Introduction
Hepatocellular carcinoma (HCC) is the sixth most
commonly diagnosed cancer globally (4.7%) and ranks as the third
leading cause of cancer-associated mortality (8.3%) in 2020
(1). Although early-stage HCC can
potentially be treated with surgical resection, liver
transplantation or ablation, the majority of patients are diagnosed
with inoperable disease, resulting in a ~18% 5-year relative
survival rate (2,3). In recent years, despite advancements
in HCC treatments, including loco-regional therapies, targeted
drugs and immunotherapy (4,5), overall survival outcomes remain
unsatisfactory. Therefore, understanding the mechanism underlying
HCC occurrence and progression and identifying effective targets
are key for improving treatment strategies.
Cytochrome P450 (CYP) enzymes constitute a
superfamily of monooxygenases notable for their extensive substrate
diversity, surpassing all other enzyme families in this regard
(6). Liver CYP enzymes serve a
crucial role in the oxidative metabolism of endogenous compounds
and xenobiotics, including drugs, carcinogens and toxins.
Typically, aging is associated with a decline in CYP enzyme
activity, which impacts the metabolism and clearance of CYP
substrates (7). Bile acids (BAs),
the end products of cholesterol catabolism, serve as amphiphilic
emulsifiers, aiding lipid absorption in the intestine and
facilitating the biliary excretion of cholesterol and phospholipids
(8,9). Numerous CYP enzymes are involved in BA
metabolism. Primary BAs are synthesized in the liver through the
classical and alternative pathways. In the classical pathway,
cholesterol is converted to 7α-hydroxycholesterol by cholesterol
7α-hydroxylase (CYP7A1), the rate-limiting enzyme, and subsequently
to cholic acid by sterol 12α-hydroxylase, also known as cytochrome
P450 8B1 (CYP8B1). In the alternative pathway, sterol
27-hydroxylase (CYP27A1) and oxysterol 7α-hydroxylase (CYP7B1)
metabolize cholesterol into chenodeoxycholic acid (10). Beyond BA production, CYP8B1 also
regulates lipid and glucose metabolism (11). CYP8B1 expression is influenced by
various factors, including nutritional status and metabolic
signaling (12). In metabolic
diseases such as diabetes and obesity, CYP8B1 expression is
increased, indicating its potential role in pathological states
(13). Moreover, CYP8B1 activity is
associated with key biological processes, including cholesterol
metabolism and lipid absorption, making it a valuable biomarker for
investigating metabolic syndrome and associated diseases (14). The formation and progression of
tumors is a complex process influenced by the interplay of internal
and external factors. The tumor microenvironment, genetic
variation, lifestyle choices and environmental influences all
contribute to tumorigenesis. Within this multifactorial framework,
the development of HCC and other cancers is strongly associated
with disruptions in BA metabolism. Alterations in BA composition
and metabolism may facilitate tumor cell proliferation and
metastasis, particularly in organs such as the liver, which are
associated with BA metabolism (15). A comprehensive understanding of the
multifactorial mechanisms underlying tumorigenesis is key for
developing new therapeutic strategies. CYP8B1 has been implicated
in various pathological conditions. For example, in patients with
ulcerative colitis, Chen et al (10) demonstrated that the CYP8B1-cholic
acid metabolic axis suppresses the renewal of leucine-rich
repeat-containing G-protein coupled receptor 5) intestinal stem
cells by inhibiting peroxisome proliferator-activated receptor
α-mediated fatty acid oxidation. This inhibition slows epithelial
barrier repair and exacerbates enteritis progression (10). Esophageal squamous cell carcinoma
(ESCC), one of the most prevalent cancers worldwide with a poor
prognosis and limited therapeutic targets, is also associated with
CYP8B1. Liu et al (15)
confirmed that CYP8B1 contributes to ESCC-associated malignancy and
serves as a potential prognostic factor. Additionally, CYP8B1
expression is associated with the severity of non-alcoholic fatty
liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH),
suggesting its potential as a biomarker for NASH (16). In bariatric surgery, hepatic CYP8B1
is a critical target for regulating glycolipid metabolism. CYP8B1
controls the proportion of 12α hydroxylated BAs in the BA pool;
changes in this ratio significantly alter gut microbiota
composition, intestinal fat absorption and various metabolic
signaling pathways, underscoring its central role in systemic
metabolic regulation (17).
Although some studies (18,19) have highlighted the association
between CYP8B1 and HCC, its precise mechanisms and prognostic value
in HCC remain insufficiently understood.
The present study aimed to explore the mechanisms
through which CYP8B1 influences HCC progression.
Materials and methods
Clinical samples
HCC and adjacent normal tissues (distance, >5cm)
were collected from five patients (age range from 51 to 73, five
patients were all males) with HCC from September to November 2024
in The First People's Hospital of Anqing (Anqing, China) during
routine surgery. The present study was approved by the Research
Ethics Committee of The First People's Hospital of Anqing (approval
no. 20240077), and written informed consent was obtained from all
participants prior to participation.
Gene expression profiles
Gene expression profiles for datasets GSE84402,
GSE26538 and GSE141090 were retrieved from the Gene Expression
Omnibus (GEO, ncbi.nlm.nih.gov.cn). The Xiantao Academic website
(20) was used for generating Venn diagrams and analyzing the
association between immune infiltration and CYP8B1 expression. The
association between CYP8B1 expression and immune cell infiltration
was analyzed based on ssGSEA algorithm provided in R package
(version 4.2.1; cran.r-project.org/). The expression of CYP8B1 and
the content of immune cells was obtained from The Cancer Genome
Atlas (TCGA) database [TCGA-liver hepatocellular carcinoma (LIHC,
the URL was: portal.gdc.cancer.gov). The association between the
expression of CYP8B1 and the content of immune cells was analyzed
by Spearman's test and to further show the relationship between
CYP8B1 and immune infiltration. The content of the immune cells can
indicate the degree of immune infiltration. The survival and
expression analyses of CYP8B1 were performed using CYP8B1
expression data and clinical information from the UALCAN
(ualcan.path.uab.edu) (21) and
Gene Expression Profiling Interactive Analysis (GEPIA) platform
(gepia.cancer-pku.cn) (22). Cox
regression analysis and Kaplan-Meier plots for TCGA datasets was
performed using RStudio software (version 4.2.1) to investigate the
association between CYP8B1 expression and cancer prognosis,
including overall survival in LIHC subclinical groups.
Additionally, the UALCAN and GEPIA database were also used to
determine the association between CYP8B1 expression and overall
survival. The heatmap of positively and negatively associated genes
with CYP8B1 in LIHC was analyzed by UALCAN database to show the top
24 associated genes. Furthermore, gene analysis of positively and
negatively associated genes with CYP8B1 in LIHC were performed
using the Metascape website (metascape.org) to show the pathway and
biological progress of these genes (23). Gene expression of CYP8B1 in HCC cell
lines was performed using the Depmap website
(depmap.org/portal/).
Cell culture
HCC cell lines [Huh7 (cat. no. SCSP-526) and Hep3b
(cat. no. SCSP-5045)] were obtained from Shanghai Institutes of the
Chinese Academy of Sciences. The cells were maintained in
high-glucose DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin and 100 µg/ml streptomycin (complete medium) at
37°C in a humidified incubator with 5% CO2.
Overexpression of CYP8B1 in HCC cell
lines
The recombinant CYP8B1 plasmid was constructed by
cloning into the pcDNA3.0 (Shanghai GeneChem Co., Ltd.) vector and
transfected into HCC cells using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.), following the manufacturer's
protocols. A total of 2 µg/well overexpressed CYP8B1 plasmid
(oe-CYP8B1) and empty vector (negative control, NC) was transfected
into HCC cells in 6-plate well for 6 h for duration in at 37°C, the
medium was replaced with complete medium. After 48 h, the cells
were collected for subsequent experimentation.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay were performed to assess cell
proliferation as previously described (20). Briefly, transfected 5000 HCC cells
were seeded into every well of 96-well plates. Cell proliferation
was measured using CCK-8 (Nanjing KeyGen Biotech Co., Ltd.)
according to the manufacturer's instructions after incubation for
1, 2, 3 and 4 days. All the experiments were performed in
triplicate.
EdU assay
EdU incorporation assay was used to measure cell
proliferation rate as previously described (20). The proliferation rate was calculated
following the manufacturer's instructions using the BeyoClick™
EdU-555 EdU kit (Beyotime Institute of Biotechnology). Each EdU
incorporation experiment was performed ≥3 times.
TUNEL assay
Cell apoptosis was assessed using the TUNEL assay
kit (Nanjing KeyGen Biotech Co., Ltd.) according to the
manufacturer's instructions, as previously described (24). The experiments were repeated ≥3
times.
Western blotting analysis
Protein samples from patient tissue and tumor cells)
were lysed in RIPA buffer supplemented with fresh protease and
phosphatase inhibitor cocktails (Beyotime Biotech Co., Ltd.). The
protein concentration was determined using the BCA assay (Pierce;
Thermo Fisher Scientific, Inc.). Equal amounts of proteins (20
µg/lane) were separated by10% SDS-PAGE and transferred onto PVDF
membranes. The membranes were blocked with 3% BSA (Beyotime Biotech
Co., Ltd.) in 10 mM Tris-HCl (pH 7.4) containing 0.05% Tween-20 to
prevent non-specific binding. Membranes were incubated with primary
antibody at 4°C for 12 h, followed by incubation with a
corresponding peroxidase-conjugated secondary antibody (Abcam;
Table SI) at room temperature for
2 h. Immunoreactive bands were visualized using SuperSignal West
Pico Chemiluminescent Substrate (Pierce; Thermo Fisher Scientific,
Inc.) and band density was quantified using a Versadoc Imaging
System Model 3000. Each experiment was conducted ≥3 times.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from HCC cell lines using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. RT-qPCR was performed as previously
described (25). Briefly, 1 µg
total RNA was reverse-transcribed using random primers and
Primescript reverse transcriptase (Vazyme Biotech Co., Ltd.). RT
protocol was as follows: 37°C for 15 min, 85°C for 5 sec, 4°C
forever. qPCR for the target genes was conducted using the
SYBR-Green qPCR kit (Vazyme Biotech Co., Ltd.) on a fluorescent
temperature cycler (ABI 7500 Real-Time PCR system; Thermo Fisher
Scientific, Inc.). The primers were as follows: CYP8B1: Forward,
5′-ACCTGAGCTTGTTCGGCTAC-3′ and reverse, 5′-CGGAGAGCATCTTGTGAAAG-3′
and β-actin: Forward, 5′-ATCGTGCGTGACATTAAGGAGAAC-3′ and reverse,
5′-AGGAAGGAAGGCTGGAAGAGTG-3′. The thermocycling conditions were as
follows: Initial denaturation at 95°C for 5 min, followed by 45
cycles of amplification at 95°C for 15 sec and annealing at 60°C
for 1 min. Each experiment was performed ≥3 three times. Relative
levels of mRNA expression were obtained via the 2−ΔΔCq
method (26).
Statistical analysis
All statistical analyses were performed using SPSS
18.0 software (SPSS Inc.). Data are presented mean ± SD, and
independent experiments repeats three times. Differences between
categorical variables were analyzed using the independent sample
unpaired or paired Student's t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
CYP8B1 expression is downregulated in
HCC
HCC gene expression profiles from GEO datasets,
including data from human, mice and rat studies, were used to
identify differentially expressed genes (DEGs) using a Venn
diagram. The analysis revealed one up- (CDK1) and four
downregulated DEGs [CYP8B1, MFSD2A (major facilitator super family
domain containing 2a), VIPR1 (Vasoactive intestinal peptide
receptor 1), THRSP (Thyroid hormone-responsive protein)] in HCC
compared with normal tissues (Fig.
1A-C). Except for CYP8B1, these DEGs have been studied in HCC
(27–29); therefore, the present study
investigated CYP8B1. In TCGA database, CYP8B1 expression was
significantly lower in HCC tumor tissues compared with normal
tissues, as shown on the UALCAN (Fig.
1D) and the GEPIA platform (Fig.
1E). Consistently, GTEx and TCGA databases demonstrated reduced
CYP8B1 expression in HCC compared with normal tissue (Fig. 1F). In the GSE84402 dataset, CYP8B1
levels were significantly lower in HCC tissues than in
corresponding normal tissue (Fig.
1G). Additionally, tumor and adjacent normal tissue samples
were collected from five patients with HCC. Immunoblotting analysis
showed that CYP8B1 expression was significantly lower in tumor
tissues compared with adjacent normal tissues (Fig. 1H and I).
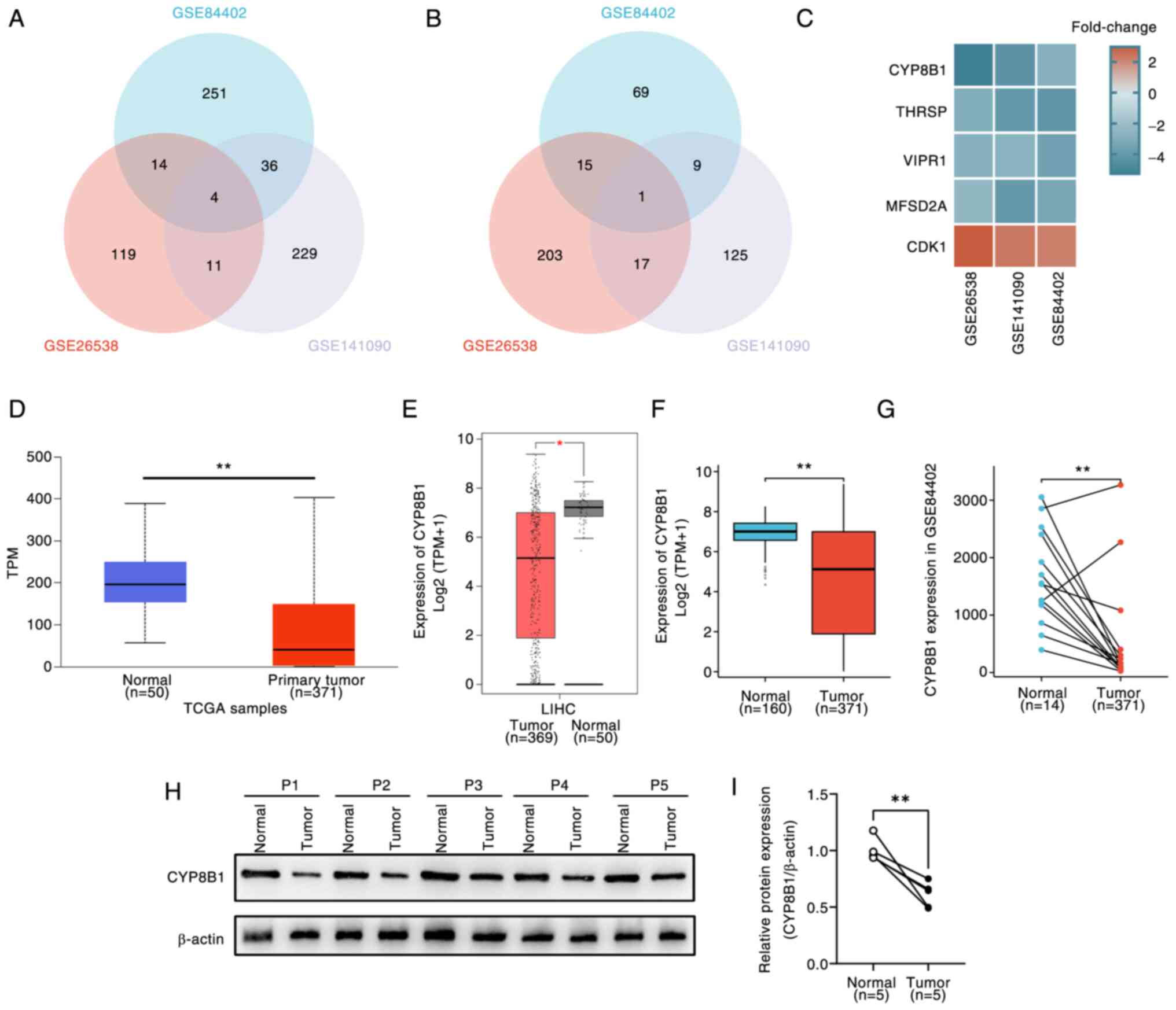 | Figure 1.CYP8B1 expression in public database
and clinical samples in LIHC. (A) Venn plots of (A) down- and (B)
upregulated overlapping DEGs. (C) Fold-change of overlapping DEGs.
CYP8B1 expression was significantly lower in tumor tissues compared
with normal tissue in LIHC in (D) University of Alabama at
Birmingham Cancer data analysis Portal), (E) Gene Expression
Profiling Interactive Analysis) and (F) TCGA and GTEx
(Genotype-Tissue Expression) database. CYP8B1 expression was
significantly lower in tumor compared with corresponding adjacent
normal tissue in (G) GSE84402 and (H) patients with hepatocellular
carcinoma. (I) Relative protein expression. *P<0.05,
**P<0.01. CYP8B1, cytochrome P450 8B1; LIHC, Liver
hepatocellular carcinoma; DEG, Differentially expressed gene; TCGA,
The Cancer Genome Atlas; THRSP, Thyroid hormone-responsive protein;
VIPR1, Vasoactive intestinal peptide receptor 1; MFSD2A, major
facilitator super family domain containing 2a; CDK,
Cyclin-dependent kinases; TPM, Transcripts per million; P,
patient. |
Prognostic value of CYP8B1 in HCC
To evaluate the prognostic significance of CYP8B1 in
HCC, UALCAN and GEPIA databases were used. Patients with HCC with
low CYP8B1 expression exhibited worse overall survival outcomes
(Fig. 2A and B). Decreased CYP8B1
expression was associated with poorer OS in specific clinical
subgroups, including those with AFP (α-fetoprotein) levels >400
ng/ml, individuals aged ≥60 years, male patients, patients taller
than 170 cm, Asian patients and patients with residual tumor (R0;
Fig. 2C-H).
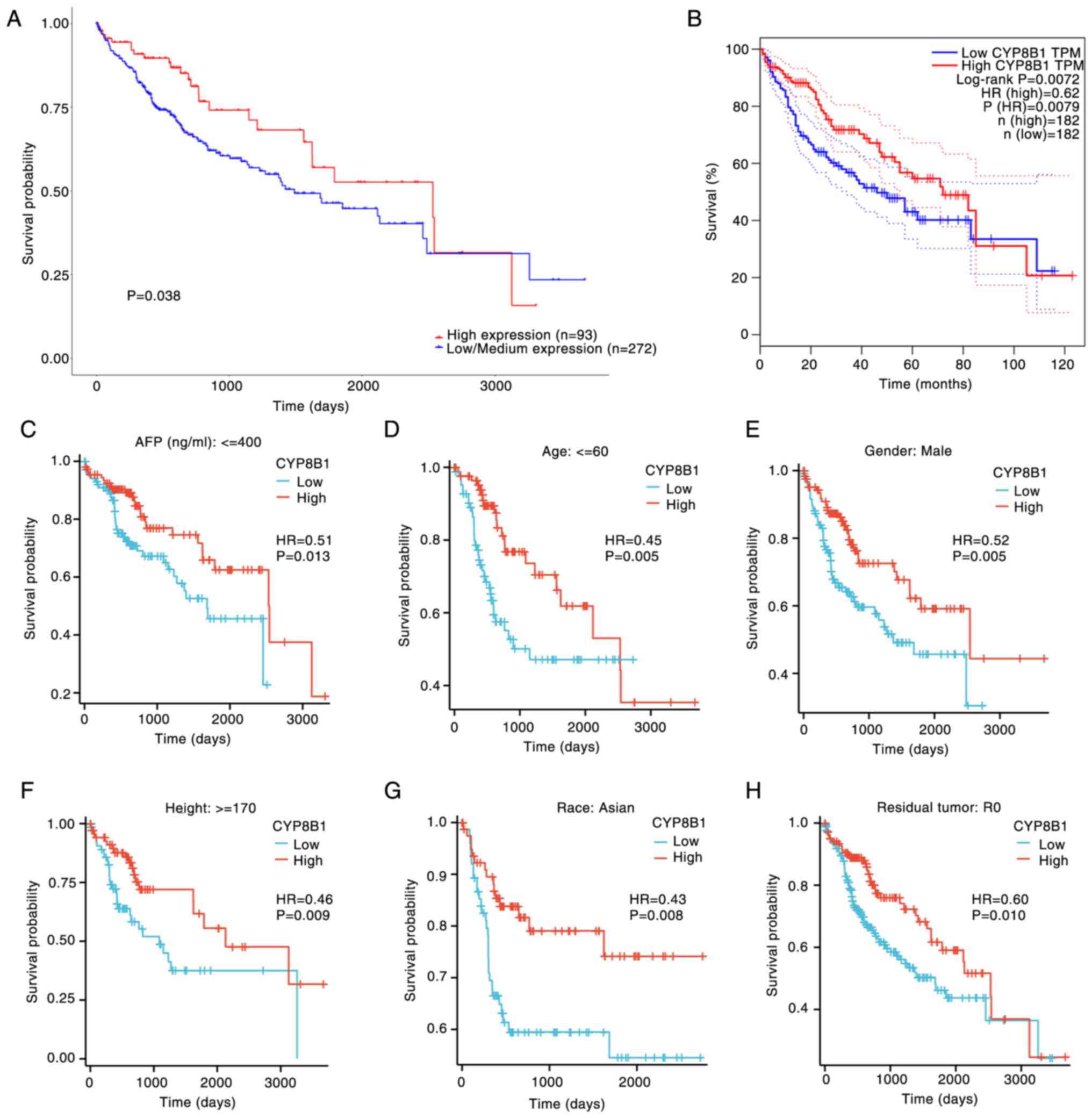 | Figure 2.Association between the OS and CYP8B1
expression in various LIHC clinical subgroups. Effect of CYP8B1 on
OS in patients with LIHC in (A) UALCAN (The University of Alabama
at Birmingham Cancer data analysis Portal) and (B) GEPIA (Gene
Expression Profiling Interactive Analysis)platform. Effect of
CYP8B1 on OS in patients with LIHC and (C) AFP (Alpha-fetoprotein)
≤400 ng/ml, (D) age ≤60 years, (E) male sex, (F) height ≥170 cm,
(G) Asian ethnicity and (H) residual tumor, R0. OS, Overall
survival; CYP8B1, Cytochrome P450 8B1; LIHC, Liver hepatocellular
carcinoma; TPM, Transcripts per million. A, low expression group
means the expression value <25%, medium expression group means
the value was 25–75%, and the high expression group means the value
was >75%. B-H, Arrange the CYP8B1 expression value from small to
larger, the low expression group means the expression value
<50%, and the high expression group means the expression
>50%. |
Association of CYP8B1 with immune cell
infiltration
Given the key role of tumor-infiltrating lymphocytes
in cancer progression and their impact on patient prognosis
(30), along with the potential
oncogenic role of CYP8B1 in HCC, the relationship between CYP8B1
expression and the degree of immune infiltration in HCC was
investigated. We detected the association between CYP8B1 expression
and different immune cells content in TCGA database and found that
CYP8B1 expression was positively associated with content of the
following immune cells such as T helper cell (Th17) cells,
neutrophils, central memory T cell) cells, Tregs (Regulatory T
cells), DC (Dendritic cells), Tgd (T γΔ) cells and eosinophils.
Conversely, CYP8B1 expression showed a significant negative
correlation with content of the following immune cells such as Th1
cells, active Dendritic cells), B cells (Bone marrow-dependent
lymphocyte), interdigitating Dendritic cells), Th (T helper cells)
and T (Thymus-dependent lymphocyte) cells, macrophages and
effective memory T cell), Th2 (T helper cell 2), TFH (Follicular
helper T cell) and NK (Nature killer) CD56 bright cells. (Fig. 3A-G). Furthermore, we selected 17
types immune cells which was significantly associated with
expression of CYP8B1 and found that the low and high expression of
CYP8B1 has significant differences in all the 16 immune cells
except for eosinophils (Fig.
3H).
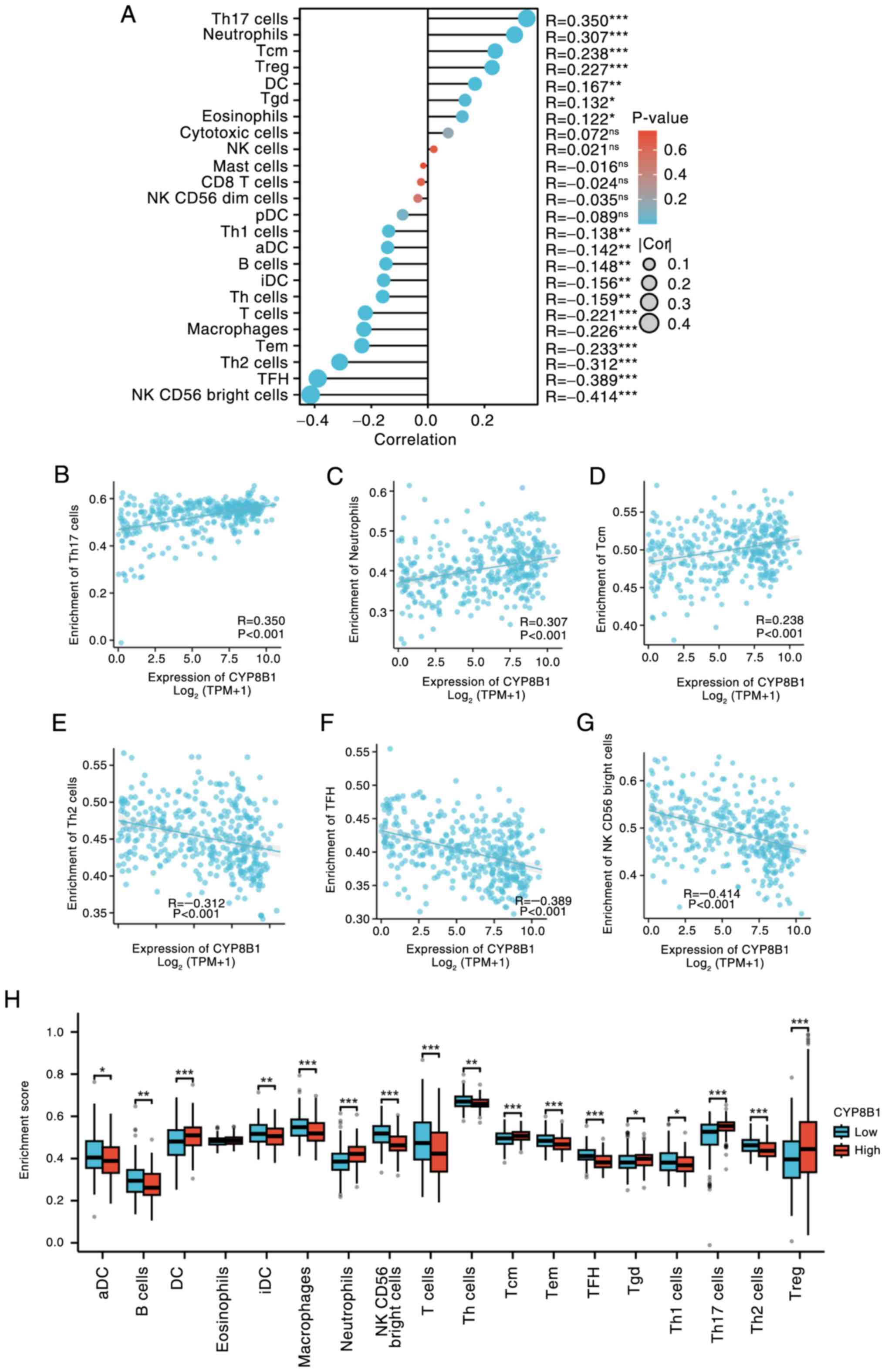 | Figure 3.Correlation analysis of CYP8B1
expression and immune cell infiltration. (A) Correlation analysis
of CYP8B1 expression and immune cell infiltration. Top three
positively correlated immune cells, (B) Th17 cells, (C) neutrophils
and (D) Tcm cells. Top three negatively correlated immune cells,
(E) Th2, (F) TFH and (G) NK CD56 bright cells. (H) CYP8B1
expression. CYP8B1, Cytochrome P450 8B1; Th, T helper cells; Tcm,
Central memory T cell; TFH, Follicular helper T cell; NK, Natural
killer; Treg, regulatory T cell; pDC, Plasmacytoid Dendritic cells;
Tgd, T gamma delta; aDC, active Dendritic cells; iDC,
interdigitating Dendritic cells; Tem, Effective memory T cell; Cor,
Correlation; TPM, Transcripts per million; ns, not significant.
*P<0.05, **P<0.01, ***P<0.001. |
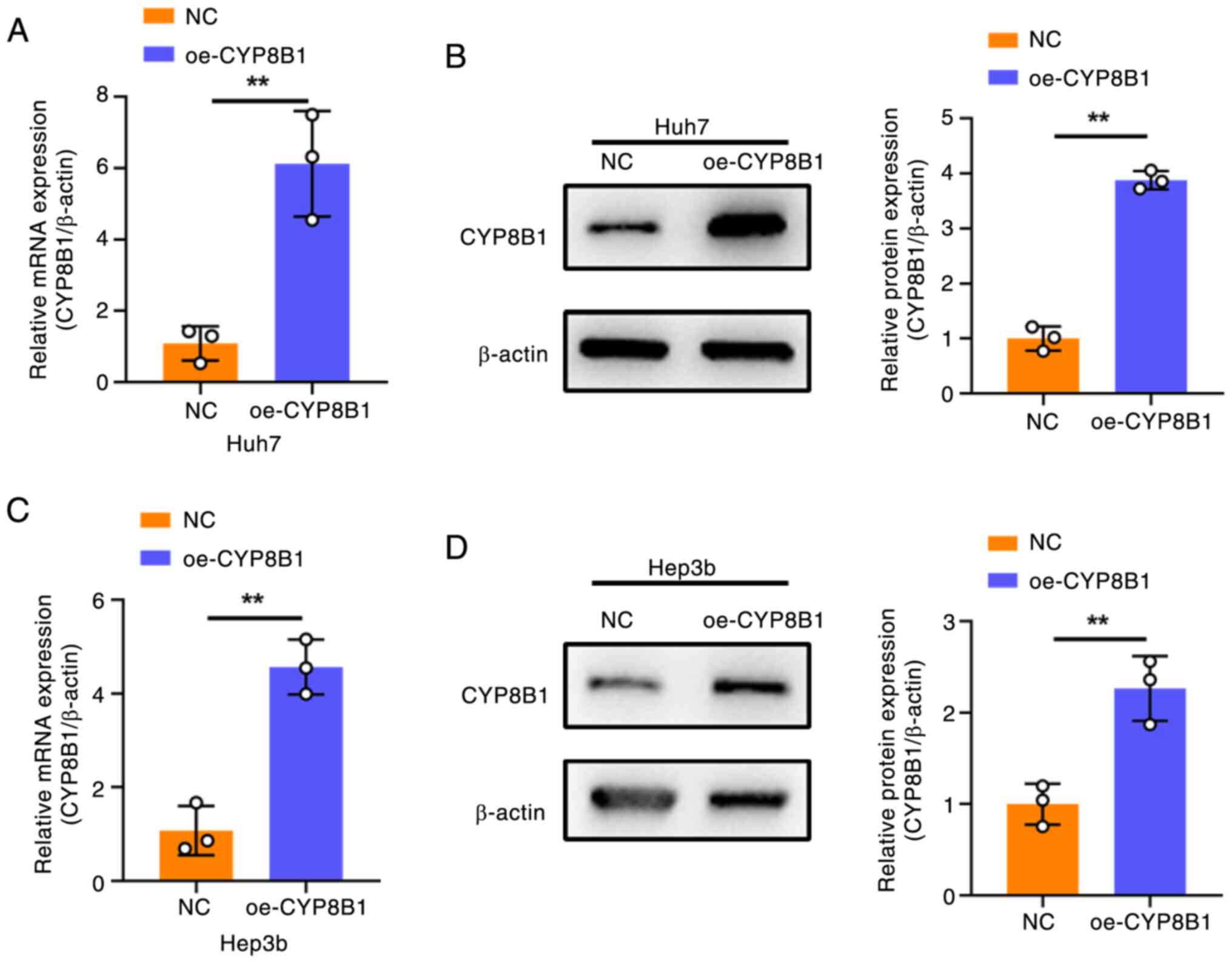
Detection of CYP8B1 overexpression
efficiency
To investigate the role of CYP8B1 in HCC, its
expression levels were searched in public datasets
(depmap.org/portal/). CYP8B1 expression was high in HepG2 and low
in Huh7 and Hep3B cells (Table
SII). To study the functional effects, CYP8B1 was overexpressed
in Hep3B and Huh7 cells by transfection with a
CYP8B1-overexpression plasmid. Following transfection, both mRNA
(Fig. 4A and C) and protein levels
(Fig. 4B and D) of CYP8B1 were
significantly increased in the two HCC cell lines.
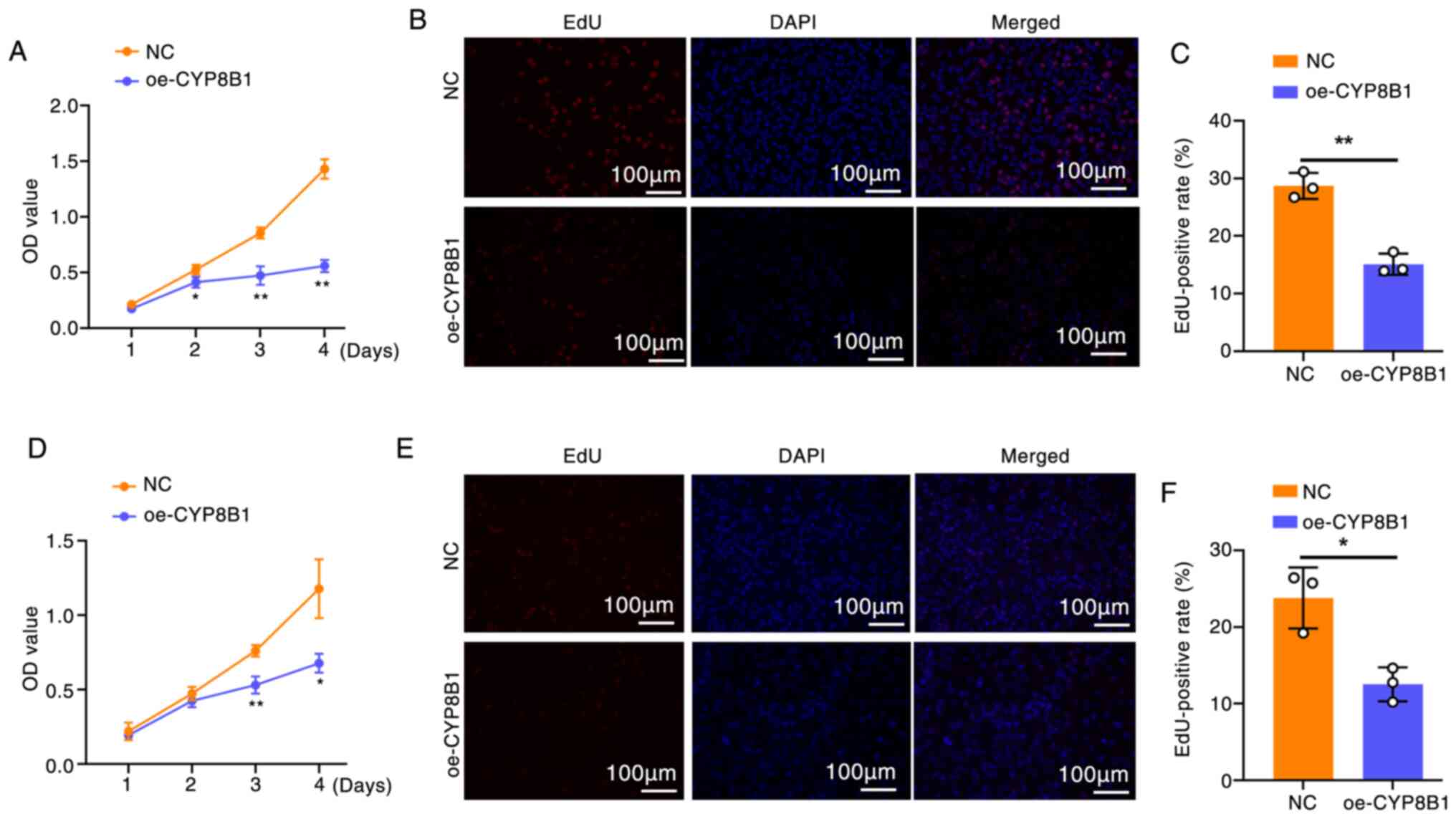
CYP8B1 overexpression inhibits cell
proliferation and promotes apoptosis
To evaluate the effect of CYP8B1 on cell
proliferation, CCK-8 (Fig. 5A) and
EdU assays (Fig. 5B and C) were
performed in cells overexpressing CYP8B1. Both assays showed that
CYP8B1 overexpression significantly decreased the proliferation of
Huh7 and Hep3b cells (Fig.
5D-F).
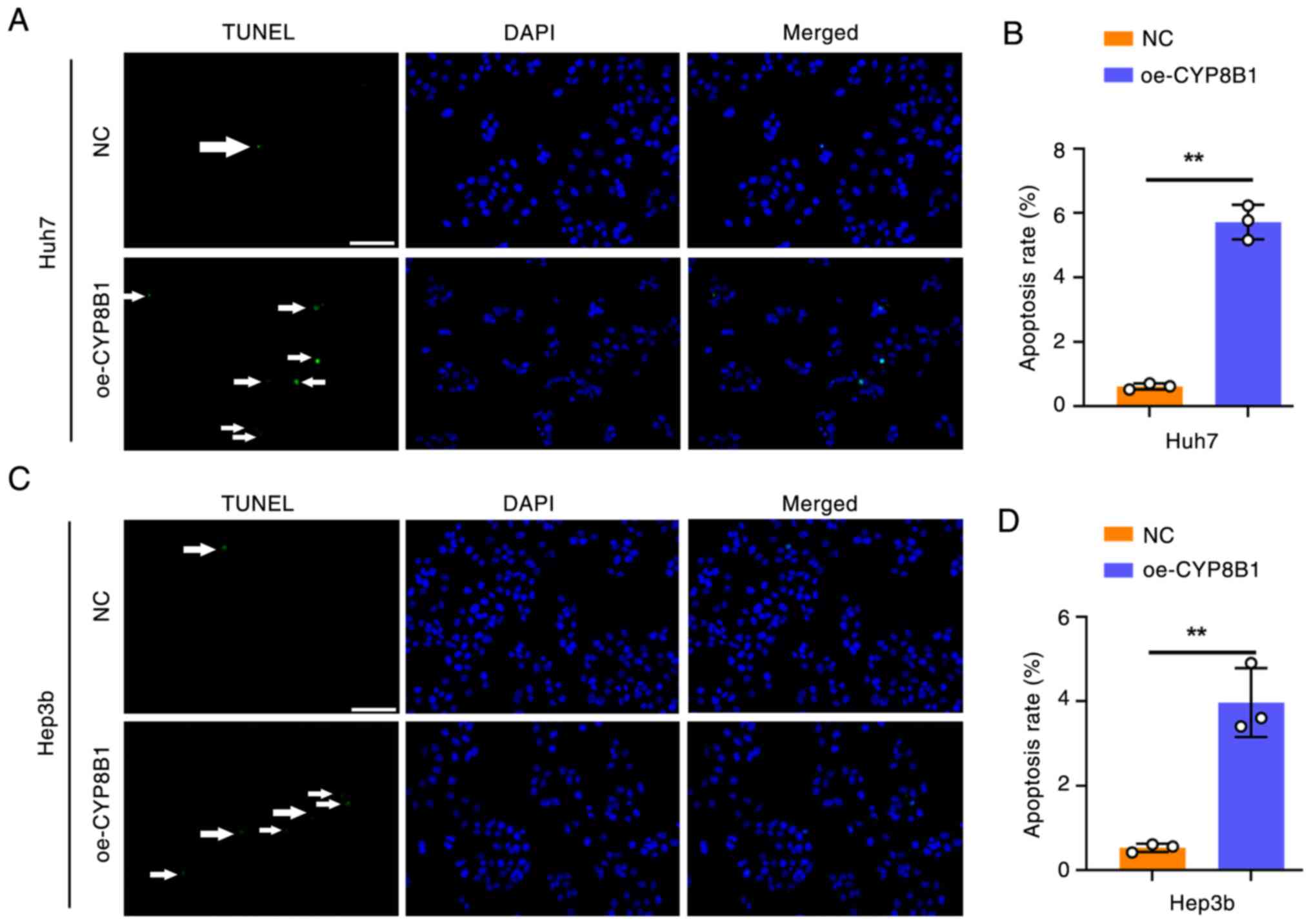
To investigate the effect of CYP8B1 on apoptosis,
TUNEL assay was performed. CYP8B1 overexpression markedly increased
the apoptosis rate in Huh7 (Fig. 6A and
B) and Hep3b cells (Fig. 6C and
D).
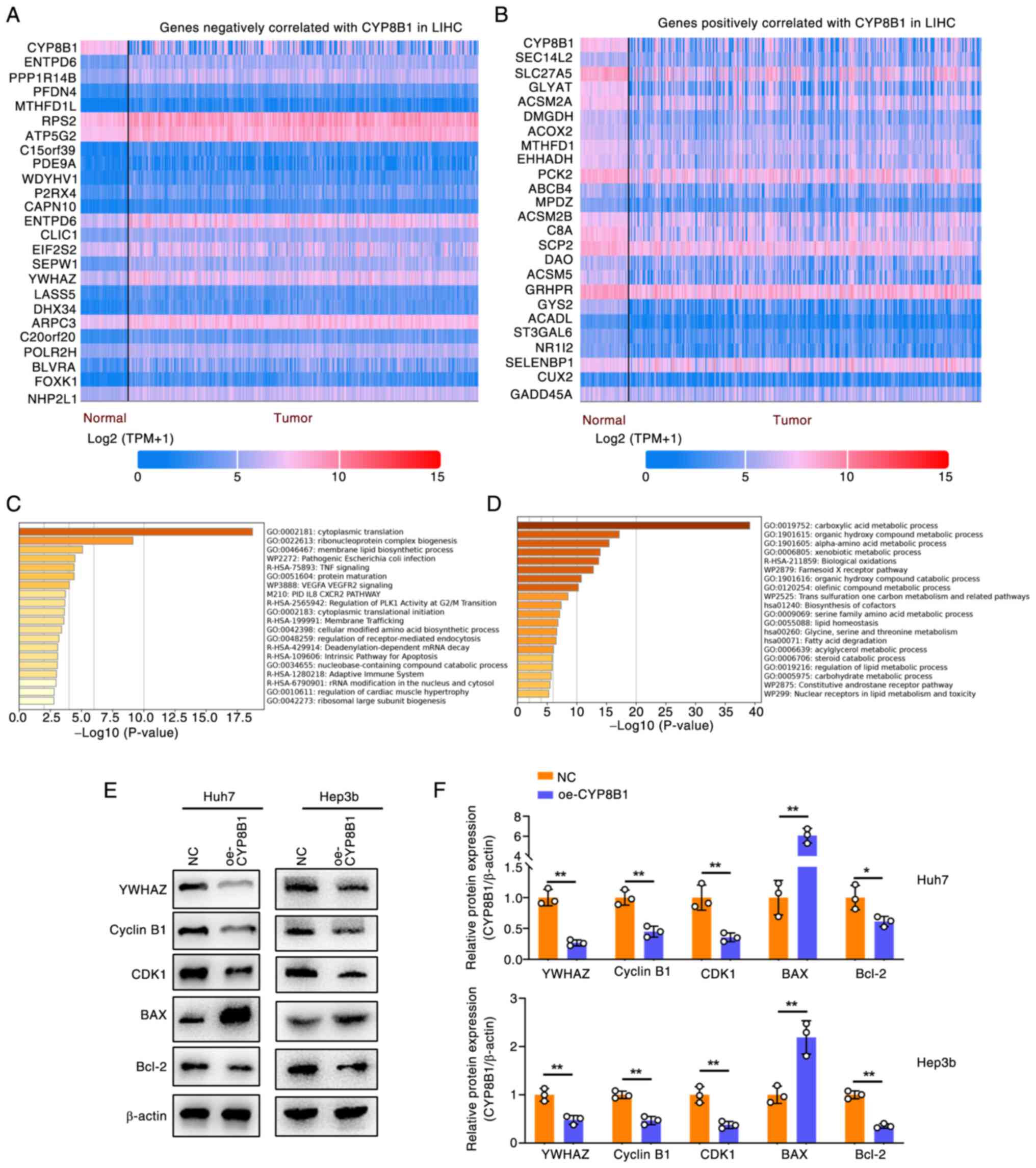
Mechanisms of CYP8B1 in regulating HCC
cell function
To explore the mechanisms of CYP8B1 in HCC, the top
24 genes positively and negatively correlated with CYP8B1
expression in LIHC were identified using the UALCAN platform
(Fig. 7A and B) and submitted to
the Metascape platform for pathway enrichment analysis (Fig. 7C and D). This revealed two key
pathways potentially regulated by CYP8B1: ‘Regulation of PLK
activity at G2/M transition’ and ‘intrinsic pathway for apoptosis’
(Fig. 7C). As CYP8B1 overexpression
inhibited cell proliferation and promoted apoptosis, the present
study identified genes from PubMed involved in the G2/M transition
and apoptosis. YWHAZ (Tyrosine 3/tryptophan 5 monooxygenase
activation protein ζ) downregulation induces G2/M transition and
apoptosis (23,24). Expression of YWHAZ was negatively
associated with CYP8B1 in LIHC (Fig.
7A). Immunoblot assay was used to clarify the expression of
YWHAZ after CYP8B1 overexpression, as well as the expression of
downstream factors of YWHAZ, including Cyclin B1, CDK1, Bax and
Bcl2 (Fig. 7E and F) in Huh7 and
Hep3b cells. YWHAZ was significantly downregulated following CYP8B1
overexpression. Downstream factors of YWHAZ, including Cyclin B1
and CDK1 which was involved in the G2/M cell cycle was
significantly downregulated. Apoptosis-related genes Bax was
upregulated and Bcl2 was downregulated after CYP8B1 overexpression.
The results suggested that CYP8B1 mediates G2/M arrest via the
YWHAZ/CyclinB1/CDK1 axis and apoptosis primarily via the
YWHAZ/Bax/Bcl2 axis.
Discussion
CYP8B1 plays a key role in the conversion of
cholesterol into BAs. It facilitates the hydroxylation of the
steroid ring at the C12 position, leading to the production of the
BA cholic acid (31). Due to its
importance in cholesterol homeostasis and lipid metabolism, CYP8B1
is as a key target for managing metabolic diseases, including NAFLD
and type 2 diabetes (32,33). CYP8B1 has been implicated in tumor
growth and apoptosis. For example, kaempferol has been shown to
upregulate CYP8B1, thereby attenuating colorectal cancer
progression (34). The present
study identified CYP8B1 as a prognostic biomarker for HCC based on
analysis from GEO datasets, with low expression correlating with
poor prognosis. The tumor microenvironment, especially the immune
microenvironment, serves a vital role in the malignant progression
and metastasis of cancer (35). The
present findings further demonstrated an association between CYP8B1
expression and immune cell infiltration in the liver cancer
microenvironment. Additionally, CYP8B1 overexpression inhibited
proliferation and promoted apoptosis in HCC cells. Through analysis
of CYP8B1 negatively correlated genes in LIHC, the present study
identified YWHAZ as a potential downstream factor regulated by
CYP8B1.
YWHAZ, a member of the 14-3-3protein family, is a
critical hub protein involved in numerous signal transduction
pathways and plays a key role in tumor progression (36). YWHAZ regulates the G2/M cell cycle
transition, thereby promoting cell proliferation in various types
of cancer such as gastric and colorectal cancer (37,38).
Silencing YWHAZ has been shown to increase apoptosis by modulating
key apoptotic markers such as Bax and Bcl2 (24). CylinB1 and CDK1 are the downstream
factors of YWHAZ that regulate G2/M transition (37), and Bax and Bcl2 are the downstream
factors of YWHAZ involved in cell apoptosis (39). Consistent with the present findings,
CYP8B1 is negatively associated with YWHAZ and G2/M cell cycle.
YWHAZ may serve as a critical downstream effector of CYP8B1.
Subsequent validation by immunoblotting confirmed this
hypothesis.
The number of clinical samples in the present study
was small; future studies require more samples to prove the present
conclusions. Secondly, the present study was based on in
vitro experiments and lacked validation in animal experiments.
Thirdly, the application of this gene in the clinical treatment of
HCC has not been explored.
In summary, by analyzing gene expression profiles
from humans, mice and rats, the present study identified CYP8B1 as
a significantly dysregulated gene in HCC. Bioinformatics and
experimental analyses demonstrated that low CYP8B1 expression
correlates with poor prognosis in HCC. Overexpression of CYP8B1
inhibited proliferation and promoted apoptosis in HCC cell lines.
Mechanistically, CYP8B1 may regulate cell proliferation and
apoptosis, at least in part, via YWHAZ. CYP8B1 could be a biomarker
for prognosis of HCC. The present study provides evidence that
CYP8B1 could serve as a promising therapeutic target for HCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by Anqing Science and Technology
Bureau Project (grant no. 2023Z2007).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
CL and FL designed the study and wrote the
manuscript. CL, FL, QD and QP performed the experiments. ZK
supplied the clinical samples. FL, QD, QP and ZK analyzed the data.
FL, DQ, ZK QP and CL confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Ethics approval was permitted by the Research Ethics
Committee of The First People's Hospital of Anqing (approval no.
20240077; Anqing, China) and written informed consent was obtained
from the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bi F, Qiu Y, Wu Z, Liu S, Zuo D, Huang Z,
Li B, Yuan Y, Niu Y and Qiu J: METTL9-SLC7A11 axis promotes
hepatocellular carcinoma progression through ferroptosis
inhibition. Cell Death Discov. 9:4282023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang C, Zhang H, Zhang L, Zhu AX, Bernards
R, Qin W and Wang C: Evolving therapeutic landscape of advanced
hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol.
20:203–222. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shokoohian B, Negahdari B, Aboulkheyr Es
H, Abedi-Valugerdi M, Baghaei K, Agarwal T, Maiti TK, Hassan M,
Najimi M and Vosough M: Advanced therapeutic modalities in
hepatocellular carcinoma: Novel insights. J Cell Mol Med.
25:8602–8614. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim HJ, Lee SH, Shim HJ, Bang HJ, Cho SH,
Chung IJ, Hwang EC, Hwang JE and Bae WK: Hepatic arterial infusion
chemotherapy versus systemic therapy for advanced hepatocellular
carcinoma: A systematic review and meta-analysis. Front Oncol.
13:12652402023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sheikholeslami B, Tootoonchi Z, Lavasani
H, Hosseinzadeh Ardakani Y and Rouini M: Investigation of MDMA
inhibitory effect on cytochromeP450 3A4 in isolated perfused rat
liver model using tramadol. Adv Pharm Bull. 11:530–536. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Haduch A, Bromek E, Kuban W,
Basińska-Ziobroń A, Danek PJ, Alenina N, Bader M and Daniel WA: The
effect of brain serotonin deficit (TPH2-KO) on the expression and
activity of liver cytochrome P450 enzymes in aging male Dark Agouti
rats. Pharmacol Rep. 75:1522–1532. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chiang JYL and Ferrell JM: Discovery of
farnesoid X receptor and its role in bile acid metabolism. Mol Cell
Endocrinol. 548:1116182022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shulpekova Y, Shirokova E, Zharkova M,
Tkachenko P, Tikhonov I, Stepanov A, Sinitsyna A, Izotov A, Butkova
T, Shulpekova N, et al: A recent ten-year perspective: Bile acid
metabolism and signaling. Molecules. 27:19832022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen L, Jiao T, Liu W, Luo Y, Wang J, Guo
X, Tong X, Lin Z, Sun C, Wang K, et al: Hepatic cytochrome P450 8B1
and cholic acid potentiate intestinal epithelial injury in colitis
by suppressing intestinal stem cell renewal. Cell Stem Cell.
29:1366–1381.e9. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kaur A, Patankar JV, de Haan W, Ruddle P,
Wijesekara N, Groen AK, Verchere CB, Singaraja RR and Hayden MR:
Loss of Cyp8b1 improves glucose homeostasis by increasing GLP-1.
Diabetes. 64:1168–1179. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pozzo L, Vornoli A, Coppola I, Croce CM,
Giorgetti L, Gervasi PG and Longo V: Effect of HFD/STZ on
expression of genes involved in lipid, cholesterol and glucose
metabolism in rats. Life Sci. 166:149–156. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pathak P and Chiang JYL: Sterol
12α-hydroxylase aggravates dyslipidemia by activating the
ceramide/mTORC1/SREBP-1C pathway via FGF21 and FGF15. Gene Expr.
19:161–173. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hoogerland JA, Lei Y, Wolters JC, de Boer
JF, Bos T, Bleeker A, Mulder NL, van Dijk TH, Kuivenhoven JA, Rajas
F, et al: Glucose-6-phosphate regulates hepatic bile acid synthesis
in mice. Hepatology. 70:2171–2184. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu X, Hong R, Du P, Yang D, He M, Wu Q,
Li L, Wang Y, Chen J, Min Q, et al: The metabolic genomic atlas
reveals potential drivers and clinically relevant insights into the
etiology of esophageal squamous cell carcinoma. Theranostics.
12:6160–6178. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu F, Yu Z, Liu Y, Du T, Yu L, Tian F,
Chen W and Zhai Q: A high-fat, high-cholesterol diet promotes
intestinal inflammation by exacerbating gut microbiome dysbiosis
and bile acid disorders in cholecystectomy. Nutrients. 15:38292023.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y, Tu J, Shi L, Fang Z, Fan M, Zhang
J, Ding L, Chen Y, Wang Y, Zhang E, et al: CYP8B1 downregulation
mediates the metabolic effects of vertical sleeve gastrectomy in
mice. Hepatology. 79:1005–1018. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang X, Liao X, Yang C, Huang K, Yu T, Yu
L, Han C, Zhu G, Zeng X, Liu Z, et al: Identification of prognostic
biomarkers for patients with hepatocellular carcinoma after
hepatectomy. Oncol Rep. 41:1586–1602. 2019.PubMed/NCBI
|
|
19
|
Zhang R, Huang M, Wang H, Wu S, Yao J, Ge
Y, Lu Y and Hu Q: Identification of potential biomarkers from
hepatocellular carcinoma with MT1 deletion. Pathol Oncol Res.
27:5975272021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vivian J, Rao AA, Nothaft FA, Ketchum C,
Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD,
Musselman-Brown A, et al: Toil enables reproducible, open source,
big biomedical data analyses. Nat Biotechnol. 35:314–316. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Li R, Guo S, Li Y, Wang Y, Wen X,
Lan T and Gong K: Bioinformatics-based identification of lipid- and
immune-related biomarkers in abdominal aortic aneurysms. Heliyon.
9:e136222023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li F, Liu CS, Wu P, Ling AS, Pan Q and Li
XN: CCT4 suppression inhibits tumor growth in hepatocellular
carcinoma by interacting with Cdc20. Chin Med J (Engl).
134:2721–2729. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu C, Li F, Li X, Cao M, Feng G, Yuan X
and Shi X: WIPI2 depletion inhibits the growth of hepatocellular
carcinoma cells through the AMPK signaling pathway. Oncol Rep.
43:1467–1478. 2020.PubMed/NCBI
|
|
26
|
Wang W, Chen H, Gao W, Wang S, Wu K, Lu C,
Luo X, Li L and Yu C: Girdin interaction with vimentin induces EMT
and promotes the growth and metastasis of pancreatic ductal
adenocarcinoma. Oncol Rep. 44:637–649. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xing S, Kan J, Su A, Liu QD, Wang K, Cai X
and Dong J: The prognostic value of major facilitator superfamily
domain-containing protein 2A in patients with hepatocellular
carcinoma. Aging (Albany NY). 11:8474–8483. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fu Y, Liu S, Rodrigues RM, Han Y, Guo C,
Zhu Z, He Y, Mackowiak B, Feng D, Gao B, et al: Activation of VIPR1
suppresses hepatocellular carcinoma progression by regulating
arginine and pyrimidine metabolism. Int J Biol Sci. 18:4341–4356.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu Q, Ma X, Li C, Zhou C, Chen J and Gu X:
Downregulation of THRSP promotes hepatocellular carcinoma
progression by triggering ZEB1 transcription in an ERK-dependent
manner. J Cancer. 12:4247–4256. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu F, Zeng G, Zhou S, He X, Sun N, Zhu X
and Hu A: Blocking Tim-3 or/and PD-1 reverses dysfunction of
tumor-infiltrating lymphocytes in HBV-related hepatocellular
carcinoma. Bull Cancer. 105:493–501. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu J, Carlson HA and Scott EE: The
structure and characterization of human cytochrome P450 8B1
supports future drug design for nonalcoholic fatty liver disease
and diabetes. J Biol Chem. 298:1023442022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Patankar JV, Wong CK, Morampudi V, Gibson
WT, Vallance B, Ioannou GN and Hayden MR: Genetic ablation of
Cyp8b1 preserves host metabolic function by repressing
steatohepatitis and altering gut microbiota composition. Am J
Physiol Endocrinol Metab. 314:E418–E432. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mao Y, Zhao Y, Luo S, Chen H, Liu X, Wu T,
Ding G, Liu X, Sheng J, Meng Y and Huang H: Advanced paternal age
increased metabolic risks in mice offspring. Biochim Biophys Acta
Mol Basis Dis. 1868:1663552022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li X, Khan I, Huang G, Lu Y, Wang L, Liu
Y, Lu L, Hsiao WLW and Liu Z: Kaempferol acts on bile acid
signaling and gut microbiota to attenuate the tumor burden in
ApcMin/+ mice. Eur J Pharmacol. 918:1747732022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mou P, Ge QH, Sheng R, Zhu TF, Liu Y and
Ding K: Research progress on the immune microenvironment and
immunotherapy in gastric cancer. Front Immunol. 14:12911172023.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gan Y, Ye F and He XX: The role of YWHAZ
in cancer: A maze of opportunities and challenges. J Cancer.
11:2252–2264. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sheng N, Yan L, Wu K, You W, Gong J, Hu L,
Tan G, Chen H and Wang Z: TRIP13 promotes tumor growth and is
associated with poor prognosis in colorectal cancer. Cell Death
Dis. 9:4022018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ma J, Chen W, Wang K, Tian K, Li Q, Zhao
T, Zhang L, Wang L, Wu Z and Zhang J: Identification of the
different roles and potential mechanisms of T isoforms in the tumor
recurrence and cell cycle of chordomas. Onco Targets Ther.
12:11777–11791. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guo F, Jiao D, Sui GQ, Sun LN, Gao YJ, Fu
QF and Jin CX: Anticancer effect of YWHAZ silencing via inducing
apoptosis and autophagy in gastric cancer cells. Neoplasma.
65:693–700. 2018. View Article : Google Scholar : PubMed/NCBI
|