Introduction
Lung cancer is a frequent malignancy and a leading
cause of cancer-related deaths worldwide (1,2). As
a common type of lung cancer, lung adenocarcinoma (LUAD) accounts
for approximately 50% of primary lung cancer cases (3,4). The
prevalence of LUAD has been markedly increased in recent years,
especially in women, which may be associated with smoking (4). At present, the treatment of LUAD
mainly includes radical resection, radiotherapy, chemotherapy,
molecular targeted therapy and immunotherapy (4). In particular, molecular targeted
therapy and immunotherapy are important treatment methods for LUAD
patients, which significantly prolong the survival time and improve
the quality of life. However, due to tumor invasion and distant
metastasis, the 5-year survival rate of LUAD remains <20%
(1,5,6).
Thus, LUAD is a great threat to global health. Hence, it is
essential to investigate the underlying mechanism of LUAD
progression and search for new therapeutic strategies.
Long non-coding RNAs (lncRNAs) are a category of
non-coding RNAs (ncRNA) with a length normally longer than 200
nucleotides (7). Although lncRNAs
cannot be translated into proteins, they effectively regulate gene
expression by acting as competing endogenous RNAs (ceRNAs) that
potently sponge microRNAs (miRNAs) (8,9).
Accumulating evidence has revealed that lncRNAs play multiple roles
in developing numerous diseases, especially in cancers (10–13).
For example, lncRNA RP11-757G1.5 has been revealed to increase the
proliferation and metastatic ability of colorectal cancer via
sponging miR-139-5p that subsequently increases the expression of
YAP1 (14). LINC00665 has been
demonstrated to facilitate breast cancer development by regulating
the expression of LIN28B via sponging miR-379-5p (15).
In LUAD, some lncRNAs play vital roles in cancer
progression (16–18). For example, lncRNA ZFPM2-AS1 was
demonstrated to promote the proliferation ability of LUAD via the
miR-18b-5p/VMA21 axis (16).
LncRNA LINC00511 accelerated the progression of LUAD through
modulation of the expression level of PKM2 by sponging miR-625-5p
(17). To provide improved
explication of the regulatory role of lncRNAs in LUAD, the
repertoire of genes dysregulated in LUAD from The Cancer Genome
Atlas (TCGA) dataset were first analyzed and it was revealed that
RP11-805J14.5 expression was significantly increased in LUAD tumor
tissues. In the present study, it was revealed that RP11-805J14.5
could sponge miR-34b-3p and miR-139-5p, thereby regulating CCND2
expression. Interestingly, in bladder cancer, miR-34b-3p could
directly bind to CCND2 and P2RY1 mRNAs to regulate their expression
and mitigate the chemotherapy resistance of bladder cancer cells
(19). However, the role of
RP11-805J14.5 in LUAD progression has not been reported. Therefore,
the present study aimed to investigate the effect of RP11-805J14.5
on LUAD progression and elucidate the underlying mechanism.
Materials and methods
Patient tissues and cell culture
Tumor tissues and adjacent normal tissues were
collected from 60 LUAD patients (age range, 41–73 years; mean age,
58±2.2 years; male/female ratio, 6:7) who underwent surgical
resection between August 2016 and June 2020 at Hwa Mei Hospital
(Ningbo, China). The inclusion criteria was as follows: None of the
patients underwent preoperative chemoradiotherapy and anticancer
treatments before surgery. All the cases were confirmed by
histological detection. All LUAD patients signed informed consent.
The present study was approved by The Ethics Committee of Hwa Mei
Hospital (approval no. IR-B-2021-3-18). Human lung epithelial cell
line BEAS-2B (CRL-9609), large cell lung carcinoma (LCLC) cell line
H460 (HTB-177), LUAD cell lines H1650 (CRL-5883), A549 (CCL-185)
and H1975 (HTB-183), and LCLC cell line H1299 (CRL-5826) were
purchased from American Type Culture Collection (ATCC). BEAS-2B
cells were cultured in Dulbecco's modified Eagle's medium (DMEM)
with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100
µg/ml streptomycin (all from Gibco; Thermo Fisher Scientific, Inc.)
at 37°C in an atmosphere containing 5% CO2. H460, H1650,
A549, H1975 and H1299 cells were maintained in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS, 100 U/ml
penicillin, and 100 µg/ml streptomycin at 37°C, in an atmosphere
containing 5% CO2. The cell lines were cultured for 6–8
passages for gene expression analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from tissues and cell lines were extracted
using TRIzol reagent (Thermo Fisher Scientific, Inc.) and reversely
transcribed to cDNA by High-Capacity cDNA Reverse Transcription Kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. SYBR Green Master Mix (Thermo Fisher Scientific,
Inc.) was employed to conduct the RT-qPCR assay. The reaction
mixture was incubated at 95°C for 2 min. The thermocycling program
consisted of holding at 94°C for 2 min, followed by 30 cycles of 30
sec at 94°C, 30 sec at 56°C, and 60 sec at 72°C. The primers of
RP11-805J14.5, miR-34b-3p, miR-139-5p and CCND2 were as follows:
RP11-805J14.5 forward, 5′-GAGTAGATGAGAGCCGGCAG-3′ and reverse,
5′-GGGGATCCCAGGACTCTTCT-3′; miR-34b-3p forward,
5′-TCTATTTGCCATCGTCTA-3′ and reverse, 5′-CAGGCAGCTCATTTGGAC-3′
(20); miR-139-5p forward,
5′-GCCTCTACAGTGCACGTGTCTC-3′ and reverse,
5′-CGCTGTTCTCATCTGTCTCGC-3′ (21);
CCND2 forward, 5′-TACACCGACAACTCCATCAAGC-3′ and reverse,
5′-GCCAGGTTCCACTTCAACTTC-3′ (19).
β-actin was used as an internal reference of RP11-805J14.5 and
CCND2, and the primers were as follows: Forward,
5′-CCTGTACGCCAACACAGTGC-3′ and reverse, 5′-ATACTCCTGCTTGCTGATCC-3′.
In addition, U6 was used as an internal reference of miR-34b-3p and
miR-139-5p. The primers for U6 were: Forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
The analysis of the relative expression levels was conducted using
the 2−ΔΔCq method (22).
Cell transfection
The siRNAs targeting RP11-805J14.5, miR-34b-3p mimic
and miR-139-5p mimic as well as miR-34b-3p inhibitor, miR-139-5p
inhibitor were separately designed by Shanghai GeneChem Co., Ltd.
The open reading frame of CCND2 was inserted into the expression
vector pcDNA 3.1(+) (Sigma-Aldrich; Merck KGaA) to overexpress
CCND2, which was also constructed by Shanghai GenePharma Co., Ltd.
An empty vector [(pcDNA3.1(+)] was used as a negative control for
CCND2 overexpression. A total of 20 nM of each construct was
transfected into cells at 37°C for 15 min using Lipofectamine 3000
(Thermo Fisher Scientific, Inc.) following the manufacturer's
protocol. After 48 h, the transfected cells were harvested to
perform subsequent experiments. The sequences were as follows:
si-NC, 5′-TTCTCCGAACGTGTCACGT-3′; si-RNA-RP11-805J14.5#1,
5′-TTGTAGTATGGAAGTTGTAAAGC-3′; si-RNA-RP11-805J14.5#2,
5′-TAGTATGGAAGTTGTAAAGCTGT-3′; mimics-NC,
5′-TTCTCCGAACGTGTCACGT-3′; miR-34b-3p mimics,
5′-UACCGUCACCUCAAUCACUAAC-3′; miR-139-5p mimics,
5′-UGACCUCUGUGCACGUGACAUCU-3′; inhibitor-NC,
5′-UUCUCCGAACGUGUCACGUTT-3′; miR-34b-3p inhibitor,
5′-AUGGCAGUGGAGUUAGUGAUUG-3′; miR-139-5p inhibitor,
5′-ACUGGAGACACGUGCACUGUAGA-3′; and CCND2,
5′-AUCUGUUCAGUGUUAAGUGAUUU-3′.
Cell Counting Kit-8 (CCK-8) assay
Transfected LUAD cells were seeded into 96-well
plates at a density of 1×103 cells/well. Subsequently,
10 µl of CCK-8 (Dojindo Laboratories, Inc.) was mixed with cells in
each well at indicated time-points (0, 24, 48 and 72 h), and
incubated for 1 h at 37°C in an atmosphere containing 5%
CO2. The absorbance at 450 nm was recorded to analyze
cell viability.
Flow cytometry
Annexin V-FITC Apoptosis Detection Kit (cat. no.
331200; Thermo Fisher Scientific, Inc.) was utilized to analyze
cell apoptosis according to the manufacturer's instructions.
Briefly, transfected 2×105 LUAD cells were collected,
washed using PBS, and suspended in 500 µl binding buffer, followed
by treatment with 5 µl Annexin V-FITC and 3 µl propidium iodide
(PI) at room temperature for 15 min in darkness. The proportion of
apoptotic cells was determined by flow cytometry (FACSCalibur; BD
Biosciences). Cell apoptosis was determined using FlowJo 7.6.1
software (FlowJo LLC).
Transwell migration assay
Transfected LUAD cells (1×105) were
seeded into the upper Transwell chamber (8-µm pore size) and
cultured in the DMEM without FBS. The complete medium (DMEM
containing 10% FBS) was added to the lower chamber. Subsequently,
cells were incubated at 37°C for 24 h, the migrating cells were
fixed in 4% paraformaldehyde for 15 min at 37°C and subjected to
0.1% crystal violet staining for 30 min at room temperature. Five
areas were randomly selected to determine the number of migrating
cells under a light microscope (Nikon Corporation) at a
magnification of ×200.
Transwell invasion assay
Transfected LUAD cells (1×105) were
seeded into the upper Transwell chamber, which was precoated with
Matrigel for 24 h at 37°C, and cultured in DMEM without FBS. DMEM
containing 10% FBS was added to the lower chamber. After cells were
incubated at 37°C for 24 h, the invading cells were fixed in 4%
paraformaldehyde for 15 min at 37°C and subjected to 0.1% crystal
violet staining for 30 min at room temperature. Five areas were
randomly selected to determine the number of invading cells under a
light microscope (Nikon Corporation) at a magnification of
×200.
Dual-luciferase reporter assay
The wild-type (WT) and mutant (MUT) sequences of
RP11-805J14.5 targeted to miR-34b-3p and miR-139-5p, and the WT and
MUT sequences of the 3′-untranslated region (UTR) of CCND2 were
subcloned into the psiCHECK-2 vector (Promega Corporation)
respectively, followed by co-transfecting with miR-34b-3p mimic or
miR-139-5p mimic into lx106 293T cells (CRL-3216; ATCC)
using Lipofectamine 3000 (Thermo Fisher Scientific, Inc.). After 48
h, the transfected cells were harvested to record the relative
luciferase activity using the Dual-luciferase reporter assay system
(Promega Corporation) and normalized to Renilla luciferase
activity.
Nuclear-cytoplasmic fractionation
Cell fractionation buffer (Thermo Fisher Scientific,
Inc.) was used to isolate nuclear and cytoplasmic fractions. Then,
the extraction of cytoplasmic and nuclear RNAs was accomplished
using PARIS™ Kit (cat. no. AM1921; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions, and was followed by
RT-qPCR to assess RP11-805J14.5 expression.
RNA pull-down assay
Biotinylated-miR-34b-3p probes and
biotinylated-miR-139-5p were obtained from Guangzhou RiboBio Co.,
Ltd. The probes were incubated at 4°C for 2 h with 50 µl C-1
magnetic beads (Thermo Fisher Scientific, Inc.) to generate
probe-coated beads and then incubated at 4°C for 4 h with 0.7 ml
RIPA lysis buffer (Thermo Fisher Scientific, Inc.) to lyse LUAD
cells. After washing, RNA complexes were subjected to
centrifugation at 11,100 × g for 10 min and then eluted by
denaturation in 1X protein loading buffer for 10 min at 100°C.
Subsequently, RT-qPCR was used to determine the expression of
absorbed RP11-805J14.5.
Western blot analysis
LUAD cell lysates were obtained utilizing RIPA lysis
buffer (Sigma-Aldrich; Merck KGaA) and quantified using the BCA
method. Lysate samples (40 µg) were resolved in 12% SDS-PAGE and
transferred to a PVDF membrane. The membrane was blocked with 5%
skim milk for nonspecific binding for 2 h at 4°C. The membrane was
then probed with anti-CCND2 (1:1,000 dilution; cat. no. 3741)
antibody and anti-GAPDH (1:1,000 dilution; cat. no. 8884; both from
Cell Signaling Technology, Inc.) antibody at 4°C for 12 h.
Subsequently, the membrane was incubated with IgG H&L
(HRP-conjugated; 1:1,000; cat. no. 7076S; Cell Signaling
Technology, Inc.) for 2 h at 24°C. GAPDH was used as the internal
control protein. The protein bands were visualized using the ECL
Western Blotting Substrate (Thermo Fisher Scientific, Inc.).
Xenograft tumor establishment
A total of 12 male BALB/c nude mice aged 4 weeks
(weight, 15–20 g) were purchased from Charles River Laboratories,
Inc. and kept under standard laboratory conditions: temperature at
22°C, humidity 55% and a 12-h light/dark cycle. Food and water were
provided ad libitum. A549 cells that were stably transfected
with the siRNA targeting RP11-805J14.5 or negative control were
selected using puromycin at 0.5 mg/ml. To establish xenograft
tumors, stably transfected A549 cells (5×106) in 100 µl
PBS were subcutaneously injected into the right flank of the mice.
The tumor size was measured based on the formula (length ×
width2/2) every 7 days until 28 days post injection. The
humane endpoint of this experiment was set to the maximum size the
tumors (2,000 mm3) allowed to grow in the mice before
euthanasia. Both the research team and the veterinary staff
monitored animals twice daily. At the end of animal experiment, the
maximum percentage of tumor weight/mice weight observed was 5.9%.
The maximum tumor diameter observed in the present study was 17.2
mm. The duration of the experiment was 4 weeks. Subsequently, 12
mice were sacrificed under anesthesia induced by mask inhalation of
vaporized isoflurane (concentration was maintained at 1.5%) and
sacrificed by cervical dislocation. Sacrifice was confirmed by
cessation of breathing and heartbeat. Then the tumor volume and
RP11-805J14.5 expression were determined. The expression of
RP11-805J14.5 was determined using RT-qPCR. All experiments abided
by the Institutional Animal Care of Hwa Mei Hospital and were
approved by the Experimental Animal Ethics Committee of Hwa Mei
Hospital (approval no. IR-D-2021-5-25).
Bioinformatics analysis
The TCGA dataset of RP11-805J14.5 expression in LUAD
patients was acquired from the Gene Expression Profiling
Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) database. The target
miRNAs that bound RP11-805J14.5 were studied by screening analysis
using the LncBase v2.0 (http://carolina.imis.athena-innovation.gr/diana_tools/web/)
database. The target mRNAs that bound miRNA were studied by
screening TargetScan Release 7.2 (http://www.targetscan.org/vert_72/).
Statistical analysis
Data were presented as the mean ± standard deviation
(SD) and analyzed using SPSS version 22.0 (IBM Corp.). The
normality test was performed by Kolmogorov-Smirnov method. The
statistical difference between two groups was analyzed by unpaired
Student's t-test and paired Student's t-test was used for Figs. 1C and 3E analysis. Pearson's correlation
analysis was performed to determine the correlation between the
expression level of RP11-805J14.5 and miR-34b-3p or miR-139-5p.
Survival analysis was conducted using Kaplan-Meier analysis with
the log-rank testing. Group differences were compared by one-way
ANOVA with Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
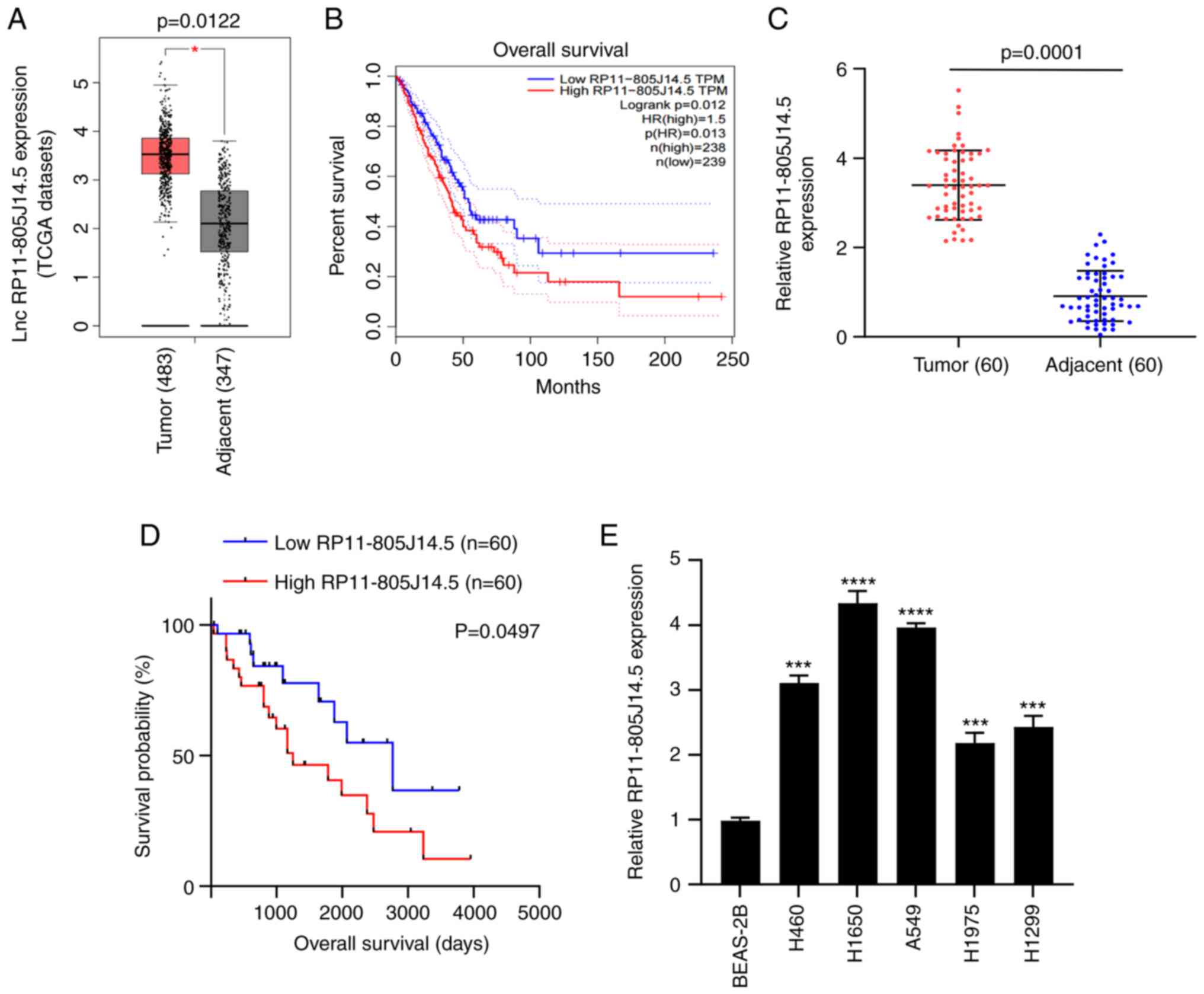
RP11-805J14.5 is highly expressed in
LUAD
To investigate the role of lncRNAs in LUAD
progression, the TCGA dataset was first analyzed using GEPIA
(23) which contains RNA
sequencing expression data of lung cancer samples of patients
worldwide. The results revealed that the expression of
RP11-805J14.5 was significantly elevated in LUAD tumor tissues
(P<0.05; Fig. 1A) and some
overlap in tumor and non-tumor tissues was revealed. The LUAD
patients with high expression of RP11-805J14.5 had poor survival
(P=0.013; Fig. 1B). Next, RT-qPCR
was used to verify the expression of RP11-805J14.5 in tumor tissues
and adjacent normal tissues from 60 LUAD patients. It was revealed
that RP11-805J14.5 expression was significantly increased in tumor
tissues (P=0.0001; Fig. 1C).
Furthermore, Kaplan-Meier analysis revealed that the high
expression of RP11-805J14.5 was associated with poor survival of
LUAD patients (P=0.0497; Fig. 1D).
Due to the amount of data, sample source and tumor heterogeneity,
the median survival time was also different, but this did not
affect the effect of RP11-805J14.5 expression on survival rate.
Subsequently, the RP11-805J14.5 expression in LUAD cells was
determined. Consistent with the expression in LUAD tissues,
RP11-805J14.5 expression was significantly increased in LUAD cell
lines H460, H1650, A549, H1975, and H1299 compared with BEAS-2B
cells (P<0.001; Fig. 1E).
Therefore, it was concluded that RP11-805J14.5 was highly expressed
in LUAD and was associated with poor overall survival of LUAD
patients.
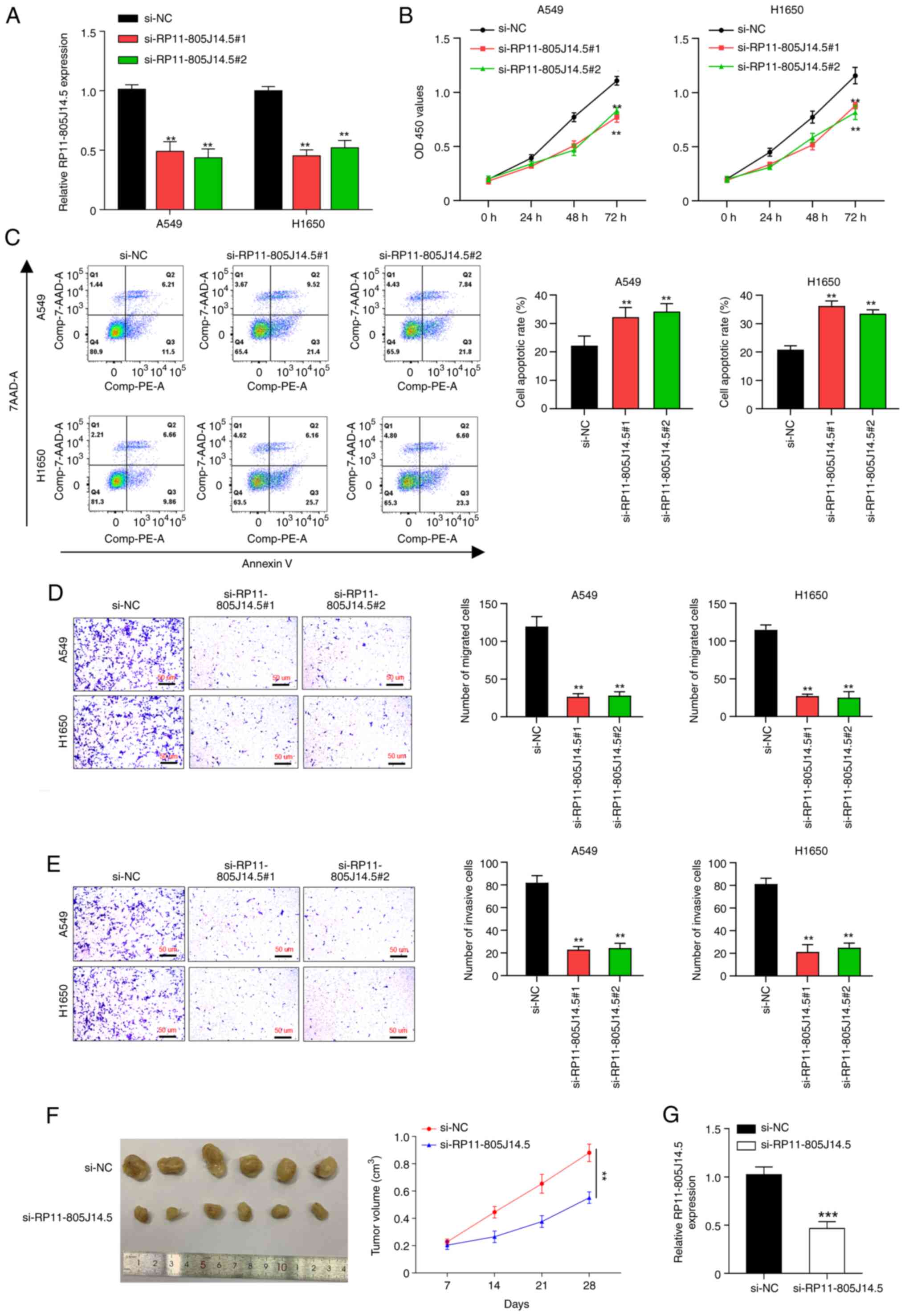
Knockdown of RP11-805J14.5 inhibits
LUAD cell growth, invasion and migration, as well as tumor
growth
To elucidate the function of RP11-805J14.5 in LUAD,
two siRNAs targeting RP11-805J14.5 were transfected into A549 and
H1650 cells that exhibited the highest expression of RP11-805J14.5.
RP11-805J14.5 was significantly downregulated by these two siRNAs
in A549 and H1650 cells (P<0.01; Fig. 2A). Then, functional assays revealed
that knockdown of RP11-805J14.5 suppressed cell viability
(P<0.01; Fig. 2B), induced cell
apoptosis (P<0.01; Fig. 2C) and
inhibited cell migration (P<0.01; Fig. 2D) and invasion abilities
(P<0.01; Fig. 2E). To study the
role of RP11-805J14.5 in tumor growth, A549 cells transfected with
siRNA targeting RP11-805J14.5 were injected into the mice to
establish a xenograft tumor model. Results revealed that
RP11-805J14.5 knockdown significantly inhibited tumor size
(P<0.01; Fig. 2F). Moreover,
the expression level of RP11-805J14.5 was significantly decreased
in mice tumor tissues (P<0.001; Fig. 2G). Thus, RP11-805J14.5 knockdown
inhibited the cell growth, invasion and migration of LUAD cells as
well as tumor growth.
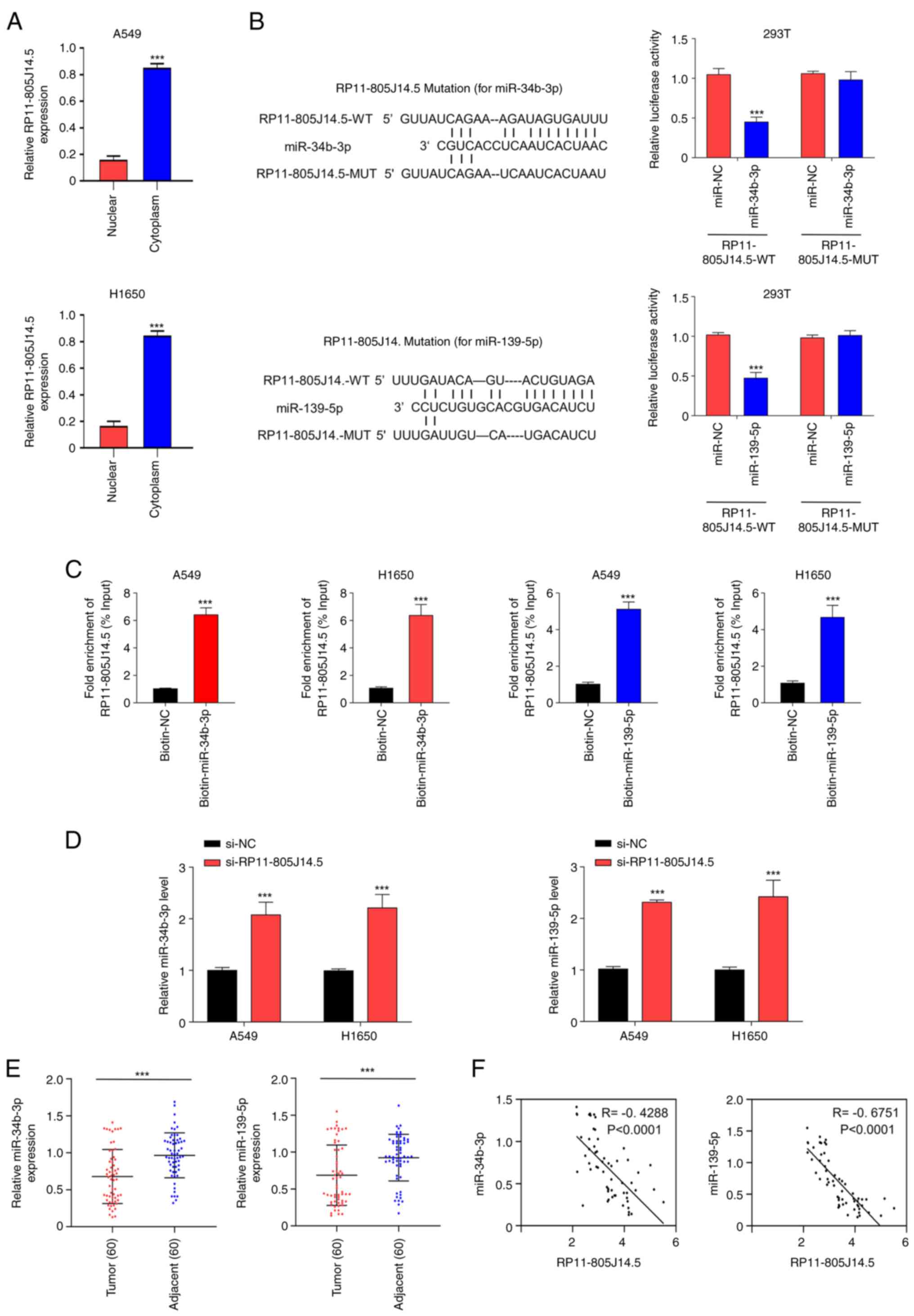
RP11-805J14.5 serves as miR-34b-3p and
miR-139-5p sponges in LUAD
To investigate the underlying mechanism of
RP11-805J14.5 in LUAD, the subcellular location of RP11-805J14.5 in
LUAD cells was identified. It was revealed that RP11-805J14.5 was
highly expressed in the cytoplasm compared with the nucleus in A549
and H1650 cells (P<0.001; Fig.
3A). Reasonably, the lncRNA was produced in the nucleus, and
subsequently transported to the cytoplasm to play its biological
function. Accumulating evidence has revealed that lncRNAs sponge
miRNAs and regulate miRNA expression (24,25).
Therefore, the target miRNAs that bound RP11-805J14.5 were studied.
The screening analysis using LncBase revealed that RP11-805J14.5
may bind to miR-34b-3p and miR-139-5p. The predicted binding motifs
of RP11-805J14.5, miR-34b-3p and miR-139-5p are presented in
Fig. 3B. To confirm the
prediction, luciferase and RNA pull-down assays were conducted.
miR-34b-3p mimics and miR-139-5p mimics significantly decreased the
activity of the WT of RP11-805J14.5 reporter while they did not
affect the activity of MUT (P<0.001; Fig. 3B). An RNA pull-down assay revealed
that biotinylated-miR-34b-3p probe and biotinylated-miR-139-5p
probe enriched more RP11-805J14.5 compared with the control probe
in A549 and H1650 cells (P<0.001; Fig. 3C). In addition, RP11-805J14.5
knockdown significantly increased the expression of miR-34b-3p and
miR-139-5p in A549 and H1650 cells (P<0.001; Fig. 3D). The expression of miR-34b-3p and
miR-139-5p was significantly decreased in tumor tissues compared
with that in adjacent normal tissues (P<0.001; Fig. 3E). Pearson's correlation analysis
revealed a negative correlation between the expression of
miR-34b-3p and RP11-805J14.5 in LUAD (P<0.0001; Fig. 3F). Moreover, a negative correlation
was also identified between the expression of miR-139-5p and
RP11-805J14.5 in LUAD (P<0.0001; Fig. 3F). Collectively, these results
indicated that RP11-805J14.5 served as a miR-34b-3p and miR-139-5p
sponge in LUAD. The expression of miR-34b-3p and miR-139-5p in A549
and H1650 cells transfected with miR-34b-3p and miR-139-5p mimics
or inhibitor was then detected (Fig.
S1A and B) using RT-qPCR. The results revealed that in cells
transfected with miR-34b-3p or miR-139-5p mimics, the level of
miR-34b-3p or miR-139-5p was significantly increased but
significantly decreased when transfected with miR-34b-3p or
miR-139-5p inhibitor, respectively.
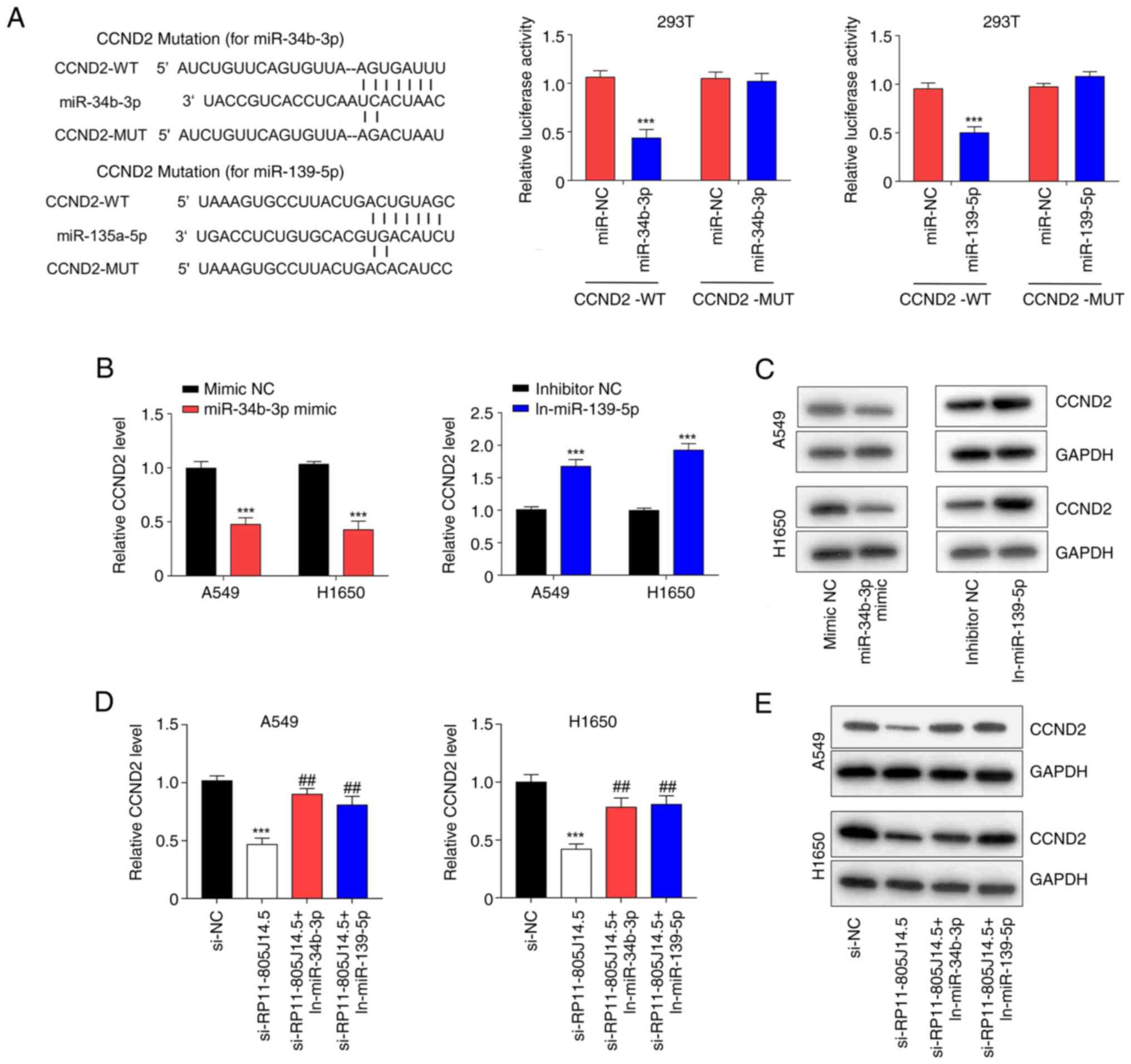
RP11-805J14.5 regulates CCND2
expression via sponging miR-34b-3p and miR-139-5p
Usually, miRNAs directly bind target mRNAs and
regulate their expression. Through screening TargetScan Release 7.2
(26), it was revealed that CCND2
was a potential target gene of miR-34b-3p and miR-139-5p. The
predicted binding sequences of CCND2, miR-34b-3p, and miR-139-5p
are presented in Fig. 4A.
Luciferase assays revealed that miR-34b-3p mimics and miR-139-5p
mimics significantly reduced the activity of the 3′-UTR of CCND2 WT
but did not affect the activity of the 3′-UTR of CCND2 MUT in 293T
cells (P<0.001; Fig. 4A).
Moreover, miR-34b-3p mimics decreased the mRNA and protein levels
of CCND2 while miR-139-5p inhibitor increased these levels in A549
and H1650 cells (P<0.001; Fig. 4B
and C). Furthermore, knockdown of RP11-805J14.5 decreased the
mRNA and protein levels of CCND2 (P<0.001), however this
decrease was abolished by either miR-34b-3p inhibitor or miR-139-5p
inhibitor in A549 and H1650 cells (P<0.01; Fig. 4D and E). Collectively, these
results indicated that RP11-805J14.5 served as a sponge of
miR-34b-3p and miR-139-5p, thereby modulating the expression of
CCND2.
RP11-805J14.5 regulates LUAD
progression by modulating CCND2 expression via sponging miR-34b-3p
and miR-139-5p
Given the demonstrated interactions between
RP11-805J14.5 and miR-34b-3p/miR-139-5p, and between these two miRs
and CCND2, it was investigated whether miR-34b-3p, miR-139-5p and
CCND2 mediated the regulation of RP11-805J14.5 on LUAD progression.
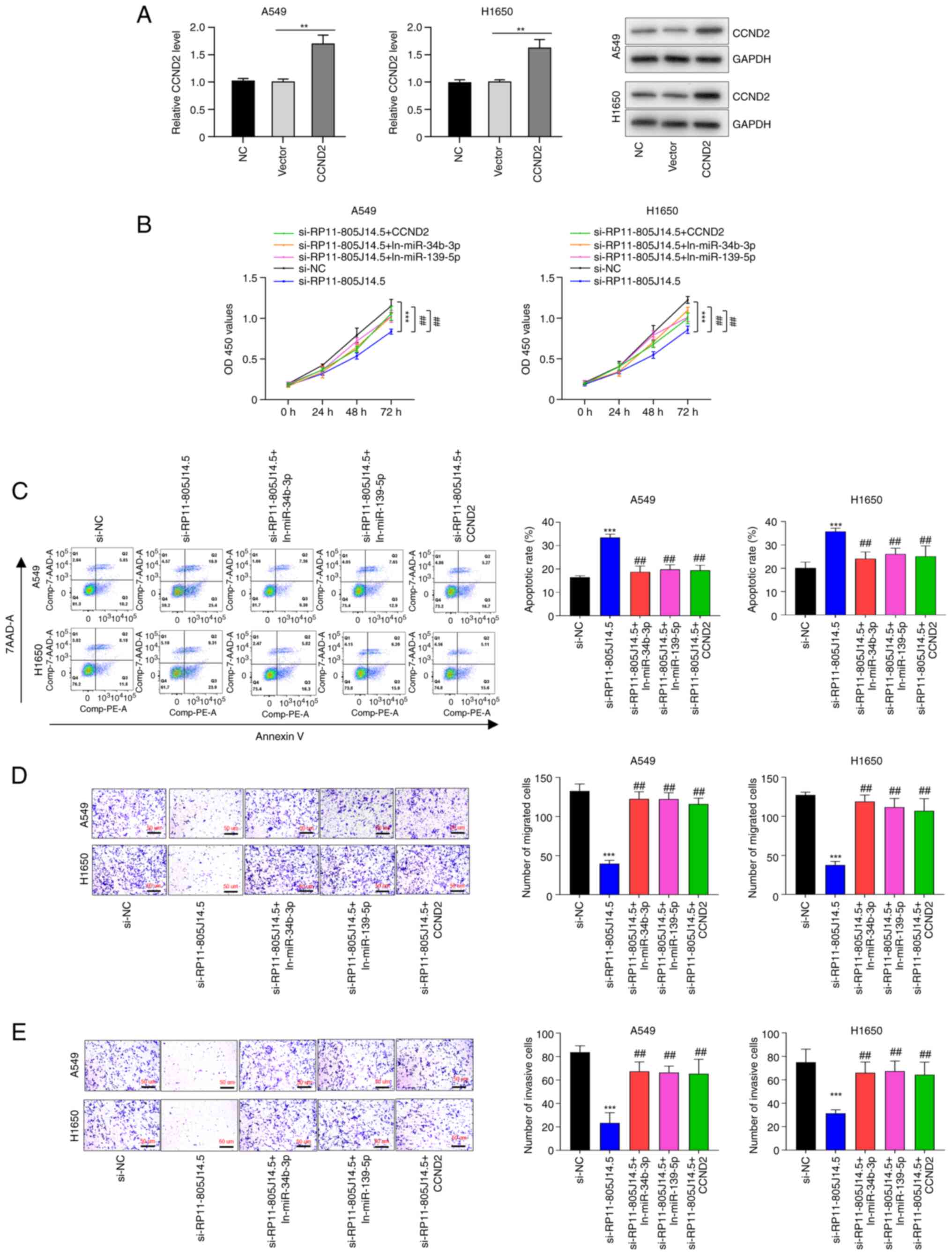
To this end, the overexpression plasmid of CCND2 was transfected
into A549 and H1650 cells, and transfection efficiency was verified
using RT-qPCR and western blotting (P<0.01; Fig. 5A). Then, the functional assays of
RP11-805J14.5, miR-34b-3p, miR-139-5p and CCND2 on LUAD progression
were conducted. Results revealed that knockdown of RP11-805J14.5
suppressed cell viability (P<0.001, Fig. 5B), but this effect was reversed by
downregulation of miR-34b-3p and miR-139-5p or overexpression of
CCND2 in A549 and H1650 cells (P<0.01; Fig. 5B). Moreover, cell apoptosis induced
by RP11-805J14.5 knockdown was inhibited by decreasing miR-34b-3p
and miR-139-5p expression or by CCND2 overexpression in A549 and
H1650 cells (P<0.01; Fig. 5C).
Furthermore, RP11-805J14.5 knockdown suppressed cell migration and
invasion (P<0.001), while this effect was abolished by either
the downregulation of miR-34b-3p or miR-139-5p or CCND2
overexpression in A549 and H1650 cells (all P<0.01; Fig. 5D and E). Thus, our results
indicated that RP11-805J14.5 regulated LUAD progression by
modulating CCND2 expression via acting as a miR-34b-3p and
miR-139-5p sponge.
Discussion
LUAD is a major type of primary lung cancer, and its
incidence has been increased in recent years (3,4).
However, the prognosis of LUAD patients is low due to the
aggressive behavior of this cancer (6). Therefore, it is imperative to
elucidate the molecular mechanism of LUAD development and explore
new therapeutic strategies. LncRNAs play a pivotal role in LUAD
progression (16–18). However, the biological function of
numerous lncRNAs has not been fully understood.
To define the role of lncRNAs in LUAD progression,
the data of LUAD from TCGA dataset were screened, which revealed
that RP11-805J14.5 expression was significantly elevated in LUAD
tumor tissues even though RP11-805J14.5 expression revealed some
overlap in tumor and non-tumor tissues. Next, the elevated
expression of RP11-805J14.5 in LUAD tissues (which were collected
from Hwa Mei Hospital) and cells was confirmed using RT-qPCR. The
results of the present study explicitly demonstrated that
RP11-805J14.5 was increased in LUAD tissues and cells. Moreover,
the increased expression of RP11-805J14.5 was associated with poor
survival of LUAD patients. Although the median survival time in
TCGA datasets was not exactly the same as that in the LUAD samples
collected, it still indicated that lncRNA RP11-805J14.5 is a
potential prognostic marker. Therefore, it is necessary to further
expand the sample size to confirm that lncRNA RP11-805J14.5 is a
valuable prognostic indicator. Furthermore, it was revealed that
knockdown of RP11-805J14.5 inhibited LUAD cell growth, invasion,
migration and tumor growth. The present study is, to the best of
our knowledge, the first study on the role of RP11-805J14.5 in LUAD
progression.
LncRNAs regulate gene expression via acting as
sponges of miRNAs, thereby modulating disease development (27). In order to elucidate the mechanism
of RP11-805J14.5 in LUAD, it was first determined that
RP11-805J14.5 mainly exists in the cytoplasm. The putative target
miRNAs were then screened and confirmed. miR-34b-3p and miR-139-5p
were identified as the miRNAs that bound to RP11-805J14.5, and
which were negatively regulated by RP11-805J14.5. Both miR-34b-3p
and miR-139-5p were decreased in LUAD tissues, which was consistent
with recent studies (28,29). Specifically, miR-34b-3p and
miR-139-5p have been revealed as key regulators in numerous
diseases. For example, miR-34b-3p inhibited the chemoresistance of
bladder cancer cells (19).
miR-34b-3p may promote antiplatelet efficiency of aspirin by
inhibiting thromboxane synthase expression (29) and miR-34b-3p impaired HUVEC
viability and migration via targeting PDK1 in an in vitro
model of gestational diabetes mellitus (30). LncRNA GAS5 silencing was revealed
to attenuate oxygen-glucose deprivation/reperfusion-induced injury
in brain microvascular endothelial cells via miR-34b-3p-dependent
regulation of EPHA4 (31).
However, miR-34b-3p played an important role in non-small cell lung
cancer (NSCLC), which suppressed cell proliferation and the cell
cycle via binding to CDK4 (32).
Piperlongumine inhibited the growth of NSCLC cells via the
miR-34b-3p/TGFBR1 pathway (33).
In addition, miR-139-5p suppressed the epithelial-mesenchymal
transition and increased the chemotherapeutic sensitivity of
colorectal cancer cells via targeting BCL2 (34). LncRNA AFAP1-AS1 suppressed
miR-139-5p and promoted cell proliferation and chemotherapy
resistance of NSCLC by competitively upregulating RRM2 (35). Furthermore, miR-139-5p expression
was decreased in lung cancer patients with lytic bone metastasis,
which indicated that miR-139-5p may be a biomarker and target in
monitoring and controlling bone metastasis of lung cancer (28). All aforementioned studies have
revealed that miR-34b-3p and miR-139-5p play an indispensable role
in NSCLC, which further supports our research.
MiRNAs can suppress the expression of target genes
via binding to the 3′-UTR of target genes (36). In the present study, it was
revealed that CCND2 could be targeted by miR-34b-3p and miR-139-5p
and negatively regulated by these two miRNAs. Furthermore, it was
also demonstrated that RP11-805J14.5 served as the sponge of
miR-34b-3p and miR-139-5p to modulate the expression of CCND2.
Previous studies have analyzed the pathogenesis and progression of
NSCLC at the molecular level. With the development of NGS
technology, RNA-seq gene expression profiles have also been drawn
(37–39). In addition, Tao et al
provided a systematic view of the functional alterations during
tumorigenesis that may help to elucidate the mechanisms of lung
cancer and lead to improved treatments for patients (37). A computational method was built to
identify new potential candidate cancer driver genes. The analyses
indicated that some of the obtained genes have the potential to
drive tumorigenesis on multiple differentiation levels. It is
hopeful that the findings of the present study will promote the
study of cancer driver genes and provide new insights into the
investigation of tumor initiation. To achieve this purpose, Chen
et al used protein information (protein-protein interaction)
to investigate cancer driver genes (40). Liu et al detected a series
of genes and the identification of the transcription factors
provided a new insight into its oncogenic role in tumor initiation
and progression, and benefited the discovery of a functional core
set that may reverse malignant transformation and reprogram cancer
cells (41). In the present study,
it was also demonstrated that the aberrant expression level of
CCND2 which was triggered by RP11-805J14.5 was the underlying cause
of LUAD. However, most of the genes in these expression profiles
have not been verified by functional experiments. CCND2 belongs to
the D-type cyclin family, and it plays a critical role in the cell
cycle (42). The function of CCND2
in cancer progression regulation has been demonstrated in prostate
(43), ovarian (44), thyroid cancer (45), and NSCLC (46). Accumulating evidence revealed that
the imbalance of circular RNAs plays a key role in multiple solid
tumors. Circ-RAD23B that is overexpressed in NSCLC, has been
revealed to be associated with lymph node invasion, poor
differentiation and short overall survival (OS). In addition,
circ-RAD23B has been demonstrated to act as an oncogene in NSCLC
cells. Mechanistically, circ-RAD23B can bind miR-593-3p and
miR-653-5p to increase the expression of CCND2 and TIAM1. Rescue
experiments revealed that circ-RAD23B promoted cell growth through
the miR-593-3p/CCND2 axis, and promoted cell invasion through the
miR-653-5p/TIAM1 pathway. In conclusion, it is considered that
circ-RAD23B is a promising biomarker and therapeutic target for
NSCLC (46). The present study
demonstrated the role of CCND2 in promoting cell growth in NSCLC.
In our study, RP11-805J14.5 regulated LUAD cell growth, invasion
and migration through modulation of CCND2 expression via sponging
miR-34b-3p and miR-139-5p, which also indicated the function of
CCND2 in LUAD. Collectively, lncRNA RP11-805J14.5 functions as a
ceRNA to regulate CCND2 expression by sponging miR-34b-3p and
miR-139-5p in LUAD. However, our research still has some
shortcomings, such as the small sample size which will be expanded
in a future study. In addition, the prognosis analysis of lung
cancer patients will be further improved in order to more
accurately determine whether lncRNAs can be used as prognostic
indicators of NSCLC. In conclusion, RP11-805J14.5 was decreased in
LUAD, and its knockdown suppressed LUAD cell growth, invasion and
migration by decreasing CCND2 via sponging miR-34b-3p and
miR-139-5p, rendering RP11-805J14.5 a prospective target for LUAD
therapy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by Natural Science Foundation of
Ningbo (grant no. 2018A610270), Ningbo Health Branding Subject Fund
(grant no. PPXK2018-05) and Hwamei Fund (grant no.
2019HMZDKY04).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HZ and GZ made substantial contributions to
conception and design of the study. XX, EZ and MY performed the
experiments and acquired the data. JN and XJ analyzed the data. HZ
and XX drafted the manuscript. GZ and XX critically revised the
study for important intellectual content. EZ and MY confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript and agree to be accountable for all aspects of the
work.
Ethics approval and consent to
participate
The present study was approved (approval no.
IR-B-2021-3-18) by The Ethics Committee of Hwa Mei Hospital
(Ningbo, China) and written informed consent was obtained from
patients in all cases. All animal experiments abided by the
National and International regulations and policies and were
approved (approval no. IR-D-2021-5-25) by the Experimental Animal
Ethics Committee of Hwa Mei Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
He F, Zhong X, Lin Z, Lin J, Qiu M, Li X
and Hu Z: Plasma exo-hsa_circRNA_0056616: A potential biomarker for
lymph node metastasis in lung adenocarcinoma. J Cancer.
11:4037–4046. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan
Y, Kong X, Bu J, Liu M and Xu S: circRNA-002178 act as a ceRNA to
promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis.
11:322020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Myers DJ and Wallen JM: Lung
Adenocarcinoma. StatPearls.edn Treasure Island, FL: 2022
|
|
5
|
Lin JJ, Cardarella S, Lydon CA, Dahlberg
SE, Jackman DM, Jänne PA and Johnson BE: Five-year survival in
EGFR-mutant metastatic lung adenocarcinoma treated with EGFR-TKIs.
J Thorac Oncol. 11:556–565. 2016. View Article : Google Scholar
|
|
6
|
Luo C, Lei M and Zhang Y, Zhang Q, Li L,
Lian J, Liu S, Wang L, Pi G and Zhang Y: Systematic construction
and validation of an immune prognostic model for lung
adenocarcinoma. J Cell Mol Med. 24:1233–1244. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Teppan J, Barth DA, Prinz F, Jonas K,
Pichler M and Klec C: Involvement of long non-coding RNAs (lncRNAs)
in tumor angiogenesis. Noncoding RNA. 6:422020.
|
|
8
|
Chan JJ and Tay Y: Noncoding RNA:RNA
regulatory networks in cancer. Int J Mol Sci. 19:13102018.
View Article : Google Scholar
|
|
9
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar
|
|
10
|
Li L, Wang M, Mei Z, Cao W, Yang Y, Wang Y
and Wen A: lncRNAs HIF1A-AS2 facilitates the up-regulation of
HIF-1α by sponging to miR-153-3p, whereby promoting angiogenesis in
HUVECs in hypoxia. Biomed Pharmacother. 96:165–172. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ye S, Lu Y, Ru Y, Wu X, Zhao M, Chen J, Xu
M, Huang Q, Wang Y, Shi S, et al: LncRNAs GACAT3 and LINC00152
regulate each other through miR-103 and are associated with
clinicopathological characteristics in colorectal cancer. J Clin
Lab Anal. 34:e233782020. View Article : Google Scholar
|
|
12
|
Feng Y, Ge Y, Wu M, Xie Y, Wang M, Chen Y
and Shi X: Long NonCoding RNAs regulate inflammation in diabetic
peripheral neuropathy by acting as ceRNAs targeting miR-146a-5p.
Diabetes Metab Syndr Obes. 13:413–422. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang T, Liu H, Mao R, Yang H, Zhang Y,
Zhang Y, Guo P, Zhan D, Xiang B and Liu Y: The lncRNA RP11-142A22.4
promotes adipogenesis by sponging miR-587 to modulate Wnt5β
expression. Cell Death Dis. 11:4752020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang L, He S, Wang Y, Zhu X, Shao W, Xu Q
and Cui Z: miRNA-20a suppressed lipopolysaccharide-induced HK-2
cells injury via NFκB and ERK1/2 signaling by targeting CXCL12. Mol
Immunol. 118:117–123. 2020. View Article : Google Scholar
|
|
15
|
Ji W, Diao YL, Qiu YR, Ge J, Cao XC and Yu
Y: LINC00665 promotes breast cancer progression through regulation
of the miR-379-5p/LIN28B axis. Cell Death Dis. 11:162020.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xue M, Tao W, Yu S, Yan Z, Peng Q, Jiang F
and Gao X: lncRNA ZFPM2-AS1 promotes proliferation via
miR-18b-5p/VMA21 axis in lung adenocarcinoma. J Cell Biochem.
121:313–321. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xue J and Zhang F: LncRNA LINC00511 plays
an oncogenic role in lung adenocarcinoma by regulating PKM2
expression via sponging miR-625-5p. Thorac Cancer. 11:2570–2579.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang X, Du L, Han J, Li X, Wang H, Zheng
G, Wang Y, Yang Y, Hu Y and Wang C: Novel long non-coding RNA
LINC02323 promotes epithelial-mesenchymal transition and metastasis
via sponging miR-1343-3p in lung adenocarcinoma. Thorac Cancer.
11:2506–2516. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tan Y, Zhang T, Zhou L, Liu S and Liang C:
MiR-34b-3p represses the multidrug-chemoresistance of bladder
cancer cells by regulating the CCND2 and P2RY1 genes. Med Sci
Monit. 25:1323–1335. 2019. View Article : Google Scholar
|
|
20
|
Zhuang XF, Zhao LX, Guo SP, Wei S, Zhai JF
and Zhou QH: miR-34b inhibits the migration/invasion and promotes
apoptosis of non-small-cell lung cancer cells by YAF2. Eur Rev Med
Pharmacol Sci. 23:2038–2046. 2019.
|
|
21
|
Cao DN, Shi JJ, Wu N and Li J: Modulation
of miR-139-5p on chronic morphine-induced, naloxone-precipitated
cAMP overshoot in vitro. Metab Brain Dis. 33:1501–1508. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao W, Geng D, Li S, Chen Z and Sun M:
LncRNA HOTAIR influences cell growth, migration, invasion, and
apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer
Med. 7:842–855. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shetty A, Venkatesh T, Kabbekodu SP,
Tsutsumi R and Suresh PS: LncRNA-miRNA-mRNA regulatory axes in
endometrial cancer: A comprehensive overview. Arch Gynecol Obstet.
Feb 18–2022.(Epub ahead of print). doi: 10.1007/s00404-022-06423-5,
2022. View Article : Google Scholar
|
|
25
|
Dastsooz H, Alizadeh A, Habibzadeh P,
Nariman A, Hosseini A, Mansoori Y and Haghi-Aminjan H:
LncRNA-miRNA-mRNA networks of gastrointestinal cancers representing
common and specific LncRNAs and mRNAs. Front Genet. 12:7919192022.
View Article : Google Scholar
|
|
26
|
Zhang Y, Hu G, Zhang Z, Jing Y, Tao F and
Ye M: CircRNA_0043691 sponges miR-873-3p to promote metastasis of
gastric cancer. Mamm Genome. 32:476–487. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Arun K, Arunkumar G, Bennet D,
Chandramohan SM, Murugan AK and Munirajan AK: Comprehensive
analysis of aberrantly expressed lncRNAs and construction of ceRNA
network in gastric cancer. Oncotarget. 9:18386–18399. 2018.
View Article : Google Scholar
|
|
28
|
Xu S, Yang F, Liu R, Li X, Fan H, Liu J,
Wei S, Chen G, Chen J and Da Y: Serum microRNA-139-5p is
downregulated in lung cancer patients with lytic bone metastasis.
Oncol Rep. 39:2376–2384. 2018.PubMed/NCBI
|
|
29
|
Liu WW, Wang H, Chen XH, Fu SW and Liu ML:
miR-34b-3p may promote antiplatelet efficiency of aspirin by
inhibiting thromboxane synthase expression. Thromb Haemost.
119:1451–1460. 2019. View Article : Google Scholar
|
|
30
|
Song F, Cai A, Ye Q, Chen X, Lin L and Hao
X: MiR-34b-3p impaired HUVECs viability and migration via targeting
PDK1 in an in vitro model of gestational diabetes mellitus. Biochem
Genet. 59:1381–1395. 2021. View Article : Google Scholar
|
|
31
|
Shen B, Wang L, Xu Y, Wang H and He S:
LncRNA GAS5 silencing attenuates oxygen-glucose
deprivation/reperfusion-induced injury in brain microvascular
endothelial cells via miR-34b-3p-dependent regulation of EPHA4.
Neuropsychiatr Dis Treat. 17:1667–1678. 2021. View Article : Google Scholar
|
|
32
|
Feng H, Ge F, Du L, Zhang Z and Liu D:
MiR-34b-3p represses cell proliferation, cell cycle progression and
cell apoptosis in non-small-cell lung cancer (NSCLC) by targeting
CDK4. J Cell Mol Med. 23:5282–5291. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu X, Xu C, Xu Z, Lu C, Yang R, Zhang F
and Zhang G: Piperlongumine inhibits the growth of non-small cell
lung cancer cells via the miR-34b-3p/TGFBR1 pathway. BMC Complement
Med Ther. 21:152021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Q, Liang X, Wang Y, Meng X, Xu Y, Cai
S, Wang Z, Liu J and Cai G: miR-139-5p inhibits the
epithelial-mesenchymal transition and enhances the chemotherapeutic
sensitivity of colorectal cancer cells by downregulating BCL2. Sci
Rep. 6:271572016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang N, Guo W, Ren K, Li W, Jiang Y, Sun
J, Dai W and Zhao W: LncRNA AFAP1-AS1 supresses miR-139-5p and
promotes cell proliferation and chemotherapy resistance of
non-small cell lung cancer by competitively upregulating RRM2.
Front Oncol. 9:11032019. View Article : Google Scholar
|
|
36
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tao X, Wu X, Huang T and Mu D:
Identification and analysis of dysfunctional genes and pathways in
CD8(+) T cells of non-small cell lung cancer based on RNA
sequencing. Front Genet. 11:3522020. View Article : Google Scholar
|
|
38
|
Wang C, Tan S, Liu WR, Lei Q, Qiao W, Wu
Y, Liu X, Cheng W, Wei YQ, Peng Y and Li W: RNA-Seq profiling of
circular RNA in human lung adenocarcinoma and squamous cell
carcinoma. Mol Cancer. 18:1342019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Seo D, Roh J, Chae Y and Kim W: Gene
expression profiling after LINC00472 overexpression in an NSCLC
cell line1. Cancer Biomark. 32:175–188. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen L, Huang T, Zhang YH, Jiang Y, Zheng
M and Cai YD: Identification of novel candidate drivers connecting
different dysfunctional levels for lung adenocarcinoma using
protein-protein interactions and a shortest path approach. Sci Rep.
6:298492016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu C, Zhang YH, Huang T and Cai Y:
Identification of transcription factors that may reprogram lung
adenocarcinoma. Artif Intell Med. 83:52–57. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang L, Cui Y, Zhang L, Sheng J, Yang Y,
Kuang G, Fan Y, Zhang Q and Jin J: The silencing of CCND2 by
promoter aberrant methylation in renal cell cancer and analysis of
the correlation between CCND2 methylation status and clinical
features. PLoS One. 11:e01618592016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dong Q, Meng P, Wang T, Qin W, Qin W, Wang
F, Yuan J, Chen Z, Yang A and Wang H: MicroRNA let-7a inhibits
proliferation of human prostate cancer cells in vitro and in vivo
by targeting E2F2 and CCND2. PLoS One. 5:e101472010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chang L, Guo R, Yuan Z, Shi H and Zhang D:
LncRNA HOTAIR regulates CCND1 and CCND2 expression by sponging
miR-206 in ovarian cancer. Cell Physiol Biochem. 49:1289–1303.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Di W, Li Q, Shen W, Guo H and Zhao S: The
long non-coding RNA HOTAIR promotes thyroid cancer cell growth,
invasion and migration through the miR-1-CCND2 axis. Am J Cancer
Res. 7:1298–1309. 2017.PubMed/NCBI
|
|
46
|
Han W, Wang L, Zhang L, Wang Y and Li Y:
Circular RNA circ-RAD23B promotes cell growth and invasion by
miR-593-3p/CCND2 and miR-653-5p/TIAM1 pathways in non-small cell
lung cancer. Biochem Biophys Res Commun. 510:462–466. 2019.
View Article : Google Scholar : PubMed/NCBI
|