Introduction
Cervical cancer is the second most common type of
cancer and one of the most common types of gynecological malignancy
in women worldwide (1–3) with ~500,000 new cases and 270,000
deaths in 2018 (4). In China, the
average age range is 30–35 years for onset of cervical cancer and
45–55 years for invasive cancer (5). Investigation of the molecular
mechanisms that promote the pathogenesis of cervical cancer is
needed.
Cancer cells produce energy by taking up glucose and
converting it to lactate, even under experimental conditions with
sufficient oxygen. This was first described by Otto Warburg in 1924
and was called the Warburg effect or aerobic glycolysis (4,6,7).
Cancer cells obtain enough energy and biosynthetic precursors (such
as nucleotides, proteins and lipids) via the Warburg effect to
achieve enhanced proliferation, survival and long-term maintenance
(8).
Accumulating evidence has demonstrated that aerobic
glycolysis is a key hallmark of cancer cells (9,10);
molecules involved in the aerobic glycolysis pathway may be
potential targets in cancer therapy, including cervical cancer
(7,11). Therefore, certain metabolic enzymes
such as Hexokinase 2, Phosphoinositide-Dependent Kinase 1 and
Lactate dehydrogenase A chain (LDHA) involved in aerobic glycolysis
have been identified as potential therapeutic targets (12–15).
LDH is a key enzyme in aerobic glycolysis that
catalyzes interconversion of lactate and pyruvate, accompanied by
interconversion of NADH and NAD+ (16). LDH is a tetramer composed of two
subunits: muscle- and heart-type, encoded by LDHA and LDHB
(17). LDHA is abnormally
expressed in multiple types of human cancer, including renal
(14), gastric (18) and pancreatic (19,20),
hepatocellular (21) and
nasopharyngeal carcinoma (22), as
well as breast cancer (23).
Furthermore, studies have indicated that inhibition of LDHA
decrease tumor proliferation in numerous types of cancer cell, such
as renal and breast cancer and human hepatocellular and
nasopharyngeal carcinoma cells (14,21,22,24,25).
LDHA inhibition suppresses migration of cancer cells and increases
radiosensitivity and chemosensitivity (19,22,26,27).
However, the underlying roles and mechanism of LDHA in cervical
cancer are largely unknown.
The present study aimed to verify expression of LDHA
in human cervical cancer. Furthermore, the effect of LDHA
inhibition by oxamate, a classic inhibitor of LDHA (28,29),
or short hairpin (sh)RNA on cervical cancer cells was investigated.
The purpose of the present study was to determine the effect of
LDHA inhibition on metabolism, proliferation and apoptosis of
cervical cancer cells and to investigate the underlying
mechanism.
Materials and methods
Cell lines, reagents and
antibodies
Human cervical squamous cell carcinoma and
endocervical adenocarcinoma (CESC; HeLa and SiHa) and 293T cell
lines were obtained from American Type Culture Collection, which
had characterized the cell lines by short tandem repeat profiling,
cell morphology and karyotyping assay. The cell lines were
maintained in RPMI-1640 supplemented with 10% fetal bovine serum
(both Gibco; Thermo Fisher Scientific, Inc.) at 37°C in 5%
CO2. Oxamate was purchased from Sigma-Aldrich (Merck
KGaA). SP600125 was purchased from Abcam. For JNK inhibitor assay,
HeLa cells were treated with 10 µM SP600125 1 h at 37°C prior to
oxamate treatment and cells were incubated for another 48 h at
37°C. LDHA-knockdown HeLa cells were treated with 10 µM SP600125
for 48 h at 37°C. The primary antibodies for LDHA, cleaved
caspase-3, caspase-9, Cyclin A, Cyclin B1, p21, cyclin-dependent
kinase (CDK)1, JNK, phosphorylated (p-)JNK, p38 and p-p38 were
obtained from Cell Signaling Technology, Inc. The antibody for
β-actin was purchased from Santa Cruz Biotechnology, Inc. The
antibodies for cytochrome c and cytochrome c oxidase
IV (COX IV) were obtained from Abcam.
Bioinformatics analysis
Gene Expression Profiling Interactive Analysis
(GEPIA; gepia.cancer-pku.cn/index.html) comprises open databases
for analysis of gene expression data from The Cancer Genome Atlas
(TCGA; cancer.gov/tcga) (30).
GEPIA was used to analyze mRNA expression levels of LDHA in human
tumor and normal samples, the correlation between LDHA and
mitochondrial pyruvate carrier 1 (MPC1) and the potential
prognostic value of LDHA expression levels. Data were extracted
from TGGA.
Cytotoxicity assay
HeLa and SiHa cells were seeded at 10,000 cells/well
in triplicate in 96-well plates for 24 h at 37°C, then treated with
10, 20, 50, 80 and 100 mM oxamate for 44 h at 37°C. Cytotoxicity of
oxamate was measured using MTT Cell Proliferation and Cytotoxicity
Assay kit (cat. no. M1020; Beijing Solarbio Science &
Technology Co., Ltd.) according to the manufacturer's protocol. MTT
solution was added to each well and incubated for 4 h at 37°C in a
humidified atmosphere. MTT formazan precipitate was dissolved in
dimethyl sulfoxide. Absorbance was measured at 570 nm. In addition,
viability of LDHA knockdown and control cells were also measured by
MTT detection kit as aforementioned.
BrdU incorporation assay
LDHA knockdown or oxamate-treated HeLa and SiHa
cells were incubated with BrdU for 6 h at 37°C. BrdU uptake was
analyzed by Cell Proliferation ELISA, BrdU (colorimetric) assay
(cat. no. 11647229001; Roche Diagnostrics) according to the
manufacturer's protocol.
Colony formation assay
For colony formation assay, 1,000 cells from LDHA
knockdown or oxamate-treated HeLa and SiHa groups were plated in
six-well plates and cultured for 7 days at 37°C. The cells were
fixed in 4% paraformaldehyde solution for 15 min at room
temperature and stained with 0.1% crystal violet solution for 10
min at room temperature. Then, colonies (>50 cells/colony) were
observed using a light microscope (magnification, ×40; Olympus
Corporation), counted by using the ImageJ Lab software (version
k1.7.9; National Institutes of Health), photographed by using a
Canon EOS600D digital camera (Canon, Inc.).
ATP detection assay
ATP levels in HeLa and SiHa cells were determined
using ATP Assay Kit (cat. no. S0026; Beyotime Institute of
Biotechnology) according to the manufacturer's protocol and as
previously described (31,32). In brief, samples were lysed with
200 µl lysis buffer, centrifuged at 6,000 g for 5 min at 4°C and
the supernatant was collected. Then, the supernatant was mixed with
100 µl ATP detection working buffer and measured by a plate-reading
luminometer.
Lactate production measurement
Lactate production was determined by Lactic Acid
assay kit (cat. no. A019-2-1; Nanjing Jiancheng Bioengineering
Institute) as previously described (33). Briefly, HeLa and SiHa cells were
harvested, washed twice with PBS and lysed in RIPA buffer (cat. no.
89900; Thermo Fisher Scientific) for 30 min on ice. Following
centrifugation at 6,000 g for 10 min at 4°C, the supernatant was
collected in microcentrifuge tubes, mixed with an equal volume of
substrate and incubated at 37°C for 10 min. Following the addition
of stop solution, each sample (volume, 200 µl) was aliquoted in the
wells of a 96-well microtiter plate. Lactate concentration was
evaluated by measuring absorbance using a microplate reader at 530
nm. The result was normalized to the total protein content,
determined by BCA assay.
Glucose assay
Glucose Assay kit (cat. no. 361500; Shanghai
Rongsheng Biotech Co., Ltd.) was used to detect the glucose uptake
according to the manufacturer's instructions and quantified by
measuring the absorption at 450 nm. HeLa and SiHa cells were
cultured in 96-well plates seeded at 10,000 cells/well in RPMI-1640
(Gibco; Thermo Fisher Scientific, Inc.) for 24 h at 37°C, then
treated with oxamate for 24 h. The cells were centrifuged at 6,000
g for 10 min at 4°C and the supernatant was collected. Glucose
uptake was calculated as follows: original glucose concentration in
medium-detected glucose concentration in culture supernatant.
Transfection of CESC cells with LDHA
shRNA lentiviral vector
shRNA sequences targeting LDHA were as follows:
shRNA1,
5′-GATCCCCACCATGATTAAGGGTCTTTCTTCCTGTCAGAAAAGACCCTTAATCATGGTGGTTTTTG-3′
and shRNA2,
5′-GATCCGCAAACTCCAAGCTGGTCATTCTTCCTGTCAGAAATGACCAGCTTGGAGTTTGCTTTTTG-3′.
The shRNA sequence of the negative control (PGreenpuro shRNA) was
5′-GATCCGTGCTCCACGGCATTTCATTACTTCCTGTCAGATAATGAAATGCCGTGGAGCACTTTTTG-3′.
pGreenPuro-LDHA-shRNA1 and pGreenPuro-LDHA-shRNA 2 were prepared by
inserting target sequences into PGreenpuro shRNA vectors.
pGreenPuro-LDHA-shRNA1, pGreenPuro-LDHA-shRNA 2 and PGreenpuro
shRNA were obtained from Chengdu Transvector Biotechnology. The
concentration of extracted plasmid was >500 ng/µl with an
A260/280 ratio of 1.8-2.0. A second generation lentivirus packaging
system was used. The plasmid pGreenPuro-LDHA-shRNA (2 µg) was
co-transfected into 293T cells together with packaging vectors
psPAX2 (2 µg) and envelop plasmids pMD2.G (1.5 µg; both Addgene,
Inc.) using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. The viral supernatant fraction was collected at 48 h
post-transfection and filtered through a 0.45 µm filter. Viral
titers were determined by QuickTiter Quantitation kit (Cell
Biolabs, Inc.) according to the manufacturer's instructions.
Multiplicity of infection of 50 and 60 were used for HeLa and SiHa
cells, respectively. HeLa and SiHa cells were co-cultured with
viral supernatant and 6 µg/ml polybrene for 16 h at 37°C. Then,
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) was replaced with
fresh medium. After 24 h, the medium was replaced with fresh
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing
1 µg/ml puromycin (Thermo Fisher Scientific, Inc.). The cells were
maintained in medium with 1 µg/ml puromycin for 2 weeks at
37°C.
Analysis of cell cycle distribution
and apoptosis
For cell cycle analysis, HeLa and SiHa cells were
collected following treatment with 20 mM oxamate for 48 h at 37°C,
then fixed with 4% paraformaldehyde at 4°C for 30 min. Following
washing with PBS three times, cells were stained with propidium
iodine (PI; 50 µg/ml; cat. no. P2667; Sigma-Aldrich; Merck KGaA) at
room temperature for 30 min in the dark according to the
manufacturer's instructions. Cell cycle distribution was analyzed
using 10,000 cells by flow cytometry analysis using a FACS Canto II
flow cytometer (Becton, Dickinson and Company) and BD FACS DIVA
software, version 6.1.3 (Becton-Dickinson and Company). For
apoptosis assay, HeLa and SiHa cells were harvested, washed with
PBS, stained with 5 µl Annexin V-FITC and 5 µl PI at room
temperature for 15 min using FITC Annexin V Apoptosis Detection kit
(cat. no. 556547; BD Biosciences) and analyzed by FACSCanto II flow
cytometer as previously described (34). Apoptotic rate was calculated as
follows: Late apoptosis cell number/total cell number ×100%.
Western blot analysis
HeLa and SiHa cells were harvested and washed with
PBS. The protein was were extracted and western blot analysis was
performed as previously described (35). Mitochondria were isolated from
cells using Mitochondria Isolation Kit for Cultured Cells (TransGen
Biotech Co., Ltd.) according to the manufacturer's protocol. Total
protein was extracted from cells using Mammalian Total Protein
Extraction kit (TransGen Biotech Co., Ltd.) according to the
manufacturer's protocol. Enhanced BCA Protein Assay kit (cat. no.
P0010; Beyotime Institute of Biotechnology) was used to determine
protein concentration. Then, 12% separation and 5% stacking gels
were prepared for electrophoresis. The proteins (20 µg/lane) were
separated and transferred onto PVDF membranes (EMD Millipore) via
wet transfer method. Next, 5% skimmed milk in 1X Tris-buffered
saline-0.05% Tween-20 (TBST; Beijing Solarbio Science &
Technology Co., Ltd.), was used to block membranes at room
temperature for 1 h. The membranes were incubated with primary
antibodies against LDHA (cat. no. 3582; 1:1,000), Cyclin A (cat.
no. 67955; 1:1,000), Cyclin B1 (cat. no. 12231; 1:1,000), p21 (cat.
no. 2947; 1:1,000), CDK1 (cat. no. 28439; 1:500), p38 (cat. no.
8690; 1:1,000), p-p38 (cat. no. 4511; 1:1,000), JNK (cat. no. 9252;
1:1,000), p-JNK (cat. no. 9255; 1:2,000), cleaved-caspase-3 (cat.
no. 9664; 1:1,000), caspase-3 (cat. no. 14220; 1:1,000; Cell
Signaling Technology, Inc.), cleaved-caspase-9 (cat. no. 20750;
1:1,000; Cell Signaling Technology, Inc.), caspase-9 (cat. no.
9508; 1:1,000), β-actin (cat. no. 4778; 1:2,000), cytochrome
c (cat. no. ab133504; 1:5,000) and COX–IV (cat. no.
ab202554; 1:2,000) overnight at 4°C. The membranes were washed with
1X TBST for 5 min (3 times). Horseradish peroxidase-conjugated goat
anti-rabbit (cat. no. 7074; 1:2,000; Cell Signaling Technology,
Inc.) and anti-mouse antibody (cat. no. 7076; 1:2,000; Cell
Signaling Technology, Inc.) were used as secondary antibodies. The
membranes were incubated with secondary antibodies at room
temperature for 1 h and washed with 1X TBST for 5 min (3 times).
The membranes were visualized using Luminata™ Crescendo Western HRP
Substrate (EMD Millipore). Protein expression was quantified using
ImageJ Lab software (version k1.7.9; National Institutes of
Health).
Statistical analysis
All data were presented as the as mean ± SD of ≥3
independent experimental repeats. The overall survival curve was
generated and assessed via Kaplan-Meier and log-rank tests.
Correlation analysis was performed using Pearson correlation
analysis. Data containing two groups were analyzed by unpaired
Student's t-test and two-tailed distribution. Data containing >2
groups was analyzed by one-way ANOVA followed by Tukey's post-hoc
test. Data were analyzed using SPSS 18.0 (SPSS, Inc.) and GraphPad
Prism 5.0 software (GraphPad Software, Inc.). *P<0.05 was
considered to indicate a statistically significant difference.
Results
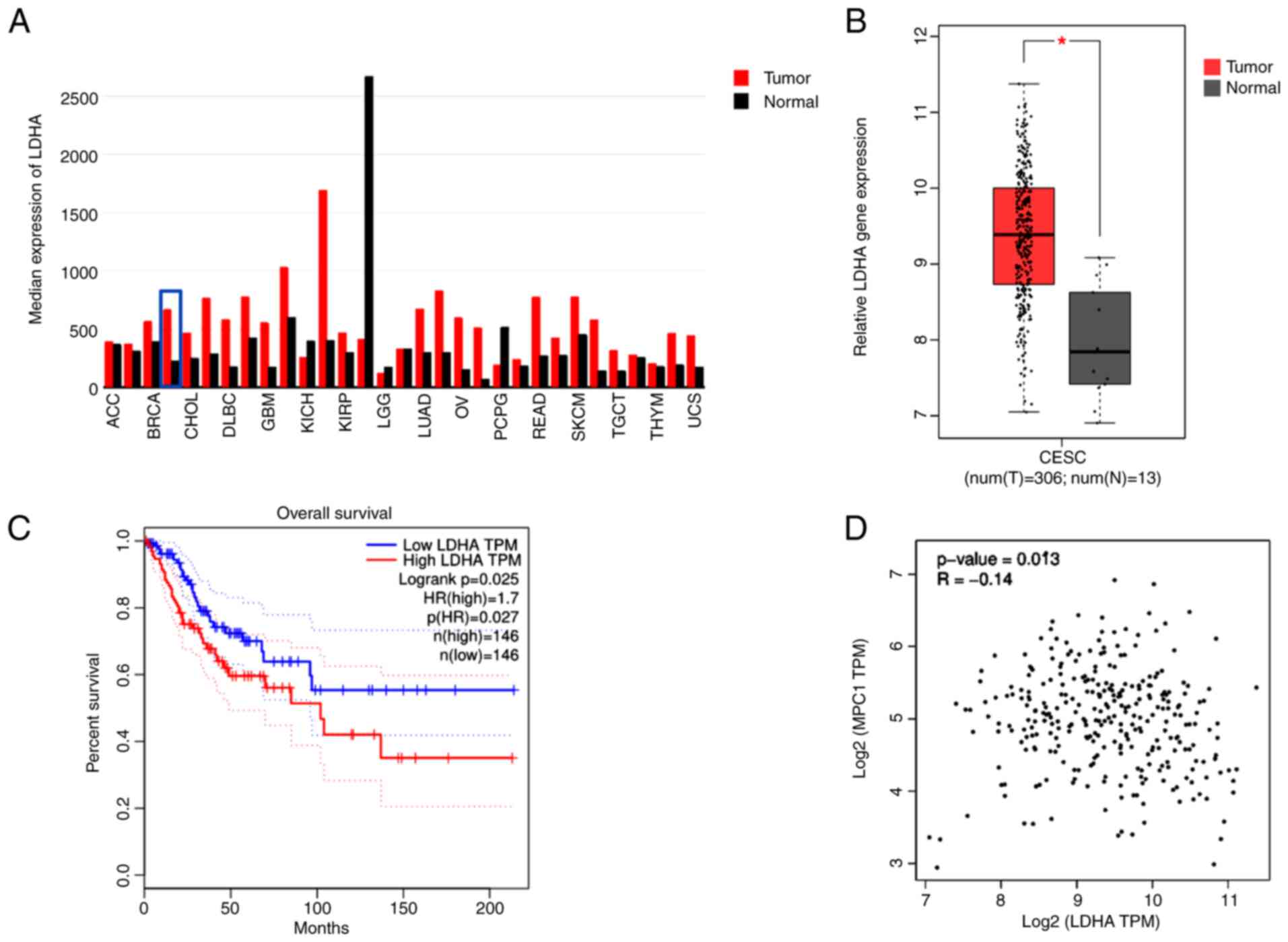
LDHA is upregulated in cervical cancer
and associated with patient survival
To determine the gene expression of LDHA in human
tumor and normal samples, TCGA datasets were analyzed via GEPIA.
LDHA gene expression levels were upregulated in most types of
tumor, including CESC, compared with normal tissue (Fig. 1A). Furthermore, analysis of TCGA
data using GEPIA showed a significantly higher LDHA gene expression
in CESC compared with normal cervical tissue (Fig. 1B). In addition, tumor samples were
separated into low and high groups based on median expression level
of LDHA. LDHA overexpression was significantly associated with
shorter overall survival in patients with cervical cancer (HR=1.7;
Fig. 1C), suggesting that high
LDHA expression was a risk factor for poor prognosis in cervical
cancer. These results suggesting that LDHA may play a critical role
in cerivical cancer progression. Because of the key role of MPC1 in
pyruvate metabolism, the association between LDHA and MPC1 was
evaluated using GEPIA. Correlation analysis (Fig. 1D) showed that LDHA levels were
negatively correlated with MPC1 (R=−0.14), supporting the
hypothesis that cervical cancer was aerobic glycolysis-dependent.
Taken together, these results suggested that LDHA was upregulated
in cervical cancer and dysregulation of LDHA served a key role in
cervical cancer.
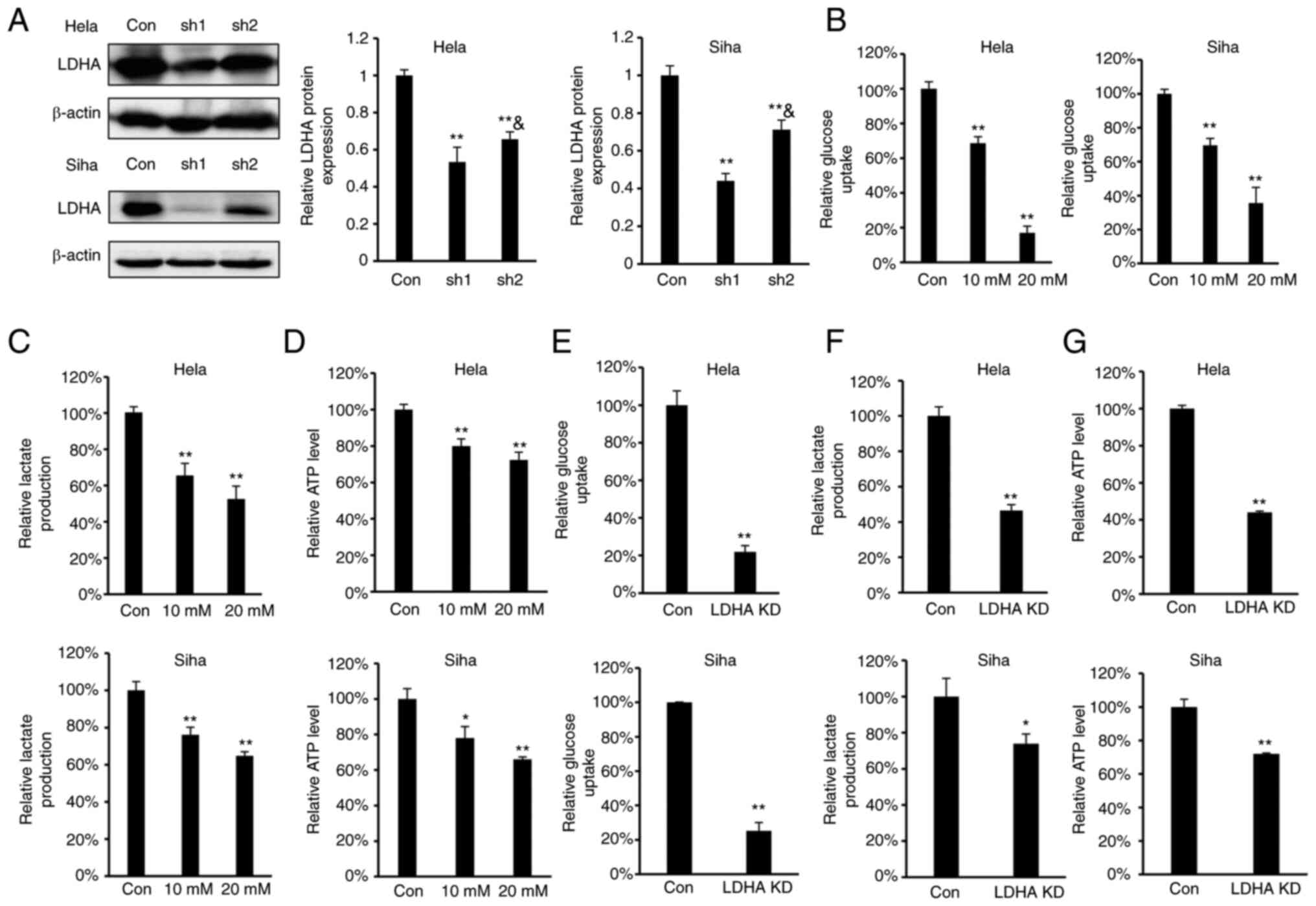
LDHA inhibition disrupts energy
metabolism
As LDHA overexpression were negatively correlated
with overall survival in patients with cervical cancer, the effect
of LDHA inhibition on glycolysis in CESC cells was investigated. To
determine the effect of LDHA inhibition on energy metabolism in
CESC cells, oxamate, a small-molecule inhibitor of LDHA (36), was used. According to the
concentrations of oxamate used in previous reports (18,20,37),
we selected 10 and 20 mM oxamate for the energy metabolism study.
Meanwhile, four stable LDHA-depleted CESC cell lines (HeLa and SiHa
sh1 and sh2) were constructed to investigate the effect of
knockdown on protein expression levels (Fig. 2A). The results indicated that sh1
and sh2 efficiently knocked down expression of LDHA compared with
the control. Considering that the knockdown effect of sh1 was
significantly greater compared with sh2 in both HeLa and SiHa
cells, HeLa and SiHa sh1 cells were selected for further study.
Intracellular biochemical indicators were detected in both HeLa and
SiHa cells. Following treatment with oxamate for 24 h, glucose
uptake, lactate production and ATP levels decreased significantly
in both HeLa and SiHa cells (Fig.
2B-D). Similarly, knockdown of LDHA in CESC cells decreased
glucose uptake, lactate production and ATP levels (Fig. 2E-G). These results demonstrated
that LDHA inhibition disrupted energy metabolism and suppressed the
Warburg effect in CESC cells.
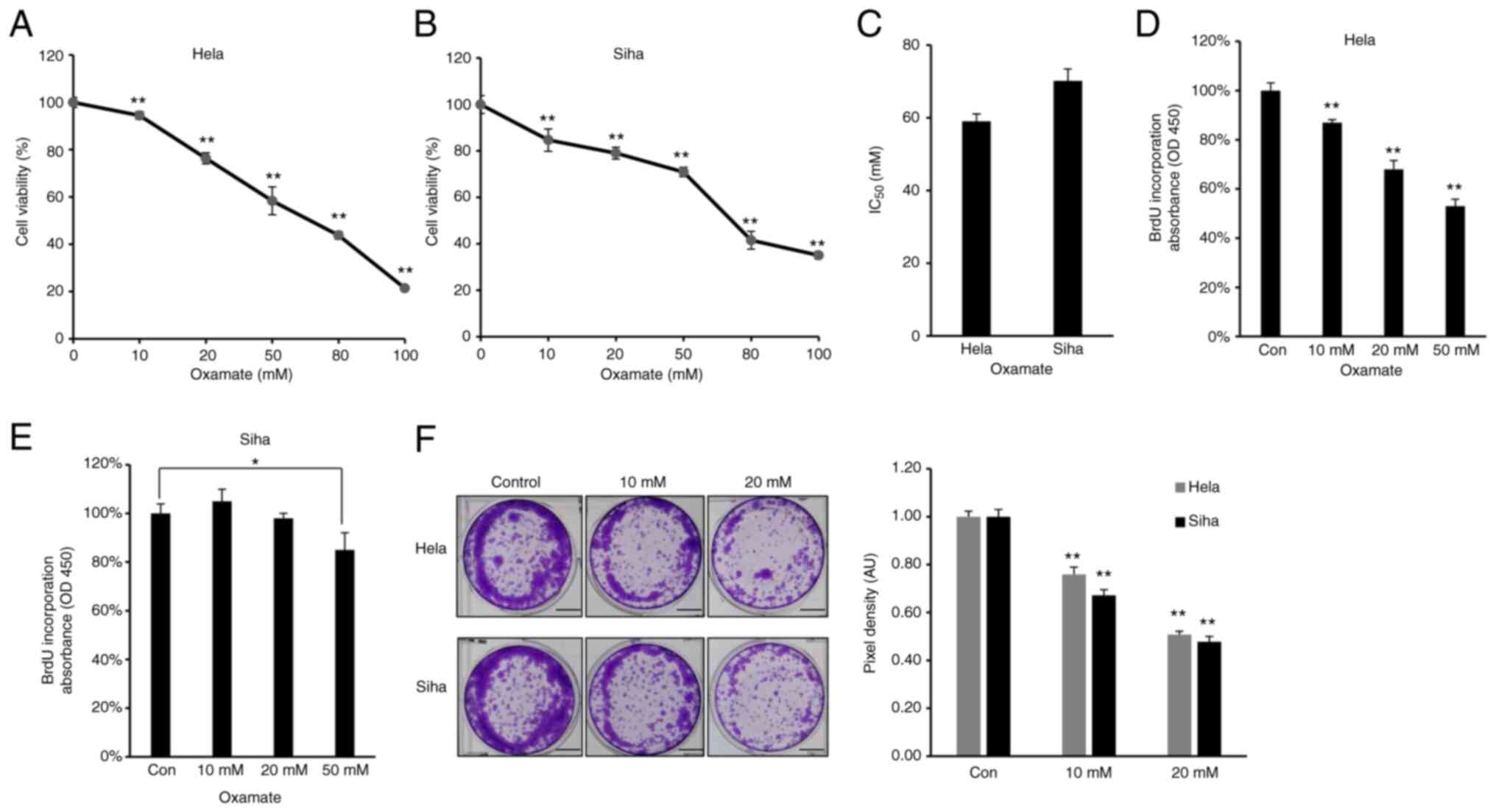
LDHA inhibition by oxamate suppresses
proliferation of CESC cells
As increased glycolysis enables cancer cells to
suppress apoptotic signaling and promotes viability and
proliferation (38), the effect of
LDHA on viability of CESC cells was assessed. Firstly, MTT assay
was performed to investigate the cytotoxic effect of oxamate on
CESC cells. HeLa and SiHa cells were treated with different doses
of oxamate for 48 h. Oxamate exhibited significant cytotoxicity
toward these two CESC cancer cell lines in a dose-dependent manner
(Fig. 3A and B). The
IC50 value of oxamate for HeLa cells was 59.05 mM, while
the IC50 value for SiHa cells was 70.19 mM (Fig. 3C). Cell proliferation was
quantified by measuring BrdU incorporation (39); oxamate significantly inhibited
proliferation of HeLa and SiHa cells (Fig. 3D and E). Clone formation assay
showed that HeLa and SiHa cells formed significantly fewer clones
following treatment with oxamate (Fig.
3F), suggesting that oxamate inhibited clone formation capacity
in both HeLa and SiHa cells. Collectively, these results suggested
that LDHA inhibition by oxamate in HeLa and SiHa cells inhibited
proliferation.
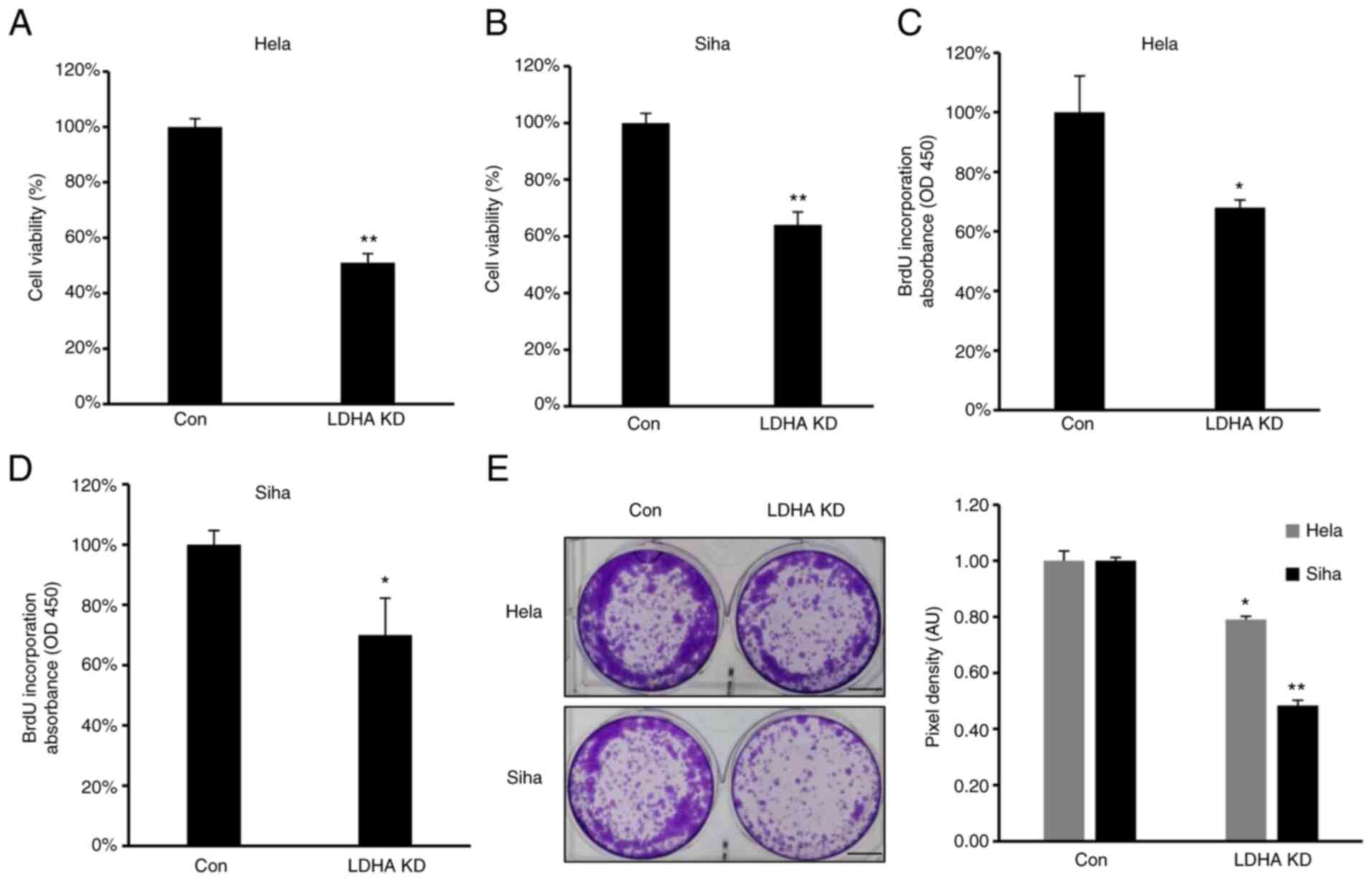
LDHA inhibition by shRNA suppresses
proliferation of CESC cells
To confirm the role of LDHA on proliferation of CESC
cells, proliferation and viability of LDHA-knockdown CESC and
control cells were investigated. Consistent with the aforementioned
results, MTT assay showed that viability was significantly
inhibited in all LDH-knockdown CESC cells (Fig. 4A and B). Furthermore, lower levels
of BrdU incorporation were detected in HeLa and SiHa #h1 cells
compared with control (Fig. 4C and
D). The clone formation assay demonstrated that LDHA knockdown
inhibited clone formation capacity of HeLa and SiHa cells (Fig. 4E). In summary, similar to LDHA
inhibition by oxamate, sh-induced LDHA knockdown suppressed
viability, proliferation and colony forming ability of CESC
cells.
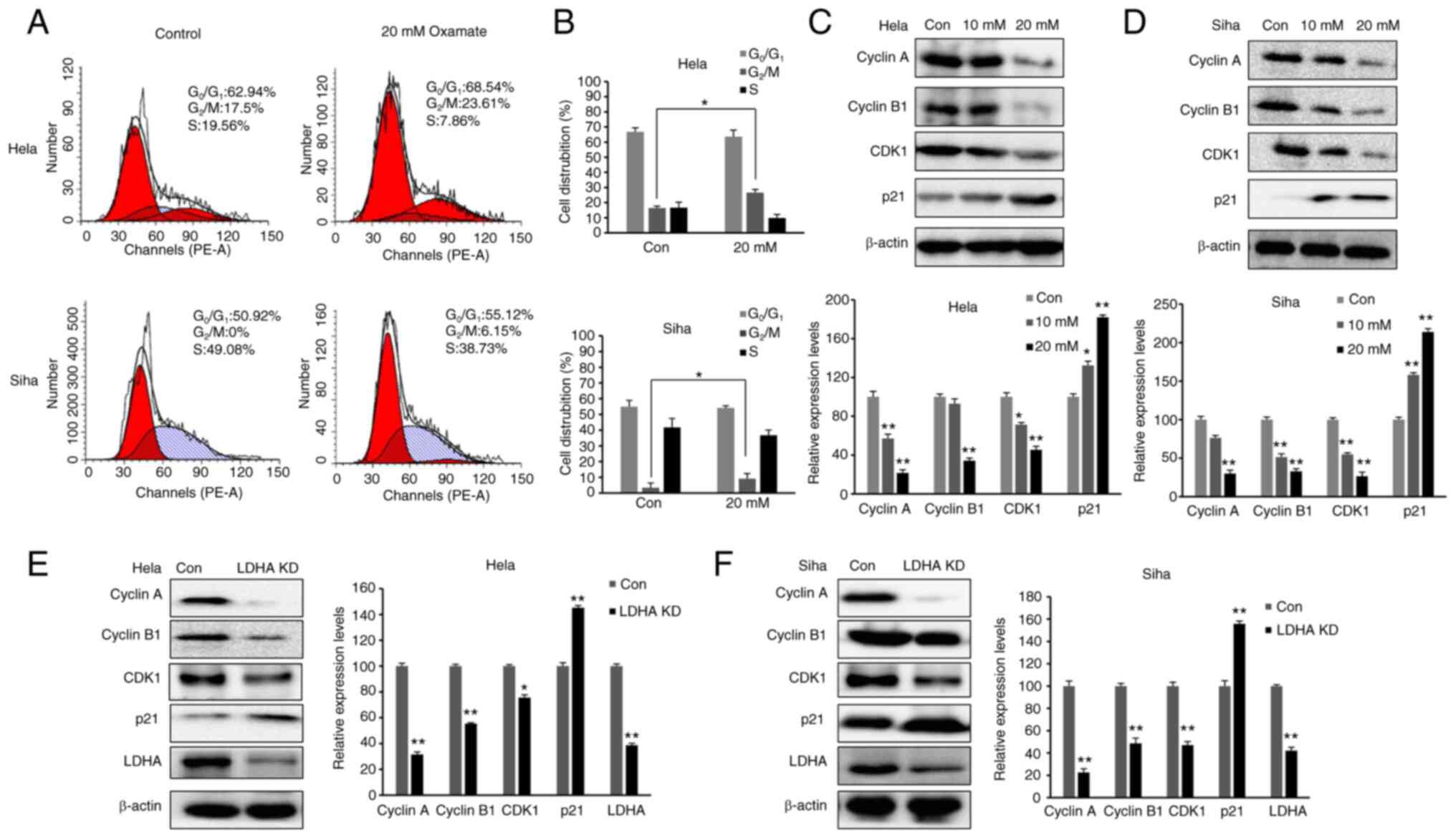
Inhibition of LDH induces cell cycle
arrest in G2/M phase
As LDHA inhibition suppressed proliferation of CESC
cells, cell cycle distribution was investigated. The aforementioned
data demonstrated that 20 mM oxamate had a more significant effect
than 10 mM oxamate on cell survival, proliferation and metabolism;
therefore, 20 mM oxamate was used for cell cycle analysis. Cells
were exposed to 20 mM oxamate for 48 h and flow cytometry was used
to analyze the cell cycle distribution following PI staining. There
was an increase in the number of HeLa and SiHa cells in
G2/M phase following treatment with oxamate (Fig. 5A and B). To verify the
G2/M arrest induced by oxamate and determine the
underlying mechanisms, western blot analysis was used to
investigate changes in expression levels of proteins associated
with G2/M transition (Fig.
5C and D). Protein levels of Cyclin B1, Cyclin A and CDK1 were
significantly decreased in both SiHa and HeLa cells following
treatment with oxamate, suggesting that oxamate induced
G2/M arrest by modulating expression of Cyclin B1,
Cyclin A and CDK1. Expression levels of proteins in upstream
signaling pathways affecting cell cycle were also assessed;
expression levels of p21 significantly increased following
treatment with oxamate. These findings were consistent with
inhibition of cell proliferation. The same results were found in
both SiHa and HeLa cells following knockdown of LDHA (Fig. 5E and F). Collectively, these data
indicated that LDHA inhibition in CESC cells induced cell cycle
arrest, suggesting a tumor-suppressive role of LDHA in CESC
cells.
LDHA inhibition induces apoptosis via
the mitochondrial pathway
The present study demonstrated that LDH inhibition
impaired proliferation and increased the G2/M fraction
in CESC cells. To determine whether LDHA inhibition induces
apoptosis, cells were exposed to oxamate for 48 h, then Annexin
V/PI staining and flow cytometric analysis were performed.
Following 48 h treatment with oxamate, the percentages of apoptotic
cells significantly increased (Fig. 6A
and B). Next, expression of apoptosis-associated proteins were
detected by western blot analysis. Expression of cleaved-caspase-3
and −9 was significantly enhanced following treatment with oxamate
for 48 h (Fig. 6C and D).
Apoptosis-associated protein expression levels were compared
between LDHA-knockdown CESC and control cells (Fig. 6E and F). Western blot results
showed that higher expression of cleaved-caspase-3 and −9 was
detected following knockdown of LDHA, supporting the results of
oxamate treatment experiments. Protein levels of cytochrome
c in the cytoplasm and mitochondria were detected;
cytochrome c was released from mitochondria into the
cytoplasm following LDHA inhibition (Fig. 6G and H). Taken together, these
results indicated that LDHA inhibition induced apoptosis via the
mitochondrial pathway in CESC cells.
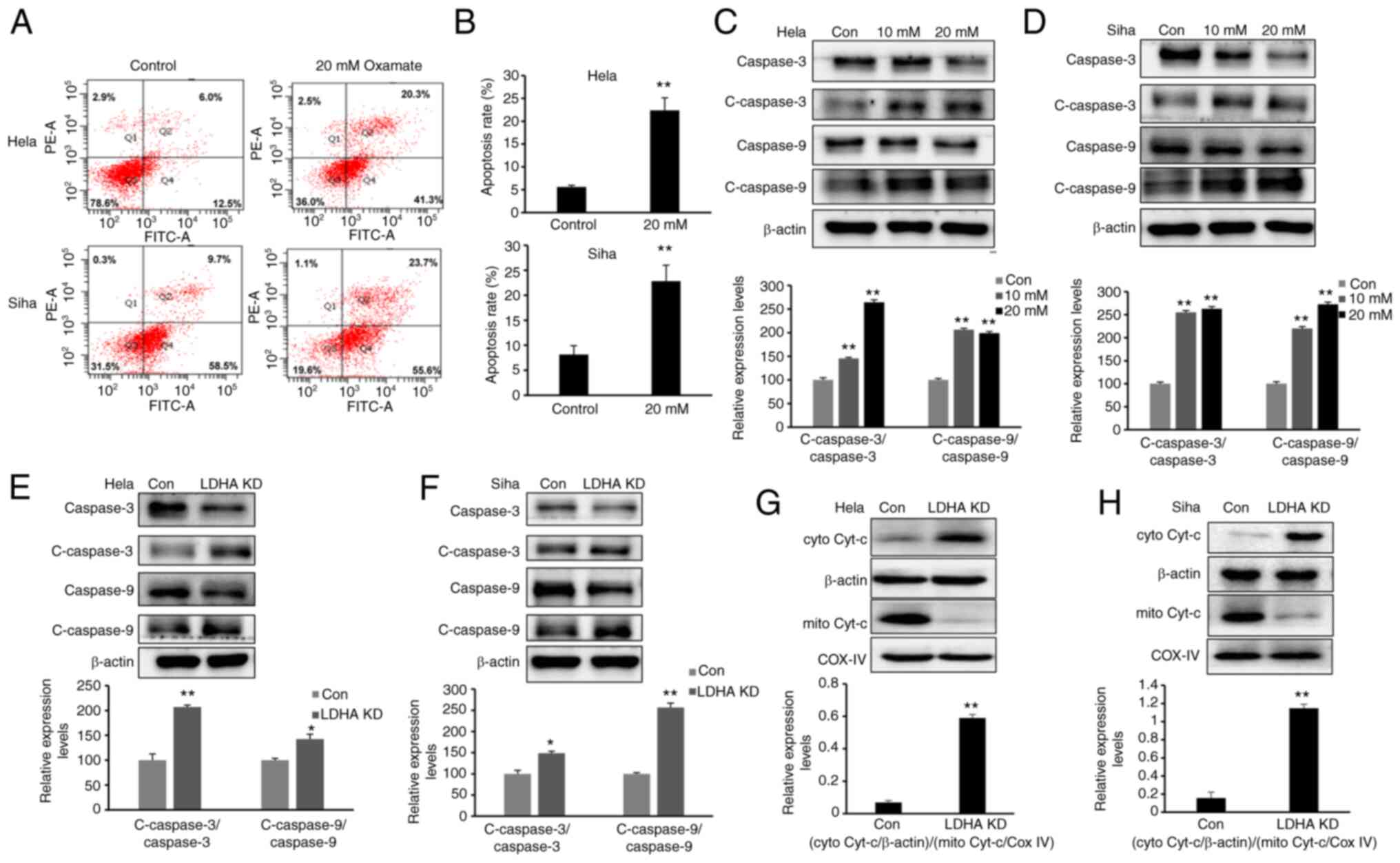 | Figure 6.LDHA inhibition induces mitochondrial
apoptosis. (A) Flow cytometric analysis was performed to detect
apoptosis of HeLa and SiHa cells following treatment with oxamate
for 48 h. (B) Apoptosis rates of HeLa and SiHa cells were analyzed
in each group. Western blot analysis was used to detect expression
levels of c-caspase-3, caspase-3, c-caspase-9 and caspase-9 in (C)
HeLa and (D) SiHa cells following treatment with oxamate for 48 h.
Western blot analysis was used to detect expression of c-caspase-3,
caspase-3, c-caspase-9 and caspase-9 in LDHA KD (E) HeLa and (F)
SiHa cells. Enhanced Cyt-c release in LDHA KD (G) HeLa and (H) SiHa
cells. Western blot analysis was used to detect levels of Cyt-c in
the cytoplasm and mitochondria. *P<0.05, **P<0.01 vs. con.
Con, control; KD, knockdown; c-, cleaved; cyto, cytosolic; mito,
mitochondrial; Cyt-c, cytochrome c; LDHA, lactate
dehydrogenase A chain; COX IV, cytochrome c oxidase IV. |
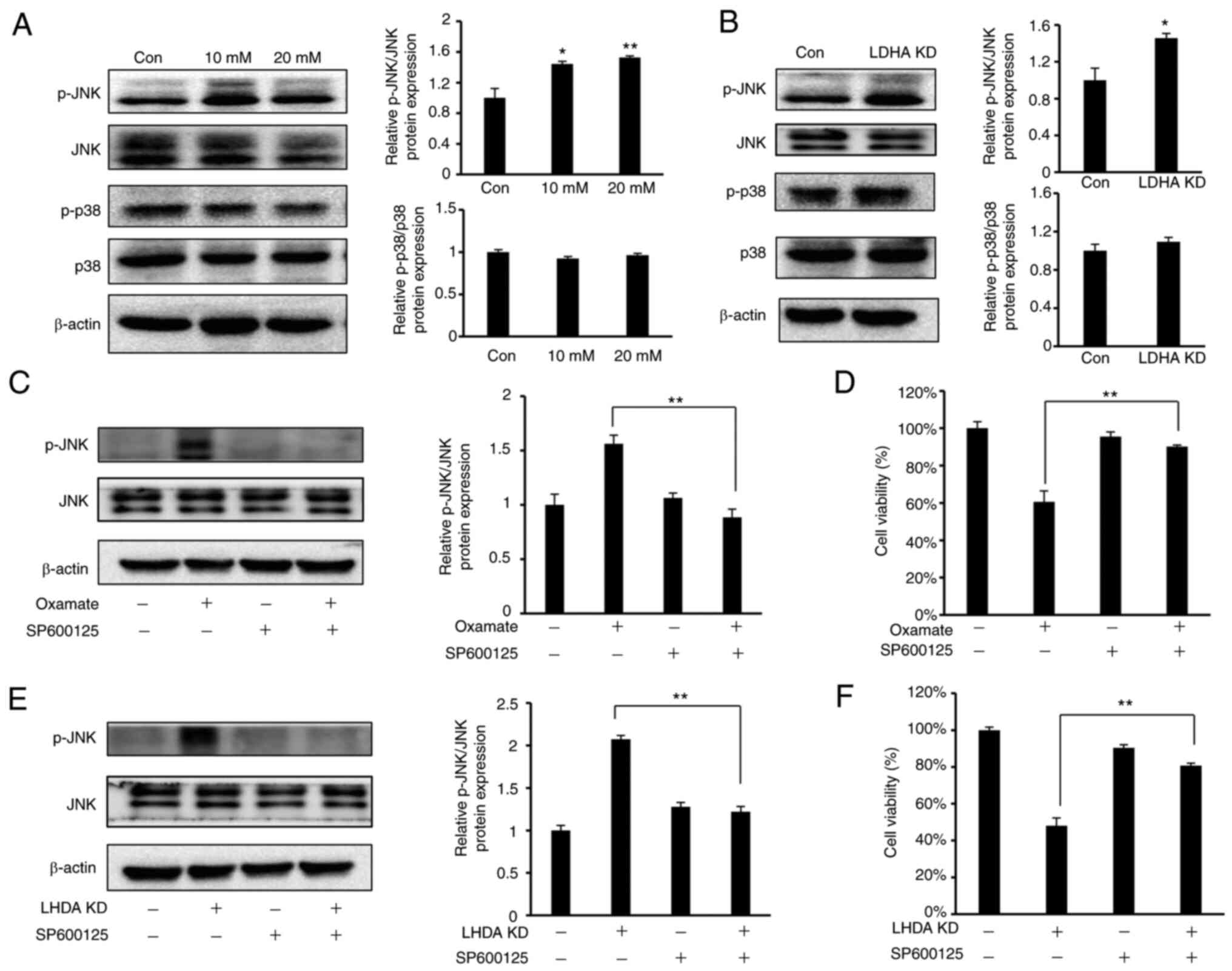
LDHA inhibition induces cell cycle
arrest and apoptosis via the JNK pathway
Multiple studies have indicated that
mitogen-activated protein kinases (MAPKs), such as p38 and JNK,
pathways are associated with cell proliferation, apoptosis and cell
cycle progression (40,41). As HeLa cells are the most commonly
used cell model for cervical cancer, the present study investigated
whether p38 or JNK pathways were involved in LDHA
inhibition-mediated cell cycle arrest and apoptosis in HeLa cells.
Western blot analysis (Fig. 7A)
showed a significant increase in JNK phosphorylation in HeLa cells
following treatment with oxamate, while there was no notable change
in phosphorylation level of p38. The same results were observed
following knockdown of LDHA (Fig.
7B).
To verify whether JNK signaling was required for
inhibition of cell proliferation following LDHA knockdown or
oxamate treatment, SP600125, a JNK inhibitor (42), was used to suppress JNK signaling
in HeLa cells. SP600125 led to a significant decrease in JNK
phosphorylation induced by oxamate treatment and LDHA knockdown
(Fig. 7C and E). The present study
investigated the regulatory effect of SP600125 on cell viability.
SP600125 treatment significantly reversed the inhibitory effect on
cell viability caused by LDHA knockdown and oxamate treatment
(Fig. 7D and F). These findings
indicated that the JNK pathway was involved in regulating apoptosis
and cell cycle arrest induced by LDHA inhibition in HeLa cells.
Discussion
Increasing evidence indicates that aerobic
glycolysis (Warburg effect) serves a key role in regulating
proliferation and cell cycle changes, especially in cancer cells
(43,44). Given the key role of LDHA in
aerobic glycolysis, the present study investigated the effect of
LDHA inhibition on cell cycle progression, proliferation and
apoptosis of cervical cancer cells and the underlying mechanism.
LDHA knockdown resulted in increased apoptosis and cell cycle
arrest and decreased in lactate production and glucose uptake,
which revealed an oncogenic role of LDHA in cervical cancer and
suggested that LDHA may be a potential therapeutic target against
cervical cancer.
Normal cells generate energy via mitochondrial
oxidative phosphorylation, while cancer cells primarily utilize
glucose via glycolysis to produce energy, even under normoxic
conditions; this phenomenon is known as the Warburg effect
(6). The Warburg effect is one of
the primary hallmarks of cancer cells. LDHA is a major subunit of
LDH, a key enzyme involved in the Warburg effect (45). Previous studies revealed that LDHA
expression was elevated in in most types of cancer cell (46,47).
Here, TCGA datasets showed that LDHA expression was upregulated in
most types of tumor and higher LDHA gene expression was found in
CESC compared with normal cervical tissue. Furthermore, LDHA was
associated with poor prognosis in patients with cervical cancer.
MPC1 is a key enzyme in mitochondrial pyruvate transport during
oxidative phosphorylation (48).
In certain types of cancer, MPC1 deficiency or inactivation
accelerates aerobic glycolysis and malignant progression (49,50).
LDHA levels were negatively correlated with MPC1, indicating that
cervical cancer was aerobic glycolysis-dependent. The present study
suggested that LDHA may be a therapeutical target for cervical
cancer. The present study investigated the effect of LDHA
inhibition by oxamate or knockdown LDHA by shRNA in cervical
cancer.
Given that LDHA serves an important role in Warburg
effect, which provides a constant supply of metabolites necessary
for cell proliferation and division, it was hypothesized that
inhibition of LDHA regulates aerobic glycolysis and impairs cell
proliferation in cervical cancer. Consistent with this hypothesis,
knocking down expression of LDHA significantly suppressed
viability, clone formation capacity and proliferation of HeLa and
SiHa cells and decreased lactate production and glucose uptake. The
same results were obtained following inhibition of LDHA activity by
oxamate both in HeLa and SiHa cells. Notably, 10 mM oxamate
significantly decreased glucose uptake and ATP production; however
the significant effect of 10 mM oxamate on cell viability was less
pronounced. This may be because abnormal metabolism is only one
factor affecting proliferation of tumor cells; other factors, such
as oncogene activation, genomic instability and inflammatory
signals, also suppress apoptosis and promote cell survival
(51). In addition, when aerobic
glycolysis of tumor cells is inhibited, cells may increase use of
other energy sources, such as glutamine, to compensate and prevent
inhibition of proliferation (52).
Both MTT and colony formation assay were used to test the effect of
LDHA inhibition on cell proliferation; the inhibitory effect of
LDHA knockdown detected by colony formation assay was greater than
that detected by MTT assay. This may be because there were only
1,000 cells/well in colony formation assay, there was insufficient
cell-cell communication to protect against damage caused by oxamate
or LDHA knockdown. Moreover, cells were exposed for longer period
(~1 week) compared with MTT assay.
Cyclins and CDKs are involved in regulating
different phases of the cell cycle (53). Here, knockdown or inhibition of
LDHA by oxamate induced G2/M cell cycle arrest,
decreased expression levels of cyclin B1, cyclin A and CDK1 and
increased expression levels of p21 in cervical cancer cells. The
present results revealed that G2/M cell cycle arrest
induced by inhibition of LDH was regulated by p21-mediated
decreased activity of the CDK1/cyclin B1 kinase complex. A previous
study also supports the hypothesis that decreased CDK1/cyclin B1
kinase complex activity triggers G2/M cell cycle arrest
(54).
When LDHA is inhibited, increased levels of
pyruvates enter the tricarboxylic acid cycle and mitochondrial
oxidative phosphorylation pathway. The present finding that
expression of LDHA was negatively correlated with MPC1, a key
enzyme involved in oxidative phosphorylation in cervical cancer,
supported this. However, in certain types of cancer cell, such as
human gastric cancer SC-M1 and hepatocellular carcinoma HepG2
cells, mitochondria are dysfunctional and oxidative phosphorylation
is impaired (55,56). Oxidative phosphorylation is
abnormally activated and more reactive oxygen species (ROS) are
generated, resulting in mitochondrial apoptosis (57). In the present study, caspase-3 and
caspase-9 activation was increased in CESC cells following
inhibition of LDHA. In mitochondrial-dependent apoptosis, apoptotic
proteins are activated by release of cytochrome c (58). Here, cytochrome c was
released from mitochondria to the cytoplasm following LDHA
knockdown. Whether caspase-3 and caspase-9 activation and
cytochrome c release induced by LDHA inhibition were due to
increased levels of ROS requires confirmation. The present data
indicated that LDH inhibition induced intrinsic mitochondrial
apoptosis in cervical cancer cells.
LDHA inhibition results in cell cycle arrest and
apoptosis. LDH inhibition by oxamate induced G2/M cell
cycle arrest via downregulation of the CDK1/Cyclin B1 pathway and
promoted apoptosis via enhancement of mitochondrial ROS generation
in nasopharyngeal carcinoma cancer cells (22). Le et al (24) reported that LDHA inhibition
resulted in decreased levels of ATP and ROS burst in lymphoma
cancer cells, which led to apoptosis and G2/M arrest.
Apoptosis and G2/M arrest were induced by LDHA
inhibition via oxamate in non-small cell lung cancer (NSCLC) H1395
cells (37). Moreover, LDHA
inhibition induced G0/G1 arrest and autophagy
in NSCLC A549 cells (37). Other
studies have found that LDHA inhibition suppressed migration and
increases chemo- and radiosensitivity in cancer cells (19,22,26,27).
Consistent with these aforementioned reports, the present results
showed that inhibition of LDHA induced G2/M cell cycle
arrest and activated the mitochondrial apoptosis pathway in CESC
cells. The MAPK pathway is a key pathway in regulating cell
survival and proliferation (59).
In the present study, JNK phosphorylation increased in HeLa cells
following knockdown or inhibition of LDHA by oxamate. JNK inhibitor
SP600125 effectively reversed cell death induced by LDH inhibition,
indicating that JNK activation was required for LDHA
inhibition-induced G2/M cell cycle arrest and apoptosis
in cervical cancer cells.
To the best of our knowledge, the present study is
the first to identify the role of LDHA in metabolism, viability,
proliferation and apoptosis of cervical cancer cells. The present
findings support the hypothesis that inhibition of LDHA may be an
effective therapeutic strategy for regulating metabolism and tumor
growth in cervical cancer. Future studies should investigate other
key enzymes involved in the Warburg effect in cervical cancer,
which may have similar effects as LDHA, and develop novel small
molecule compounds specifically targeting LDHA.
Acknowledgements
Not applicable.
Funding
The present study was supported by Youth Foundation of The First
Affiliated Hospital of Zhengzhou University (grant no. 71251).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LM conceived the study and designed experiments. WZ
performed experiments, analyzed data and wrote the manuscript. CW
and YL performed experiments and analyzed data. XH and CD designed
the experiments and wrote and revised the manuscript. WZ and CW
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Petrelli F, Ghidini A, Pedersini R,
Cabiddu M, Borgonovo K, Parati MC, Ghilardi M, Amoroso V, Berruti A
and Barni S: Comparative efficacy of palbociclib, ribociclib and
abemaciclib for ER+ metastatic breast cancer: An
adjusted indirect analysis of randomized controlled trials. Breast
Cancer Res Treat. 174:597–604. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Cicco P, Catani MV, Gasperi V, Sibilano
M, Quaglietta M and Savini I: Nutrition and breast cancer: A
literature review on prevention, treatment and recurrence.
Nutrients. 11:15142019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen Q, Cao HZ and Zheng PS: LGR5 promotes
the proliferation and tumor formation of cervical cancer cells
through the Wnt/β-catenin signaling pathway. Oncotarget.
5:9092–9105. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li XQ, Bai YL, Zhang DL, Jiao HS and He
RX: Euphornin reduces proliferation of human cervical
adenocarcinoma HeLa cells through induction of apoptosis and G2/M
cell cycle arrest. Onco Targets Ther. 11:4395–4405. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zu XL and Guppy M: Cancer metabolism:
Facts, fantasy, and fiction. Biochem Biophys Res Commun.
313:459–465. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pavlova NN and Thompson CB: The emerging
hallmarks of cancer metabolism. Cell Metab. 23:27–47. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Potter M, Newport E and Morten KJ: The
Warburg effect: 80 Years on. Biochem Soc Trans. 44:1499–1505. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nicholas JA, Electricwala B, Lee LK and
Johnson KM: Burden of relapsing-remitting multiple sclerosis on
workers in the US: A cross-sectional analysis of survey data. BMC
Neurol. 19:2582019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Moreno-Sánchez R, Rodríguez-Enríquez S,
Marín-Hernández A and Saavedra E: Energy metabolism in tumor cells.
FEBS J. 274:1393–1418. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu W, Yu X, Zhou L, Li J, Li M, Li W and
Gao F: Sinomenine inhibits non-small cell lung cancer via
downregulation of hexokinases II-mediated aerobic glycolysis. Onco
Targets Ther. 13:3209–3221. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tamgüney T, Zhang C, Fiedler D, Shokat K
and Stokoe D: Analysis of 3-phosphoinositide-dependent kinase-1
signaling and function in ES cells. Exp Cell Res. 314:2299–2312.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie H, Valera VA, Merino MJ, Amato AM,
Signoretti S, Linehan WM, Sukhatme VP and Seth P: LDH-A inhibition,
a therapeutic strategy for treatment of hereditary leiomyomatosis
and renal cell cancer. Mol Cancer Ther. 8:626–635. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pelicano H, Martin DS, Xu RH and Huang P:
Glycolysis inhibition for anticancer treatment. Oncogene.
25:4633–4646. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Choi YJ, Jeon JH and Oh JW: Critical
combination of initial markers for predicting refractory Mycoplasma
pneumoniae pneumonia in children: A case control study. Respir Res.
20:1932019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Drent M, Cobben NA, Henderson RF, Wouters
EF and van Dieijen-Visser M: Usefulness of lactate dehydrogenase
and its isoenzymes as indicators of lung damage or inflammation.
Eur Respir J. 9:1736–1742. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang W, Zhou F, Li N, Li Q and Wang L:
FOXM1-LDHA signaling promoted gastric cancer glycolytic phenotype
and progression. Int J Clin Exp Pathol. 8:6756–6763.
2015.PubMed/NCBI
|
|
19
|
Maftouh M, Avan A, Sciarrillo R, Granchi
C, Leon LG, Rani R, Funel N, Smid K, Honeywell R, Boggi U, et al:
Synergistic interaction of novel lactate dehydrogenase inhibitors
with gemcitabine against pancreatic cancer cells in hypoxia. Br J
Cancer. 110:172–182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi M, Cui J, Du J, Wei D, Jia Z, Zhang J,
Zhu Z, Gao Y and Xie K: A novel KLF4/LDHA signaling pathway
regulates aerobic glycolysis in and progression of pancreatic
cancer. Clin Cancer Res. 20:4370–4380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sheng SL, Liu JJ, Dai YH, Sun XG, Xiong XP
and Huang G: Knockdown of lactate dehydrogenase A suppresses tumor
growth and metastasis of human hepatocellular carcinoma. FEBS J.
279:3898–3910. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhai X, Yang Y, Wan J, Zhu R and Wu Y:
Inhibition of LDH-A by oxamate induces G2/M arrest, apoptosis and
increases radiosensitivity in nasopharyngeal carcinoma cells. Oncol
Rep. 30:2983–2991. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao YH, Zhou M, Liu H, Ding Y, Khong HT,
Yu D, Fodstad O and Tan M: Upregulation of lactate dehydrogenase A
by ErbB2 through heat shock factor 1 promotes breast cancer cell
glycolysis and growth. Oncogene. 28:3689–3701. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Le A, Cooper CR, Gouw AM, Dinavahi R,
Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL and Dang
CV: Inhibition of lactate dehydrogenase A induces oxidative stress
and inhibits tumor progression. Proc Natl Acad Sci USA.
107:2037–2042. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang ZY, Loo TY, Shen JG, Wang N, Wang DM,
Yang DP, Mo SL, Guan XY and Chen JP: LDH-A silencing suppresses
breast cancer tumorigenicity through induction of oxidative stress
mediated mitochondrial pathway apoptosis. Breast Cancer Res Treat.
131:791–800. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou M, Zhao Y, Ding Y, Liu H, Liu Z,
Fodstad O, Riker AI, Kamarajugadda S, Lu J, Owen LB, et al: Warburg
effect in chemosensitivity: Targeting lactate dehydrogenase-A
re-sensitizes taxol-resistant cancer cells to taxol. Mol Cancer.
9:332010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao Y, Liu H, Liu Z, Ding Y, Ledoux SP,
Wilson GL, Voellmy R, Lin Y, Lin W, Nahta R, et al: Overcoming
trastuzumab resistance in breast cancer by targeting dysregulated
glucose metabolism. Cancer Res. 71:4585–4597. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Novoa WB, Winer AD, Glaid AJ and Schwert
GW: Lactic dehydrogenase. V. Inhibition by oxamate and by oxalate.
J Biol Chem. 234:1143–1148. 1959. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ramanathan A, Wang C and Schreiber SL:
Perturbational profiling of a cell-line model of tumorigenesis by
using metabolic measurements. Proc Natl Acad Sci USA.
102:5992–5997. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dong Y, Stewart T, Bai L, Li X, Xu T,
Iliff J, Shi M, Zheng D, Yuan L, Wei T, et al: Coniferaldehyde
attenuates Alzheimer's pathology via activation of Nrf2 and its
targets. Theranostics. 10:179–200. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang W, Gong J, Ding L, Zhang Z, Pan X,
Chen X, Guo W, Zhang X, Yang X, Peng G, et al: Functional
validation of a human GLUD2 variant in a murine model of
Parkinson's disease. Cell Death Dis. 11:8972020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma WQ, Sun XJ, Zhu Y and Liu NF: PDK4
promotes vascular calcification by interfering with autophagic
activity and metabolic reprogramming. Cell Death Dis. 11:9912020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang WJ, Song ZB, Bao YL, Li WL, Yang XG,
Wang Q, Yu CL, Sun LG, Huang YX and Li YX: Periplogenin induces
necroptotic cell death through oxidative stress in HaCaT cells and
ameliorates skin lesions in the TPA- and IMQ-induced psoriasis-like
mouse models. Biochem Pharmacol. 105:66–79. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tan WX, Xu TM, Zhou ZL, Lv XJ, Liu J,
Zhang WJ and Cui MH: TRP14 promotes resistance to cisplatin by
inducing autophagy in ovarian cancer. Oncol Rep. 42:1343–1354.
2019.PubMed/NCBI
|
|
36
|
An J, Zhang Y, He J, Zang Z, Zhou Z, Pei
X, Zheng X, Zhang W, Yang H and Li S: Lactate dehydrogenase A
promotes the invasion and proliferation of pituitary adenoma. Sci
Rep. 7:47342017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang Y, Su D, Zhao L, Zhang D, Xu J, Wan
J, Fan S and Chen M: Different effects of LDH-A inhibition by
oxamate in non-small cell lung cancer cells. Oncotarget.
5:11886–11896. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dayton TL, Jacks T and Vander Heiden MG:
PKM2, cancer metabolism, and the road ahead. EMBO Rep.
17:1721–1730. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Malsy M, Gebhardt K, Gruber M, Wiese C,
Graf B and Bundscherer A: Effects of ketamine, s-ketamine, and MK
801 on proliferation, apoptosis, and necrosis in pancreatic cancer
cells. BMC Anesthesiol. 15:1112015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yim NH, Kim A, Liang C, Cho WK and Ma JY:
Guibitang, a traditional herbal medicine, induces apoptotic death
in A431 cells by regulating the activities of mitogen-activated
protein kinases. BMC Complement Altern Med. 14:3442014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xie P, Horio F, Fujii I, Zhao J, Shinohara
M and Matsukura M: A novel polysaccharide derived from algae
extract inhibits cancer progression via JNK, not via the p38 MAPK
signaling pathway. Int J Oncol. 52:1380–1390. 2018.PubMed/NCBI
|
|
42
|
Hao D, Li Y, Shi J and Jiang J: Baicalin
alleviates chronic obstructive pulmonary disease through regulation
of HSP72-mediated JNK pathway. Mol Med. 27:532021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Netea-Maier RT, Smit JWA and Netea MG:
Metabolic changes in tumor cells and tumor-associated macrophages:
A mutual relationship. Cancer Lett. 413:102–109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lu Y, Cheng J, Cai W, Zhuo H, Wu G and Cai
J: Inhibition of circRNA circVPS33B reduces warburg effect and
tumor growth through regulating the miR-873-5p/HNRNPK axis in
infiltrative gastric cancer. Onco Targets Ther. 14:3095–3108. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ždralević M, Brand A, Di Ianni L, Dettmer
K, Reinders J, Singer K, Peter K, Schnell A, Bruss C, Decking SM,
et al: Double genetic disruption of lactate dehydrogenases A and B
is required to ablate the ‘Warburg effect’ restricting tumor growth
to oxidative metabolism. J Biol Chem. 293:15947–15961. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Miao P, Sheng S, Sun X, Liu J and Huang G:
Lactate dehydrogenase A in cancer: A promising target for diagnosis
and therapy. IUBMB Life. 65:904–910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lu QY, Zhang L, Yee JK, Go VW and Lee WN:
Metabolic consequences of LDHA inhibition by epigallocatechin
gallate and oxamate in MIA PaCa-2 pancreatic cancer cells.
Metabolomics. 11:71–80. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
McCommis KS, Hodges WT, Bricker DK,
Wisidagama DR, Compan V, Remedi MS, Thummel CS and Finck BN: An
ancestral role for the mitochondrial pyruvate carrier in
glucose-stimulated insulin secretion. Mol Metab. 5:602–614. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li X, Han G, Li X, Kan Q, Fan Z, Li Y, Ji
Y, Zhao J, Zhang M, Grigalavicius M, et al: Mitochondrial pyruvate
carrier function determines cell stemness and metabolic
reprogramming in cancer cells. Oncotarget. 8:46363–46380. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li Y, Li X, Kan Q, Zhang M, Li X, Xu R,
Wang J, Yu D, Goscinski MA, Wen JG, et al: Mitochondrial pyruvate
carrier function is negatively linked to Warburg phenotype in vitro
and malignant features in esophageal squamous cell carcinomas.
Oncotarget. 8:1058–1073. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Nossol C, Landgraf P, Kahlert S, Oster M,
Isermann B, Dieterich DC, Wimmers K, Dänicke S and Rothkötter HJ:
Deoxynivalenol affects cell metabolism and increases protein
biosynthesis in intestinal porcine epithelial cells (IPEC-J2): DON
increases protein biosynthesis. Toxins (Basel). 10:4642018.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pal HC and Katiyar SK: Cryptolepine, a
plant alkaloid, inhibits the growth of non-melanoma skin cancer
cells through inhibition of topoisomerase and induction of DNA
damage. Molecules. 21:17582016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lindqvist A, van Zon W, Karlsson Rosenthal
C and Wolthuis RM: Cyclin B1-Cdk1 activation continues after
centrosome separation to control mitotic progression. PLoS Biol.
5:e1232007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hung WY, Huang KH, Wu CW, Chi CW, Kao HL,
Li AF, Yin PH and Lee HC: Mitochondrial dysfunction promotes cell
migration via reactive oxygen species-enhanced β5-integrin
expression in human gastric cancer SC-M1 cells. Biochim Biophys
Acta. 1820:1102–1110. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Guo Y, Zhang W, Yan YY, Ma CG, Wang X,
Wang C and Zhao JL: Triterpenoid pristimerin induced HepG2 cells
apoptosis through ROS-mediated mitochondrial dysfunction. J BUON.
18:477–485. 2013.PubMed/NCBI
|
|
57
|
Bartz RR, Suliman HB and Piantadosi CA:
Redox mechanisms of cardiomyocyte mitochondrial protection. Front
Physiol. 6:2912015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jiang Q, Liu G, Wang X, Hou Y, Duan Y, Wu
G, Yin Y and Yao K: Mitochondrial pathway is involved in the
protective effects of alpha-ketoglutarate on hydrogen peroxide
induced damage to intestinal cells. Oncotarget. 8:74820–74835.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ades F and Metzger-Filho O: Targeting the
cellular signaling: BRAF inhibition and beyond for the treatment of
metastatic malignant melanoma. Dermatol Res Pract. 2012:2591702012.
View Article : Google Scholar : PubMed/NCBI
|