Introduction
Cervical cancer is a very lethal type of cancer with
a high incidence and mortality (1),
~80% of them are cervical squamous cell carcinoma (2). Although in recent years, with the
ongoing developments of treatment technologies and the broad
availability of the HPV vaccine the incidence and mortality rate of
cervical cancer have decreased (3),
the therapeutic efficacy remains limited, and the main cause of the
mortality of cervical cancer patients is the invasive metastasis of
the tumor. Therefore, the mining of molecular markers that can
effectively modulate the invasive metastasis of cervical cancer may
be beneficial in order to reduce the mortality rate of patients
with cervical cancer.
TFAP2A is a crucial mediator of embryonic
development that is aberrantly overexpressed in various types of
cancer, including basal-squamous bladder cancer (4), pancreatic cancer (5), gastric cancer (6), lung adenocarcinoma (7), nasopharyngeal carcinoma (8) and ovarian cancer (9); TFAP2A mainly functions as a
transcription factor that can promote the expression of particular
genes, including direct upregulation of bone morphogenetic protein
4 (BMP4) promoting angiogenesis and leads to drug resistance
(10) and induction of keratin 16
and transforming growth factor-β (TGF-β) overexpression promoting
the epithelial-mesenchymal transition of tumors (11,12).
Accumulating evidence has demonstrated that TFAP2A has potential
use as a diagnostic marker for specific tumors, and it is involved
in cancer cell proliferation, invasion and migration (13). A high TFAP2A expression has been
shown to be associated with a poor tumor prognosis and an increased
risk of recurrence (14). It is
also involved in tumor adaptability to hypoxia and food deprivation
(8,15). The aforementioned findings suggested
that TFAP2A is involved in cancer genesis and progression. However,
the role of TFAP2A in cervical cancer remains unknown.
Programmed death-ligand 1 (PD-L1) is mainly
expressed on the surface of tumor cells, and an increased PD-L1
expression is associated with the poor survival of cancer patients
(16). In addition, its
immunosuppressive signal activation results in the immunological
tolerance in tumor cells (17) and
it promotes tumor stem cell proliferation (18), indicating that PD-L1 is associated
with cancer progression. Previously, immunosuppressants that target
PD-L1 have exhibited exceptional efficacy in the treatment of
cervical cancer (19). However,
certain patients do not benefit from immunotherapy due to
resistance. The response rate of PD-1 antibodies for cervical
cancer has been reported to be only ~20% (20). Therefore, there is a need for the
understanding of the mechanisms regulating PD-L1 in tumors in order
to address the limitations of current treatments. PD-L1 regulation
is also diverse (21). At present,
the association between TFAP2A and PD-L1 in cervical cancer is not
clear and it would thus be of interest to investigate this.
In the present study, the expression of TFAP2A was
first evaluated and it was found that it was highly expressed in
cervical cancer and was associated with a poor survival rate of
patients with cervical cancer. Furthermore, it was identified that
TFAP2A functions as the transcription factor of PD-L1 and PD-L1 can
also elevate the expression of TFAP2A, which indicates a positive
feedback loop between them. In vivo experiments demonstrated
that the decreased expression of TFAP2A inhibited cervical tumor
growth. The present study demonstrated that TFAP2A may function as
a molecular marker for cervical cancer and revealed a positive
feedback loop between TFAP2A and PD-L1.
Materials and methods
Data collection
In the present study, four GSE datasets were used:
GSE39001 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39001),
GSE63514 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63514),
GSE9750 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE9750)
and GSE52903 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE52903).
These datasets were utilized to evaluate the mRNA expression of
TFAP2A between clinical cancer specimens and normal specimens.
GSE52903 and The Cancer Genome Atlas (https://xenabrowser.net/) were used to analyze the
association between TFAP2A and the clinicopathological
characteristics of patients with cervical cancer, as well as to
examine the link between TFAP2A and PDL1.
Tissue sample collection
Clinical samples, including 91 cervical cancer
tissues and 30 normal cervical tissues, were obtained from the
Zhongnan Hospital of Wuhan University (Wuhan, China) from January
2019 to December 2021. All procedures were approved (approval no.
2022120K) by the Ethical board of Zhongnan Hospital of Wuhan
University (Wuhan, China). Of the 91 cervical cancer tissues, 80
were squamous carcinomas and 11 were adenocarcinomas.
Cells and cell culture
For the purposes of the present study, three human
cervical cancer cell lines (HeLa, SiHa and C33a) and normal
cervical epithelial cell lines (END1/E6E7) were selected. The cell
lines were purchased from the American Type Culture Collection
(ATCC). The cell culture medium consisted of 90% DMEM or RPMI-1640,
10% fresh inactivated fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin-streptomycin solution (Thermo
Fisher Scientific, Inc.). The culture conditions were set in an
incubator with 5% CO2, 37°C and saturated humidity.
Cell transfection
PD-L1 and TFAP2A overexpression plasmids were
constructed by inserting full-length cDNA (482 ng/µl) into target
vectors (GV238 and GV710; Shanghai GeneChem Co., Ltd.). PD-L1 (591
ng/µl), TFAP2A (377 ng/µl), and HPV 16 E6 (483 ng/µl) short hairpin
RNA were synthesized and cloned into pLKO.1 TRC (Shanghai GeneChem
Co., Ltd.) vector to knock down the target genes. The
aforementioned overexpression plasmids and gene knockdown plasmid
were transfected into cervical cancer cells using Lipofectamine
2000® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The effects of plasmid transfection were detected after 48 h
using western blot analysis. The sequences used are presented in
Table SI.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to collect and extract total RNA
from the cervical cancer cells. Following 15 min (50°C, 10 min;
85°C, 5 min) of RNA reverse transcription (Prime ScriptTM RT Master
Mix; used according to the manufacturer's protocol), the Ct values
of the samples were monitored by qPCR. The thermocycling conditions
were as follows: i) Pre-denaturation at 95°C for 30 sec; ii) cycle
reaction (40 times) at 95°C for 10 sec and at 60°C for 30 sec; and
dissolution curve: 95°C for 15 sec, 60°C for 60 sec, 15°C for 4
sec. SYBR-Green Master mix (Thermo Fisher Scientific, Inc.) was
used for qPCR and data were calculated using the 2−ΔΔCq
method (22). GAPDH was used as the
reference gene. The primers used are listed in Table SI.
Western blot analysis
After washing the cells with ice-cold PBS, protein
lysate (RIPA lysate: PMSF ratio, 100:1; Beyotime Institute of
Biotechnology) was added and collected. The BCA method (Tiangen
Biotech Co., Ltd.) was used to measure the protein concentration.
Protein loading buffer was added and boiled at 100°C and stored at
−80°C. The exact amount of protein (30 µg) was separated with 12%
SDS-PAGE. The electrophoresis time was 1–1.5 h, and the proteins
were then transferred onto PVDF membranes (Bio-Rad Laboratories,
Inc.) and blocked with 5% skimmed milk at room temperature for 2 h.
Corresponding diluted primary antibodies [PD-L1 (1:1,000), TFAP2A
(1:1,000), GAPDH (1:5,000) and HPV 16 E6 (1:1,000) were then added
followed by overnight incubation at 4°C. An HRP-labeled secondary
antibody (1:10,000) was then added and incubated at room
temperature for 1 h. ECL chemiluminescence imaging system
(Tanon-5200; Tanon Science and Technology Co., Ltd.) was used for
imaging. The antibodies used for western blot analysis are listed
in Table SII.
Cell Counting Kit-8 (CCK-8) assay
The transfected HeLa and SiHa cells were transferred
into 96-well plates (5×103 cells/well) and 10 µl/well of
CCK-8 solution [Multisciences (Lianke) Biotech Co., Ltd] was added
at 0, 24, 48, 72 and 96 h. The enzyme marker (PerkinElmer, Inc.)
was used for detection of the absorbance at 450 nm after 2 h of
incubation at 37°C.
Wound healing assay
The transfected HeLa and SiHa cells were laid flat
in six-well plates (1×105 cells/well), and when the
density reached ~60%, a wound was created in the cell monolayer
using a 200-µl pipette tip and marked and cultured with medium
containing 2% FBS, followed by observation of the cells under a
light microscope (Olympus Corporation) for 0 and 48 h and images
were captured.
Transwell assay
Cellular migration and invasion were examined using
Transwell assay. Cell invasion assays were performed in advance.
The diluted Matrigel gel (BD Biosciences) was added to the
Transwell chamber (Corning, Inc.) at 70 µl/well and incubated at
37°C until set. Subsequently, 200 µl cell dilution containing 1%
FBS medium (1×105/ml) were added to the upper chamber,
while 800 µl medium containing 10% FBS were added to the bottom
chamber. Following 48 h of incubation at 37°C, the cells were
subjected at room temperature to fixation 30 min with formaldehyde
and crystal violet (Beyotime Institute of Biotechnology) staining
20 min and placed under a fluorescent microscope (Olympus
Corporation) to observe and obtain images for counting.
Clonal formation assay
The transfected cells were spread flat in six-well
plates at a concentration of 300 cells/well, observed and cultured
for 2 weeks. After the number of cells in the minimal clonal cell
mass was >50, formaldehyde fixation for 30 min and 0.1% crystal
violet solution staining for 20 min were performed and images were
obtained by a fluorescent microscope (Olympus Corporation).
Apoptosis detection
The transfected cells were digested, resuspended and
stained according to the instructions provided with the FITC
Annexin V apoptosis detection kit [cat. no. AP104; Multisciences
(Lianke) Biotech Co., Ltd.]. The number of apoptotic cells was
immediately detected and counted using a flow cytometer (cut flex;
Beckman Coulter, Inc.).
Dual-luciferase reporter assays
An overexpression plasmid (pGL3 plasmid) containing
PD-L1 gene promoters of different lengths (P, P1, P2, P3, P4 and
P5) and TFAP2A overexpression plasmid (pCDNA3.1 plasmid) were
constructed. In 24-well plates, 0.5 µg target plasmids and 0.01 µg
pRL-TK plasmid (Shanghai GeneChem Co., Ltd.) per well were
co-transfected into 293T cells (ATCC) using Lipofectamine
2000® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The cells were analyzed to determine the activity of
luciferase using a Dual-Luciferase® Reporter Assay
System (Promega Corporation) following 48 h of transfection.
Normalization was performed by comparison with Renilla
luciferase activity.
Chromatin immunoprecipitation
(ChIP)
ChIP-qPCR primers containing the binding sequences
of the PD-L1 promoter and TFAP2A gene were designed, and the cells
were cross-linked with 1% formaldehyde, sonicated to fragment DNA,
and the corresponding 4 µg Ab or IgG and 50 µl protein A/G-agarose
(cat. no. 20421; MilliporeSigma) were then added for
immunoprecipitation. The samples were washed and uncross-linked
after rotating and mixing overnight at 4°C, and the PCR products
were recovered using the PCR product recovery kit (the IP products
were purified using the PCR product recovery kit; Tiangen Biotech
Co., Ltd.), and finally RT-qPCR was performed using the designed
primers, and positive controls were set to analyze the enriched DNA
fragments (23). The primers used
are listed in Table SI.
Tumor xenograft model
The animal experiment was approved (approval no.
ZN2021257) by the Experimental Animal Ethics Committee of Zhongnan
Hospital of Wuhan University (Wuhan, China). A total of 12 BALB/C
nude mice (female, weighing 18–20 g, aged 4–6 weeks) were housed in
an SPF-grade environment at the Animal Testing Center of the
Central South Hospital of Wuhan University in Vital River, Beijing,
China. The housing temperature was maintained at 22±2°C, with a
relative humidity of 40–60%. A 12-h diurnal cycle was applied, and
the mice were fed ad libitum with standard chow diet and water. All
mice were experimented on after 1 week of acclimatization. The
sh-TFAP2A and sh-NCs HeLa cells (5×106 cells, 100 µl
DMEM-containing medium) were stably transfected with lentivirus and
injected subcutaneously into the axilla of the nude mice. Tumor
volume and mouse body weight were recorded every other day for ~18
days until tumor volume did not exceed 2,000 mm3. The
mice were anesthetized with 1–1.5% isoflurane and euthanized by
cervical dislocation. Tumors were excised from each mouse and their
weights were calculated for statistical analysis. The tumor volume
was calculated as follows: Volume=(length ×
width)2/2.
Immunohistochemistry (IHC) and
hematoxylin-eosin (H&E) staining
The collected tumor tissues were fixed with 4%
paraformaldehyde for 24 h at room temperature, alcohol-gradient
dehydrated and paraffin-embedded and sectioned strictly according
to the IHC staining and H&E staining procedures. The 4-µm
sections were then stained with antibodies against TFAP2A (1:450;
cat. no. 67076-1-Ig; Proteintech Group, Inc.) and PD-L1 (1:150;
cat. no. 66248-1-Ig; Proteintech Group, Inc.). Incubation was
performed overnight at 4°C, and secondary antibodies [Goat
Anti-Rabbit IgG H&L (HRP; 1:1,000; cat. no. ab6721; Abcam)]
were used for re-staining at room temperature for 1 h followed by
counterstaining (10–20 sec) with 1X hematoxylin solution (cat. no.
DA1010; Beijing Solarbio Science & Technology Co., Ltd.) and
mounting, followed by staining analysis, a procedure independently
analyzed by two experienced pathologists. H&E staining
solidifies the data to identify whether the tissue type is cervical
cancer. The data of IHC staining were scored using a
semi-quantitative scoring system. The average gray value of
positive cells (staining intensity) and the percentage of positive
areas (stained area) were used together as a measure of IHC
staining, resulting in four scores: High positive (3+), positive
(2+), low positive (1+) and negative (0). Finally, the scores of
the three replicates were summed to obtain the final score; the
median was used as a cut-off to distinguish between high and low
protein expression.
Statistical analysis
SPSS 25.0 statistical software (IBM Corp.) was used
for statistical analysis, GraphPad Version 5.0 Software (Dotmatics)
for analysis and graphing, and ImageJ-win32 software (National
Institutes of Health) for image analysis. Kaplan-Meier plotter
followed by the log-rank test was used for survival analysis. Data
are expressed as the mean ± SD, and the unpaired t-test was used
for statistical analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
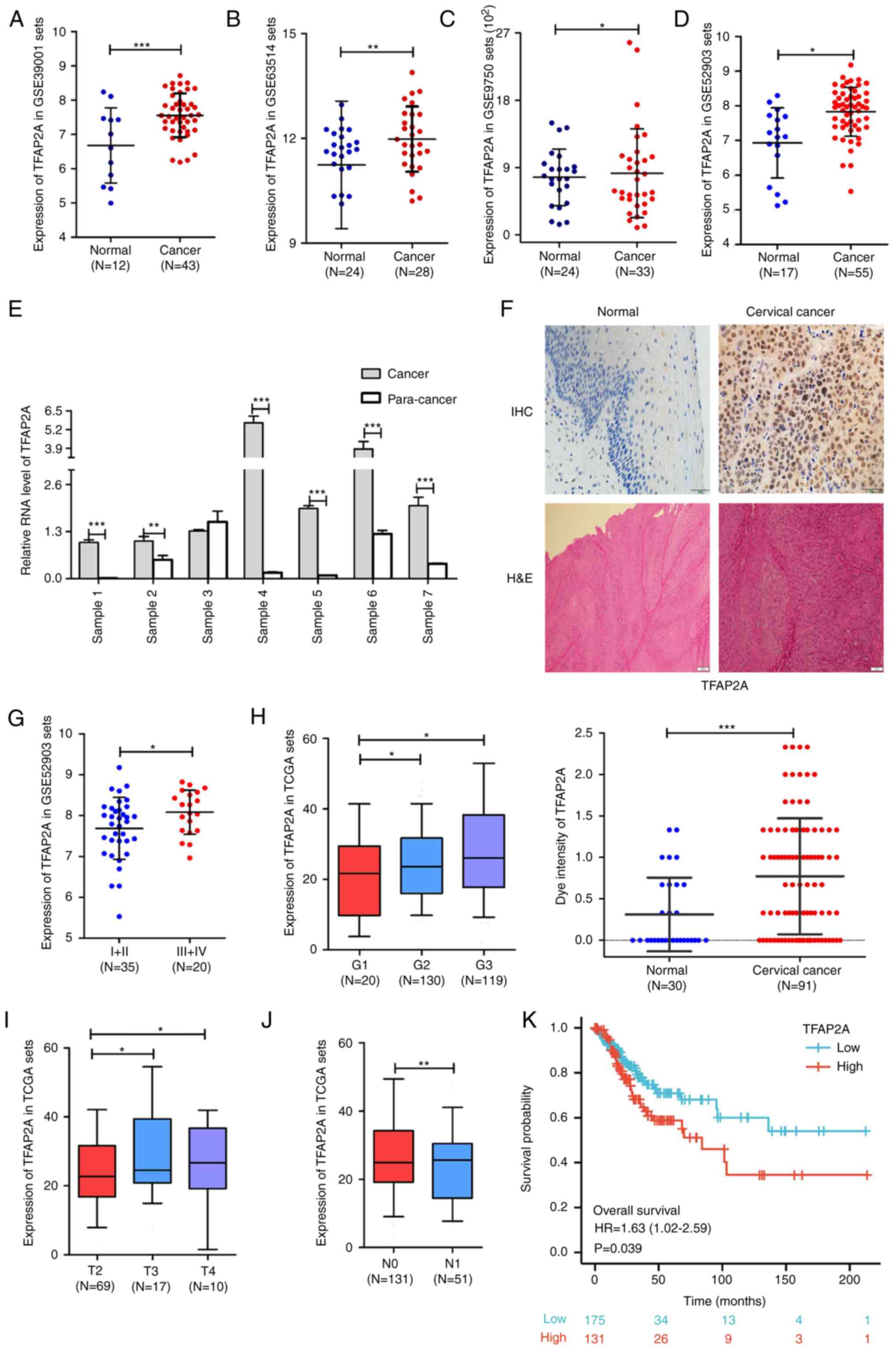
TFAP2A is highly expressed in cervical
cancer tissues and is associated with the clinicopathological
features of patients
The analysis results of multiple cervical cancer
RNA-sequencing datasets in the Gene Expression Omnibus database
suggested that the expression level of TFAP2A was significantly
higher in cervical cancer tissues than in normal cervical tissues
(Fig. 1A-D). In addition, seven
sets of paired cancer-paraneoplastic tissues were collected and
TFAP2A gene expression was evaluated. The results from RT-qPCR
further supported the data mining results (Fig. 1E). Furthermore, 30 normal cervical
tissues and 91 cervical cancer tissues were collected for IHC and
H&E staining to further assess TFAP2A gene expression. The
results suggested that the positive staining of TFAP2A was
significantly higher in the cancerous tissues than in normal
tissues (Fig. 1F). At the same
time, the analysis of the database cervical cancer samples revealed
that patients with high levels of TFAP2A mRNA expression had a
higher FIGO grade (Fig. 1G), a
higher pathological stage (Fig. 1H)
and a larger tumor size (Fig. 1I),
as well as more lymph node metastases when compared with those with
a low TFAP2A expression (Fig. 1J).
Kaplan-Meier survival analysis revealed that a high expression of
TFAP2A was positively associated with a poor overall survival of
patients with cervical cancer (Fig.
1K). It was also validated in clinical samples, as shown in
Table I, that patients with a high
TFAP2A expression exhibited fewer births, higher human epididymis
protein 4 levels, a higher FIGO grade, more HPV infections, and
were more prone to vascular invasion. These findings suggested that
TFAP2A is highly expressed in cervical cancer tissues and is
associated with the clinical characteristics and prognosis of
patients with cervical cancer.
 | Table I.Association between TFAP2A and the
clinicopathological parameters of patients with cervical
cancer. |
Table I.
Association between TFAP2A and the
clinicopathological parameters of patients with cervical
cancer.
|
| Expression level of
TFAP2A |
|
|---|
|
|
|
|
|---|
| Clinicopathological
characteristics | Low | High | P-value |
|---|
| Total cases, n
(%) | 46 | 45 |
|
| Age, years |
|
| 0.159 |
|
≤55 | 24 (26.4%) | 30 (33.0%) |
|
|
>55 | 22 (24.2%) | 15 (16.5%) |
|
| Underlying
diseases |
|
| 0.763 |
|
Yes | 35 (38.5%) | 33 (36.3%) |
|
| No | 11 (12.1%) | 12 (13.2%) |
|
| Pregnancy |
|
| 0.599 |
| ≤3 | 22 (24.1%) | 24 (26.4%) |
|
|
>3 | 24 (26.4%) | 21 (23.1%) |
|
| Childbirth |
|
| 0.014 |
| ≤2 | 27 (30.0%) | 37 (40.7%) |
|
|
>2 | 19 (20.9%) | 8 (8.8%) |
|
| CEA (ng/ml) |
|
| 0.526 |
| ≤5 | 40 (45.0%) | 39 (43.9%) |
|
|
>5 | 4 (4.5%) | 6 (6.7%) |
|
| CA125 (U/ml) |
|
| 0.080 |
|
≤35 | 38 (43.2%) | 44 (50.0%) |
|
|
>35 | 5 (5.7%) | 1 (1.1%) |
|
| HE4 (pmol/ml) |
|
| 0.044 |
|
≤60.5 | 32 (41.6%) | 25 (50.6%) |
|
|
>60.5 | 6 (7.8%) | 14 (18.2%) |
|
| SCC (ng/ml) |
|
| 0.193 |
|
≤1.5 | 6 (8.0%) | 12 (16.0%) |
|
|
>1.5 | 29 (38.7%) | 28 (37.3%) |
|
| FIGO stage |
|
| 0.043 |
|
Ib-IIa | 39 (43.3%) | 31 (34.4%) |
|
|
IIb-IV | 6 (6.7%) | 14 (15.6%) |
|
| HPV infection |
|
| 0.013 |
|
Negative | 12 (13.2%) | 3 (3.3%) |
|
|
Positive | 34 (37.4%) | 42 (46.2%) |
|
| Lymph node
metastasis |
|
| 0.534 |
|
Yes | 5 (5.8%) | 7 (8.1%) |
|
| No | 38 (44.2%) | 36 (41.9%) |
|
| Vascular
invasion |
|
| 0.006 |
|
Yes | 18 (28.1%) | 8 (12.5%) |
|
| No | 13 (20.3%) | 25 (39.1%) |
|
| Nerve
Violation |
|
| 0.772 |
|
Yes | 18 (34.6%) | 20 (38.5%) |
|
| No | 6 (11.5%) | 8 (15.4%) |
|
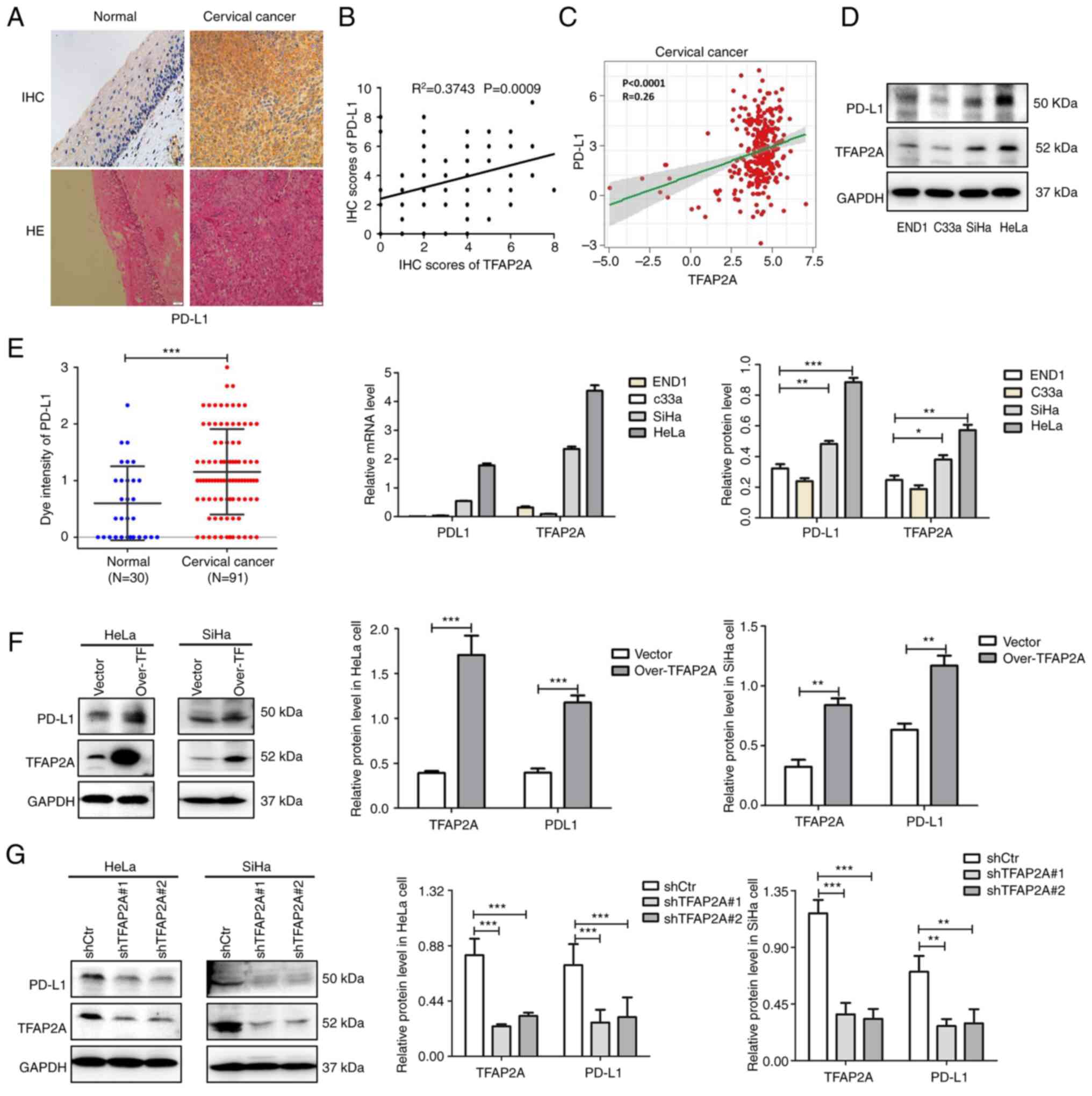
TFAP2A positively regulates PD-L1
expression in cervical cancer tissues and cell lines
The IHC staining results of 30 normal cervical
tissues and 91 cervical cancerous tissues revealed that positive
PD-L1 staining was significantly higher in cancerous tissues than
in normal tissues (Fig. 2A).
Combined with the database analysis, a significant positive
association was identified between PD-L1 and TFAP2A protein
expression (Fig. 2B and C).
In addition, the expression of TFAP2A and PD-L1 was
examined in three cervical cancer cell lines and cervical
epithelial cells. It was found that TFAP2A and PD-L1 expression was
higher in the HeLa and SiHa cervical cancer cell lines than in the
END1 cells (Fig. 2D and E). To
further validate the association between TFAP2A and PD-L1 in
cervical cancer, TFAP2A was stably knocked down and overexpressed
in HeLa and SiHa cells using lentiviral transfection. Notably, the
protein level of PD-L1 was significantly increased following TFAP2A
overexpression (Fig. 2F), whereas
the protein level of PD-L1 was significantly decreased following
TFAP2A knockdown (Fig. 2G),
indicating that TFAP2A positively regulated PD-L1 expression.
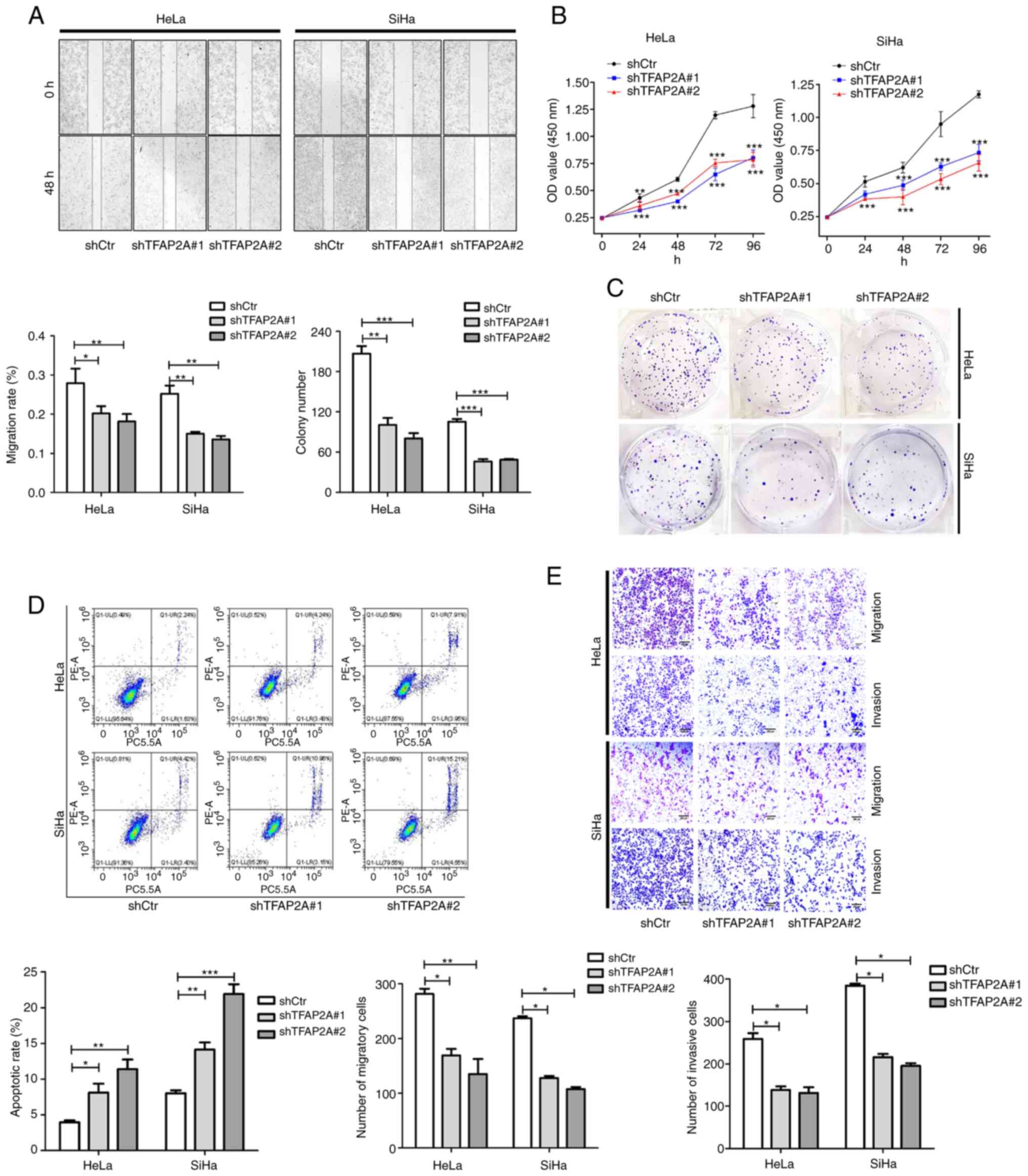
TFAP2A knockdown inhibits cervical
cancer cell proliferation, migration and invasion, and promotes
apoptosis
In HeLa and SiHa cells in which TFAP2A was knocked
down, the role of TFAP2A in cervical cancer was investigated. As
shown by the results of wound healing assay, cell migration was
significantly reduced following TFAP2A knockdown (Fig. 3A). Cell proliferation and colony
formation were also inhibited after TFAP2A knockdown (Fig. 3B and C). Furthermore, TFAP2A
knockdown increased apoptosis (Fig.
3D). Of note, TFAP2A knockdown inhibited cell migration and
invasion (Fig. 3E). These findings
indicated that tumor metastasis was reduced and apoptosis was
promoted following TFAP2A knockdown. These findings indicated that
TFAP2A has potential oncogenic properties.
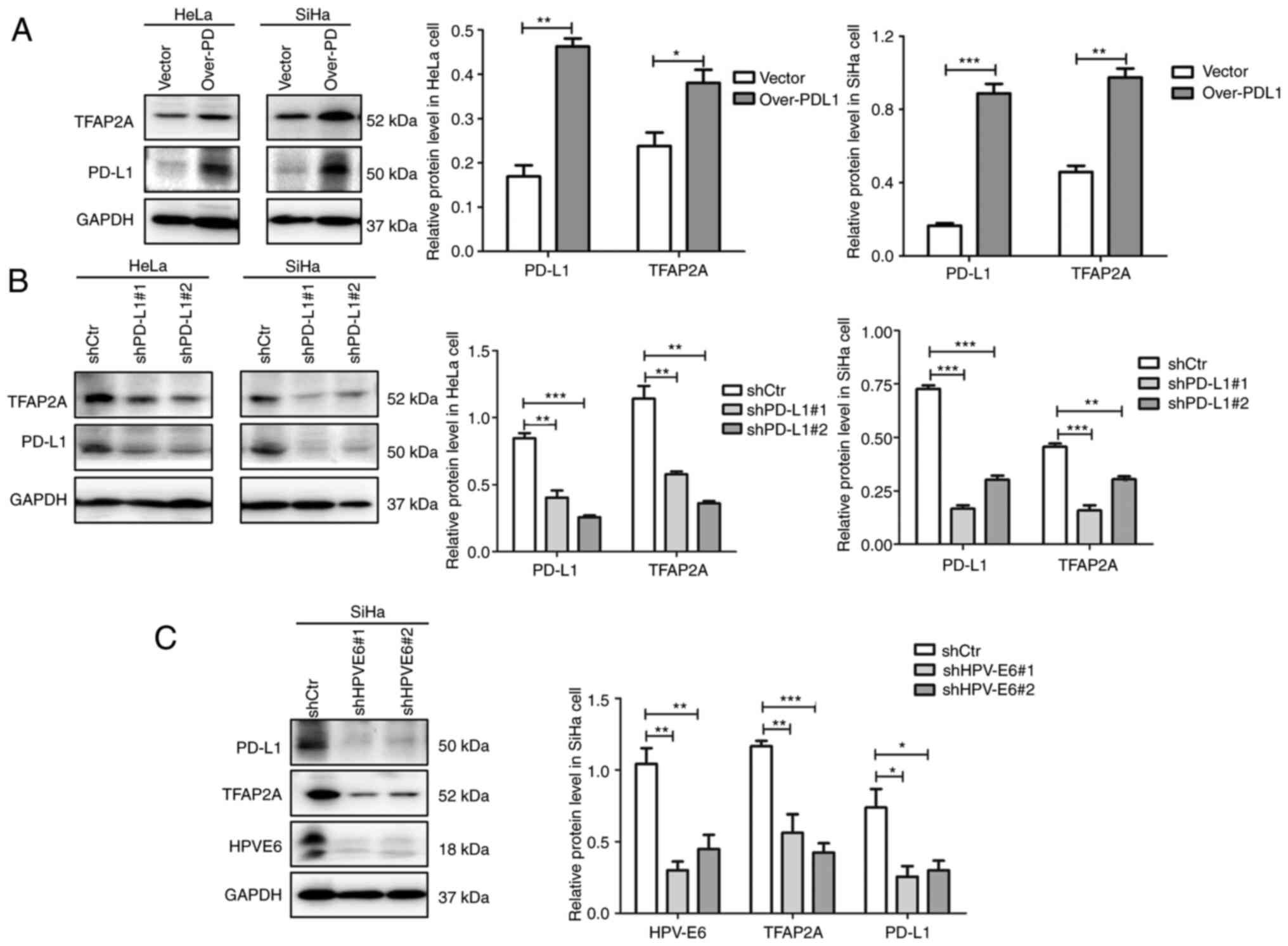
TFAP2A and PD-L1 form a positive
feedback loop that may be influenced by the HPV E6 protein
To further investigate the association between
TFAP2A and PD-L1, PD-L1 was overexpressed and knocked down in the
HeLa and SiHa cervical cancer cell lines using plasmid
transfection. Notably, it was found that the overexpression of
PD-L1 resulted in an elevated protein level of TFAP2A (Fig. 4A), whereas the knockdown of PD-L1
resulted in a decrease in its expression (Fig. 4B). Such consistent changes suggest
that TFAP2A and PD-L1 regulate each other, forming a positive
feedback loop. The results based on Table I suggested that TFAP2A expression is
linked to HPV infection. In the present study, HPV 16 E6 was
knocked down in SiHa cells to further investigate this link. The
results revealed that the knockdown of E6 protein led to a
significant decrease in TFAP2A and PD-L1 protein expression
(Fig. 4C), suggesting that HPV E6
protein regulates TFAP2A and PD-L1 protein expression. A positive
feedback regulates TFAP2A and PD-L1, according to these findings.
HPV E6 protein may upregulate TFAP2A expression, resulting in an
increased PD-L1 protein expression.
TFAP2A promotes the proliferation and
migration, and inhibits the apoptosis of cervical cancer cells via
PD-L1
To investigate the functional axis between TFAP2A
and PD-L1 in cervical cancer, sh-TFAP2A stable cell lines were
generated using the SiHa and HeLa cells, which were then
transiently transfected with a PD-L1 overexpression plasmid
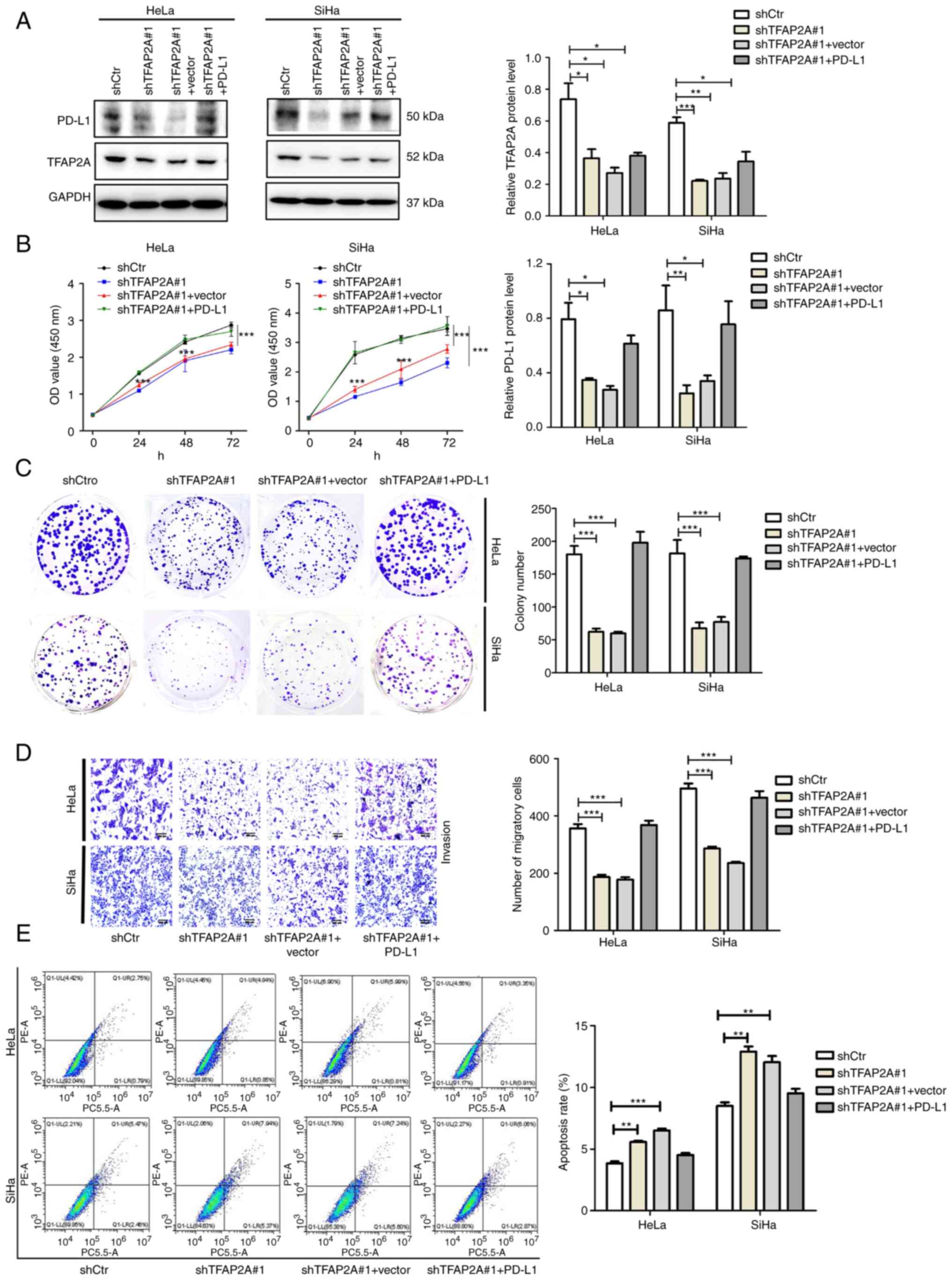
(sh-TFAP2A#1 + PD-L1). The results of western blot analysis
revealed that the expression of PD-L1 was also significantly
decreased following the knockdown of TFAP2A, and the expression of
PD-L1 was partially restored following the overexpression of PD-L1
in the sh-TFAP2A group (Fig. 5A).
In the sh-TFAP2A#1 + PD-L1 group, cell proliferation was increased
(Fig. 5B), colony formation was
restored (Fig. 5C) and cell
migration was also significantly promoted (Fig. 5D) compared with the sh-TFAP2A#1 +
Vector group. At the same time, the proportion of apoptotic cells
decreased (Fig. 5E). These results
demonstrated that TFAP2A relies on the PD-L1 pathway to regulate
the proliferation, migration and apoptosis of cervical cancer
cells.
TFAP2A suppresses the progression of
cervical cancer by targeting PD-L1
TFAP2A and PD-L1 are closely related and regulate
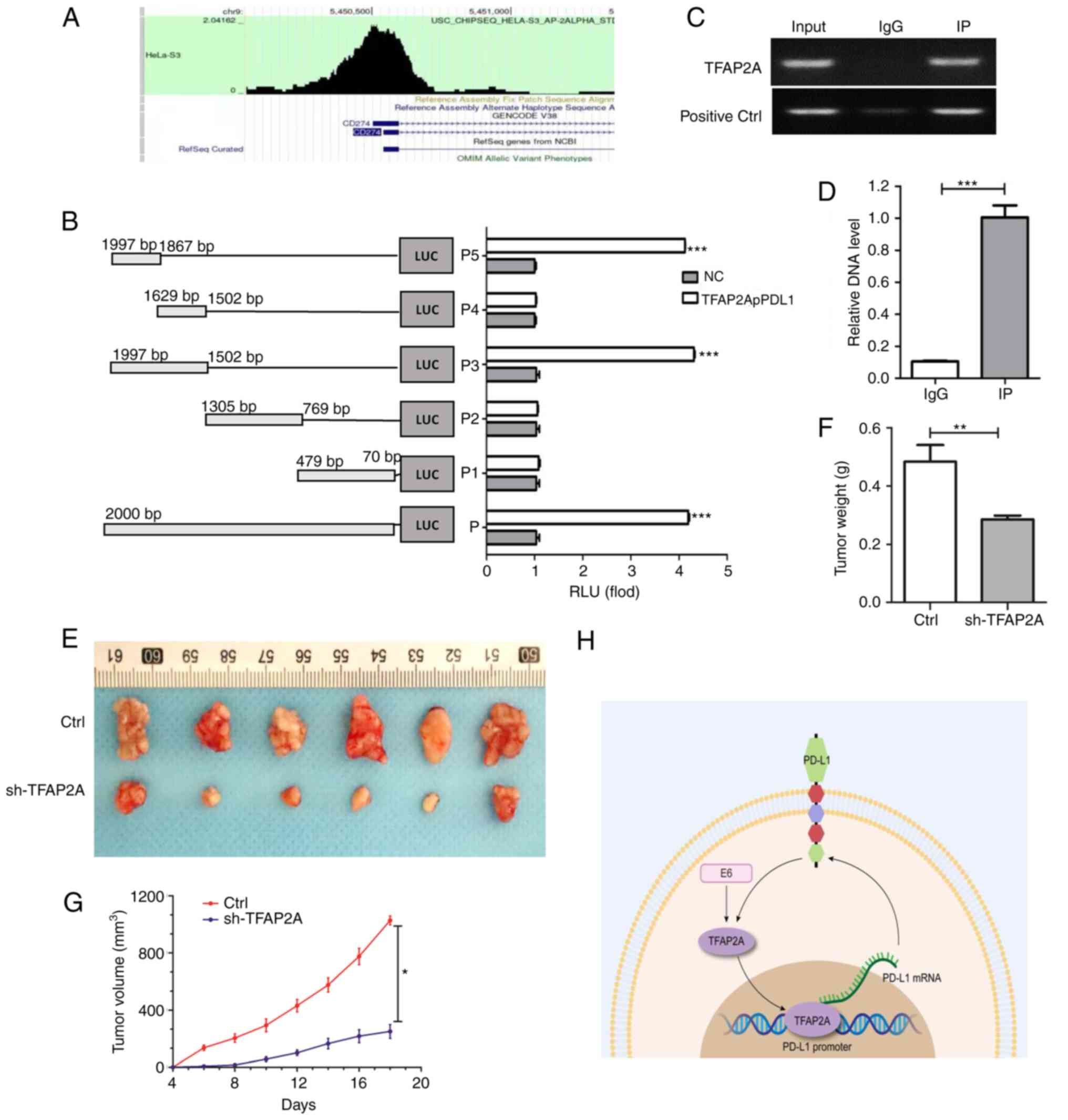
each other. Herein, to investigate the binding between them, the
enrichment of TFAP2A in the PD-L1 promoter region was first
predicted using bioinformatics (Fig.
6A). A subsequent dual luciferase reporter gene assay (Fig. 6B) confirmed that a TFAP2A binding
site was located in the PD-L1 promoter region. RT-qPCR was also
used to examine the immune complexes. The findings revealed that
TFAP2A physically interacted with PD-L1 protein (Fig. 6C and D). Finally, to improve the
understanding of the role of TFAP2A in the development of cervical
cancer in vivo, a mouse cervical cancer transplantation
tumor model was generated using HeLa cells. The results revealed
that the group subjected to knockdown TFAP2A exhibited a slower
tumor growth and a significantly lower final tumor weight and
volume (Fig. 6E-G). TFAP2A
knockdown significantly inhibited the growth of HeLa-derived tumors
in vivo. On the whole, it was found that TFAP2A can promote
the transcription of PD-L1 promoter by binding to it and affect the
expression of HPV 16 E6, thus promoting the progression of cervical
cancer (Fig. 6H).
Discussion
In the present study, it was found that TFAP2A was
highly expressed in cervical cancer tissues and cells, and was
associated with a more advanced tumor stage and distant metastasis.
The silencing of TFAP2A expression significantly inhibited the
proliferation and migration of cervical cancer cells, and promoted
apoptosis. A mouse xenograft tumor model was used to confirm the
tumor-promoting function of TFAP2A. In addition, it was found that
TFAP2A directly targeted PD-L1 to promote cervical cancer cell
growth, and HPV E6 may be a potential influencing factor of this
pathway.
The transcription factor TFAP2A, a‧ AP-2 family
member, has been demonstrated to be involved in essential life
processes, including embryonic ectodermal development, stem cell
differentiation and cell growth and proliferation (24). The role of TFAP2A in carcinogenesis
and development has only recently been recognized. It is highly
expressed in a variety of cancer types, including bladder cancer,
melanoma, head and neck squamous carcinoma (25), gastric cancer (6), ovarian cancer (9), lung adenocarcinoma (7) and nasopharyngeal carcinoma (8). The overexpression of TFAP2A predicts a
very poor prognosis. In head and neck squamous carcinoma, TFAP2A
downregulation has been shown to be associated with a reduced
proliferation (26,27). The high expression of TFAP2A
promotes cancer cell proliferation and inhibits apoptosis (13). In nasopharyngeal carcinoma, TFAP2A
overexpression has been shown to be positively associated with
tumor stage, local infiltration and a decreased overall survival,
and TFAP2 silencing results in a slower proliferation of cancer
cells (8). The silencing of TFAP2A
has been shown to inhibit the proliferation, migration and
invasiveness of esophageal cancer cells, and to enhance apoptosis
(28). The increased expression of
TFAP2A has been demonstrated in gallbladder cancer, and its
inhibition reduces the proliferation, migration and invasion of
gallbladder cancer cells (29).
However, TFAP2A has been revealed to be a tumor suppressor and to
be expressed in low levels in a few malignancies, including
hepatocellular carcinoma, glioma and colon cancer (30). This suggests that TFAP2A has a dual
identity and can serve as an oncogene or tumor suppressor gene,
depending on the tumor type. As a result, the present study aimed
to examine its expression profile in cervical cancer. It was
identified that TFAP2A was highly expressed in cervical cancer and
was associated with tumor stage and lymph node metastasis. It
should be noted that due to the extremely low incidence of cervical
adenocarcinoma (2), the present
study mainly focused on squamous carcinoma of the cervix. More than
80% of the selected public data sets and follow-up clinical
specimens were also cervical squamous carcinoma, and the selected
cell line SiHa was also derived from squamous carcinoma of the
cervix (31). In addition, it was
confirmed that TFAP2A knockdown inhibited the proliferation,
invasion and migration of cervical cancer cells, while promoting
apoptosis, indicating oncogenic potential.
TFAP2A was found to act as a transcription factor to
regulate the level and activity of downstream signaling proteins,
acting in various cancers or different types of cell lines by
activating different signaling pathways. For example, in melanoma,
TFAP2 promotes tumor metastasis by activating the Enhancer of Zeste
Homolog 2 (EZH2). Gene (25).
TFAP2A promotes the growth and survival of nasopharyngeal carcinoma
by targeting the HIF-1 and VEGF pathways in nasopharyngeal
carcinoma cells (8). In lung
cancer, TFAP2A directly upregulates BMP4 to promote angiogenesis
and lead to acquired resistance to anlotinib (32). TFAP2A activates heme oxygenase-1
(HO-1) to promote tumor growth (15), upregulates telomerase to resist
apoptosis (33), induces Keratin 16
(KRT16) and Inositol-Trisphosphate 3-Kinase A (ITPKA)
overexpression, and promotes lung cancer tumorigenicity through
epithelial-mesenchymal transition (7,11). In
addition, several studies have found that certain long non-coding
RNAs or microRNAs (miRs) regulate the post-transcriptional levels
of TFAP2A which affect the biological functions of tumors. For
example, Linc00467 can regulate miR-1285-3p/TFAP2A expression,
thereby facilitating the invasion of head and neck squamous cell
carcinoma cells and inhibiting apoptosis (13). Long-stranded non-coding RNA
TFAP2A-AS1 produces oncogenic effects in oral squamous cell
carcinoma by regulating the miR-1297/TFAP2A axis (34). miR-876-5p inhibits breast cancer
progression by regulating cell proliferation, migration and
invasion in a TFAP2A-dependent manner (35). TFAP2A enhances lung adenocarcinoma
metastasis via miR-16 family/TFAP2A/PSG9/TGF-β signaling pathway
(12). Silencing of lymphocytic
leukemia deletion gene1 (DLEU1) inhibited ovarian cancer cell
proliferation, migration and invasion by regulating the
miR-429/TFAP2A axis (9).
Zinc-finger protein 471 (ZNF471) acts as a tumor suppressor in
gastric cancer by transcriptionally repressing the downstream
targets TFAP2A and Plastin 3 (PLS3) (6). Of note, the present study found that
TFAP2A interacted directly with PD-L1 and produced a hitherto
unknown positive feedback signaling loop in cervical cancer.
PD-L1 is an immunosuppressive signaling protein that
induces tumor immune escape (36).
Previous studies have examined the expression of PD-L1 in cervical
cancer and have reported that PD-L1 is more commonly expressed in
cervical cancer than in normal cervical tissues, ranging from
34.4-96% (37,38). PD-L1 expression has been observed to
be more widespread in cervical squamous cell carcinoma, with a
percentage of tissue-positive samples exceeding 37% (39,40).
These findings demonstrated that PD-L1 is widely expressed in
cervical cancer tumor cells. These results support prior results
regarding the elevated expression of PD-L1 in cervical cancer.
Furthermore, PD-L1 protein is substantially connected with cervical
cancer prognosis, with a higher PD-L1 expression being positively
associated with tumor metastasis (41), tumor progression (42) and a poor prognosis of patients with
cervical cancer (43). In the
present study, TFAP2A was found to promote the proliferation and
migration of cervical cancer cells through PD-L1, while inhibiting
cell apoptosis However, certain researchers have concluded that
PD-L1 expression is not associated with progression-free or overall
survival in patients with cervical cancer (37), and that borderline PD-L1 expression
is associated with an improved prognosis than diffuse PD-L1
expression or no PD-L1 expression (44). The explanation for the contradictory
prior research may be due to the degree and pattern of PD-L1
expression, in addition to the diverse tissue subtypes with varied
PD-L1 expression and function, which can influence the different
prognoses of cervical cancer.
The regulatory mechanisms of PD-L1 expression in
malignancies are complex and diverse, including genomic
abnormalities, transcriptional control, mRNA stability, oncogenic
signaling and protein stability (21). The current study demonstrated that
TFAP2A promoted the expression of PD-L1 by binding to the promoter
region of PD-L1, and its binding site was 1876–1997 bp in the PD-L1
promoter region. But how TFAP2A upregulate PD-L1 remains unknown.
Earlier studies have found that the C-terminal region of AP-2
family proteins contains a helix-turn-helix domain that mediates
dimerization and site-specific DNA sequence GCCNNGGC binding,
thereby inducing transcriptional activation (45). It has also been confirmed that the
activator of human AP-2α is located in amino acids 52–108 (46). In other words, TFAP2A may activate
PD-L1 transcription by specifically recognizing and binding to the
GCCNNNGGC sequence in the 1876–1997 bp region of the PD-L1
promoter. In addition, the transcription process of PD-L1 in the
nucleus is also very complex. Studies have found that under hypoxia
(47), p-Stat3 physically interacts
with PD-L1 to promote its nuclear translocation. Another study
reported that PD-L1 in the nucleus further regulates Gas6
expression and activates the MerTK signaling pathway to promote
proliferation of NSCLC cells (48).
In addition, it was also identified that PD-L1 can
affect the expression of TFAP2A. However, there are no relevant
studies on how PD-L1 affects TFAP2A, but studies have found the
intrinsic function of PD-L1 in tumors and its interaction with
other carcinogenic pathways. The high expression of PD-L1 interacts
with mTOR signaling pathway to promote the growth and autophagy of
ovarian cancer cells and melanoma cells (49). PD-L1 binds to ITGB4 in cervical
cancer (50), triggering AKT/GSK3β
and SNAI1/SIRT3 signaling pathways. PD-L1/ITGB6/STAT3 signaling
axis regulates bladder cancer cell proliferation, glucose
metabolism and chemotherapy resistance (51). PD-L1 regulates the proliferation and
apoptosis of AML cell lines through the PI3K/AKT signaling pathway
(52). However, PI3K-AKT pathway
can affect the expression of TFAP2A (53), suggesting that PI3K-AKT pathway may
be a key mediator in PD-L1 regulation of TFAP2A, which is
interesting and worthy of further exploration.
Finally, the signaling upstream of TFAP2A was
actively investigated and it was identified that the oncogenic
protein HPV E6 may be a potential factor in regulating the
TFAP2A/PD-L1 signaling pathway. Studies have found that the PD-L1
expression was linked to promoting HPV-induced cervical
carcinogenesis. The HPV E5/E6/E7 oncogene stimulates numerous
signaling pathways, including PI3K/AKT, MAPK, hypoxia-inducible
factor 1, STAT3/NF-κB and microRNAs, which regulate the PD-L1/PD-1
axis (54). It has also been
reported that the AP-2 transcription factor family, in which TFAP2A
is located, was found to have binding sites in the transcriptional
control region (upstream regulatory region, URR) of several HPV
types (55–58), including HPV11 and HPV18, suggesting
that AP-2 may be a key activator of viral E6/E7 oncogene
expression, but it was further found to have only a slight effect
on the activity of the E6/E7 oncogene promoter (59). Suggesting that AP-2 itself is not
critical for the E6/E7 promoter, it is also possible that
additional AP-2 binding sites may be present in the viral URRs and
affect the activity of the E6/E7 promoter, as AP-2 is an epithelial
transcription factor and therefore may contribute to the
preferential transcriptional activity of HPV URRs. However, there
is no study on the functional significance of TFAP2A on HPV E6/E7.
The present study found a potential association between TFAP2A and
E6, and also provided a little idea about the regulatory mechanism
of E6 in cervical cancer.
In conclusion, the present study demonstrated the
high expression profile of TFAP2A in cervical cancer and that it is
associated with a poorer prognosis. TFAP2A plays a key role in
promoting cervical cancer progression by directly interacting with
PD-L1. Thus, the present study revealed a possible molecular
mechanism of TFAP2A in cervical cancer. The inhibition of TFAP2A
may therefore provide some theoretical basis for the treatment of
patients with cervical cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 8197103302).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
JY and YG designed the experiments. JY and SW mainly
conducted cell experiments. YG and SW undertook all animal
experiments. JY, YG, SY and HC collated and analyzed the data. JY,
HC and SY drafted the manuscript. YG and HC performed language
correction. JY and YG confirm the authenticity of all the raw data,
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
All procedures regarding human tissue studies were
approved (approval no. 2022120K) by the Ethical board of Zhongnan
Hospital of Wuhan University (Wuhan, China). The waiver of informed
consent has been approved by the ethics Committee. The animal
experiments were performed according to the Animal Care and Use
Committee guidelines at the Zhongnan Hospital of Wuhan University
(approval no. ZN2021257).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Martínez-Rodríguez F, Limones-González JE,
Mendoza-Almanza B, Esparza-Ibarra EL, Gallegos-Flores PI,
Ayala-Luján JL, Ayala-Luján JL, Godina-González S, Salinas E and
Mendoza-Almanza G: Understanding cervical cancer through
proteomics. Cells. 10:18542021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Revathidevi S, Murugan AK, Nakaoka H,
Inoue I and Munirajan AK: APOBEC: A molecular driver in cervical
cancer pathogenesis. Cancer Lett. 496:104–116. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yamashita H, Kawasawa YI, Shuman L, Zheng
Z, Tran T, Walter V, Warrick JI, Chen G, Al-Ahmadie H, Kaag M, et
al: Repression of transcription factor AP-2 alpha by PPARγ reveals
a novel transcriptional circuit in basal-squamous bladder cancer.
Oncogenesis. 8:692019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu J: Pancreatic cancer-derived exosomes
promote the proliferation, invasion, and metastasis of pancreatic
cancer by the miR-3960/TFAP2A Axis. J Oncol. 2022:35903262022.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cao L, Wang S, Zhang Y, Wong KC, Nakatsu
G, Wang X, Wong S, Ji J and Yu J: Zinc-finger protein 471
suppresses gastric cancer through transcriptionally repressing
downstream oncogenic PLS3 and TFAP2A. Oncogene. 37:3601–3616. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guoren Z, Zhaohui F, Wei Z, Mei W, Yuan W,
Lin S, Xiaoyue X, Xiaomei Z and Bo S: TFAP2A Induced ITPKA Serves
as an oncogene and interacts with DBN1 in lung adenocarcinoma. Int
J Biol Sci. 16:504–514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi D, Xie F, Zhang Y, Tian Y, Chen W, Fu
L, Wang J, Guo W, Kang T, Huang W and Deng W: TFAP2A regulates
nasopharyngeal carcinoma growth and survival by targeting HIF-1α
signaling pathway. Cancer Prev Res (Phila). 7:266–277. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu H, Wang L and Jiang X: Silencing of
lncRNA DLEU1 inhibits tumorigenesis of ovarian cancer via
regulating miR-429/TFAP2A axis. Mol Cell Biochem. 476:1051–1061.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen S, Li H, Li X, Chen W, Zhang X, Yang
Z, Chen Z, Chen J, Zhang Y, Shi D and Song M: High SOX8 expression
promotes tumor growth and predicts poor prognosis through GOLPH3
signaling in tongue squamous cell carcinoma. Cancer Med.
9:4274–4289. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yuanhua L, Pudong Q, Wei Z, Yuan W, Delin
L, Yan Z, Geyu L and Bo S: TFAP2A Induced KRT16 as an oncogene in
lung adenocarcinoma via EMT. Int J Biol Sci. 15:1419–1428. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiong Y, Feng Y, Zhao J, Lei J, Qiao T,
Zhou Y, Lu Q, Jiang T, Jia L and Han Y: TFAP2A potentiates lung
adenocarcinoma metastasis by a novel miR-16
family/TFAP2A/PSG9/TGF-β signaling pathway. Cell Death Dis.
12:3522021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang Y, Cheng G, Huang D and Yuan F:
Linc00467 promotes invasion and inhibits apoptosis of head and neck
squamous cell carcinoma by regulating miR-1285-3p/TFAP2A. Am J
Transl Res. 13:6248–6259. 2021.PubMed/NCBI
|
|
14
|
Hao H, Xiao D, Pan J, Qu J, Egger M,
Waigel S, Sanders MA, Zacharias W, Rai SN and McMasters KM:
Sentinel lymph node genes to predict prognosis in node-positive
melanoma patients. Ann Surg Oncol. 24:108–116. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pu M, Li C, Qi X, Chen J, Wang Y, Gao L,
Miao L and Ren J: MiR-1254 suppresses HO-1 expression through seed
region-dependent silencing and non-seed interaction with TFAP2A
transcript to attenuate NSCLC growth. PLoS Genet. 13:e10068962017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brody R, Zhang Y, Ballas M, Siddiqui MK,
Gupta P, Barker C, Midha A and Walker J: PD-L1 expression in
advanced NSCLC: Insights into risk stratification and treatment
selection from a systematic literature review. Lung Cancer.
112:200–215. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang Y, Chen M, Nie H and Yuan Y: PD-1
and PD-L1 in cancer immunotherapy: Clinical implications and future
considerations. Hum Vaccin Immunother. 15:1111–1122. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei F, Zhang T, Deng SC, Wei JC, Yang P,
Wang Q, Chen ZP, Li WL, Chen HC, Hu H and Cao J: PD-L1 promotes
colorectal cancer stem cell expansion by activating HMGA1-dependent
signaling pathways. Cancer Lett. 450:1–13. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Darvin P, Toor SM, Sasidharan Nair V and
Elkord E: Immune checkpoint inhibitors: Recent progress and
potential biomarkers. Exp Mol Med. 50:1–11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Frenel JS, Le Tourneau C, O'Neil B, Ott
PA, Piha-Paul SA, Gomez-Roca C, van Brummelen EMJ, Rugo HS, Thomas
S, Saraf S, et al: Safety and efficacy of pembrolizumab in
advanced, programmed death ligand 1-positive cervical cancer:
Results From the Phase Ib KEYNOTE-028 Trial. J Clin Oncol.
35:4035–4041. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun C, Mezzadra R and Schumacher TN:
Regulation and Function of the PD-L1 Checkpoint. Immunity.
48:434–452. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Orso F, Corà D, Ubezio B, Provero P,
Caselle M and Taverna D: Identification of functional TFAP2A and
SP1 binding sites in new TFAP2A-modulated genes. BMC Genomics.
11:3552010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tchieu J, Zimmer B, Fattahi F, Amin S,
Zeltner N, Chen S and Studer L: A modular platform for
differentiation of human PSCs into all major ectodermal lineages.
Cell Stem Cell. 21:399–410.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
White JR, Thompson DT, Koch KE, Kiriazov
BS, Beck AC, van der Heide DM, Grimm BG, Kulak MV and Weigel RJ:
AP-2α-Mediated Activation of E2F and EZH2 drives melanoma
metastasis. Cancer Res. 81:4455–4470. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bennett KL, Romigh T and Eng C: AP-2alpha
induces epigenetic silencing of tumor suppressive genes and
microsatellite instability in head and neck squamous cell
carcinoma. PLoS One. 4:e69312009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kagohara LT, Zamuner F, Davis-Marcisak EF,
Sharma G, Considine M, Allen J, Yegnasubramanian S, Gaykalova DA
and Fertig EJ: Integrated single-cell and bulk gene expression and
ATAC-seq reveals heterogeneity and early changes in pathways
associated with resistance to cetuximab in HNSCC-sensitive cell
lines. Br J Cancer. 123:101–113. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhu JL, Xue WB, Jiang ZB, Feng W, Liu YC,
Nie XY and Jin LY: Long noncoding RNA CDKN2B-AS1 silencing protects
against esophageal cancer cell invasion and migration by
inactivating the TFAP2A/FSCN1 axis. Kaohsiung J Med Sci.
38:1144–1154. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang HX, Yang G, Yang Y, Yan J, Tang XY
and Pan Q: TFAP2A is a novel regulator that modulates ferroptosis
in gallbladder carcinoma cells via the Nrf2 signalling axis. Eur
Rev Med Pharmacol Sci. 24:4745–4755. 2020.PubMed/NCBI
|
|
30
|
Kołat D, Kałuzińska Ż, Bednarek AK and
Płuciennik E: The biological characteristics of transcription
factors AP-2α and AP-2γ and their importance in various types of
cancers. Biosci Rep. 39:BSR201819282019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan L, Liu Z, Zhang Y, Zhu H, Yu H, Yang
F, Yang R and Wu F: MiRNA373 induces cervical squamous cell
carcinoma SiHa cell apoptosis. Cancer Biomark. 21:455–460. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Ll, Lu J, Liu RQ, Hu MJ, Zhao YM,
Tan S, Wang SY, Zhang B, Nie W, Dong Y, et al: Chromatin
accessibility analysis reveals that TFAP2A promotes angiogenesis in
acquired resistance to anlotinib in lung cancer cells. Acta
Pharmacol Sin. 41:1357–1365. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wei CW, Lin CC, Yu YL, Lin CY, Lin PC, Wu
MT, Chen CJ, Chang W, Lin SZ, Chen YL and Harn HJ:
n-Butylidenephthalide induced apoptosis in the A549 human lung
adenocarcinoma cell line by coupled down-regulation of AP-2alpha
and telomerase activity. Acta Pharmacol Sin. 30:1297–1306. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang K, Niu Y, Cui Z, Jin L, Peng S and
Dong Z: Long noncoding RNA TFAP2A-AS1 promotes oral squamous cell
carcinoma cell growth and movement via competitively binding
miR-1297 and regulating TFAP2A expression. Mol Carcinog.
61:865–875. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu J, Zheng J, Wang J and Shao J:
miR-876-5p suppresses breast cancer progression through targeting
TFAP2A. Exp Ther Med. 18:1458–1464. 2019.PubMed/NCBI
|
|
36
|
Kluger HM, Zito CR, Turcu G, Baine MK,
Zhang H, Adeniran A, Sznol M, Rimm DL, Kluger Y, Chen L, et al:
PD-L1 studies across tumor types, its differential expression and
predictive value in patients treated with immune checkpoint
inhibitors. Clin Cancer Res. 23:4270–4279. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Enwere EK, Kornaga EN, Dean M, Koulis TA,
Phan T, Kalantarian M, Köbel M, Ghatage P, Magliocco AM,
Lees-Miller SP and Doll CM: Expression of PD-L1 and presence of
CD8-positive T cells in pre-treatment specimens of locally advanced
cervical cancer. Mod Pathol. 30:577–586. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Meng Y, Liang H, Hu J, Liu S, Hao X, Wong
MSK, Li X and Hu L: PD-L1 expression correlates with tumor
infiltrating lymphocytes and response to neoadjuvant chemotherapy
in cervical cancer. J Cancer. 9:2938–2945. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mezache L, Paniccia B, Nyinawabera A and
Nuovo GJ: Enhanced expression of PD L1 in cervical intraepithelial
neoplasia and cervical cancers. Mod Pathol. 28:1594–1602. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang RSP, Haberberger J, Murugesan K,
Danziger N, Hiemenz M, Severson E, Duncan DL, Ramkissoon SH, Ross
JS, Elvin JA and Lin DI: Clinicopathologic and genomic
characterization of PD-L1-positive uterine cervical carcinoma. Mod
Pathol. 34:1425–1433. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang W, Lu YP, Yang YZ, Kang JR, Jin YD
and Wang HW: Expressions of programmed death (PD)-1 and PD-1 ligand
(PD-L1) in cervical intraepithelial neoplasia and cervical squamous
cell carcinomas are of prognostic value and associated with human
papillomavirus status. J Obstet Gynaecol Res. 43:1602–1612. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hsu PC, Li SH and Yang CT: Recurrent
Pneumonitis Induced by Atezolizumab (Anti-Programmed Death Ligand
1) in NSCLC patients who previously experienced anti-programmed
death 1 immunotherapy-related pneumonitis. J Thorac Oncol.
13:e227–e230. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Heeren AM, Punt S, Bleeker MC,
Gaarenstroom KN, van der Velden J, Kenter GG, de Gruijl TD and
Jordanova ES: Prognostic effect of different PD-L1 expression
patterns in squamous cell carcinoma and adenocarcinoma of the
cervix. Mod Pathol. 29:753–763. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Karim R, Jordanova ES, Piersma SJ, Kenter
GG, Chen L, Boer JM, Melief CJ and van der Burg SH: Tumor-expressed
B7-H1 and B7-DC in relation to PD-1+ T-cell infiltration and
survival of patients with cervical carcinoma. Clin Cancer Res.
15:6341–6347. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lambert SA, Jolma A, Campitelli LF, Das
PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR and Weirauch MT:
The human transcription factors. Cell. 172:650–665. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wankhade S, Yu Y, Weinberg J, Tainsky MA
and Kannan P: Characterization of the activation domains of AP-2
family transcription factors. J Biol Chem. 275:29701–29708. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hou J, Zhao R, Xia W, Chang CW, You Y, Hsu
JM, Nie L, Chen Y, Wang YC, Liu C, et al: PD-L1-mediated gasdermin
C expression switches apoptosis to pyroptosis in cancer cells and
facilitates tumour necrosis. Nat Cell Biol. 22:1264–1275. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Du W, Zhu J, Zeng Y, Liu T, Zhang Y, Cai
T, Fu Y, Zhang W, Zhang R, Liu Z and Huang JA: KPNB1-mediated
nuclear translocation of PD-L1 promotes non-small cell lung cancer
cell proliferation via the Gas6/MerTK signaling pathway. Cell Death
Differ. 28:1284–1300. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Clark CA, Gupta HB, Sareddy G, Pandeswara
S, Lao S, Yuan B, Drerup JM, Padron A, Conejo-Garcia J, Murthy K,
et al: Tumor-Intrinsic PD-L1 Signals Regulate Cell Growth,
Pathogenesis, and Autophagy in Ovarian Cancer and Melanoma. Cancer
Res. 76:6964–6974. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang S, Li J, Xie J, Liu F, Duan Y, Wu Y,
Huang S, He X, Wang Z and Wu X: Programmed death ligand 1 promotes
lymph node metastasis and glucose metabolism in cervical cancer by
activating integrin β4/SNAI1/SIRT3 signaling pathway. Oncogene.
37:4164–4180. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cao D, Qi Z, Pang Y, Li H, Xie H, Wu J,
Huang Y, Zhu Y, Shen Y, Zhu Y, et al: Retinoic acid-related orphan
receptor C regulates proliferation, glycolysis, and chemoresistance
via the PD-L1/ITGB6/STAT3 signaling axis in bladder cancer. Cancer
Res. 79:2604–2618. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang F, Yang L, Xiao M, Zhang Z, Shen J,
Anuchapreeda S, Tima S, Chiampanichayakul S and Xiao Z: PD-L1
regulates cell proliferation and apoptosis in acute myeloid
leukemia by activating PI3K-AKT signaling pathway. Sci Rep.
12:114442022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Beck AC, Cho E, White JR, Paemka L, Li T,
Gu VW, Thompson DT, Koch KE, Franke C, Gosse M, et al: AP-2α
Regulates S-Phase and is a marker for sensitivity to PI3K inhibitor
buparlisib in colon cancer. Mol Cancer Res. 19:1156–1167. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang L, Zhao Y, Tu Q, Xue X, Zhu X and
Zhao KN: The roles of programmed cell death ligand-1/programmed
cell death-1 (PD-L1/PD-1) in HPV-induced cervical cancer and
potential for their use in blockade therapy. Curr Med Chem.
28:893–909. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Royer HD, Freyaldenhoven MP, Napierski I,
Spitkovsky DD, Bauknecht T and Dathan N: Delineation of human
papillomavirus type 18 enhancer binding proteins: The intracellular
distribution of a novel octamer binding protein p92 is cell cycle
regulated. Nucleic Acids Res. 19:2363–2371. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Turek LP: The structure, function, and
regulation of papillomaviral genes in infection and cervical
cancer. Adv Virus Res. 44:305–356. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Parker JN, Zhao W, Askins KJ, Broker TR
and Chow LT: Mutational analyses of differentiation-dependent human
papillomavirus type 18 enhancer elements in epithelial raft
cultures of neonatal foreskin keratinocytes. Cell Growth Differ.
8:751–762. 1997.PubMed/NCBI
|
|
58
|
Zhao W, Chow LT and Broker TR: A distal
element in the HPV-11 upstream regulatory region contributes to
promoter repression in basal keratinocytes in squamous epithelium.
Virology. 253:219–229. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Beger M, Butz K, Denk C, Williams T, Hurst
HC and Hoppe-Seyler F: Expression pattern of AP-2 transcription
factors in cervical cancer cells and analysis of their influence on
human papillomavirus oncogene transcription. J Mol Med (Berl).
79:314–320. 2001. View Article : Google Scholar : PubMed/NCBI
|