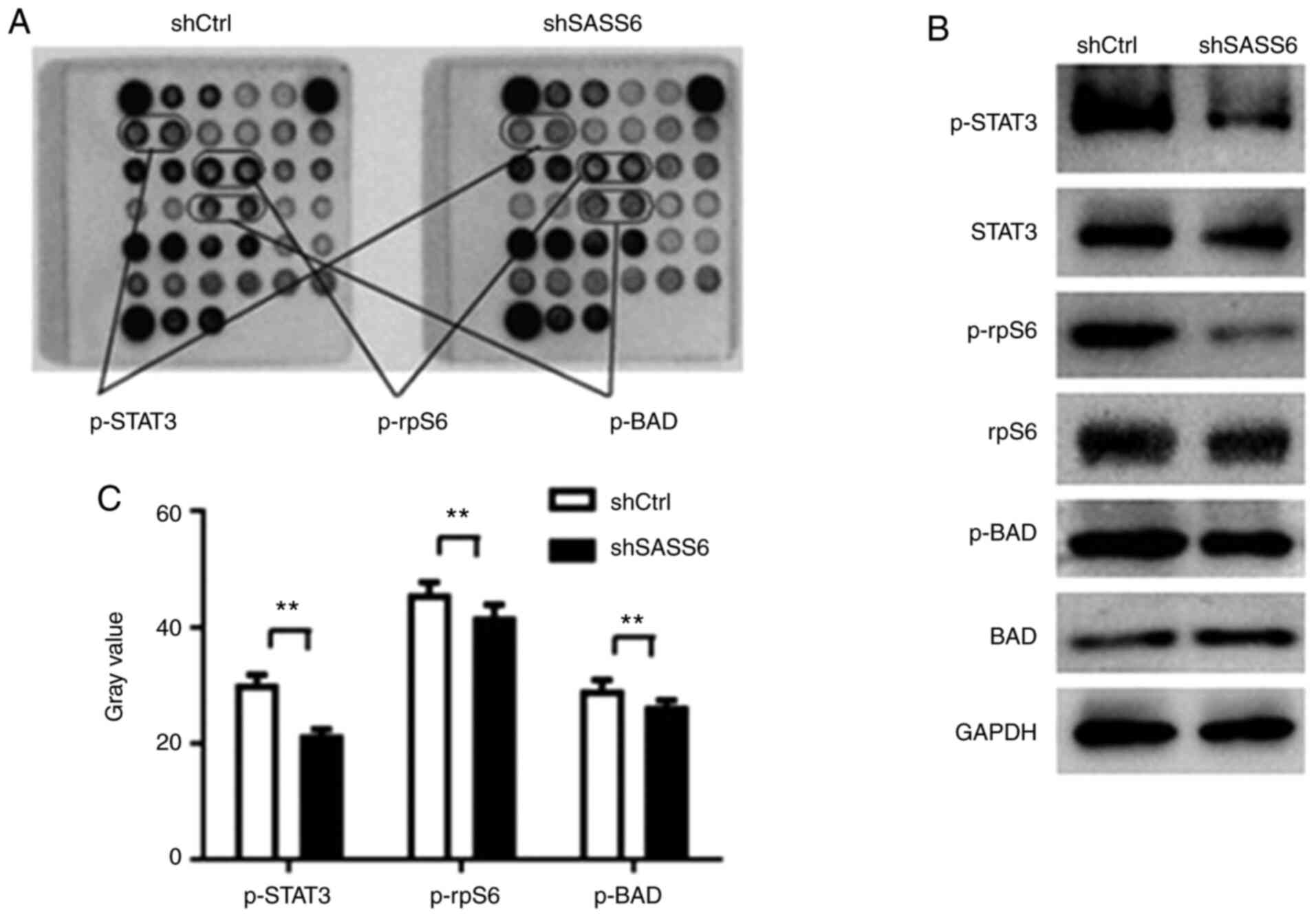 | Figure 5.Effects of shSASS6 on the expression
of multiple signaling molecules in MDA-MB-231 cells. (A)
Representative images of intracellular signaling arrays are
presented for shCtrl- and shSASS6-treated cells. (B) Quantitative
analysis in the arrays revealed that the p-STAT3, p-rpS6 and p-BAD
expression levels were decreased following SASS6 knockdown. n=3.
**P<0.01. (C) Western blot analysis of p-STAT3, STAT3, p-rpS6,
rpS6, p-BAD and BAD expression in MDA-MB-231 cells transfected with
shCtrl and shSASS6. SASS6, spindle assembly abnormal protein 6
homolog; sh, short hairpin; Ctrl, control; p-, phosphorylated;
rpS6, S6 ribosomal protein. |
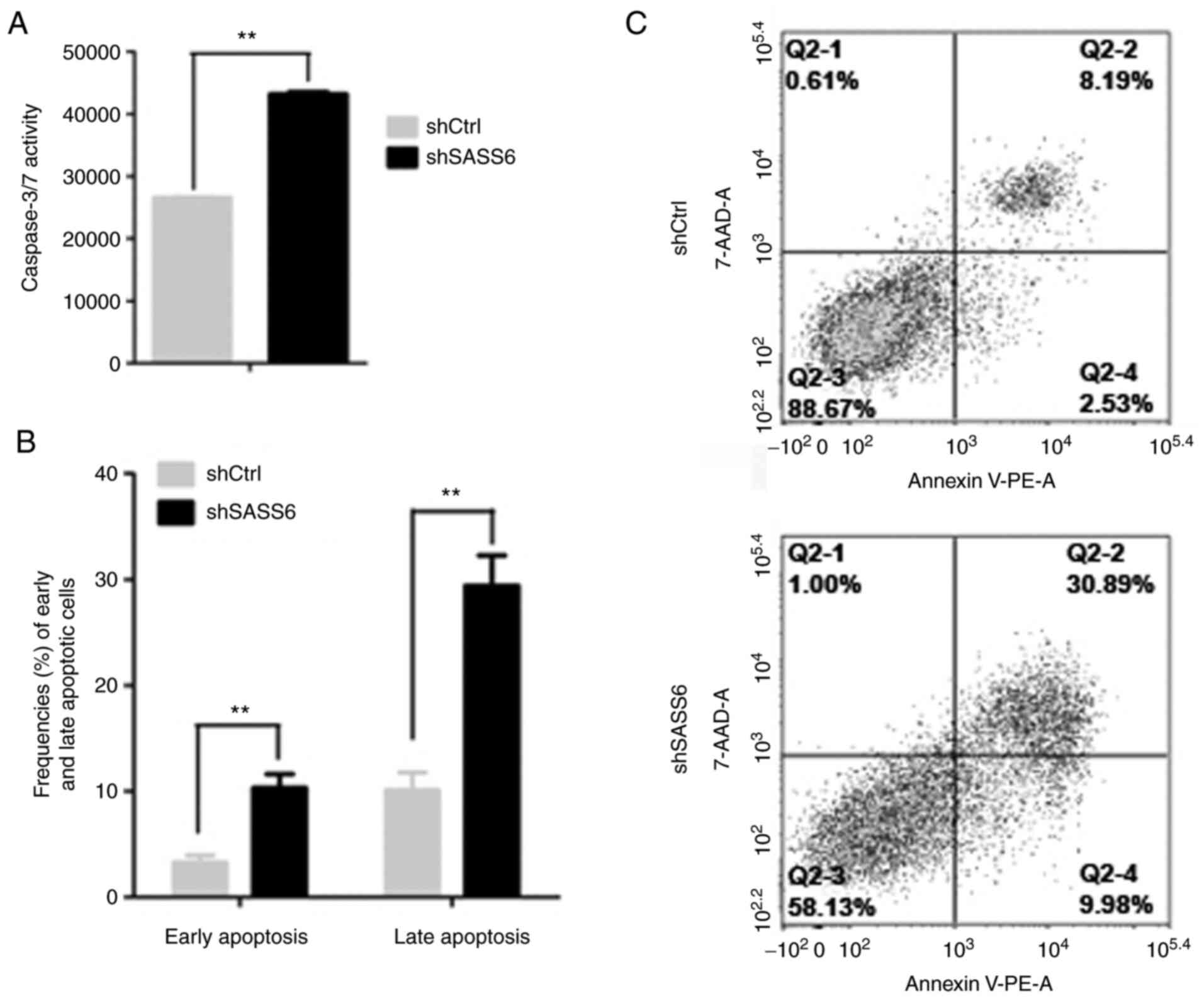
Knockdown of SASS6 induces apoptosis
of MDA-MB-231 cells
BAD, a proapoptotic Bcl-2 family member, serves an
important role in apoptosis (16).
Knockdown of SASS6 inhibited the phosphorylation of BAD, therefore,
whether SASS6 affected apoptosis was next determined. Caspase3/7
analysis was performed following knockdown of SASS6 in MDA-MB-231
cells to determine this. The results revealed that the Caspase3/7
activity in MDA-MB-231 cells infected with shSASS6 lentivirus was
significantly higher compared with the control group (P<0.01;
Fig. 6A).
Knockdown of SASS6-induced apoptosis was further
confirmed by staining cells with Annexin-V and propidium iodide.
Consistent with the Caspase3/7 analysis, the results revealed that
shSASS6-treated cells exhibited a significantly increased
proportion of cells in early (lower right) and late (upper right)
apoptosis (P<0.01; Fig. 6B and C).
These results indicated that knockdown of SASS6 induced apoptosis
in MDA-MB-231 cells.
Discussion
Ectopic expression of SASS6 induces centrosome
amplification, mitotic abnormalities, and chromosomal instability
(9,10,12,17). SASS6
upregulation has been revealed in several different types of cancer
(12). Despite extensive research on
its role in centriole formation, the regulatory mechanism of SASS6
in the proliferation of human cancer cells remains to be
elucidated. In the present study, SASS6 expression was upregulated
in TNBC tissues and cells. To determine the effect of SASS6 in TNBC
cells, SASS6 expression was knocked down in MDA-MB-231 TNBC cells,
and the results revealed that SASS6 knockdown significantly
inhibited proliferation of the MDA-MB-231 cells and arrested cell
cycle progression at the G2/M phase.
During physiological cell proliferation, SASS6
serves an important role in the regulation of the centrosome
numbers along with other proteins (17–22).
Proliferating cells have a single centrosome, which is replicated
once per cell cycle producing two centrosomes, and this guides the
assembly of bipolar spindles during mitosis, thus ensuring accurate
faithful separation of genetic material. Failure of centrosome
replication can lead to the assembly of unipolar spindles. SASS6
levels must be limited to restrict procentriole formation to a
single event per cell cycle (11);
thus, overexpression of SASS6 in cancer may result in the formation
of more than one procentriole per cell cycle leading to abnormal
cell proliferation, whereas downregulated expression of SASS6
levels may result in monopolar spindles, thus preventing cell
proliferation. The results of the present study revealed that
knockdown of SASS6 inhibited the proliferation of TNBC cells,
suggesting that SASS6 may be a potential target for treatment of
TNBC. Furthermore, the results revealed that the proportion of
cells in the G1 phase of the SASS6-knockdown group was decreased,
whereas the proportion of cells in the G2/M phase was significantly
increased. Following knockdown of SASS6 in HeLa cells using siRNA,
53% of cells contained a single centrin focus; and 66% of this
subset of cells assembled a monopolar spindle single centrin focus
(11). Inhibition of SASS6 protein
may result in the formation of a single centrin focus and thus
inhibit spindle formation, leading to G2/M blockade, resulting in 4
or more chromosomes at the G2/M phase. It has been reported that
SASS6 expression was detected at centrioles and in the cytoplasm of
S-phase cells, and accumulated further in the cytoplasm as cells
progressed through the G2 phase and until mitosis occurred
(11). Knockdown of SASS6 in the
present study resulted in G2/M phase arrest, further supporting the
role of SASS6 as a mediator during the G2 phase of the cell cycle.
Furthermore, due to G2/M blockade, cell proliferation in the
experimental group was reduced, resulting in the proportion of
cells in the G1 phase being significantly lower than the control
group. The experimental results revealed that the SASS6 gene
acts in the mitosis phase of the cell cycle, which is consistent
with the results of a previous study (11). The molecular mechanism of G2/M cell
cycle arrest induced by knockdown of SASS6 in MDA-MB-231 cells was
further assessed. The results revealed that the protein expression
levels of CDK1 and cyclin B1 were downregulated following knockdown
of SASS6. As CDK1 and cyclin B1 are two specific regulators that
act during the G2/M phase, their expression was assessed. The
results revealed that CDK1 and cyclin B1 may have been involved in
the G2/M cell cycle arrest following knockdown of SASS6 in
MDA-MB-231 cells. Moreover, shSASS6-treated cells exhibited
downregulated expression of PCNA. PCNA serves an important role in
the replisome by accommodating multiple processes at the
replication fork in the G1-S phase (23). SASS6 expression was detected at
centrioles and in the cytoplasm of S-phase cells as well, thus
knockdown of SASS6 may regulate PCNA expression; however, the
specific mechanism requires further study.
To further clarify the molecular mechanisms by which
SASS6 suppressed breast cancer cell growth, phosphorylation of
intracellular signal molecules in MDA-MB-231 cells was assessed
following SASS6 knockdown. STAT3 is an oncogene and its
constitutive activity can mediate the transformation of oncogenes
(24). Constitutive activation of
STAT3 signaling also confers resistance to apoptosis in cancer
cells (25,26). Inhibition of the STAT3 signaling
pathway can suppress the proliferation of ovarian and breast cancer
cells which possess active STAT3 (27). It has been reported that STAT3 is
constitutively activated in several different types of cancer,
including breast cancer (28),
although the mechanism underlying its activation requires further
study. In the present study, it was revealed that knockdown of
SASS6 decreased the levels of STAT3-Thr705 in human TNBC cells,
suggesting that SASS6 is involved in activation of STAT3.
Inhibition of STAT3 phosphorylation has been revealed to be
associated with downregulation of CDK1 and cyclin B1 expression
(29), thus it was concluded that
knockdown of SASS6 may inhibit phosphorylation of STAT3, and this
resulted in downregulation of CDK1 and cyclin B1, ultimately
leading to G2/M blockade. In addition, phosphorylation of rpS6 can
regulate translation initiation of certain mRNAs (30). Phosphorylation of rpS6 serves critical
roles in neoplastic transformation and initiation of pancreatic
cancer (31), dephosphorylation of
rpS6 at Ser235/236 can decrease proliferation and cell motility,
and invasion of breast cancer cells and pancreatic cancer cells
(32); however, its function requires
further study. The results of the present suggested that SASS6
knockdown significantly inhibited the growth of breast cancer cells
by blocking the activation of STAT3 and rpS6 proteins.
BAD is a proapoptotic Bcl-2 family member, and the
first cell death component identified as a survival signal
regulatory target. Dephosphorylated BAD forms a heterodimer with
Bcl-2 and Bcl-xL, inactivating them and thus allowing Bax and Bak
proapoptotic members to form complexes, resulting in the release of
cytochrome c, caspase activation and initiation of apoptosis
(33–35). In the present study, SASS6 knockdown
resulted in dephosphorylation of BAD.
The present study has some limitations. The possible
effects of inhibition of SASS6 in animal models were not assessed
to observe whether it exhibited the same effects. Additionally, the
use of cell lines may not be representative, as different cell line
models may provide differing results.
In summary, the results of the present study
revealed for the first time that SASS6 protein expression was
upregulated in TNBC tumor tissues. Additionally, SASS6 knockdown
inhibited TNBC cell growth and G2/M phase arrest through cyclin
B1/CDK1 signaling and regulation of phosphorylation of STAT3, BAD
and rpS6 protein. These data indicated that SASS6 may serve as a
potential therapeutic target and merits further investigation in
animal models or preclinical and clinical studies.
Acknowledgements
The authors would like to thank Shanghai GeneChem
Co., Ltd. for technical support.
Funding
The present study was funded in part by the National
Natural Science Foundation of China (grant no. 81573005), the
Science and Technology Program of Shanxi Provincial Health
Commission (grant no. 2017083), the Research Foundation of Capital
Institute of Pediatrics (grant no. GZ-2021-11) and the Capital
Institute of Pediatrics funds (grant no. FX-2016-03).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LD, JJ and HC conceived and designed the study, and
participated in writing the manuscript. LD, YW, WW and XX performed
the molecular genetics studies, and participated in the sequence
alignment. YS and BT performed the cell culture. TS, CH and XZ
analyzed and interpreted the data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shanxi Cancer Hospital (Shanxi, China). All patients
provided informed consent for publication of their data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sharma P: Update on the treatment of
early-stage triple-negative breast cancer. Curr Treat Options
Oncol. 19:222018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Laurentiis M, Cianniello D, Caputo R,
Stanzione B, Arpino G, Cinieri S, Lorusso V and De Placido S:
Treatment of triple negative breast cancer (TNBC): Current options
and future perspectives. Cancer Treat Rev. 36 (Suppl 3):S80–S86.
2010. View Article : Google Scholar
|
|
3
|
Rivera-Rivera Y and Saavedra HI:
Centrosome-a promising anti-cancer target. Biologics. 10:167–176.
2016.PubMed/NCBI
|
|
4
|
Raff JW: Centrosomes and cancer: Lessons
from a TACC. Trends Cell Biol. 12:222–225. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Salisbury JL: The contribution of
epigenetic changes to abnormal centrosomes and genomic instability
in breast cancer. J Mammary Gland Biol Neoplasia. 6:203–212. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Korzeniewski N, Hohenfellner M and
Duensing S: The centrosome as potential target for cancer therapy
and prevention. Expert Opin Ther Targets. 17:43–52. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chan JY: A clinical overview of centrosome
amplification in human cancers. Int J Biol Sci. 7:1122–1144. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pihan GA, Purohit A, Wallace J, Knecht H,
Woda B, Quesenberry P and Doxsey SJ: Centrosome defects and genetic
instability in malignant tumors. Cancer Res. 58:3974–3985.
1998.PubMed/NCBI
|
|
9
|
Leidel S, Delattre M, Cerutti L, Baumer K
and Gönczy P: SAS-6 defines a protein family required for
centrosome duplication in C. elegans and in human cells. Nat Cell
Biol. 7:115–125. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yoshiba S, Tsuchiya Y, Ohta M, Gupta A,
Shiratsuchi G, Nozaki Y, Ashikawa T, Fujiwara T, Natsume T,
Kanemaki MT and Kitagawa D: HsSAS-6-dependent cartwheel assembly
ensures stabilization of centriole intermediates. J Cell Sci.
132:jcs2175212019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Strnad P, Leidel S, Vinogradova T,
Euteneuer U, Khodjakov A and Gönczy P: Regulated HsSAS-6 levels
ensure formation of a single procentriole per centriole during the
centrosome duplication cycle. Dev Cell. 13:203–213. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shinmura K, Kato H, Kawanishi Y, Nagura K,
Kamo T, Okubo Y, Inoue Y, Kurabe N, Du C, Iwaizumi M, et al: SASS6
overexpression is associated with mitotic chromosomal abnormalities
and a poor prognosis in patients with colorectal cancer. Oncol Rep.
34:727–38. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Annika W, Anja MB, Kereshmeh T, Justus PB,
Paul DD, Michael S, Raymund EH, Matthias WB, Pamela LS and Reiner
S: Selective isolation and characterization of primary cells from
normal breast and tumors reveal plasticity of adipose derived stem
cells. Breast Cancer Res. 18:322016. View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Borsotti C, Borroni E and Follenzi A:
Lentiviral vector interactions with the host cell. Curr Opin Virol.
21:102–108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zha J, Harada H, Osipov K, Jockel J,
Waksman G and Korsmeyer SJ: BH3 domain of BAD is required for
heterodimerization with BCL-XL and pro-apoptotic activity. J Biol
Chem. 26(272): 24101–24104. 1997. View Article : Google Scholar
|
|
17
|
Comartin D, Gupta GD, Fussner E, Coyaud É,
hasegan M, Archinti M, Cheung SW, Pinchev D, Lawo S, Raught B, et
al: CEP120 and SPICE1 cooperate with CPAP in centriole elongation.
Curr Biol. 23:1360–1366. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu X, Huang S, Zhang B, Huang F, Chi W, Fu
J, Wang G, Li S, Jiang Q and Zhang C: DNA replication licensing
factor Cdc6 and Plk4 kinase antagonistically regulate centrosome
duplication via Sas-6. Nat Commun. 8:151642017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Arquint C, Sonnen KF, Stierhof YD and Nigg
EA: Cell-cycle-regulated expression of STIL controls centriole
number in human cells. J Cell Sci. 125:1342–1352. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin YC, Chang CW, Hsu WB, Tang CJ, Lin YN,
Chou EJ, Wu CT and Tang TK: Human microcephaly protein CEP135 binds
to hSAS-6 and CPAP, and is required for centriole assembly. EMOJ.
32:1141–1154. 2013.
|
|
21
|
Puklowski A, Homsi Y, Keller D, May M,
Chauhan S, Kossatz U, Grünwald V, Kubicka S, Pich A, Manns MP, et
al: The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and
targets HsSAS-6 to control centrosome duplication. Nat Cell Biol.
13:1004–1009. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Keller D, Orpinell M, Olivier N, Wachsmuth
M, Mahen R, Wyss R, Hachet V, Ellenberg J, Manley S and Gönczy P:
Mechanisms of HsSAS-6 assembly promoing centriole formation in
human cells. J Cell Biol. 204:697–712. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Johnson A and O'Donnell M: Cellular DNA
replicases: Components and dynamics at the replication fork. Annu.
Rev. Biochem. 74:283–315. 2005.
|
|
24
|
Bromberg JF, Wrzeszcznska MH, Devgan G,
Zhao Y, Pestell RG, Albanese C and Darnell JE Jr: Stat3 as an
oncogene. Cell. 98:295–303. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
William MB, Jin XH, Lin HJ, Huang M, Liu
R, Reynolds RK and Lin J: Inhibition of constitutively active Stat3
suppresses growth of human ovarian and breast cancer cells.
Oncogene. 20:7925–7934. 2001. View Article : Google Scholar
|
|
26
|
Catlett-Falcone R, Landowski T, Oshiro M,
Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L,
Fernández-Luna JL, Nuñez G, et al: Constitutive activation of Stat3
signaling confers resistance to apoptosis in human U266 myeloma
cells. Immunity. 10:105–115. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shen Y, Devgan G, Darnell JE Jr and
Bromberg JF: Constitutively activated Stat3 protects fibroblasts
from serum withdrawal and UV induced apoptosis and antagonizes the
proapoptotic effects of activated Stat1. Proc Natl Acad Sci USA.
98:1543–1548. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Garcia R, Yu CL, Hudnall A, Catlett R,
Nelson KL, Smithgall T, Fujita DJ, Ethier SP and Jove R:
Constitutive activation of Stat3 in fibroblasts transformed by
diverseoncoproteins and in breast carcinoma cells. Cell Growth
Differ. 8:1267–1276. 1997.PubMed/NCBI
|
|
29
|
Roshan S, Liu YY, Banafa A, Chen HJ, li
KX, Yang GX, He GY and Chen MJ: Fucoidan induces apoptosis of HepG2
cells by down-regulating p-Stat3. J Huazhong Univ Sci Technolog Med
Sci. 34:330–336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Roux PP, Shahbazian D, Vu H, Holz MK,
Cohen MS, Taunton J, Sonenberg N and Blenis J: RAS/ERK signaling
promotes site-specific ribosomal protein S6 phosphorylation via RSK
and stimulates cap-dependent translation. J Biol Chem.
282:14056–14064. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meyuhas O: Ribosomal protein S6
phosphorylation: Four decades of research. Int Rev Cell Mol Biol.
320:41–73. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Akar U, Ozpolat B, Mehta K,
Lopez-Berestein G, Zhang D, Ueno NT, Hortobagyi GN and Arun B:
Targeting p70S6K prevented lung metastasis in a breast cancer
xenograft model. Mol Cancer Ther. 9:1180–1187. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zha J, Harada H, Yang E, Jockel J and
Korsmeyer SJ: Serine phosphorylation of death agonist BAD in
response to survival factor results in binding to 14-3-3 not
BCL-X(L). Cell. 87:619–628. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang E, Zha J, Jockel J, Boise LH,
Thompson CB and Korsmeyer SJ: Bad, a heterodimeric partner for
Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell.
80:285–291. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wei MC, Zong WX, Cheng EH, Lindsten T,
Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB and
Korsmeyer SJ: Proapoptotic BAX and BAK: A requisite gateway to
mitochondrial dysfunction and death. Science. 292:727–730. 2001.
View Article : Google Scholar : PubMed/NCBI
|