Introduction
Ovarian cancer (OC) can be referred to as the
malignancy growth that originates from the ovaries. In 2018, this
invasive cancer has claimed the lives of approximately 185,000
individuals worldwide (1). In the
last decade, there has been a significant increase in individuals
succumbing to OC in Asia, including China (2). The current five-year survival rate of OC
varies from 29 to 49%, depending on the severity of the tumor
(3). Owing to the complex
histological classification of OC, the molecular pathogenesis is
somewhat complicated. Findings have shown that the functional
mutations of the TP53 gene account for the occurrence of OC,
especially high-grade ovarian carcinoma (3,4).
Additionally, the recurrent somatic mutations in the gene locus of
NF1, BRCA1, BRCA2, RB1, and CDK12 were found to be associated with
OC pathogenesis (5). Although
treatment methods such as surgery, chemotherapy and radiation
therapy have been used to combat the spread of OC, OC patients are
encumbered with poor prognosis. Therefore, the identification of
new biomarkers for the diagnosis and treatment of OC is
imperative.
Several reports in the literature have confirmed
that long non-coding RNAs (lncRNAs) contribute to the growth of
malignant tumors (6). Also known as
C8orf51, RHPN1-AS1 is located at chromosome 8q24.3, and it contains
1 exon (7). This RNA was found to be
over-expressed in uveal melanoma in 2017 (7), and it has been verified as a significant
tumor promoter in a number of malignant cancers such as gastric
cancer, hepatocellular carcinoma, breast cancer, colorectal cancer,
glioma, and cancer of the head and neck (8–14).
However, research has not explored the impact of RHPN1-AS1 in OC
development. The present study aimed to investigate the impact and
the underlying mechanism of RHPN1-AS1 on OC.
In the last decade, evidence has demonstrated that a
large number of small RNAs play a considerable role in
carcinogenesis (15). More
specifically, miRNAs are widely reported to be associated with the
tumorigenesis of multitype malignancies (16–18). A
member of miRNAs, miR-485-5p was found to play tumor-inhibitory
roles in breast cancer, hepatocellular carcinoma, cervical cancer,
melanoma, lung cancer, oral tongue squamous cell carcinoma,
osteosarcoma, glioblastoma, colorectal cancer, esophageal cancer,
and thyroid carcinoma (19–31). Another study confirmed that miR-485-5p
could inhibit the spread of OC by regulating UCA1 (32). It was also reported that miR-485-5p
could serve as a sponging target of RHPN1-AS1 in the pathogenesis
of hepatocellular carcinoma (33,34).
Findings of those studies confirmed the effects of
RHPN1-AS1/miR-485-5p on OC progression. Nevertheless, to the best
of our knowledge, no study has explored the upstream regulator of
miR-485-5p in OC or investigated whether miR-485-5p could be
regulated by RHPN1-AS1 in OC cells.
Located on human chromosome 20q11.21 with an exon
count of 18, targeting protein for Xklp2 (TPX2) can encode a
microtubule-related protein (35). In
the early stage of mitosis, TPX2 is the downstream of
Ran-GTP, and it participates in spindle formation (36). TPX2 is also regulated at all
stages of the cell cycle, and TPX2 was downregulated at the
G1-S transition boundary and upregulated as the cell cycle
progressed into S and G2 phases (37). Therefore, TPX2 may provide
insights into tumor cell proliferation. Evidence documented in the
literature suggested that TPX2 was upregulated in such
tumors as cervical cancer, lung cancer, pancreatic ductal
adenocarcinomas, bladder cancer, OC and colon cancer (38–44). More
importantly, a recent study revealed that miR-485-3p could suppress
colorectal cancer by targeting TPX2 (45). The abovementioned results emphasize
the significance of miR-485 and TPX2 in cancer development.
However, no studies have confirmed whether TPX2 could be
regulated by miR-485-5p in OC cells and whether the
RHPN1-AS1/miR-485-5p/TPX2 axis could contribute to OC
pathogenesis.
The aim of the current study was to demonstrate the
effect of the RHPN1-AS1/miR-485-5p/TPX2 axis on OC. It was
hypothesized that RHPN1-AS1 acted as a tumor promoter in OC by
interacting with miR-485-5p to increase TPX2. The results of
this study may provide insights into OC diagnoses and
treatments.
Materials and methods
Bioinformatics analysis
GSE119056 and GSE23392 downloaded from the GEO
DataSets were the mRNA expression profiles involving ovarian
cancer. GEPIA database (http://gepia.cancer-pku.cn/index.html) is a public
database showing the differentially expressed genes (DEGs) or
lncRNAs in ovarian cancer. With P-value <0.01 and
|log2 fold change| (|logFC|)≥1.5, DEGs were screened out
from GEPIA, GSE119056 and GSE23392. The STRING database (https://string-db.org/) was then used to construct the
interaction network for the screened DEGs with the medium
confidence (0.400) of interaction score. TargetScan and ENCORI
Starbase were finally employed to predict the miRNAs targeting
TPX2 and the miRNAs sponged by RHPN1-AS1, respectively.
Patients
OC tissues and adjacent normal ovarian tissues were
collected from 37 OC patients at the Yantai Affiliated Hospital of
Binzhou Medical University (China). The relevant characteristics
are shown in Table I. The collection
and use of the clinical tissues were performed based on the ethical
standards set out in the Helsinki Declaration. All participants
signed the informed consent forms, and this study was approved by
the Ethics Committee of the Yantai Affiliated Hospital of Binzhou
Medical University.
 | Table I.Baseline characteristic of 37
patients with ovarian cancer. |
Table I.
Baseline characteristic of 37
patients with ovarian cancer.
| Total no. of
patients=37 | No. (%) |
|---|
| Age at diagnosis
(years) |
|
>55 | 20 (54.05) |
|
≤55 | 17 (45.95) |
| Tumor size
(cm) |
|
≤10 | 23 (62.16) |
|
>10 | 14 (37.84) |
| Tumor type |
|
Invasive | 17 (45.95) |
|
Borderline | 15 (40.54) |
|
Unknown | 5 (13.51) |
| FIGO stage |
|
I/II | 24 (64.86) |
|
III/IV | 13 (35.14) |
| Pathological
grade |
|
G1+G2 | 21 (56.76) |
| G3 | 16 (43.24) |
RNA isolation and RT-qPCR
RNAs were extracted from tissues and cells with the
miRcute miRNA Isolation Kit (Tiangen). This extraction was carried
out according to the manufacturer's instructions. The miRcute miRNA
First-strand cDNA Synthesis Kit (Tiangen) and the PrimeScript™ RT
reagent Kit (Takara) were used to perform miRNA reverse
transcription and lncRNA and mRNA reverse transcription,
respectively. Then, the expression of miR-485-5p, RHPN1-AS1 and
mRNA of TPX2 in OC samples was analyzed using TB Green
Premix Ex Taq II (Takara) with 95°C 30 sec denaturation, followed
by 40 cycles at 95°C for 5 sec and 60°C for 30 sec. The reference
gene for RHPN1-AS1 and TPX2 mRNA was GAPDH, while the
reference gene for miR-485-5p was U6. Primers were obtained
from GeneCopoeia, and the corresponding sequences are listed in
Table II. The 2−ΔΔCt
method (46) was used to estimate
LncRNA, miRNA and mRNA expressions. This experiment was repeated
three times.
 | Table II.Primer sequences. |
Table II.
Primer sequences.
| Gene name | Forward primer
5′-3′ | Reverse primer
5′-3′ |
|---|
|
RHPN1-AS1 |
TGTGAGTCCTCCGACAATGC |
AACTTGATGACCAGGAGCCG |
|
miR-485-5p |
ACTTGGAGAGAGGCTGGC |
AAAAGAGAGGAGAGCCGTGT |
| U6 |
AGTAAGCCCTTGCTGTCAGTG |
CCTGGGTCTGATAATGCTGGG |
| TPX2 |
ATGGAACTGGAGGGCTTTTTC |
TGTTGTCAACTGGTTTCAAAGGT |
| GAPDH |
GGAGCGAGATCCCTCCAAAAT |
GGCTGTTGTCATACTTCTCATGG |
Cell culture
Biological materials were purchased from the Bena
Culture Collection (Beijing), such as CaOV4, OVCAR3, CaOV3, SKOV3,
and HOSEpiC (human ovarian surface epithelial cells) cell lines. A
mixture containing fetal bovine serum (10%) and DMEM (Dulbecco's
modified Eagle's medium) was utilized to culture CaOV3. DMEM
containing 4 mM L-glutamine and sodium pyruvate and 10% FBS was
used to culture OVCAR3 cells. SKOV3 cells were cultured in the
McCoy's 5A medium, which contained NaHCO3 (2.2 g/l) and
10% FBS. CaOV4 cells were cultured in L15 medium, which contained
10% FBS. HOSEpiC was kept in the RPMI-1640 medium, which contained
10% FBS. The temperature of the cells was sustained at 37°C in a
humidified air containing 5% CO2.
Subcellular fractionation
The PARIS™ Kit (Invitrogen; Thermo Fisher
Scientific) was used to separate and isolate the RNA from
cytoplasmic and nuclear SKOV3 and OVCAR3 cells. The fractionation
buffer was first added to the cells. Then, the cells were
centrifuged at 10,000 × g, for 5 min at 4°C. The cell supernatant
was then obtained, followed by the lysis of the pellet with a
disruption buffer. The RNAs in the cell supernatant containing
cytoplasmic lysate and nuclear lysate were later isolated with
Lysis/Binding Solution. Subsequently, the cell lines were treated
with 100% ethanol. The expression level of RHPN1-AS1, U2 (served as
a nuclear control) and GAPDH (served as a cytoplasmic control) was
determined using reverse transcription-quantitative PCR (RT-qPCR).
This experiment was repeated three times.
Cell transfection
Genomics products were purchased from GeneCopoeia
(Guangzhou), including miRNA mimic negative control, miRNA
inhibitor negative control and small interference RNA negative
control, pcDNA3.1 empty vector, pcDNA3.1-TPX2 overexpression
vector, si-TPX2 (TPX2 small interference RNA),
miR-485-5p inhibitors, miR-485-5p mimics, and si-RHPN1-AS1
(RHPN1-AS1 small interference RNA). Then, 6×104 OVCAR3
and SKOV3 cell lines were seeded into 6-well plates and cultured
overnight at 37°C before transfection. Lipofectamine 2000 reagent
(cat. no. 11668027; Thermo Fisher Scientific) was applied for the
transfection of siRNAs, miRNA mimic and miRNA inhibitors into
target cells based on the user manual. After 48-h transfection, the
cells were collected and the transfection efficiency was detected
three times.
Cell viability assessment
Cell viability at 96, 72, 48, and 24 h was measured
with CCK-8 (Cell Counting Kit-8). A total of 4,800 cells were
subsequently seeded into each well of the 96 plates and cultured at
37°C after transfection for 24, 48, 72, and 96 h. Next, 10 µl CCK-8
solution was added to each well, followed by incubation at 37°C for
2 h. Finally, the optical density at 450 nm was read with the aid
of a Multiskan FC microplate reader (Thermo Fisher Scientific).
This experiment was repeated three times.
Cell proliferation assessment
BrdU assay was utilized to evaluate cell
proliferation. After the transfected SKOV3 and OVCAR3 cells
(3×104/ml) were plated into each well of 96-well plates
for 24 h, 10 µl/well 5-bromo-2′-deoxyuridine (BrdU) was added to
the cells. The cells were then incubated for 4 h. Thymidine analog
(Abcam, ab142567) was then added to cells, and the mixture was
incubated at 22°C for 15 min. Finally, the optical density at 450
nm was measured with a Multiskan FC microplate reader (Thermo
Fisher Scientific). This experiment was repeated three times.
Cell apoptosis assessment
The transfected cells were collected and rinsed with
PBS three times. Following that, 3×105 cells were fixed
in cold methanol at 4°C for 30 min. After washing the cells with
PBS three times, 100 µl 1X binding buffer diluted with Annexin
V-FITC was added to the fixed cells in the dark for 10 min at 37°C.
Prior to subjecting the cells to flow cytometer (Cytoflex, Beckman
Coulter), the cells were stained with 5 µl PI and washed twice with
PBS. The flow cytometry (FCM) data were then obtained and analyzed
using FlowJo version 7.6.5 software (Tree Star). The rate of cell
apoptosis was calculated as the sum of ratio of the top-right
(Annexin V+/PI+) and the bottom-right
(Annexin V+/PI−). This experiment was
repeated three times.
Wound healing assay
Cells (2×105/well were plated into
12-well plates until the cells reached 90% confluency. The fused
monolayer cells were then scratched with a pipette tip (100 µl),
and the exfoliated cells were washed gently with PBS. Subsequently,
the cells were cultured in a serum-free medium for 24 h. Using an
optic microscope (Leica), the images at 0 and 24 h were captured
with ×100 magnification to evaluate cell migration. This experiment
was repeated three times.
Cell adhesion assessment
Transfected SKOV3 and OVCAR3 cell lines
(5×103 cells) were plated into 96-well plates coated
with type I collagen (10 µg/ml). After culturing for 1 h at 37°C,
the culture medium was removed, and the cell wells were rinsed with
PBS to remove the floating cells. The adherent cells underwent 4%
paraformaldehyde fixation, 0.5% crystal violet staining and dye
extraction with sodium citrate methanol solution. The optical
density (OD) at 570 nm was measured using a microplate reader. The
relative adhesion ability of the blank group was calculated and
subjected to statistical analysis. This experiment was repeated
three times.
Luciferase reporter assay
The genomics materials for this assay were obtained
from GeneCopoeia, such as SEAP (secreted alkaline phosphatase, the
internal control) and Gaussia Luciferase (GLuc) reporter gene
pEZX-MT05 with wild-type or mutant RHPN1-AS1-3′UTR or
wild-type or mutant TPX2−3′UTR. Next, the negative control
and miR-485-5p mimic were co-transfected into OVCAR3 and SKOV3 cell
lines along with SEAP, and the above Gluc reporter plasmids using
Lipofectamine 2000 reagent. The cells collected were lysed with
lysis buffer and then transfected for 48 h. The GeneCopoeia's
Secrete-Pair™ Dual Luminescence Assay Kit was later used to analyze
the relative luciferase activity. The analysis was performed with a
standard microplate reader. The activity ratio of GLuc/SEAP in each
group was calculated and compared with other groups. This
experiment was repeated three times.
RNA immunoprecipitation (RIP)
assay
The EZ-Magna RIP™ RNA-Binding Protein
Immunoprecipitation Kit RNA Immunoprecipitation (RIP) Kit was used
to perform RIP immunoprecipitation based on the manual protocol.
OVCAR3 and SKOV3 cell lines with miR-485-5p mimic transfection or
negative-control transfection were lysed in a standard RIP buffer.
The cell lysates were then incubated with magnetic beads conjugated
with AGO2 (anti-Argonaute2) or anti-IgG (anti-Immunoglobulin G)
antibodies served as the negative control for 12 h at 4°C. After
washing the beads with the RIP wash buffer, Proteinase K was used
to digest the precipitate by incubating the mixture at 55°C for 30
min. Subsequently, the total RNAs in the digested supernatant were
isolated with phenol: Chloroform: Isoamyl alcohol, which was then
reverse-transcribed into cDNA using the kit. Finally, RT-qPCR was
used to measure the relative indication of RHPN1-AS1. This
experiment was repeated three times.
RNA pull-down assay
This assay was conducted based on the methodology
used in the previous report (47).
SKOV3 and OVCAR3 cells transfected were seeded into 6-well plates
at a concentration of 6×105/well. Subsequently, the
cells were incubated for 12 h at 37°C in humid air filled with 5%
CO2. Then, biotinylated-miR-485-5p mimics and
biotinylated-negative controls purchased from RiboBio were
transfected into the cultured cells using Lipofectamine 2000
Reagent (Invitrogen; Thermo Fisher Scientific). After 2 days, the
cell lysates were collected, sonicated and incubated with
streptavidin beads (Life Technologies) for 3 h at 4°C. The cells
were then washed three times with PBS. Subsequently, the RNeasy
Mini Kit (Qiagen) was used to elute the bound RNAs. The eluted RNAs
were then reverse-transcribed into cDNA, which was then subjected
to RT-qPCR to estimate the relative expression of TPX2. This
experiment was repeated three times.
Western blot assay
The proteins in SKOV3 and OVCAR3 cells were
extracted with the RIPA lysis buffer. Next, the Thermo Fisher
Scientific Pierce™ BCA Protein Assay Kit was utilized to quantify
the concentration of protein in different groups. Then, 20 µg
protein was then loaded into 10% SDS-PAGE gel and separated using
gel electrophoresis. The separated protein was then transferred
onto a PVDF membrane, which was then blocked with 5.0% BSA at 37°C
for 60 min. Subsequently, the primary antibodies against
TPX2 (cat. no. ab252944; Abcam) and GAPDH (cat. no.
ab181602, Abcam) were used to incubate the PVDF membranes at 37°C
for 1 h. Subsequently, the cell lines were incubated for 12 h at
4°C. The Goat Anti-Rabbit IgG H&L secondary antibodies (cat.
no. ab205718; Abcam) were used to incubate the PVDF membrane for 1
h at room temperature. Lastly, the ECL Substrate Kit (Abcam) was
used for protein blot visualization, which was analyzed with
Image-Pro Plus 6.0 produced by Media Cybernetics. This experiment
was repeated three times.
Statistical analysis
Three biological repeats were carried out for each
experiment. Statistical data were evaluated with GraphPad Prism 8.0
(GraphPad Prism Inc.) and they were presented as mean ± standard
deviation (SD). Two-tailed unpaired t-test and one-way or two-way
ANOVA with Dunnett's or Tukey's post hoc test were employed for
statistical difference analyses between two groups and among
multiple groups, respectively. It was assumed that variables with
P-values <0.05 were statistically significant.
Results
mRNA and miRNA identification
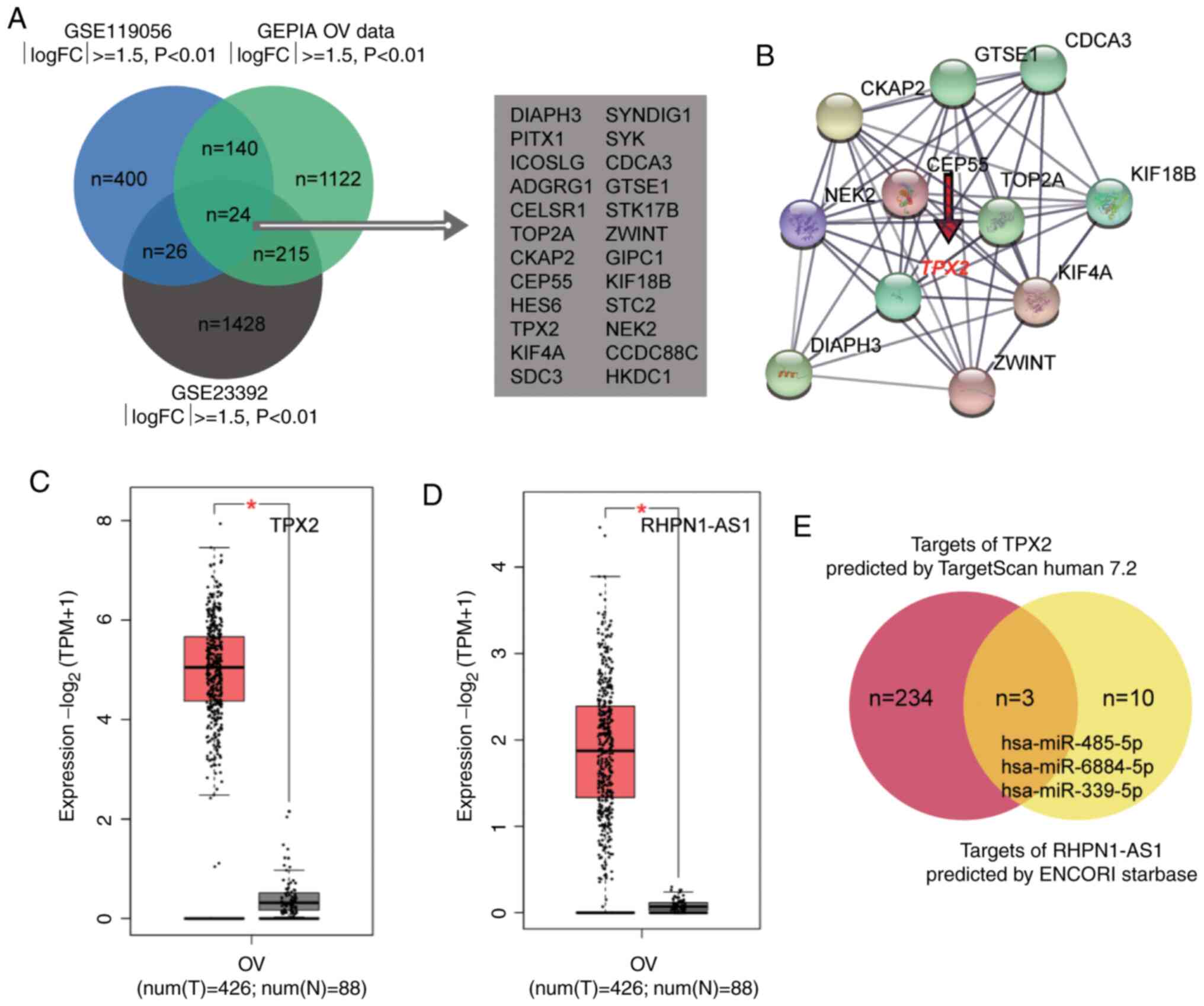
To identify the most significant genes involved in
OC, the GSE119056 and GSE23392 were downloaded from the GEO
DataSets. A total of 24 genes were found to be overlapped between
the three datasets (Fig. 1A). The 24
genes were then uploaded into the STRING database and an
interaction network analysis was constructed. A total of 11 genes
were found to be closely associated with each other in the network.
Within the network, it was observed that TPX2 (Fig. 1B) was significantly upregulated
(Fig. 1C) in OC and was partly
responsible for cancerous growth in OC (44,48,49).
However, researchers are yet to study its effect on OC cells. In
this study, lncRNA RHPN1-AS1 was evaluated and found to be
significantly upregulated in OC according to the data from GEPIA
(Fig. 1D) and was regarded as a tumor
enhancer in the OC ceRNA system (50,51). Next,
TargetScan predicted the miRNAs bound to TPX2 (Table SI), while ENCORI Starbase predicted
the miRNAs sponged by RHPN1-AS1 (Table
SII). After intersecting the target miRNAs of RHPN1-AS1 and the
target miRNAs of TPX2, findings revealed three common miRNAs
that could be sponged by RHPN1-AS1 and target TPX2 mRNA.
They included miR-485-5p, miR-6884-5p, and miR-339-5p (Fig. 1E). The effect of miR-485-5p on OC
remains unclear, and it was hypothesized that the novel
interactome, RHPN1-AS1-miR-485-5p-TPX2, may influence OC
progression.
RHPN1-AS1 enhanced OC development
The OC tumor tissues obtained from the participants
were used to observe the level of RHPN1-AS1. Experimental results
confirmed that RHPN1-AS1 upregulated OC tissues by 3-fold in
contrast with normal adjacent tissues (Fig. 2A), meaning RHPN1-AS1 is a potential
biomarker of OC. To further explore the impact of RHPN1-AS1 on OC,
RHPN1-AS1 expression was detected in a normal human ovarian
epithelial cell (HOSEpiC) and four typical OC cell lines (SKOV3,
CaOV3, OVCAR3 and CaOV4). Findings indicated that the degree of
RHPN1-AS1 expression was higher in OC cell lines than in HOSEpiC
cell lines (Fig. 2B). The subcellular
fractionation location assay was then employed to observe the
subcellular location of RHPN1-AS1 in SKOV3 and OVCAR3 cell lines.
According to the results of GAPDH and U2, RHPN1-AS1 was found
mainly in the cytoplasm (Fig. 2C).
Additionally, si-RHPN1-AS1, negative control (NC) and blank control
(blank) were transfected into SKOV3 and OVCAR3 cell lines to
evaluate the regulatory role of RHPN1-AS1 in OC. To examine the
transfection efficiency, RT-qPCR was employed. Data analyses
revealed that RHPN1-AS1 in the si-RHPN1-AS1 group was downregulated
by 70% compared to the blank group (Fig.
2D). The results of CCK-8 and BrdU assays indicated that cell
proliferation decreased in the si-RHPN1-AS1 group in contrast to
the blank groups and that there was no difference between NC and
blank groups (Fig. 2E and F). After
FCM was performed to observe cell apoptosis in the three groups,
the results indicated that the number of apoptotic cells increased
in the si-RHPN1-AS1 group compared with the blank group (Fig. 2G). Moreover, the wound-healing assay
results indicated that silencing RHPN1-AS1 impaired the migration
of SKOV3 and OVCAR3 cells (Fig. 2H and
I). According to the outcome of the adhesion assay, the
adherent cell number in the si-RHPN1-AS1 group was downregulated in
contrast to the NC group or blank group in SKOV3 and OVCAR3 cell
lines (Fig. 2J). Collectively, these
results suggested that RHPN1-AS1 could enhance malignant growth in
OC cells.
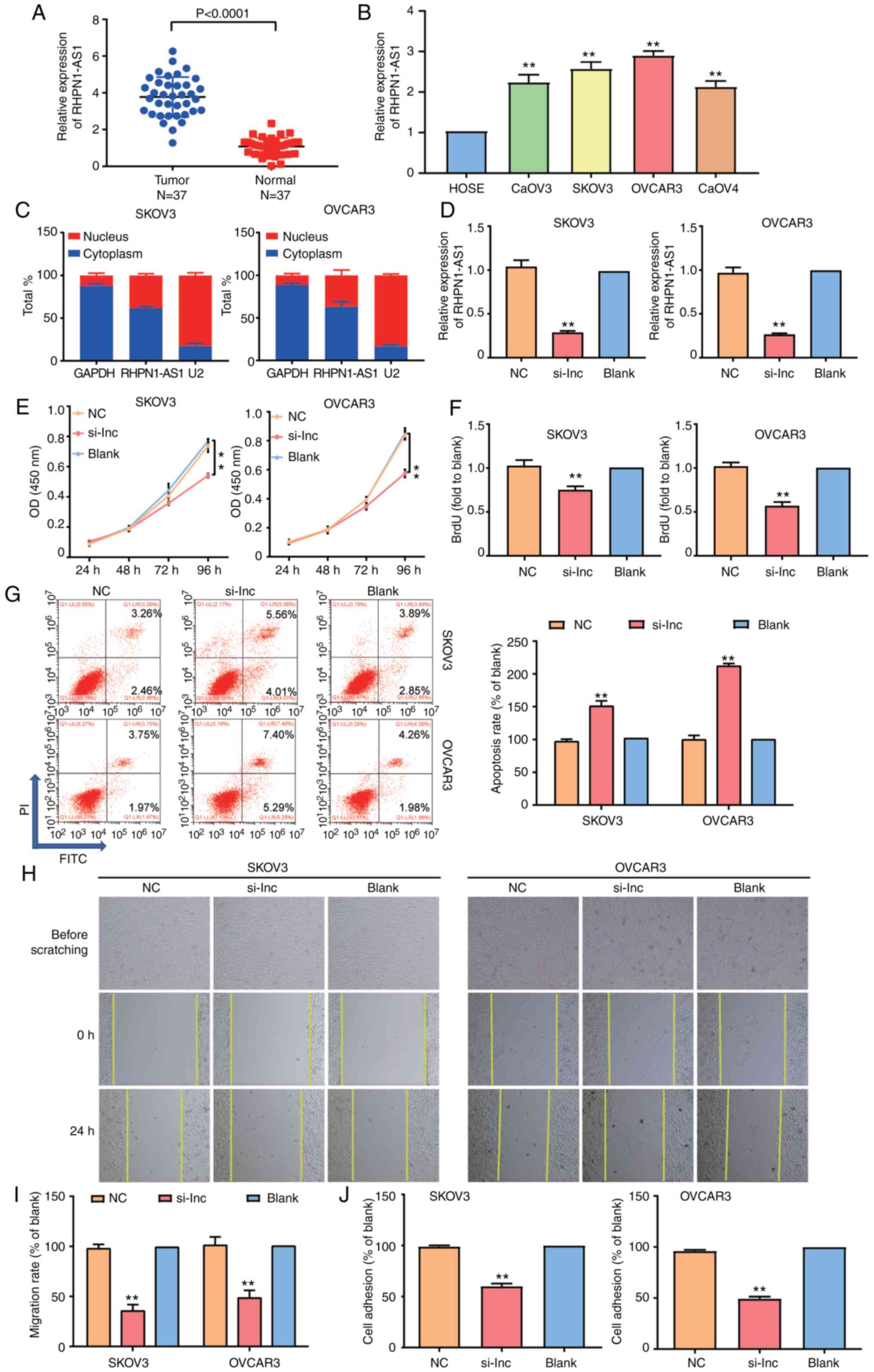 | Figure 2.The function of RHPN1-AS1 in ovarian
cancer. (A) RT-qPCR analysis revealed that the expression of
RHPN1-AS1 was increased in OC tissues compared with adjacent
healthy tissues. (B) RT-qPCR analysis revealed that the expression
of RHPN1-AS1 was higher in OC cell lines than that in normal
ovarian epithelial cells. (C) The location of RHPN1-AS1 in SKOV3
and OVCAR3 cell lines was analyzed by subcellular fractionation.
(D) Transfection efficiency of SKOV3 and OVCAR3 cell lines with the
transfection of si-RHPN1-AS1 (si-lnc group) was analyzed by
RT-qPCR. (E) CCK-8 assay was used to observe the cell proliferation
in the si-lnc, NC and blank groups. (F) BrdU assay was used to
observe the cell proliferation in the si-lnc, NC and blank groups.
NC, si-RHPN1-AS1 negative control; blank: Blank control. (G) Flow
cytometry was employed to measure the cell apoptosis in si-lnc
group, NC group and blank group. NC, si-RHPN1-AS1 negative control;
blank: Blank control. (H and I) Wound healing assay was employed to
measure the cell migration in si-lnc group, NC group and blank
group. Original magnification, ×100. (J) Cell adhesion assay was
employed to measure the cell adhesion in si-lnc, NC and blank
groups. NC, si-RHPN1-AS1 negative control; blank: Blank control.
The cellular experiςments were biologically repeated three times,
and the data were presented as mean ± standard deviation (SD).
**P<0.001 in contrast to blank group. NC, si-RHPN1-AS1 negative
control; blank, blank control. |
Effects of miR-485-5p on
RHPN1-AS1
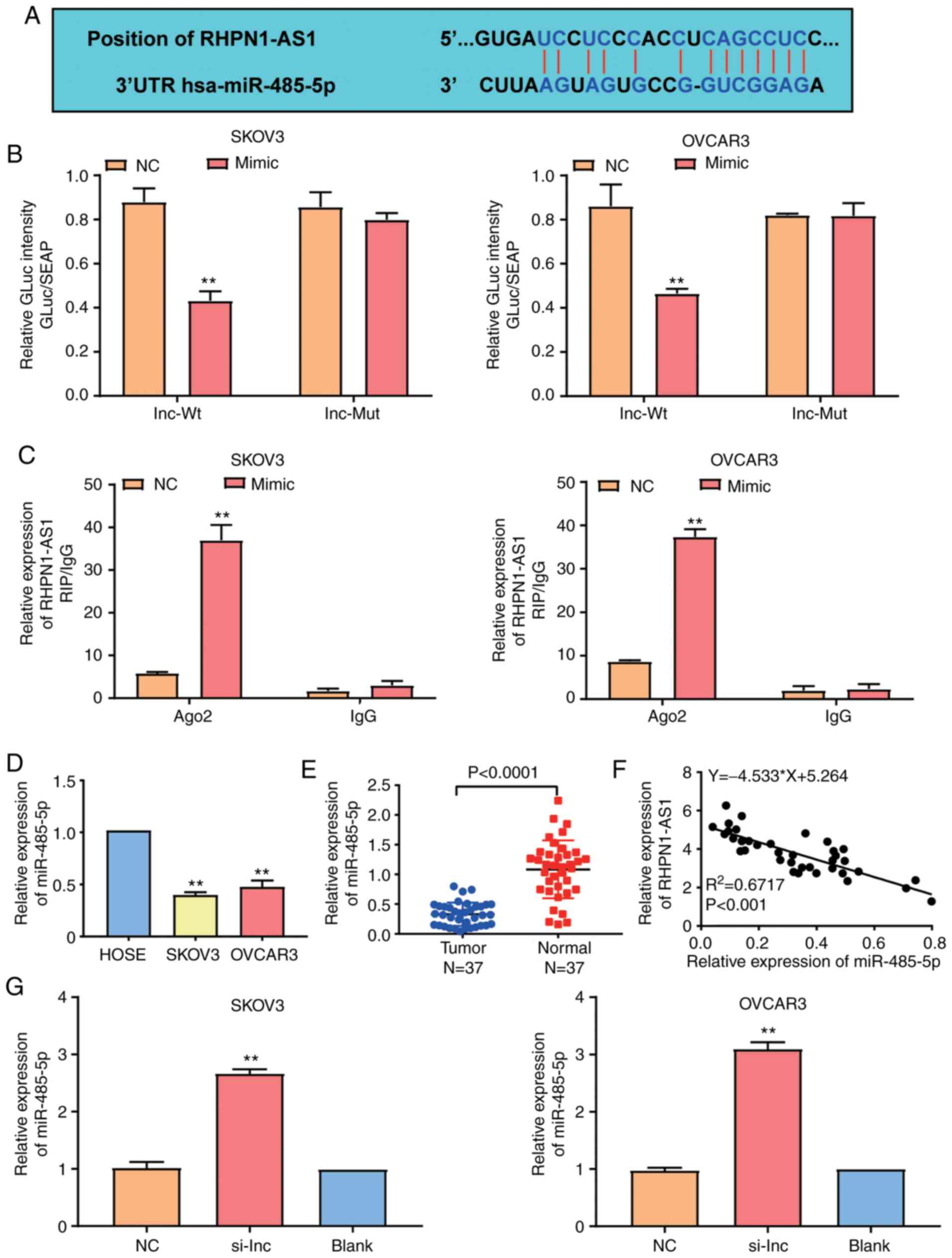
StarBase was employed to identify the binding
sequences and the relationship between RHPN1-AS1 and miR-485-5p in
OC cells (Fig. 3A). After performing
luciferase reporter assay, it was observed that luciferase
activities were reduced by 50% in SKOV3 and OVCAR3 cells
co-transfected with the wild-type RHPN1-AS1-3′UTR plasmid and
miR-485-5p mimics compared to the cells co-transfected with the
mutated RHPN1-AS1-3′UTR plasmid and negative control. On the other
hand, the luciferase activity in the cells co-transfected with
mutant RHPN1-AS1-3′UTR plasmid and miR-485-5p mimics or negative
control showed no statistical difference (Fig. 3B). The RIP assay results further
confirmed that RHPN1-AS1 could merge with miR-485-5p (Fig. 3C). It was also found that miR-485-5p
was downregulated by 60% in SKOV3 and OVCAR3 cell lines compared to
the HOSEpiC cell line (Fig. 3D).
Similarly, miR-485-5p decreased OC tissues by 60% compared to the
adjacent normal tissues (Fig. 3E).
Moreover, the correlation analysis revealed that RHPN1-AS1 had a
negatively correlated expression pattern with miR-485-5p in OC
tissues (Fig. 3F). Furthermore, to
assess whether RHPN1-AS1 could regulate miR-485-5p expression,
RHPN1-AS1 siRNAs were transfected into SKOV3 and OVCAR3 cell lines.
The RT-qPCR results indicated that the expression of miR-485-5p in
the si-RHPN1-AS1 group increased by 3-fold than that of the blank
group (Fig. 3G). Taken together, the
results revealed that a direct relationship existed between
RHPN1-AS1 and miR-485-5p in OC cells.
Association between miR-485-5p
andRHPN1-AS1
Several assays were performed to explore the
regulatory association between RHPN1-AS1 and miR-485-5p in OC
progression. Before the assay, the transfection efficiency was
evaluated by RT-qPCR, and the results showed that transfection of
si-RHPN1-AS1 significantly reduced the level of RHPN1-AS1 and
miR-485-5p, while transfection of miR-485-5p inhibitor markedly
decreased the level of miR-485-5p but had no effect on RHPN1-AS1
(Fig. S1). With the high
transfection efficiency, the functional assays were then performed.
The CCK-8 assay results showed that the miR-485-5p inhibitor
enhanced cell viability, while si-RHPN1-AS1 impaired cell
viability. When miR-485-5p inhibitor and si-RHPN1-AS1 were
co-transfected, cell viability decreased considerably compared to
the miR-485-5p inhibitor group (Fig.
4A). The BrdU assay outcome was similar to that of the CCK-8
assay in that miR-485-5p inhibitor increased the proliferation of
SKOV3 and OVCAR3 cell lines, while si-RHPN1-AS1 weakened cell
proliferation. When miR-485-5p inhibitor and si-RHPN1-AS1 were
co-transfected, the cell proliferation promotive effect of
miR-485-5p inhibitor was completely reversed (Fig. 4B). In addition, the FCM results
revealed that the cell apoptosis rate in the miR-485-5p inhibitor
group decreased by 50% in contrast to the blank group but that the
cell apoptosis rate increased by 2-fold in the si-RHPN1-AS1 group
in contrast to the blank group in SKOV3 and OVCAR3 cell lines.
After miR-485-5p inhibitor and si-RHPN1-AS1 were co-transfected,
the suppression of cell apoptosis by miR-485-5p inhibitor was
completely reversed (Fig. 4C).
Furthermore, it was observed that the exogenous inhibition of
miR-485-5p strengthened cell migration, whereas that of RHPN1-AS1
weakened the migration of SKOV3 and OVCAR3 cells. When both
RHPN1-AS1and miR-485-5p were inhibited, the enhancement of cell
migration by the miR-485-5p inhibitor was completely reversed
(Fig. 4D). After cell adhesion assay
was performed to measure the adhesion changes in SKOV3 and OVCAR3
cell lines, the results indicated that in the miR-485-5p inhibitor
group, the adherent cell number was upregulated but that in the
si-RHPN1-AS1 group, the adherent cell number was downregulated
compared to the blank group. After miR-485-5p inhibitor and
si-RHPN1-AS1 were co-transfected, the increase in the adherent cell
number by miR-485-5p inhibitor was completely reversed (Fig. 4E). These data revealed that RHPN1-AS1
could act on OC progression by negatively regulating
miR-485-5p.
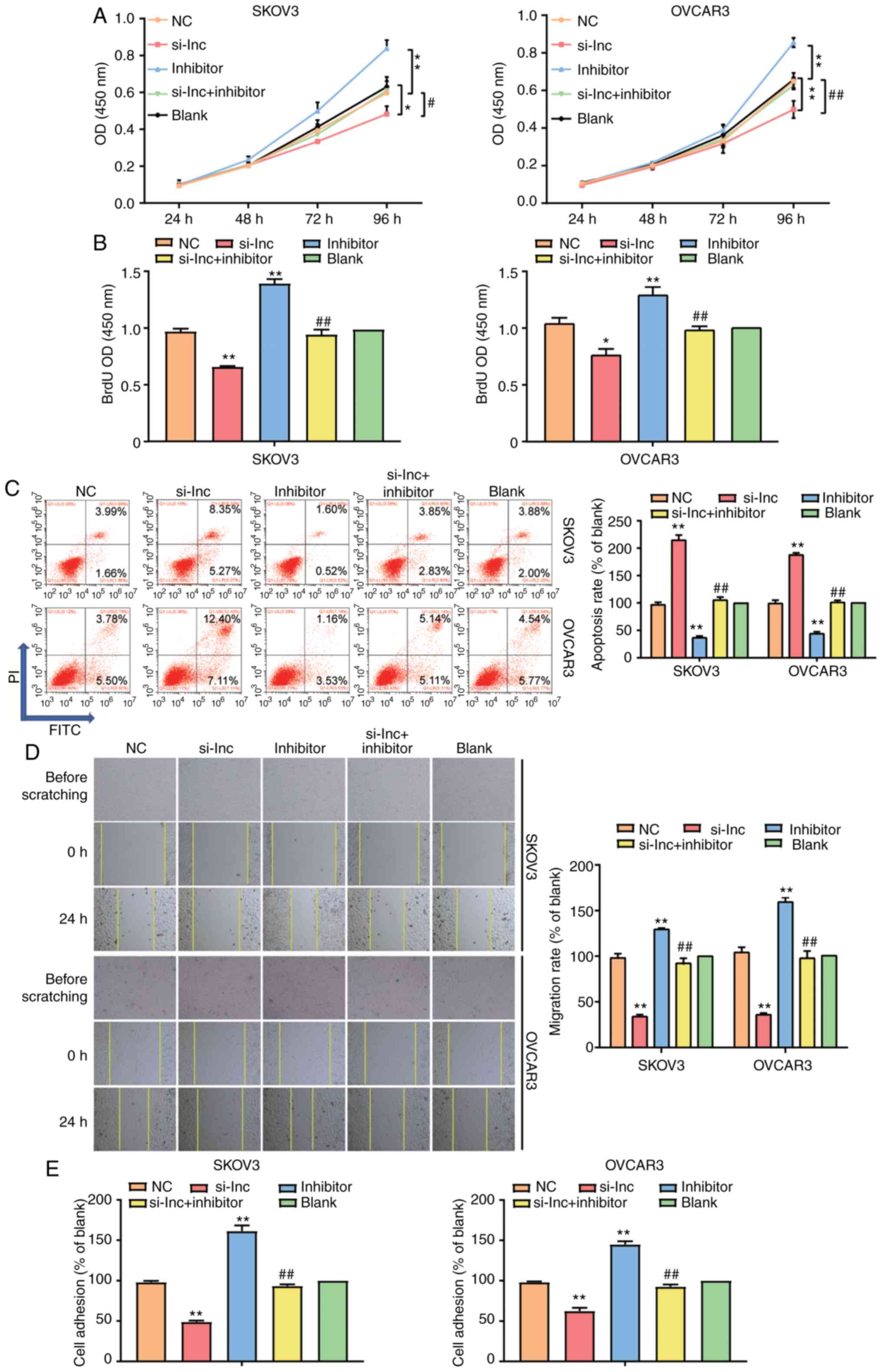 | Figure 4.miR-485-5p weakened cell
proliferation, migration and invasion while strengthened cell
apoptosis in ovarian cancer cells which was regulated by RHPN1-AS1.
(A) CCK-8 assay was used to observe the viability of SKOV3 and
OVCAR3 cell lines after transfecting miR-485-5p inhibitor
(inhibitor group), si-RHPN1-AS1 (si-lnc group), negative control
(NC group, siRNA NC+inhibitor-NC) and co-transfecting miR-485-5p
inhibitor and si-RHPN1-AS1 (si-lnc+inhibitor group) and untreated
cells (blank group). (B) BrdU assay was used to observe the cell
proliferation in the si-lnc, inhibitor, si-lnc+inhibitor, NC and
blank groups. (C) Flow cytometry was employed to measure the cell
apoptosis in the si-lnc, inhibitor, si-lnc+inhibitor, NC and blank
groups. (D) Wound healing assay was employed to measure the cell
migration in the si-lnc, inhibitor, si-lnc+inhibitor, NC and blank
groups. Original magnification: ×100. (E) Cell adhesion assay was
employed to measure the cell adhesion in the si-lnc, inhibitor,
si-lnc+inhibitor, NC and blank groups. The cellular experiments
were biologically repeated for three times, and the data were
presented as mean ± standard deviation (SD). *P<0.05,
**P<0.001 contrast to blank group. #P<0.05,
##P<0.001 in contrast to si-lnc group. |
TPX2: The downstream target gene of
miR-485-5p
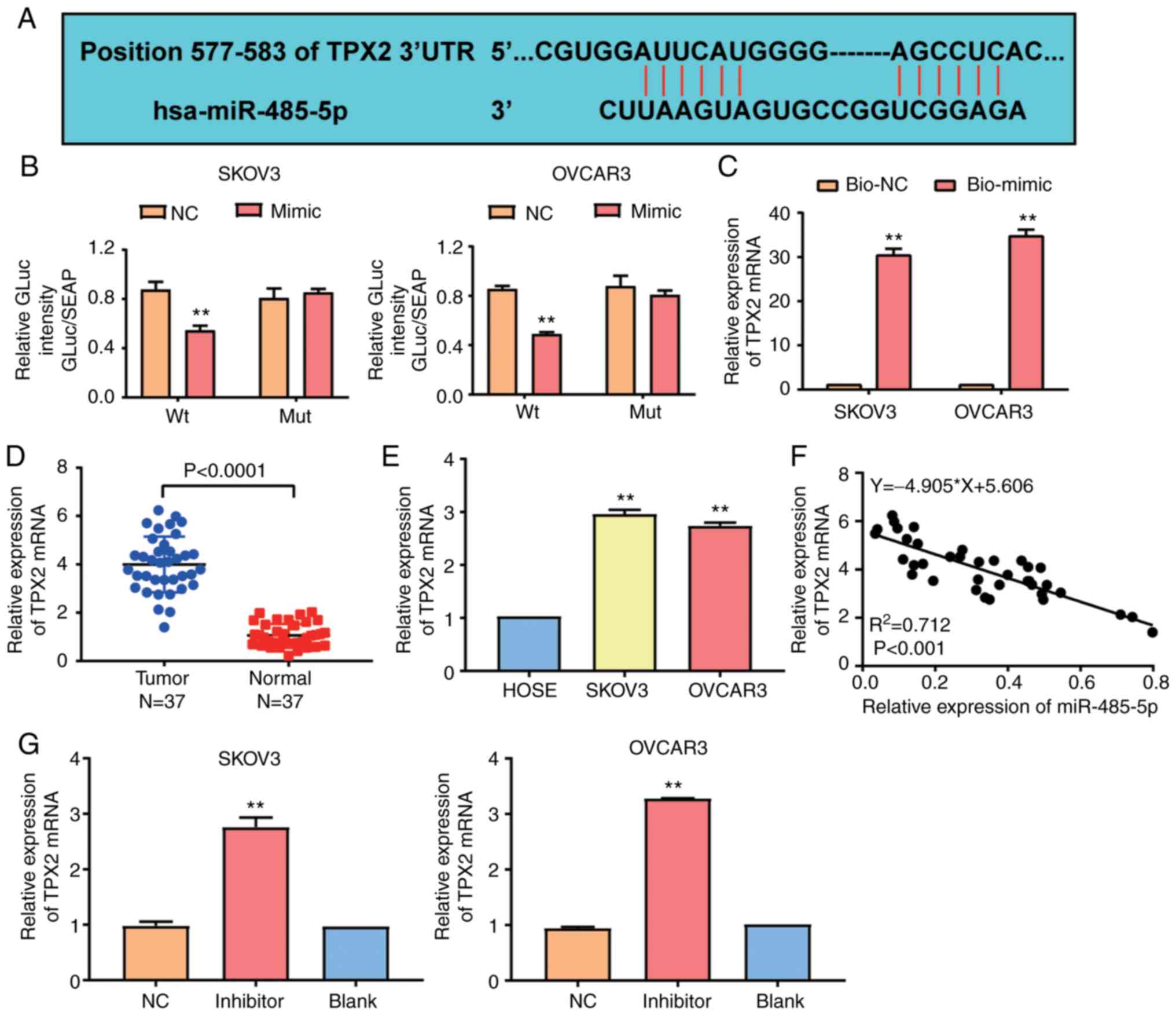
TargetScan 7.2 was employed to identify the gene
targeting miR-485-5p. The scanning results indicated that
TPX2 was the potential gene targeting miR-485-5p. The
predicted competitive region sequences between miR-485-5p and
TPX2 3′UTR are shown in Fig.
5A. Furthermore, the results of the luciferase reporter assay
revealed that the luciferase activity was reduced by 40% in SKOV3
and OVCAR3 cells co-transfected with the wild-type
TPX2−3′UTR plasmid and miR-485-5p mimic compared to cells
co-transfected with the wild-type TPX2−3′UTR plasmid and
negative control. Nonetheless, no considerable difference was
observed in cells co-transfected with mutant TPX2−3′UTR
plasmid and negative control or miR-485-5p mimics (Fig. 5B). Similarly, the RNA-pull down assay
findings indicated that miR-485-5p could bind to and interact with
TPX2 (Fig. 5C). With RT-qPCR,
it was observed that the TPX2 mRNA level in OC tissues was
almost 4-fold higher than that in adjacent normal tissues (Fig. 5D). Similarly, the TPX2 mRNA
level was increased in SKOV3 and OVCAR3 cell lines compared to the
HOSEpiC cell line (Fig. 5E). The
correlation analysis also demonstrated a negatively correlated
expression pattern between miR-485-5p and TPX2 (Fig. 5F). To explore whether miR-485-5p could
regulate TPX2 expression, miR-485-5p inhibitor was
transfected into SKOV3 and OVCAR3 cell lines, and the RT-qPCR
results indicated that the TPX2 mRNA level was upregulated
in the miR-485-5p inhibitor group in contrast to the blank group
(Fig. 5G). Overall, these data
suggested that miR-485-5p could negatively regulate
TPX2.
TPX2 strengthening ovarian malignancy
was regulated by miR-485-5p
To explore the mechanism of TPX2 related to
miR-485-5p in depth, SKOV3 and OVCAR3 cell lines were transfected
with miR-485-5p inhibitor, TPX2 siRNA, negative control or
co-transfected with miR-485-5p inhibitor and TPX2 siRNA.
After the protein level of TPX2 in different grouped cells
was evaluated using western blot analysis, the results revealed a
1.4-fold increase of TPX2 in the miR-485-5p inhibitor group
and at least a 40% decrease of TPX2 in the si-TPX2
group compared to the blank group. By contrast, it showed
comparable TPX2 expression in cells co-transfected with
miR-485-5p inhibitor and TPX2 siRNA and cells in the NC and
blank groups (Figs. 6A and S2). The CCK-8 assay data showed that
silencing TPX2 reduced the viability of SKOV3 and OVCAR3
cells, while co-transfecting the miR-485-5p inhibitor completely
reversed the changes caused by TPX2 silencing (Fig. 6B). Similarly, the BrdU assay results
showed that in contrast to blank groups, the cells in the
si-TPX2 group suppressed cell proliferation, which could be
completely reversed by co-transfecting miR-485-5p (Fig. 6C). In addition, FCM findings revealed
that the cell apoptosis rate in the si-TPX2 group was
upregulated by 2-fold; however, this effect could be completely
reversed by co-transfecting miR-485-5p (Fig. 6D). Silencing TPX2 was later
found to decrease the migration capacity of SKOV3 and OVCAR3 cells;
however, the miR-485-5p inhibitor could reverse the decrease
(Fig. 6E). The adhesion assay
results, on the other hand, showed that in the miR-485-5p inhibitor
group, the adherent cell number was upregulated by 1.5-fold,
whereas in the si-TPX2 group, the adherent cell number was
downregulated by 50%. After transfecting miR-485-5p inhibitor into
the si-TPX2 group, it was found that the increased adherent
cell number caused by silencing TPX2 was completely reversed
(Fig. 6F). In sum, these results
unveiled that after targeting TPX2, miR-485-5p could promote
the proliferation, migration and invasion of OC cells and inhibit
the apoptosis of OC cells.
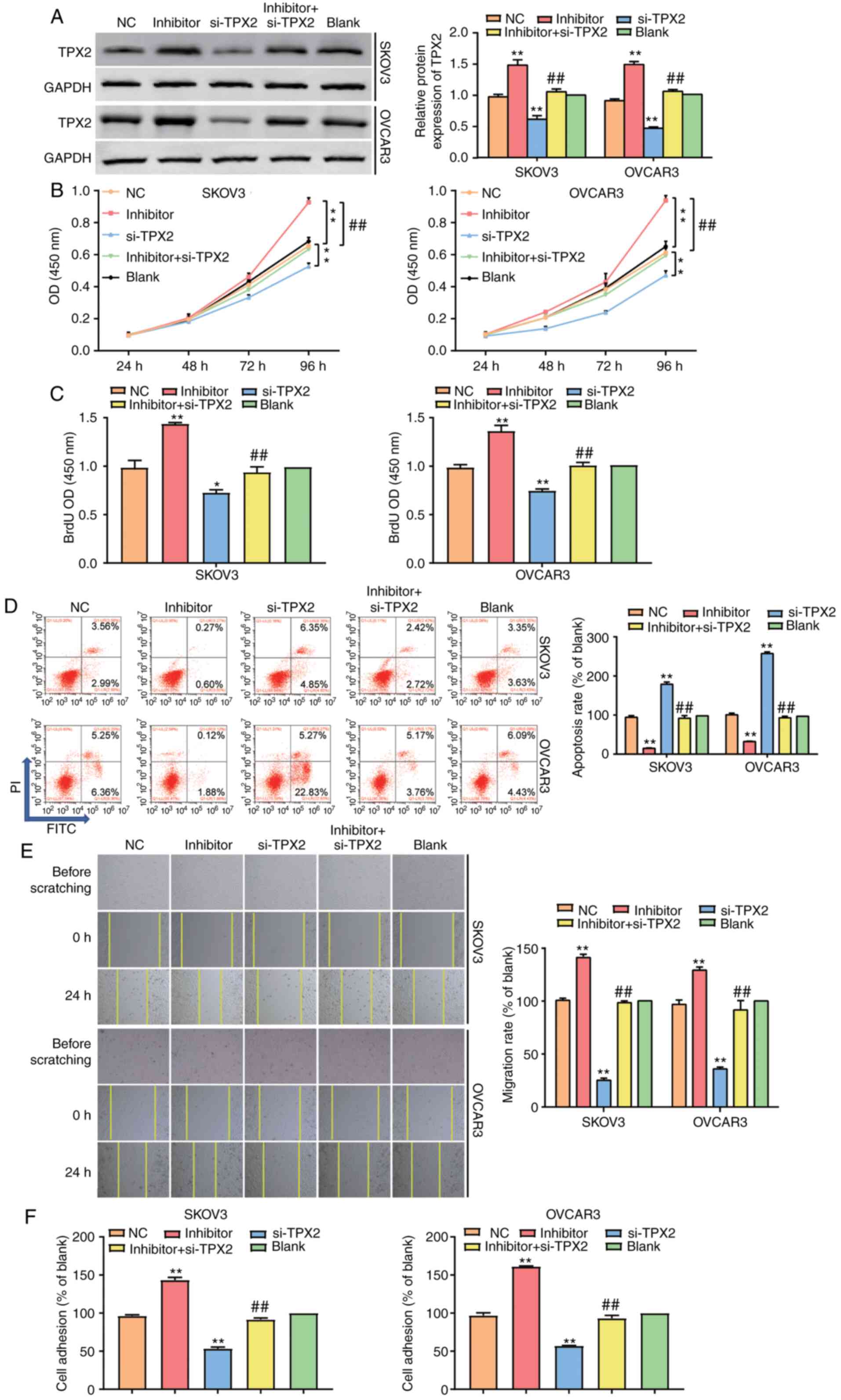 | Figure 6.TPX2 strengthened cell
propagation, cell migration and invasion while weakened cell
apoptosis in ovarian cancer cells which was regulated by
miR-485-5p. (A) Western blot was employed to detect the protein
level of TPX2 in SKOV3 and OVCAR3 cell lines after
transfecting miR-485-5p inhibitor (inhibitor group), si-TPX2
(si-TPX2 group), negative control (NC group, si-TPX2
NC +inhibitor-NC) and co-transfecting miR-485-5p inhibitor and
si-TPX2 (inhibitor+si-TPX2 group) and untreated cells
(blank group). (B) CCK-8 assay was used to observe the cell
proliferation in the si-TPX2, inhibitor,
inhibitor+si-TPX2, NC and blank groups. (C) BrdU assay was
used to observe the cell proliferation in the si-TPX2,
inhibitor, inhibitor+si-TPX2, NC and blank groups. (D) Flow
cytometry was employed to measure the cell apoptosis in the
si-TPX2, inhibitor, inhibitor+si-TPX2, NC and blank
groups. (E) Wound healing assay was employed to measure the cell
migration in the si-TPX2, inhibitor,
inhibitor+si-TPX2, NC and blank groups. Original
magnification: ×100. (F) Cell adhesion assay was employed to
measure the cell adhesion in the si-TPX2, inhibitor,
inhibitor+si-TPX2, NC and blank groups. The cellular
experiments were biologically repeated for three times, and the
data were presented as mean ± standard deviation (SD). *P<0.05,
**P<0.001 in contrast to blank group. ##P<0.001 in
contrast to inhibitor group. |
Effect of RHPN1-AS1 on OC progression
depended on TPX2
Western blot assay was performed to detect the
protein level of TPX2 in SKOV3 and OVCAR3 cells transfected
with si-RHPN1-AS1 and/or miR-485-5p inhibitor. The results showed
that TPX2 decreased by 60% in OC cells transfected with
si-RHPN1-AS1 compared to the control cells. However, this decrease
was completely reversed by co-transfecting the miR-485-5p inhibitor
(Fig. 7A). Subsequently, western blot
assay was employed to examine the protein level of TPX2 in
SKOV3 and OVCAR3 cells transfected with si-RHPN1-AS1 and/or
OE-TPX2, and the results showed that TPX2
overexpression could significantly upregulate the protein level of
TPX2 and also effectively reversed the suppressive effect of
si-RHPN1-AS1 on TPX2 expression (Figs. 7B and S3). CCK-8 and BrdU assays were also
performed to confirm the effect of RHPN1-AS1 on OC progression. The
results of CCK8 assays revealed that silencing RHPN1-AS1 reduced
the viability of SKOV3 and OVCAR3 cells; nevertheless,
co-transfecting OE-TPX2 reversed the reduction (Fig. 7C). Similarly, the BrdU assay results
demonstrated that the weakened cell-proliferation ability induced
by si-RHPN1-AS1 could be completely reversed by co-transfecting
OE-TPX2 (Fig. 7D).
Collectively, these results revealed that the effect of RHPN1-AS1
on OC progression was dependent on TPX2.
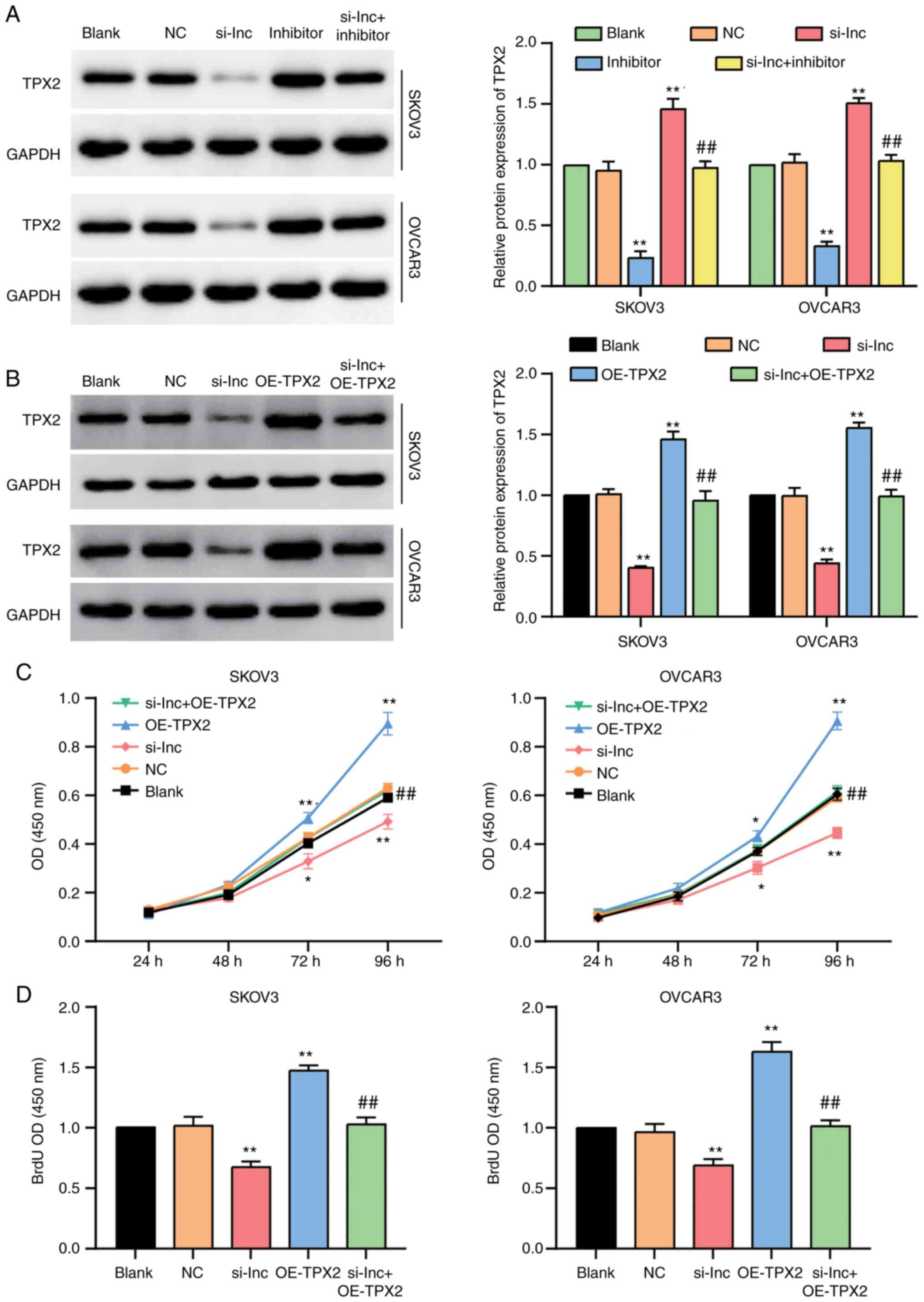 | Figure 7.The regulation of RHPN1-AS1 to OC
progression was dependent on TPX2. (A) Western blot was
employed to detect the protein level of TPX2 in SKOV3 and
OVCAR3 cell lines with the transfection of si-RHPN1-AS1 (si-lnc
group), miR-485-5p inhibitor (inhibitor group), co-transfecting
si-RHPN1-AS1 and inhibitor (si-lnc+inhibitor group), negative
control (NC group, siRNA NC+inhibitor-NC) and untreated cells
(blank group). (B) Western blot was employed to detect the protein
level of TPX2 in SKOV3 and OVCAR3 cell lines with the
transfection of si-RHPN1-AS1 (si-lnc group), OE-TPX2
(OE-TPX2 group), co-transfecting si-RHPN1-AS1 and
OE-TPX2 (si-lnc+OE-TPX2 group), negative control (NC
group, siRNA NC+empty vector) and untreated cells (blank group).
(C) CCK-8 assay was used to observe the cell proliferation in
si-lnc group, OE-TPX2 group, si-lnc+OE-TPX2 group, NC
group and blank group. (D) BrdU assay was used to observe the cell
proliferation in si-lnc group, OE-TPX2 group,
si-lnc+OE-TPX2 group, NC group and blank group. The cellular
experiments were biologically repeated for three times, and the
data were presented as mean ± standard deviation (SD). *P<0.05,
**P<0.001 in contrast to blank group. ##P<0.001 in
contrast to si-lnc group. |
Discussion
Our findings revealed that RHPN1-AS1 dominantly
located in the cell cytoplasm was highly expressed in OC tissues
and cell lines. In addition, RHPN1-AS1 enhanced the proliferation,
migration and adhesion of OC cells but suppressed the apoptosis of
OC cells. Apart from that, findings revealed that RHPN1-AS1
facilitated the tumorigenesis of OC by sponging miR-485-5p, which
could regulate the proliferation, migration, adhesion and apoptosis
of OC cells by targeting TPX2.
Previous research indicated that lncRNA RHPN1-AS1
was expressed in various types of cancer and was associated with
tumorigenesis. For instance, in uveal melanoma, RHPN1-AS1 was
overexpressed, thus promoting cell propagation, clone formation,
cell migration and cell invasion (7).
In another study, RHPN1-AS1 was upregulated in glioma, and it
promoted cell propagation, migration and invasion by regulating
miR-625-5p/REG3A directly (9). It was
also previously reported that by targeting FGF2, RHPN1-AS1
influenced cervical carcinoma and exhibited a significantly
negative relation with miR-299-3p to enhance cell propagation,
migration, and invasion (10).
Furthermore, RHPN1-AS1 accelerated cell proliferation and clone
formation by sponging miR-4261 and targeting c-Myc in breast cancer
(52). RHPN1-AS1 was utilized to
predict poor prognosis in breast cancer patients due to its ability
to enhance the growth of breast cancer cells by targeting the
miR-6884-5p/ANXA11 pathway (53).
Similarly, the upregulated RHPN1-AS1 in cell lines with colorectal
carcinoma strengthened the propagation, migration and invasion of
the tumor but weakened the apoptosis of the cancer by combining
with miR-7-5p to stabilize O-GlcNAcylation transferase (OGT)
(12).
In addition, the expression level of RHPN1-AS1 was
higher in tissues with hepatocellular carcinoma than that in normal
adjacent tissues (13). Findings of
that study predicted shorter survival times in patients. RHPN1-AS1
overexpression also enhanced the proliferation and metastasis of
hepatocellular carcinoma (13).
Another research reported that the high expression of RHPN1-AS1 had
a significant correlation with advanced tumor metastasis stage,
histologic grade and poor prognosis in hepatocellular carcinoma,
which mainly resulted from its promotive action on cell
proliferation, migration, and invasion through the miR-485/CDCA5
pathway (34). The sponging effect of
RHPN1-AS1 on miR-485-5p has also been proved in another research
that focused on hepatocellular carcinoma (33). Similar to the studies mentioned, our
study verified that RHPN1-AS1 was overexpressed in OC, and this RNA
may become a new biomarker in OC. Our research also showed that
RHPN1-AS1 could not only promote cell proliferation, adhesion and
migration but also suppress cell apoptosis in OC.
The potential role of miR-485-5p in the metastasis
of multiple malignancies has been reported in the literature. For
example, miR-485-5p was found to restrain mitochondrial
respiration, cell invasion, cell migration and cell proliferation
of breast cancer by directly suppressing PGC-1α (20). Similarly, miR-485-5p was downregulated
in malignant melanoma cells and tissues compared to their
corresponding controls (23). In
non-small cell lung cancer, miR-485-5p could prevent cell growth,
G0/G1 cell-cycle arrest, invasion and epithelial mesenchymal
transformation (EMT) by downregulating IGF2BP2 (24). Moreover, lncRNA DSCR8 could reduce
miR-485-5p by targeting the downstream molecule FZD7 to enhance the
progression of hepatocellular carcinoma (21). In papillary thyroid cancer, the
overexpression of LINC00460 and the downexpression of miR-485-5p
were observed. An increase in LINC00460 expression enhanced cell
proliferation, migration, invasion and EMT by targeting the
miR-485-5p/Raf1 pathway (53).
Similar to the results of previous research, the results of the
present study showed a decrease in the expression of miR-485-5p in
cells with OC. We also observed that miR-485-5p restricted cell
proliferation, migration and adhesion, but facilitated cell
apoptosis. Overall, these results offered more insights into the
mechanism of OC development.
TPX2 is associated with aberrant expression
and is highly expressed in various malignant tumors (55). In OC, TPX2 facilitated cell
proliferation, invasion and migration but weakened cell apoptosis
via the AKT signaling pathway (44).
TPX2 was also overexpressed in pancreatic cancer tissues and
cell lines compared with normal controls; however, the exogenous
silence of TPX2 suppressed cancerous growth (38). In addition, a report documented that
TPX2 was highly expressed in tissues with gastric carcinoma
compared to normal adjacent tissues and that it could strengthen
cell proliferation, invasion and migration by enhancing EMT-related
proteins (cdk2, cyclin D1, slug, MMP-9 and N-cadherin) and
restraining E-cadherin (56).
Findings of another study showed that, TPX2 was overexpressed in
cervical carcinoma, thereby enhancing cell migration, invasion, and
proliferation but inhibiting cell apoptosis and S-phase cell cycle
arrest (57). In addition, this
protein was found to be the downstream target gene of miR-8075, and
it enhanced the proliferation, migration, and invasion of cervical
cancer (42). The findings of this
research were the same as those of previous studies: TPX2 was
regulated by miR-485-5p, and it enhaned OC progression. This
finding could aid the understanding of TPX2 in OC and thus
provide new ways of increasing the survival rate of OC
patients.
Nevertheless, the current study has some
limitations. We only designed in vitro cell experiments to
determine the role of RHPN1-AS1, miR-485-5p and TPX2 in OC
progression. Put simply, in vivo experiments were not
performed to verify our findings. Furthermore, we did not reveal
how TPX2 participated in OC development. Thus, we recommend
that future research should verify our conclusion in vivo
and investigate how TPX2 influences OC tumorigenesis via
potential signaling pathways such as AKT signaling pathway and EMT.
In addition, due to the limitation of the microarray datasets used
in this study, the DEGs at different stages were unable to be
analyzed, which impedes the exploration of the more specific
molecular basis of RHPN1-AS1 functioning in OC. In future work, we
aim to identify other datasets or perform the microarray in
different stages of OC to explore and analyze the DEGs, providing
clues for the identification of specific biological significance of
RHPN1-AS1 in OC.
In summary, our study suggested that RHPN1-AS1 could
enhance OC progression by downregulating miR-485-5p and boosting
TPX2 expression. For this reason, we consider that
RHPN1-AS1, miR-485-5p and TPX2 may become new therapy
targets for OC treatments.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Yantai Affiliated Hospital of Binzhou Medical University. All
procedures performed in this study were in accordance with the 1964
Helsinki declaration. All the patients provided written informed
consent.
Authors' contributions
SC was responsible for the conceptualization,
methodology, investigation, data analysis, and manuscript
preparation. CL was involved in methodology, investigation,
visualization, resources, and manuscript preparation. CL and SC
confirmed the authenticity of the data shown in the present
manuscript. Both authors approved the final version of the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lheureux S, Gourley C, Vergote I and Oza
AM: Epithelial ovarian cancer. Lancet. 393:1240–1253. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oza AM, Tinker AV, Oaknin A,
Shapira-Frommer R, McNeish IA, Swisher EM, Ray-Coquard I,
Bell-McGuinn K, Coleman RL, O'Malley DM, et al: Antitumor activity
and safety of the PARP inhibitor rucaparib in patients with
high-grade ovarian carcinoma and a germline or somatic BRCA1 or
BRCA2 mutation: Integrated analysis of data from Study 10 and
ARIEL2. Gynecol Oncol. 147:267–275. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim G, Ison G, McKee AE, Zhang H, Tang S,
Gwise T, Sridhara R, Lee E, Tzou A, Philip R, et al: FDA approval
summary: Olaparib Monotherapy in patients with Deleterious Germline
BRCA-Mutated advanced ovarian cancer treated with three or more
lines of chemotherapy. Clin Cancer Res. 21:4257–4261. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu L, Yu X, Zhang L, Ding X, Pan H, Wen X,
Xu S, Xing Y, Fan J, Ge S, et al: The long non-coding RNA RHPN1-AS1
promotes uveal melanoma progression. Int J Mol Sci. 18:2262017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qiu X, Lei Z, Wang Z, Xu Y, Liu C, Li P,
Wu H and Gong Z: Knockdown of LncRNA RHPN1-AS1 inhibits cell
migration, invasion and proliferation in head and neck squamous
cell carcinoma. J Cancer. 10:4000–4008. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cui P, Su J, Li Q, Xu G and Zhu N: LncRNA
RHPN1-AS1 Targeting miR-625/REG3A promotes cell proliferation and
invasion of glioma cells. Onco Targets Ther. 12:7911–7921. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Duan H, Li X, Chen Y, Wang Y and Li Z:
LncRNA RHPN1-AS1 promoted cell proliferation, invasion and
migration in cervical cancer via the modulation of miR-299-3p/FGF2
axis. Life Sci. 239:1168562019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zheng S, Lv P, Su J, Miao K, Xu H and Li
M: Silencing of the long non-coding RNA RHPN1-AS1 suppresses the
epithelial-to-mesenchymal transition and inhibits breast cancer
progression. Am J Transl Res. 11:3505–3517. 2019.PubMed/NCBI
|
|
12
|
Zheng W, Li H, Zhang H, Zhang C, Zhu Z,
Liang H and Zhou Y: Long noncoding RNA RHPN1-AS1 promotes
colorectal cancer progression via targeting miR-7-5p/OGT axis.
Cancer Cell Int. 20:542020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fen H, Hongmin Z, Wei W, Chao Y, Yang Y,
Bei L and Zhihua S: RHPN1-AS1 drives the progression of
hepatocellular carcinoma via regulating miR-596/IGF2BP2 Axis. Curr
Pharm Des. 25:4630–4640. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ding L, Wang L, Li Z, Jiang X, Xu Y and
Han N: The positive feedback loop of RHPN1-AS1/miR-1299/ETS1
accelerates the deterioration of gastric cancer. Biomed
Pharmacother. 124:1098482020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mattick JS: Non-coding RNAs: The
architects of eukaryotic complexity. EMBO Rep. 2:986–991. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Z, Xu R and Li N: MicroRNAs from plants
to animals, do they define a new messenger for communication? Nutr
Metab (Lond). 15:682018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Seok H, Ham J, Jang ES and Chi SW:
MicroRNA target recognition: Insights from Transcriptome-wide
non-canonical interactions. Mol Cells. 39:375–381. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shao L, He Q, Liu Y, Liu X, Zheng J, Ma J,
Liu L, Li H, Li Z and Xue Y: UPF1 regulates the malignant
biological behaviors of glioblastoma cells via enhancing the
stability of Linc-00313. Cell Death Dis. 10:6292019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lou C, Xiao M, Cheng S, Lu X, Jia S, Ren Y
and Li Z: miR-485-3p and miR-485-5p suppress breast cancer cell
metastasis by inhibiting PGC-1α expression. Cell Death Dis.
7:e21592016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Sun L, Wang L, Liu Z, Li Q, Yao B,
Wang C, Chen T, Tu K and Liu Q: Long non-coding RNA DSCR8 acts as a
molecular sponge for miR-485-5p to activate Wnt/β-catenin signal
pathway in hepatocellular carcinoma. Cell Death Dis. 9:8512018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ou R, Lv J, Zhang Q, Lin F, Zhu L, Huang
F, Li X, Li T, Zhao L, Ren Y and Xu Y: circAMOTL1 Motivates AMOTL1
expression to facilitate cervical cancer growth. Mol Ther Nucleic
Acids. 19:50–60. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu J, Li J, Ren J and Zhang D:
MicroRNA-485-5p represses melanoma cell invasion and proliferation
by suppressing Frizzled7. Biomed Pharmacother. 90:303–310. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang RS, Zheng YL, Li C, Ding C, Xu C and
Zhao J: MicroRNA-485-5p suppresses growth and metastasis in
non-small cell lung cancer cells by targeting IGF2BP2. Life Sci.
199:104–111. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Hu J, Zhou W and Gao H: LncRNA
FOXD2-AS1 accelerates the papillary thyroid cancer progression
through regulating the miR-485-5p/KLK7 axis. J Cell Biochem.
2018.(Epub ahead of print).
|
|
26
|
Duan J, Zhang H, Li S, Wang X, Yang H,
Jiao S and Ba Y: The role of miR-485-5p/NUDT1 axis in gastric
cancer. Cancer Cell Int. 17:922017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chai Y, Du Y, Zhang S, Xiao J, Luo Z, He F
and Huang K: MicroRNA-485-5p reduces O-GlcNAcylation of Bmi-1 and
inhibits colorectal cancer proliferation. Exp Cell Res.
368:111–118. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin XJ, He CL, Sun T, Duan XJ, Sun Y and
Xiong SJ: hsa-miR-485-5p reverses epithelial to mesenchymal
transition and promotes cisplatin-induced cell death by targeting
PAK1 in oral tongue squamous cell carcinoma. Int J Mol Med.
40:83–89. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han DL, Wang LL, Zhang GF, Yang WF, Chai
J, Lin HM, Fu Z and Yu JM: MiRNA-485-5p, inhibits esophageal cancer
cells proliferation and invasion by down-regulating O-linked
N-acetylglucosamine transferase. Eur Rev Med Pharmacol Sci.
23:2809–2816. 2019.PubMed/NCBI
|
|
30
|
Wang FR, Xu SH, Wang BM and Wang F:
miR-485-5p inhibits metastasis and proliferation of osteosarcoma by
targeting CX3CL1. Eur Rev Med Pharmacol Sci. 22:7197–7204.
2018.PubMed/NCBI
|
|
31
|
Gao F, Wu H, Wang R, Guo Y, Zhang Z, Wang
T, Zhang G, Liu C and Liu J: MicroRNA-485-5p suppresses the
proliferation, migration and invasion of small cell lung cancer
cells by targeting flotillin-2. Bioengineered. 10:1–12. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang Y, Jiang Y, Wan Y, Zhang L, Qiu J,
Zhou S and Cheng W: UCA1 functions as a competing endogenous RNA to
suppress epithelial ovarian cancer metastasis. Tumour Biol.
37:10633–10641. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang W, Han L, Xing P, Xing P, Liu B, Sun
Z, Zhou W and Dong J: LncRNA RHPN1-AS1 accelerates proliferation,
migration, and invasion via regulating miR-485-5p/BSG axis in
hepatocellular carcinoma. Naunyn Schmiedebergs Arch Pharmacol.
393:2543–2551. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang X, Yan Z, Wang L, Zhang S and Gao M:
STAT1-induced upregulation of lncRNA RHPN1-AS1 predicts a poor
prognosis of hepatocellular carcinoma and contributes to tumor
progression via the miR-485/CDCA5 axis. J Cell Biochem. 2020.(Epub
ahead of print).
|
|
35
|
Wittmann T, Boleti H, Antony C, Karsenti E
and Vernos I: Localization of the kinesin-like protein Xklp2 to
spindle poles requires a leucine zipper, a microtubule-associated
protein, and dynein. J Cell Biol. 143:673–685. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gruss OJ and Vernos I: The mechanism of
spindle assembly: Functions of Ran and its target TPX2. J Cell
Biol. 166:949–955. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stewart S and Fang G: Anaphase-promoting
complex/cyclosome controls the stability of TPX2 during mitotic
exit. Mol Cell Biol. 25:10516–10527. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Warner SL, Stephens BJ, Nwokenkwo S,
Hostetter G, Sugeng A, Hidalgo M, Trent JM, Han H and Von Hoff DD:
Validation of TPX2 as a potential therapeutic target in pancreatic
cancer cells. Clin Cancer Res. 15:6519–6528. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tonon G, Wong KK, Maulik G, Brennan C,
Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, et
al: High-resolution genomic profiles of human lung cancer. Proc
Natl Acad Sci USA. 102:9625–9630. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Smith LT, Mayerson J, Nowak NJ, Suster D,
Mohammed N, Long S, Auer H, Jones S, McKeegan C, Young G, et al:
20q11.1 amplification in giant-cell tumor of bone: Array CGH, FISH,
and association with outcome. Genes Chromosomes Cancer. 45:957–966.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wei P, Zhang N, Xu Y, Li X, Shi D, Wang Y,
Li D and Cai S: TPX2 is a novel prognostic marker for the growth
and metastasis of colon cancer. J Transl Med. 11:3132013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Song T, Xu A, Zhang Z, Gao F, Zhao L, Chen
X, Gao J and Kong X: CircRNA hsa_circRNA_101996 increases cervical
cancer proliferation and invasion through activating TPX2
expression by restraining miR-8075. J Cell Physiol.
234:14296–14305. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yan L, Li Q, Yang J and Qiao B:
TPX2-p53-GLIPR1 regulatory circuitry in cell proliferation,
invasion, and tumor growth of bladder cancer. J Cell Biochem.
119:1791–1803. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tian Y, Liu LL, Guo DM, Wang Y, Zha WH, Li
Y and Wu FJ: TPX2 gene silencing inhibits cell proliferation and
promotes apoptosis through negative regulation of AKT signaling
pathway in ovarian cancer. J Cell Biochem. 119:7540–7555. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Taherdangkoo K, Kazemi Nezhad SR, Hajjari
MR and Tahmasebi Birgani M: miR-485-3p suppresses colorectal cancer
via targeting TPX2. Bratisl Lek Listy. 121:302–307. 2020.PubMed/NCBI
|
|
46
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang
C, Liu D, Wang M, Wang L, Zeng F and Jiang G: CircHIPK3 sponges
miR-558 to suppress heparanase expression in bladder cancer cells.
EMBO Rep. 18:1646–1659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ma S, Rong X, Gao F, Yang Y and Wei L:
TPX2 promotes cell proliferation and migration via PLK1 in OC.
Cancer Biomark. 22:443–451. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang D, Chen J, Yang C and Wang M: TPX2
silencing mediated by joint action of microvesicles and ultrasonic
radiation inhibits the migration and invasion of SKOV3 cells. Mol
Med Rep. 17:7627–7635. 2018.PubMed/NCBI
|
|
50
|
Zhao J, Yang T, Ji J, Zhao F, Li C and Han
X: RHPN1-AS1 promotes cell proliferation and migration via
miR-665/Akt3 in ovarian cancer. Cancer Gene Ther. 28:33–41. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang J, Ding W, Xu Y, Tao E, Mo M, Xu W,
Cai X, Chen X, Yuan J and Wu X: Long non-coding RNA RHPN1-AS1
promotes tumorigenesis and metastasis of ovarian cancer by acting
as a ceRNA against miR-596 and upregulating LETM1. Aging (Albany
NY). 12:4558–4572. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhu P, Li Y, Li P, Zhang Y and Wang X:
c-Myc induced the regulation of long non-coding RNA RHPN1-AS1 on
breast cancer cell proliferation via inhibiting P53. Mol Genet
Genomics. 294:1219–1229. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liang D, Liu H, Yang Q, He Y, Yan Y, Li N
and You W: Long noncoding RNA RHPN1-AS1, induced by KDM5B, is
involved in breast cancer via sponging miR-6884-5p. J Cell Biochem.
2020.(Epub ahead of print). View Article : Google Scholar
|
|
54
|
Huang S, Zou C, Tang Y, Wa Q, Peng X, Chen
X, Yang C, Ren D, Huang Y, Liao Z, et al: miR-582-3p and miR-582-5p
suppress prostate cancer metastasis to bone by repressing TGF-β
Signaling. Mol Ther Nucleic Acids. 16:91–104. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Morgan-Lappe SE, Tucker LA, Huang X, Zhang
Q, Sarthy AV, Zakula D, Vernetti L, Schurdak M, Wang J and Fesik
SW: Identification of Ras-related nuclear protein, targeting
protein for xenopus kinesin-like protein 2, and stearoyl-CoA
desaturase 1 as promising cancer targets from an RNAi-based screen.
Cancer Res. 67:4390–4398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liang B, Zheng W, Fang L, Wu L, Zhou F,
Yin X, Yu X and Zou Z: Overexpressed targeting protein for Xklp2
(TPX2) serves as a promising prognostic marker and therapeutic
target for gastric cancer. Cancer Biol Ther. 17:824–832. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chang H, Wang J, Tian Y, Xu J, Gou X and
Cheng J: The TPX2 gene is a promising diagnostic and therapeutic
target for cervical cancer. Oncol Rep. 27:1353–1359.
2012.PubMed/NCBI
|