Introduction
Colon adenocarcinoma (COAD) is the third most common
type of cancer worldwide (1).
Although great progress has been made in terms of the treatment of
COAD, the high recurrence rates associated with this type of cancer
remain a major clinical challenge (2,3). There
are only limited methods available to effectively inhibit COAD
metastasis and there are no effective therapies for patients with
distant metastases (4). Numerous
studies have investigated the underlying molecular mechanisms of
COAD, which may solve clinical problems associated with this type
of cancer (5,6). The complex function and regulation of
the immune system offers more diverse strategies for cancer
treatment. These strategies include adoptive T-cell transfer,
cytokine therapy, and administration of ligands and monoclonal
antibodies. Immunotherapy provides important leads for potential
therapies for the treatment of patients with advanced colon cancer;
therefore, further scientific research is required to improve the
efficacy of immunotherapy against colon cancer (7).
Gα-interacting protein (GAIP) C-terminus (GIPC)
PDZ-domain-containing family member 2 (GIPC2) is a member of the
GIPC family of proteins, which can activate the Wnt signaling
pathway by binding to the GTP-coupled proteins, RCSI9 and RCSUGAIP,
and TGF-β type 3 receptor proteins through their PDZ domain
(8). The expression levels of GIPC2
vary among human tissues, with high expression levels being
reported in digestive organs, such as the small intestine, colon,
stomach, esophagus, liver and other tissues, whereas tissues such
as the bone marrow, thymus, retina, smooth muscle and placenta have
negligible levels of expression (9). In addition, GIPC2 is expressed in
certain glands, such as breast, adrenal, salivary and thyroid
glands (10). Previous studies have
demonstrated an important role for GIPC2 in embryonic development
(11,12) and the occurrence of digestive tract
tumors (13,14); however, the role of GIPC2 in COAD
has yet to be elucidated.
In the present study, database analysis was used to
explore the expression levels of GIPC2 in different types of cancer
tissue, and to determine the association between its expression and
prognosis, the level of infiltrating immune cells and expression of
immune checkpoint-associated genes in COAD.
Materials and methods
GIPC2 expression analyses
The Xiantao bioinformatics analysis tool (https://www.xiantao.love/products) is an online
comprehensive bioinformatics tool platform based on visual R
language programming (15). This
tool was first employed to analyze the mRNA expression levels of
GIPC2 in 11,093 samples of 33 types of cancer based on data
retrieved from The Cancer Genome Atlas (TCGA; http://portal.gdc.cancer.gov/). The expression
levels of GIPC2 were then compared between normal and tumor tissues
obtained from 478 patients with COAD, which included tumor tissues
from all patients, and 41 normal tissues adjacent to the cancer
from 41 patients.
Patient samples
A total of 22 pairs of COAD samples and adjacent
normal colon tissues were collected from surgical samples at the
People's Hospital of Tongling City (Tongling, China). Among the 22
patients who underwent surgery between July 2019 and June 2021, 15
were male and seven were female, with a mean age of 69 years. Of
these patients, 16 underwent laparoscopic right hemicolectomy, five
underwent laparoscopic sigmoidectomy and one underwent laparoscopic
left hemicolectomy. None of the patients were treated with
preoperative therapy. All patients provided written informed
consent. The present study was approved by the Ethics Committee of
the People's Hospital of Tongling City (approval no. 2022002).
Immunohistochemistry (IHC)
GIPC2 protein detection was performed using IHC with
horseradish peroxidase. The concentrated rabbit polyclonal antibody
against human GIPC2 protein was purchased from BIOSS (cat no.
KT22301; 1:200). Normal adult kidney tissue was used as a positive
control and PBS was used as a negative control. The normal adult
kidney tissue was derived from the same patients with kidney
cancer, with normal tissue taken >5 cm away from cancer tissue.
IHC kits were purchased from Fuzhou Maixin Biotech Co., Ltd. and
were performed according to the manufacturer's protocol. Briefly,
the ex vivo tissue was immediately fixed in 10% neutral
formalin fixative for 24 h at 20–25°C, embedded in paraffin,
continuously sectioned (4 µm) and mounted on slides at 60°C for 2
h. Subsequently, the tissue was conventionally dewaxed using xylene
and hydrated in a gradient series of alcohol. The endogenous
peroxidase was blocked with 3% hydrogen peroxide for 15 min at room
temperature and antigen retrieval was performed in a pressure
cooker with 1% citric acid antigen repair solution (pH 6.0) for 2
min. Tissues were then incubated with 50 µl primary antibody at 4°C
for ~12 h, and with 50 µl secondary antibody (ready to use; cat.
no. KIT-5010; Fuzhou Maixin Biotech Co., Ltd.) at room temperature
for 30 min. All sections were counterstained with hematoxylin after
the reaction. Two senior pathologists performed double-blind
evaluations by examining the sections under a light microscope.
According to a previous study (16), 10 high-power fields were randomly
selected from each section. A semi-quantitative score based on the
intensity of positively stained cells and the staining area was
used to evaluate the results of IHC; the comprehensive score of
staining intensity was multiplied by the staining area. Staining
intensity was scored as follows: 0, colorless areas (no staining);
1, light yellow staining; 2, brown-yellow staining; 3, brown
staining. The numbers of positive cells were evaluated as follows:
<5%, 0; 5–25%, 1; 26–50%, 2; 51–75%, 3; >75%, 4. The staining
intensity score was subsequently multiplied by the positive cell
number score; a score of 0–3 was classified as negative, whereas a
score >3 was classified as positive.
Association between GIPC2 and
clinicopathological features
Differential expression of GIPC2 according to
pathological stage, tumor stage, lymph node status, metastasis,
sex, age, lymphatic invasion and perineural invasion in patients
with COAD based on TCGA database was assessed using box plots. The
same variables were used in multiple logistic regression analyses
to determine the factors associated with GIPC2 expression.
Survival analyses based on GIPC2
expression
The effect of GIPC2 expression on the survival of
patients with COAD was determined by performing a Kaplan-Meier
analysis and log-rank test, based on data retrieved from TCGA
database. Data for the patients with COAD for whom the relevant
prognostic information was known were used to analyze the
association between GIPC2 expression (based on median levels) and
overall survival (OS), disease-specific survival (DSS) and
progression-free interval (PFI). Cox regression analysis was
subsequently used to determine the risk factors for OS.
Analyses of genes co-expressed with
GIPC2
The Gene Expression Profiling Interactive Analysis 2
(GEPIA2) server (http://gepia.cancer-pku.cn/index.html) is an online
server for the analysis of TCGA data. GEPIA2 was used to estimate
the top 100 genes co-expressed with GIPC2. Enrichment analysis was
subsequently performed using the clusterProfiler package in R
(version 4.0.3) on the Xiantao bioinformatics analysis tool
(https://www.xiantao.love/) to perform
Gene Ontology (GO) biological process and Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathway analysis. Finally, Spearman rank
correlation test was used to determine the correlation between
GIPC2 and the top five co-expressed genes.
Gene set enrichment analysis
(GSEA)
To explore the potential pathological processes
associated with GIPC2, GSEA was conducted for TCGA-COAD data using
GSEA v4.3.0 software (https://www.gsea-msigdb.org/gsea/index.jsp). The gene
set c2.cp.kegg.v7.1.symbols.gmt was selected for further analysis.
The number of permutations was set as 5,000. Normalized enrichment
scores >1, false discovery rate q-values <0.05 and adjusted
P-values <0.05 were set as the cut-off values for significant
enrichment.
Evaluation of tumor-infiltrating
immune cells
The immunedeconv package in R (https://www.aclbi.com/static/index.html#/immunoassay),
which integrates CIBERSORT (17),
is a deconvolution algorithm based on gene expression that is able
to evaluate changes in the expression of one set of genes relative
to all other genes in the sample. This package was used to analyze
the levels of tumor-infiltrating immune cells. Among 478 COAD
samples based on TCGA-COAD data, samples with the top 25% and the
lowest 25% levels of GIPC2 expression were classified into the
high- and low-expression groups, respectively. The abundance of 22
types of immune cells [naïve B cells, memory B cells, plasma B
cells, CD8+ T cells, naïve CD4+ T cells,
resting CD4+ memory T cells, activated CD4+
memory T cells, follicular helper T cells, regulatory T cells, γδ T
cells, resting natural killer (NK) cells, activated NK cells,
monocytes, M0 macrophages, M1 macrophages, M2 macrophages, resting
myeloid dendritic cells, activated myeloid dendritic cells,
activated mast cells, resting mast cells, eosinophils and
neutrophils] were estimated using the CIBERSORT algorithm. Briefly,
gene expression datasets from TCGA were uploaded to the Xiantao
bioinformatics analysis tool, and after standard annotation, the
immunedeconv R package was used to estimate the P-values for
deconvolution via the CIBERSORT algorithm. This tool was then used
to compare the expression of immune checkpoint-associated genes,
including CD274, CTLA4, HAVCR2, LAG3, PDCD1, PDCD1LG2, TIGIT and
SIGLEC15, between patients with COAD in the high and low GIPC2
expression groups, respectively. The aforementioned analyses and R
package were implemented using R foundation for statistical
computing (2020) version 4.0.3 (18) and the software packages ggplot2
(https://cran.r-project.org/web/packages/ggplot2/index.html)
and pheatmap (https://cran.r-project.org/web/packages/pheatmap/index.html)
were used for generating images.
Statistical analysis
SPSS 19.0 software (IBM Corp.) was used to perform
the statistical analyses. Comparisons between or among groups were
performed using unpaired χ2 test, Student's t-test,
paired Student's t-test, Mann-Whitney U-test or one-way ANOVA.
Tukey's HSD was used as a post hoc test following ANOVA. As
aforementioned, a Kaplan-Meier analysis and log-rank test was
performed for survival analysis using R language package (version
4.0.3). Spearman rank correlation test was used for the correlation
analysis between GIPC2 and co-expressed genes. Logistic multiple
regression analysis was used to determine the risk factors
associated with GIPC2 expression, and Cox regression analysis was
performed to determine the risk factors for OS. P<0.05 was
considered to indicate a statistically significant difference.
Results
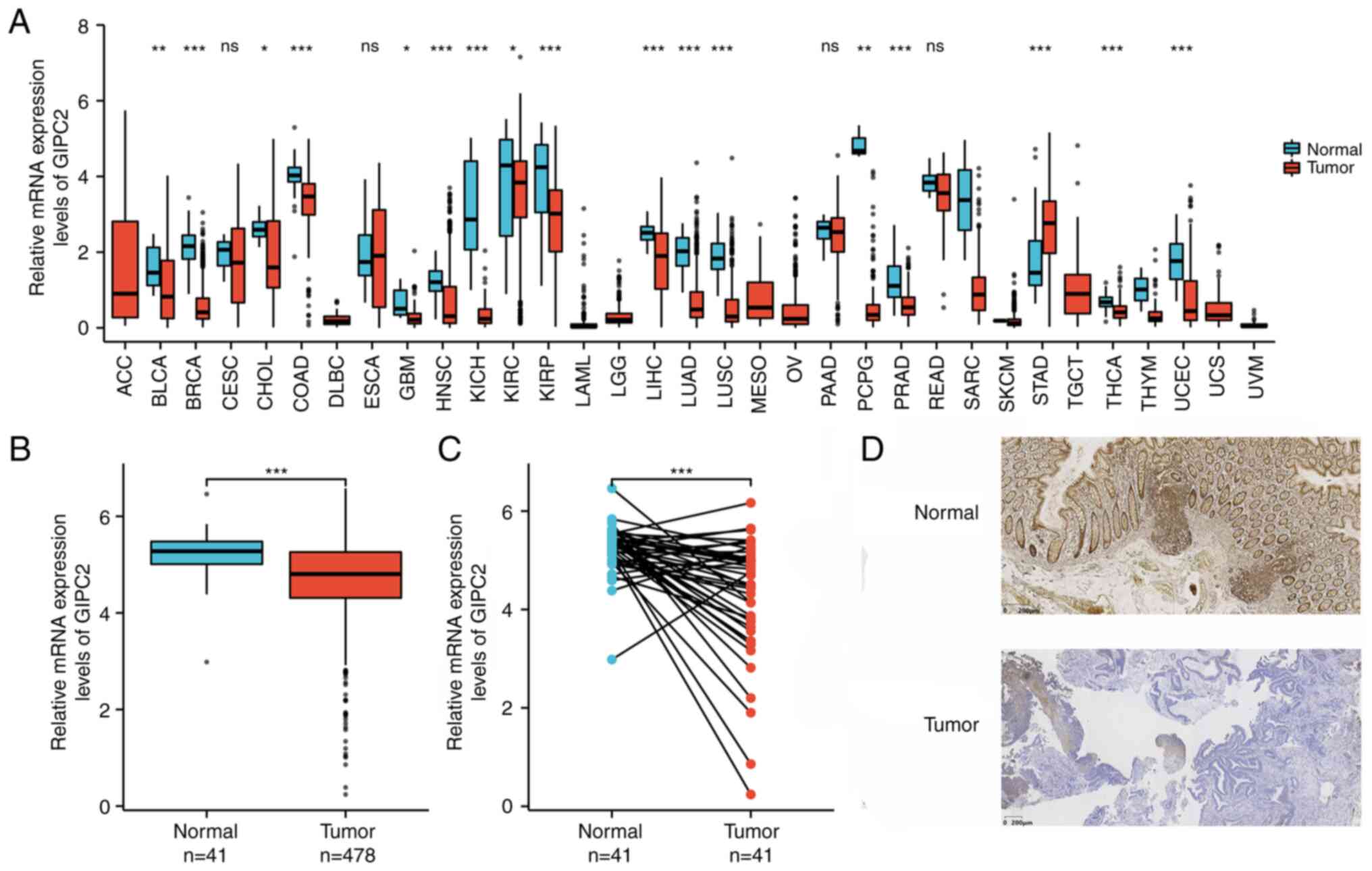
Pan-cancer analysis of the expression levels of
GIPC2. The expression levels of GIPC2 in 33 types of cancer based
on TCGA data were analyzed. GIPC2 expression was revealed to be low
in bladder cancer, breast cancer, bile duct cancer, COAD,
glioblastoma, head and neck cancer, chromophobe renal cell
carcinoma, kidney clear cell carcinoma, kidney papillary cell
carcinoma, liver cancer, lung adenocarcinoma, lung squamous cell
carcinoma, pheochromocytoma and paraganglioma, prostate cancer,
thyroid cancer, thymoma and endometrioid cancer, whereas it was
high in stomach cancer compared with the tissue adjacent to the
cancer (Fig. 1A). The results of
unpaired (Fig. 1B) and paired
(Fig. 1C) analyses of COAD
confirmed that the expression levels of GIPC2 were significantly
higher in normal tissues compared with those in tumor tissues. The
results of IHC also confirmed that the protein expression levels of
GIPC2 were lower in COAD tissues compared with those in normal
tissue samples (Fig. 1D). The
positive rate in the normal intestinal mucosa group (18/22,81.82%)
was significantly higher than that in the COAD group (3/22, 13.64%;
χ2=20.497; P<0.001) (data not shown).
Association between GIPC2 expression
and clinicopathological variables
R version 4.0.3 was used to assess the association
of GIPC2 with the relevant clinical information from 478 cases of
COAD obtained from TCGA. Differential expression of GIPC2 according
to the pathological stage (Fig.
2A), tumor stage (Fig. 2B),
lymph node status (Fig. 2C),
metastasis status (Fig. 2D), sex
(Fig. 2E), age (Fig. 2F), lymphatic invasion (Fig. 2G) and perineural invasion (Fig. 2H) was analyzed. The results of
univariate analysis revealed that GIPC2 expression (based on the
median expression value) was markedly associated with pathological
stage, tumor stage, lymph node status and lymphatic invasion.
Multivariate analysis using logistic regression showed that tumor
stage, lymph node status and lymphatic invasion were significantly
associated with GIPC2 expression (Table
I).
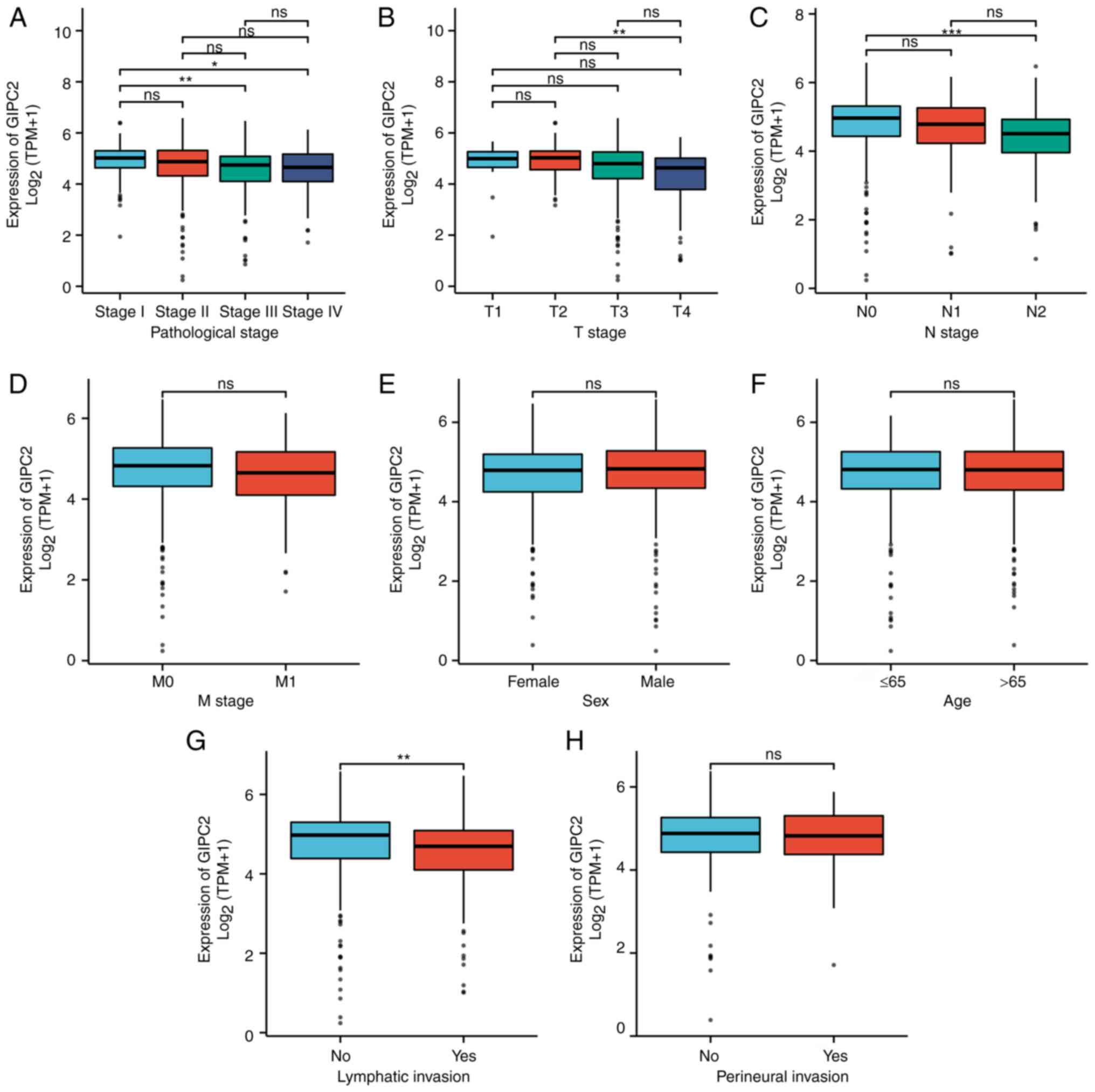 | Figure 2.Association between GIPC2 expression
levels and clinicopathological variables. Association of GIPC2 with
(A) pathological stage, (B) T stage, (C) N stage, (D) M stage, (E)
sex, (F) age, (G) lymphatic invasion and (H) perineural invasion.
Data are presented as the mean ± SD. *P<0.05, **P<0.01,
***P<0.001. GIPC2, Gα-interacting protein C-terminus
PDZ-domain-containing family member 2; ns, not significant. |
 | Table I.Relationship between Gα-interacting
protein C-terminus PDZ-domain-containing family member 2 expression
and clinicopathological variables. |
Table I.
Relationship between Gα-interacting
protein C-terminus PDZ-domain-containing family member 2 expression
and clinicopathological variables.
| Characteristic | Odds ratio (95%
CI) | P-value |
|---|
| T stage (T3 and T4
vs. T1 and T2) | 0.439
(0.271–0.698) |
<0.001a |
| N stage (N1 and N2
vs. N0) | 0.496
(0.341–0.717) |
<0.001a |
| M stage (M1 vs.
M0) | 0.530
(0.304–0.907) | 0.022 |
| Sex (Male vs.
Female) | 1.106
(0.772–1.585) | 0.583 |
| Age (>65 vs. ≤65
years) | 0.966
(0.670–1.392) | 0.852 |
| Perineural invasion
(Yes vs. No) | 1.011
(0.517–1.991) | 0.974 |
| Lymphatic invasion
(Yes vs. No) | 0.452
(0.304–0.669) |
<0.001a |
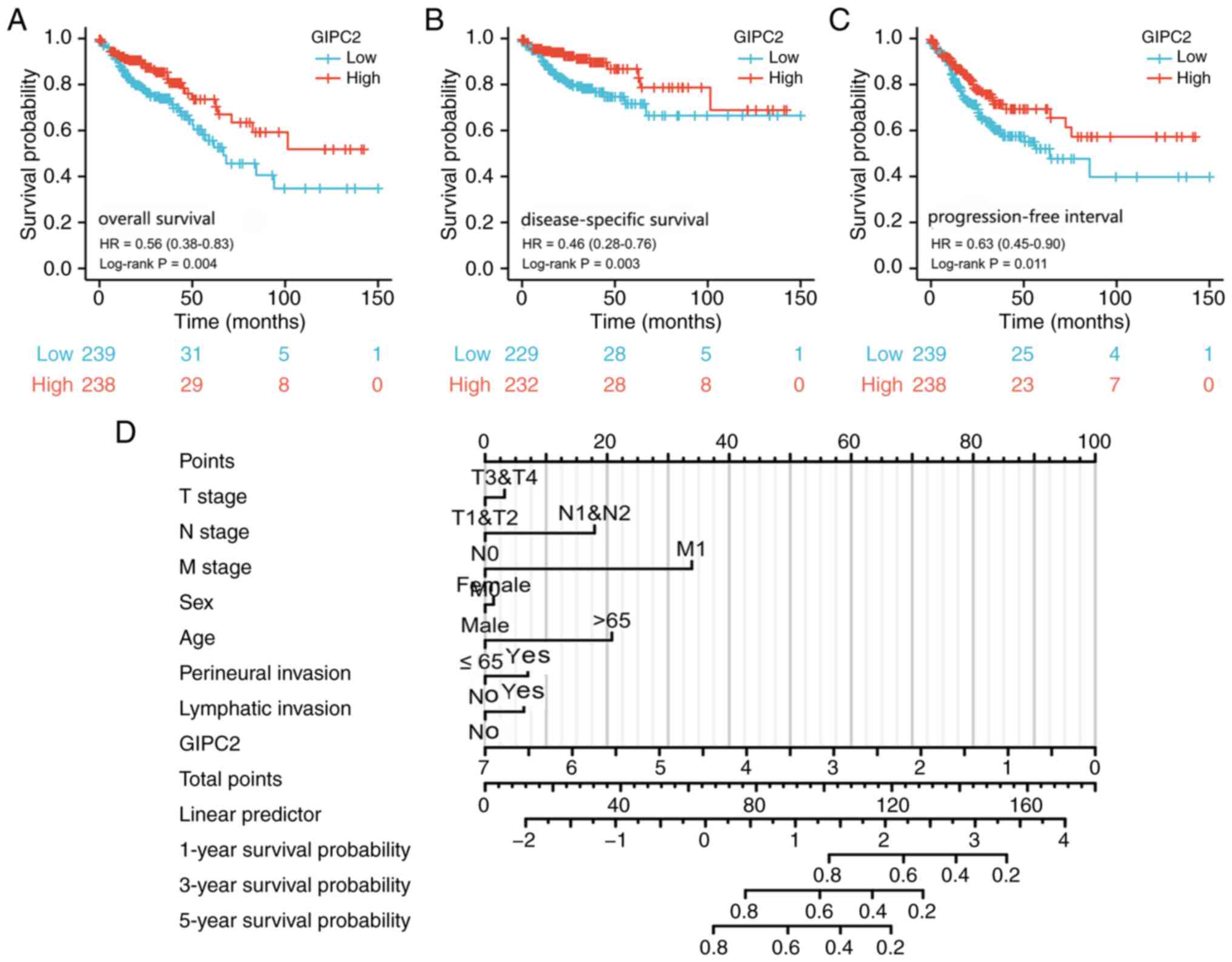
Low GIPC2 expression is associated
with poor prognosis in patients with COAD
As shown in Fig. 3,
low expression levels of GIPC2 (based on the median expression
value) was significantly associated with poor OS (Fig. 3A), DSS (Fig. 3B) and PFI (Fig. 3C). Furthermore, the results of the
Cox multivariate analysis revealed that low expression levels of
GIPC2 and positive distant metastasis were independent prognostic
factors (Fig. 3D; Table II).
 | Table II.Cox regression analysis for overall
survival. |
Table II.
Cox regression analysis for overall
survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Characteristic | Hazard ratio (95%
CI) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| T stage |
|
|
|
|
| T1 and
T2 | Reference |
|
|
|
| T3 and
T4 | 3.072
(1.423–6.631) | 0.004a | 1.120
(0.218–5.747) | 0.892 |
| N stage |
|
|
|
|
| N0 | Reference |
|
|
|
| N1 and
N2 | 2.592
(1.743–3.855) |
<0.001a | 1.973
(0.600–6.491) | 0.263 |
| M stage |
|
|
|
|
| M0 | Reference |
|
|
|
| M1 | 4.193
(2.683–6.554) |
<0.001a | 3.593
(1.300–9.930) | 0.014a |
| Sex |
|
|
|
|
|
Female | Reference |
|
|
|
|
Male | 1.101
(0.746–1.625) | 0.627 |
|
|
| Age |
|
|
|
|
| ≤65
years | Reference |
|
|
|
| >65
years | 1.610
(1.052–2.463) | 0.028a | 2.157
(0.947–4.913) | 0.067 |
| Perineural
invasion |
|
|
|
|
| No | Reference |
|
|
|
|
Yes | 1.940
(0.982–3.832) | 0.056 | 1.286
(0.502–3.294) | 0.600 |
| Lymphatic
invasion |
|
|
|
|
| No | Reference |
|
|
|
|
Yes | 2.450
(1.614–3.720) | <0.001 | 1.269
(0.523–3.079) | 0.598 |
| GIPC2, log2+1 | 0.832
(0.695–0.997) | 0.046a | 0.582
(0.393–0.862) | 0.007a |
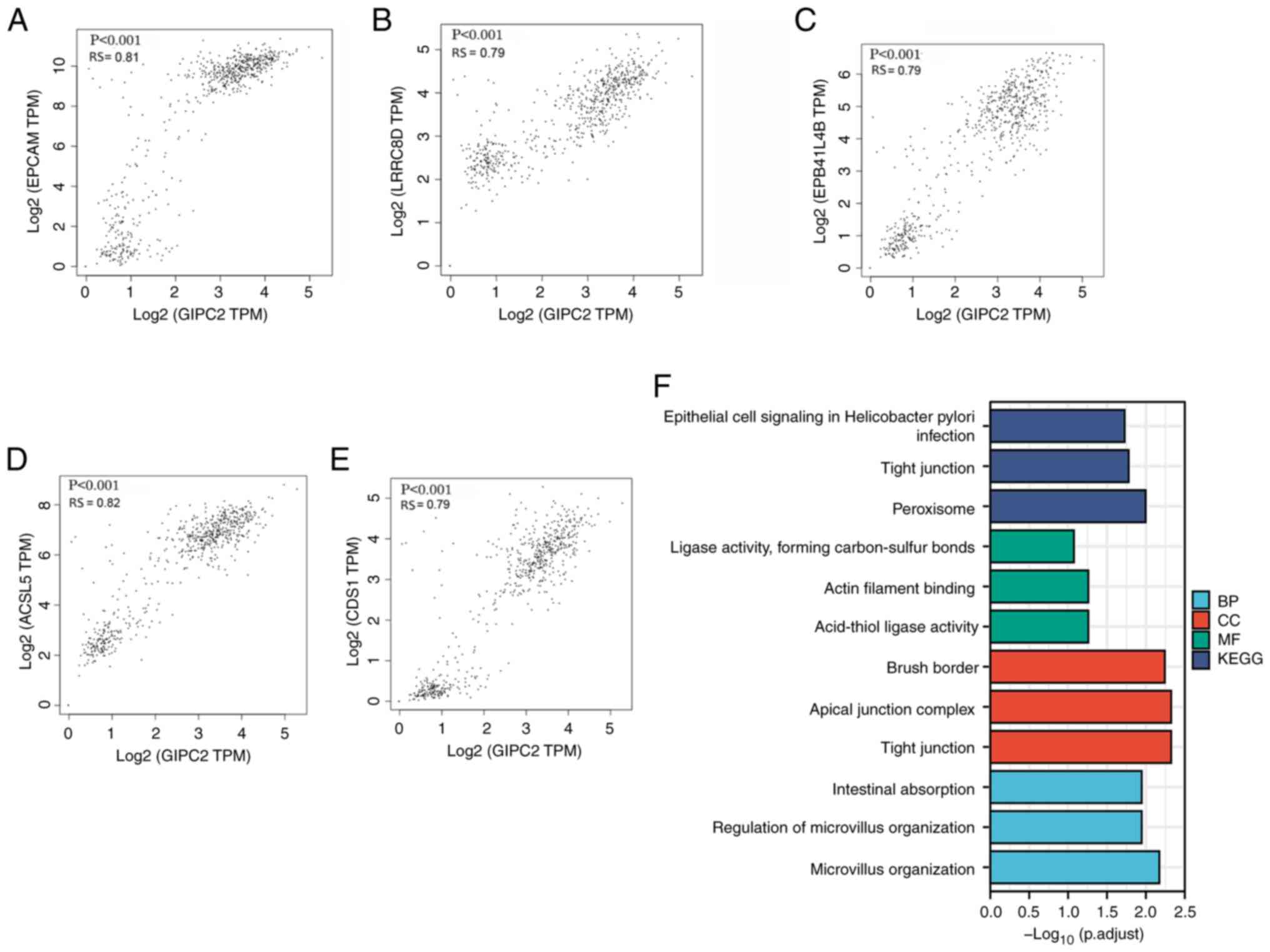
Analysis of genes co-expressed with
GIPC2 in COAD
The top five genes that exhibited a significant
positive correlation with GIPC2 were EPCAM (Fig. 4A), LRRC8D (Fig. 4B), EPB41L4B (Fig. 4C), ACSL5 (Fig. 4D) and CDS1 (Fig. 4E). The top 100 genes co-expressed
with GIPC2 were subsequently selected for enrichment analysis. The
terms ‘epithelial cell signaling in Helicobacter pylori
infection’, ‘tight junction’ and ‘peroxisome’ were significantly
enriched in the GO biological process analysis (Fig. 4F). The terms ‘ligase activity,
forming carbon-sulfur bonds’, ‘actin filament binding’ and
‘acid-thiol ligase activity’ were significantly enriched in the GO
cellular component analysis (Fig.
4F). In the GO molecular function analysis, the terms ‘brush
border’, ‘apical junction complex’ and ‘tight junction’ were highly
enriched (Fig. 4F). Finally, the
KEGG pathway analysis indicated that the pathways ‘intestinal
absorption’, ‘regulation of microvillus organization’ and
‘microvillus organization’ were significantly enriched (Fig. 4F).
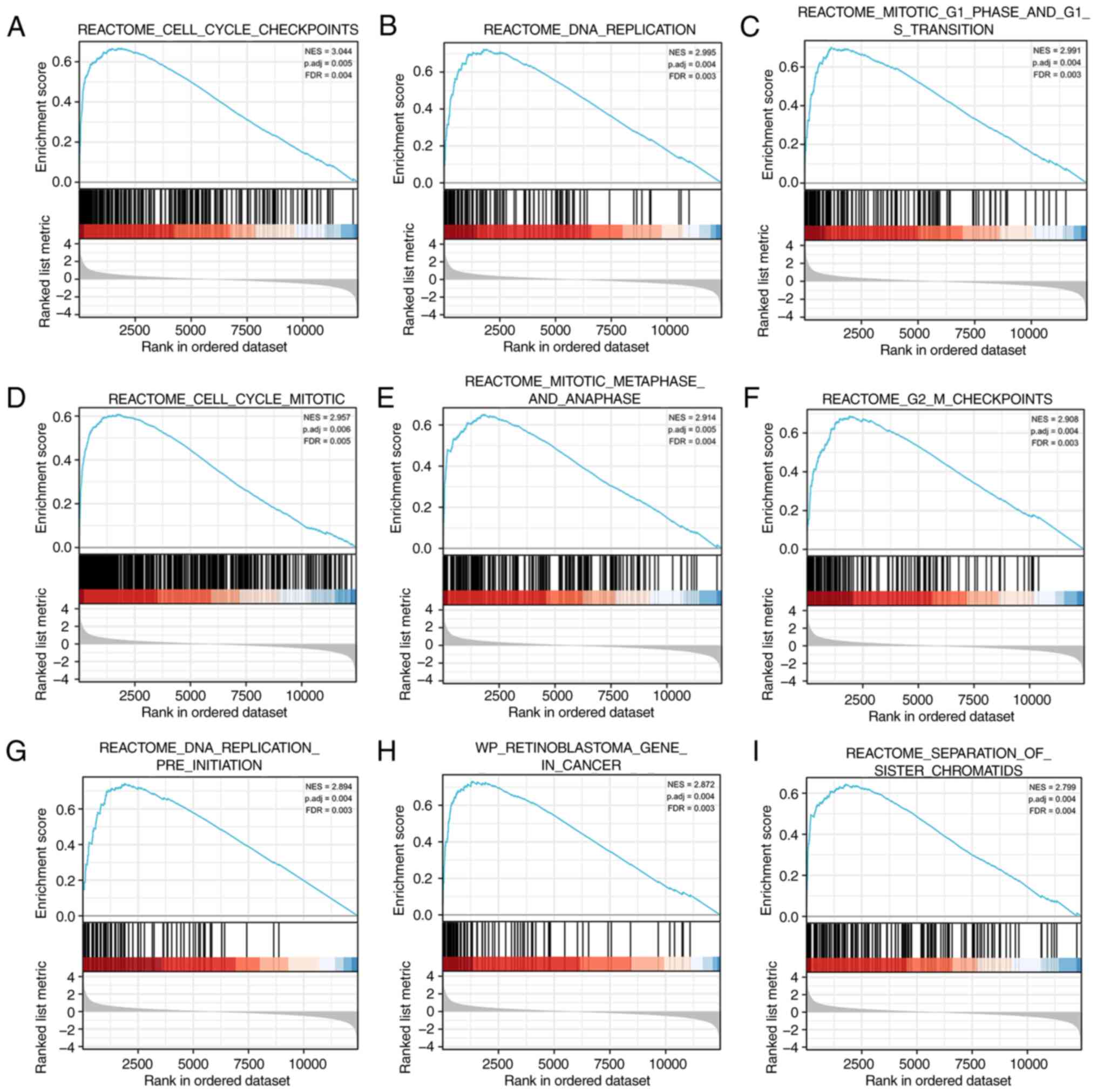
Identification of GIPC2-associated
pathways by GSEA
GSEA was performed using COAD data from TCGA, and
the results were compared between tissues with high and low GIPC2
expression to identify the possible biological pathways regulated
by GIPC2. A total of 316 pathways were significantly enriched in
the GIPC2 high expression group. The results showed that the top
nine significantly enriched terms comprised ‘cell cycle
checkpoints’ (Fig. 5A), ‘DNA
replication’ (Fig. 5B), ‘mitotic G1
phase and G1-S transition’ (Fig.
5C), ‘cell cycle mitotic’ (Fig.
5D), ‘mitotic metaphase and anaphase’ (Fig. 5E), ‘G2 M checkpoints’ (Fig. 5F), ‘DNA replication pre-initiation’
(Fig. 5G), ‘retinoblastoma gene in
cancer’ (Fig. 5H) and ‘separation
of sister chromatids’ (Fig.
5I).
Association between GIPC2 expression
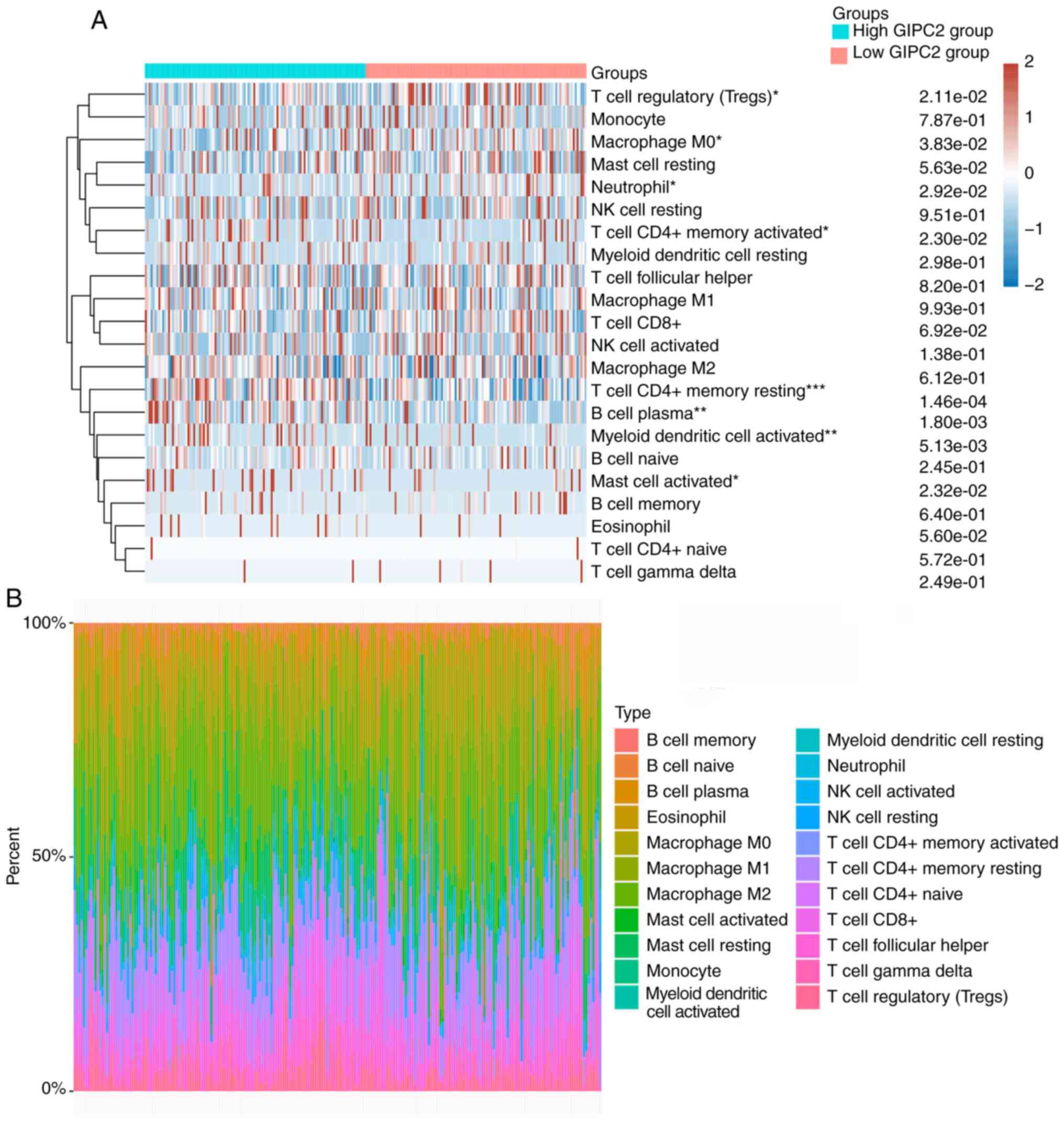
and tumor-infiltrating immune cells
The results showed that the numbers of plasma B
cells (P=0.018), resting CD4+ memory T cells (P=0.015),
activated CD4+ memory T cells (P=0.023), activated
myeloid dendritic cells (P=0.005) and activated mast cells
(P=0.023) were significantly higher, whereas the numbers of
regulatory T cells (P=0.021), M0 macrophages (P=0.038) and
neutrophils (P=0.029) were significantly lower in the high GIPC2
expression group compared with the low expression group (Fig. 6A and B). The expression levels of
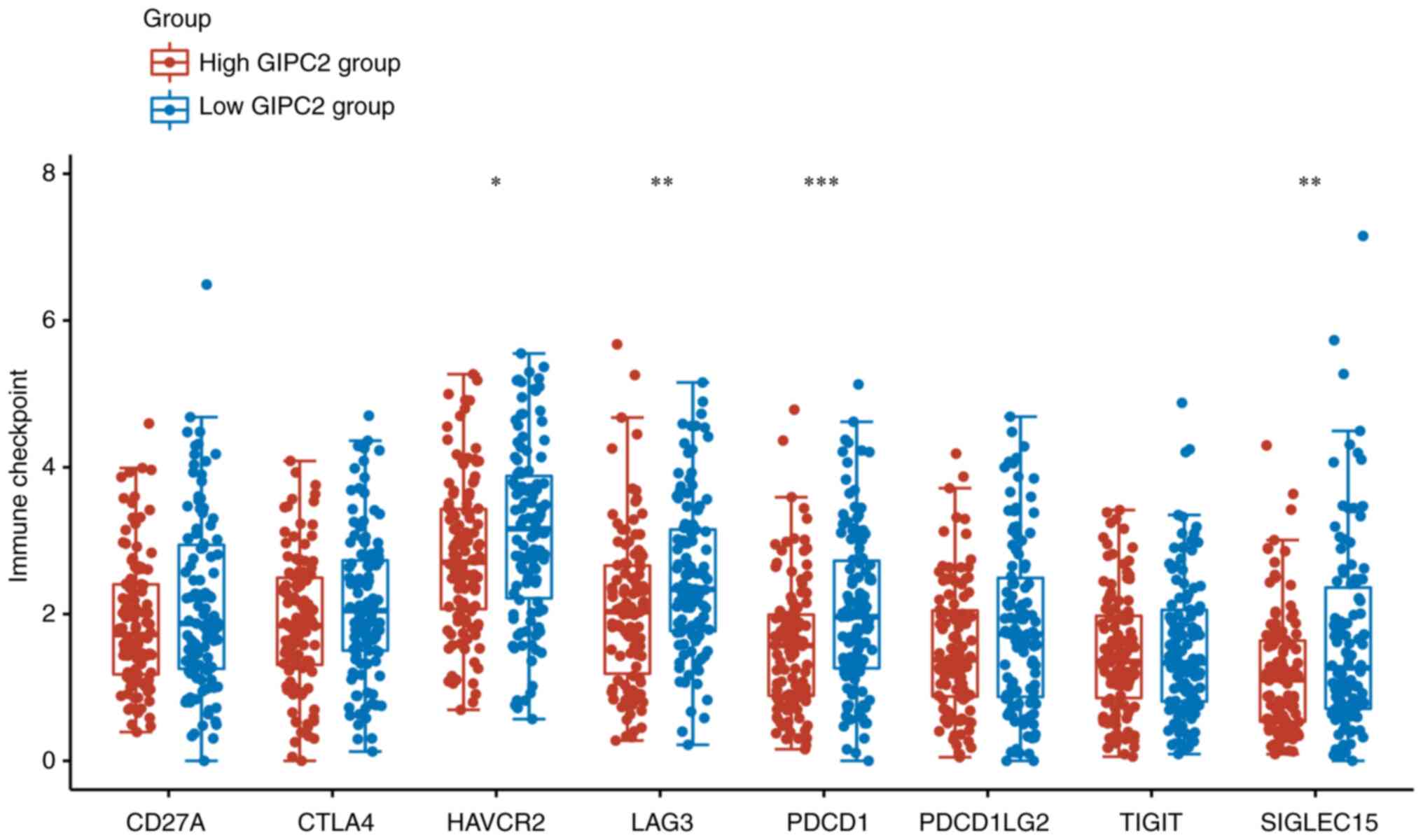
immune checkpoint-associated genes, including HAVCR2, LAG3, PDCD1
and SIGLEC15 were significantly higher in the low GIPC2 expression
group compared with in the GIPC2 high expression group (Fig. 7).
Discussion
GIPC2 is an important member of the PDZ domain
family, and its abnormal expression has previously been reported to
be associated with the development of tumors and abnormal embryonic
development (19–22). Notably, GIPC2 expression is
significantly increased in diffuse gastric cancer cell lines,
including the OKAJIMA, TMK1, MKN45 and KATO-I cell lines; however,
the expression of GIPC2 has been shown to be negligible in the
HL-60 leukemia cell line, HeLaS3 cervical cancer cell line, K-562
chronic myeloid leukemia cell line, Burkitt lymphoma, SW480 colon
cancer cell line and A549 lung cancer cell line (19).
The PDZ domain is the main functional domain of the
GIPC2 protein, which interacts with FZD3-type Wnt receptor,
insulin-like growth factor receptor, receptor tyrosine kinase A
receptor, TGF-β R type II receptor and the RGS19 protein of the RGS
family (23–25). The RGS19 protein is an important
protein that regulates heterotrimers in the G-protein signaling
pathway. Therefore, GIPC2 may have an important role in
tumorigenesis and embryonic development through promoting the
interaction between G-protein heterotrimers and Wnt receptors or
receptor tyrosine kinases (26,27).
Somatic mutations of GIPC2 in different types of cancer have been
detected by whole-genome or whole-exome sequencing. Cancer genomic
testing of ovarian cancer cases identified a G102E missense
mutation in GIPC2 (28). D125N and
E288K missense mutations of GIPC2 have also been identified in
malignant melanoma (9).
Furthermore, the F74Y and R312Q missense mutations, and E216X
nonsense mutation of GIPC2 have been identified upon performing a
colorectal cancer genome-level analysis (29). The E216X nonsense mutation is a
deleterious mutation that causes the loss of the GH2 domain, which
enables GIPC2 to bind to MY06 (30). Collectively, these data suggested
that GIPC2 serves certain biological functional roles in different
diseases.
To the best of our knowledge, the present study is
the first to investigate the role of GIPC2 in COAD. The results
demonstrated that the expression of GIPC2 was reduced in COAD
tissues compared with that in normal tissues. Moreover, the
expression levels of GIPC2 were negatively associated with COAD
tumor stage, lymph node status and lymphatic invasion.
Additionally, the survival analysis revealed that high expression
of GIPC2 was significantly associated with favorable OS, DSS and
PFI in patients with COAD. The results of the regression analysis
also suggested that GIPC2 was an independent prognostic factor for
COAD. Taken together, these findings suggested that GIPC2 may act
as a prognostic biomarker for COAD.
Co-expressed genes often have similar functions. The
results of the co-expression analysis performed in the present
study identified a significant positive correlation between GIPC2
and EPCAM, LRRC8D, EPB41L4B, ACSL5 and CDS1. Several of these genes
have been reported to serve important roles in maintaining normal
intestinal mucosal function and cancer resistance (31–35),
thus indicating that GIPC2 and its co-expressed genes may serve as
potential prognostic markers for COAD. At present, a large number
of studies have shown that members of the GIPC family are able to
fully exert their role as adaptors and interact with a variety of
proteins, including RGS19/GAIP, MY06 and type III TGF-P receptors,
and subsequently participate in the regulation of a variety of
biological processes, including cell signaling, transmembrane
protein transport, cell movement and endocytosis (36,37).
To explore the underlying biological mechanism of GIPC2, GO and
KEGG analyses, and GSEA were performed on genes co-expressed with
GIPC2 in the present study. The enrichment analysis of the top 100
co-expressed genes and the GSEA revealed that ‘intestinal
absorption’, ‘regulation of microvillus organization’, ‘microvillus
organization’, ‘cell cycle checkpoints’, ‘DNA replication’ and
‘mitosis-associated’ pathways were significantly enriched. Although
certain pathways, including cell cycle checkpoints, DNA replication
and mitosis, have been verified in the occurrence and development
of cancer (38), further
mechanistic studies are required to fully elucidate their roles in
the association between GIPC2 and COAD.
An important finding in the present study was
identifying the association between GIPC2 expression and the level
of immune cell infiltration in COAD. Analysis of the results
obtained using the CIBERSORT algorithm demonstrated that plasma B
cells, resting CD4+ memory T cells, activated
CD4+ memory T cells, activated myeloid dendritic cells
and activated mast cells were present in significantly higher
proportions in the GIPC2 high expression group in COAD. Immune
checkpoints are a class of immunosuppressive molecules that are
expressed on immune cells and are able to regulate the degree of
immune activation (39,40). These checkpoints have an important
role in preventing the occurrence of autoimmunity. Immunotherapy
through immune checkpoints is a treatment method that modulates
T-cell activity to kill tumor cells through a series of pathways,
including co-suppression or co-stimulatory signaling (41,42).
The present study also revealed that there were significant
differences in the expression levels of immune
checkpoint-associated genes, including HAVCR2, LAG3, PDCD1 and
SIGLEC15, between the high and low GIPC2 expression groups. Taken
together, these findings indicated that GIPC2 may have an important
role in regulating tumor-infiltrating immune cells in COAD, and may
be considered a biomarker for immune therapy.
In conclusion, the present study revealed that GIPC2
expression was significantly downregulated in COAD and that it was
associated with malignant progression in patients with COAD.
Furthermore, increased expression levels of GIPC2 may regulate the
level of infiltrating immune cells and proteins involved in various
pathways during COAD progression. With a deeper understanding of
its function, GIPC2 may serve as an independent prognostic factor
for COAD, and therefore may be a target for the diagnosis and
treatment of COAD.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated during and/or analyzed during
the current study are available from the corresponding author on
reasonable request.
Authors' contributions
SZ and KM designed the study, collected data and
performed the analysis. SZ drafted the manuscript. KM performed
immunohistochemistry. Both authors read and approved the final
manuscript. SZ and KM confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
All patients provided written informed consent. The
present study was approved by the Ethics Committee of the People's
Hospital of Tongling City (approval no. 2022002).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GIPC2
|
Gα-interacting protein C-terminus
PDZ-domain-containing family member 2
|
|
COAD
|
colon adenocarcinoma
|
|
IHC
|
immunohistochemistry
|
|
TCGA
|
The Cancer Genome Atlas
|
|
OS
|
overall survival
|
|
DSS
|
disease-specific survival
|
|
PFI
|
progression-free interval
|
|
GEPIA2
|
Gene Expression Profiling Interactive
Analysis 2
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shawki S, Ashburn J, Signs SA and Huang E:
Colon cancer: Inflammation-associated cancer. Surg Oncol Clin N Am.
27:269–287. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu C: Systemic therapy for colon cancer.
Surg Oncol Clin N Am. 27:235–242. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mody K and Bekaii-Saab T: Clinical trials
and progress in metastatic colon cancer. Surg Oncol Clin N Am.
27:349–365. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bao X, Zhang H, Wu W, Cheng S, Dai X, Zhu
X, Fu Q, Tong Z, Liu L, Zheng Y, et al: Analysis of the molecular
nature associated with microsatellite status in colon cancer
identifies clinical implications for immunotherapy. J Immunother
Cancer. 8:e0014372020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ruan H, Leibowitz BJ, Zhang L and Yu J:
Immunogenic cell death in colon cancer prevention and therapy. Mol
Carcinog. 59:783–793. 2020. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lichtenstern CR, Ngu RK, Shalapour S and
Karin M: Immunotherapy, inflammation and colorectal cancer. Cells.
9:6182020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Katoh M: GIPC gene family (review). Int J
Mol Med. 9:585–589. 2002.PubMed/NCBI
|
|
9
|
Katoh M: Functional proteomics, human
genetics and cancer biology of GIPC family members. Exp Mol Med.
45:e262013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Saitoh T, Mine T and Katoh M: Molecular
cloning and characterization of human GIPC3, a novel gene
homologous to human GIPC1 and GIPC2. Int J Oncol. 20:577–582.
2002.PubMed/NCBI
|
|
11
|
De Marco N, Tussellino M, Carotenuto R,
Ronca R, Rizzolio S, Biffo S and Campanella C: Eukaryotic
initiation factor eIF6 modulates the expression of Kermit 2/XGIPC
in IGF-regulated eye development. Dev Biol. 427:148–154. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tussellino M, De Marco N, Campanella C and
Carotenuto R: Involvement of the eukaryotic initiation factor 6 and
kermit2/gipc2 in Xenopus laevis pronephros formation. Int J Dev
Biol. 56:357–362. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Comijn J, Berx G, Vermassen P, Verschueren
K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D and van Roy
F: The two-handed E box binding zinc finger protein SIP1
downregulates E-cadherin and induces invasion. Mol Cell.
7:1267–1278. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang LH, Kalb RG and Strittmatter SM: A
PDZ protein regulates the distribution of the transmembrane
semaphorin, M-SemF. J Biol Chem. 274:14137–14146. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang C, He Y, You Z and Chen X: TMSB10
promotes progression of clear cell renal cell carcinoma via JUN
transcription regulation. Ann Clin Lab Sci. 52:230–239.
2022.PubMed/NCBI
|
|
16
|
Peng W, Li W, Zhang X, Cen W and Liu Y:
The intercorrelation among CCT6A, CDC20, CCNB1, and PLK1
expressions and their clinical value in papillary thyroid carcinoma
prognostication. J Clin Lab Anal. 36:e246092022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sturm G, Finotello F, Petitprez F, Zhang
JD, Baumbach J, Fridman WH, List M and Aneichyk T: Comprehensive
evaluation of transcriptome-based cell-type quantification methods
for immuno-oncology. Bioinformatics. 35:i436–i445. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
R Core Team, . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: 2020, https://www.r-project.org/
|
|
19
|
Kirikoshi H and Katoh M: Up-regulation of
GIPC2 in human gastric cancer. Int J Oncol. 20:1183–1187.
2020.PubMed/NCBI
|
|
20
|
Wang L, Wang J, Yin X, Guan X, Li Y, Xin C
and Liu J: GIPC2 interacts with Fzd7 to promote prostate cancer
metastasis by activating WNT signaling. Oncogene. 41:2609–2623.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu Z, Wan Y, Yang M, Qi X, Dong Z, Huang
J and Xu J: Identification of methylation-driven genes related to
the prognosis of papillary renal cell carcinoma: A study based on
the cancer genome atlas. Cancer Cell Int. 20:2352020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dong Y, Huang Y, Fan C, Wang L, Zhang R,
Li W, Guo Z, Wang D and Zheng Z: GIPC2 is an endocrine-specific
tumor suppressor gene for both sporadic and hereditary tumors of
RET- and SDHB-, but not VHL-associated clusters of
pheochromocytoma/paraganglioma. Cell Death Dis. 12:4442021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu X and Fuentes EJ: Emerging themes in
PDZ domain signaling: Structure, function, and inhibition. Int Rev
Cell Mol Biol. 343:129–218. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Khan Z and Lafon M: PDZ domain-mediated
protein interactions: Therapeutic targets in neurological
disorders. Curr Med Chem. 21:2632–2641. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tso PH, Wang Y, Wong SY, Poon LS, Chan AS
and Wong YH: RGS19 enhances cell proliferation through its
C-terminal PDZ motif. Cell Signal. 22:1700–1707. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu Y, Lou W, Chen G, Ding B, Kuang J,
Zhang Y, Wang C, Duan S, Deng Y and Lu X: Genome-wide screening for
the G-protein-coupled receptor (GPCR) pathway-related therapeutic
gene RGS19 (regulator of G protein signaling 19) in bladder cancer.
Bioengineered. 12:5892–5903. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ji YR, Kim MO, Kim SH, Yu DH, Shin MJ, Kim
HJ, Yuh HS, Bae KB, Kim JY, Park HD, et al: Effects of regulator of
G protein signaling 19 (RGS19) on heart development and function. J
Biol Chem. 285:28627–28634. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cancer Genome Atlas Research Network, .
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Berger MF, Hodis E, Heffernan TP, Deribe
YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E,
Ghosh P, et al: Melanoma genome sequencing reveals frequent PREX2
mutations. Nature. 485:502–506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cancer Genome Atlas Network, .
Comprehensive molecular characterization of human colon and rectal
cancer. Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McGowan CH: Checking in on Cds1 (Chk2): A
checkpoint kinase and tumor suppressor. Bioessays. 24:502–511.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li S, Ma J, Si Y, Cheng S, Hu M, Zhi X, Li
B, Yu H and Jiang WG: Differential expression and functions of Ehm2
transcript variants in lung adenocarcinoma. Int J Oncol.
54:1747–1758. 2019.PubMed/NCBI
|
|
33
|
Quan J, Bode AM and Luo X: ACSL family:
The regulatory mechanisms and therapeutic implications in cancer.
Eur J Pharmacol. 909:1743972021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma W, Li T, Wu S, Li J, Wang X and Li H:
LOX and ACSL5 as potential relapse markers for pancreatic cancer
patients. Cancer Biol Ther. 20:787–798. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hartmann F, Sparla D, Tute E, Tamm M,
Schneider U, Jeon MK, Kasperk R, Gassler N and Kaemmerer E: Low
acyl-CoA synthetase 5 expression in colorectal carcinomas is
prognostic for early tumour recurrence. Pathol Res Pract.
213:261–266. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jeanneteau F, Guillin O, Diaz J, Griffon N
and Sokoloff P: GIPC recruits GAIP (RGS19) to attenuate dopamine D2
receptor signaling. Mol Biol Cell. 15:4926–4937. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ding X, Philip S, Martin BK, Pang Y,
Burkett S, Swing DA, Pamala C, Ritt DA, Zhou M, Morrison DK, et al:
Survival of BRCA2-deficient cells is promoted by GIPC3, a novel
genetic interactor of BRCA2. Genetics. 207:1335–1345. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Williams GH and Stoeber K: The cell cycle
and cancer. J Pathol. 226:352–364. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Topalian SL, Taube JM, Anders RA and
Pardoll DM: Mechanism-driven biomarkers to guide immune checkpoint
blockade in cancer therapy. Nat Rev Cancer. 16:275–287. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Galluzzi L, Humeau J, Buqué A, Zitvogel L
and Kroemer G: Immunostimulation with chemotherapy in the era of
immune checkpoint inhibitors. Nat Rev Clin Oncol. 17:725–741. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Funes SC, Manrique de Lara A,
Altamirano-Lagos MJ, Mackern-Oberti JP, Escobar-Vera J and Kalergis
AM: Immune checkpoints and the regulation of tolerogenicity in
dendritic cells: Implications for autoimmunity and immunotherapy.
Autoimmun Rev. 18:359–368. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang Y and Zheng J: Functions of immune
checkpoint molecules beyond immune evasion. Adv Exp Med Biol.
1248:201–226. 2020. View Article : Google Scholar : PubMed/NCBI
|