Introduction
Intrahepatic cholangiocarcinoma (ICC) is a highly
aggressive and malignant liver cancer that accounts for 10–20% of
primary liver cancers and is the second most common type after
hepatocellular carcinoma (HCC) (1,2). The
incidence of ICC has been increasing globally, with an average
annual growth rate of 4.4% over the past decade (3). Unfortunately, most patients are
diagnosed at advanced stages because of the asymptomatic nature of
early ICC, leading to limited treatment options and poor clinical
outcomes (4). The 5-year survival
rate remains dismal, at ~25-40% even after curative resection, with
a high recurrence rate (5).
Surgical resection is currently the only potentially curative
treatment for ICC, but it is suitable for only a minority of
patients. The molecular mechanisms underlying ICC growth and
metastasis are not well understood, which hinders the development
of new therapies. Factors such as chronic inflammation, genetic
mutations in genes such as isocitrate dehydrogenase
(NADP+ IDH 1/2), epidermal growth factor receptors,
fibroblast growth factor receptors and ROS proto-oncogene 1, as
well as aberrant signaling pathways, contribute to ICC development
(6–8). However, the clear mechanisms of ICC
have not been fully elucidated, and there is an urgent need to
investigate the molecular pathogenesis of ICC to identify new
therapeutic targets and approaches.
PTP4A1, also known as protein tyrosine phosphatase
4A1, is an enzyme that belongs to the protein tyrosine phosphatase
(PTP) family. It plays significant roles in various cellular
processes, including cell proliferation, differentiation and
migration (9–11). PTP4A1 is involved in the
dephosphorylation of tyrosine residues on proteins, which can
influence the PI3K/AKT, ERK and transforming growth factor beta 1
(TGFβ) signaling pathways (12–15).
PTP4A1 has also been implicated in the development and progression
of several types of cancer, including non-small cell lung cancer
(NSCLC) (9), HCC (16), cervical cancer (13) and colon cancer (17), where it may promote cell
proliferation, survival and metastasis. High levels of PTP4A1
expression are associated with poor prognosis in various types of
cancer (9,18,19),
suggesting its potential as a therapeutic target and biomarker. A
previous study by the authors revealed that the lncRNA NEAT1
promoted cell proliferation, migration and invasion via the
miR-186-5p/PTP4A1 axis in cholangiocarcinoma (20). However, the function and mechanism
of PTP4A1 in ICC remain largely unclear.
In the present study, the potential role and
underlying molecular mechanism of PTP4A1 in ICC were explored. Our
results revealed that PTP4A1 was upregulated in ICC and associated
with lymph node metastasis, vascular invasion, tumor node
metastasis (TNM) stage and differentiation of ICC. Furthermore, it
was demonstrated that PTP4A1 promoted the proliferation, metastasis
and invasion of ICC. In terms of mechanism, PTP4A1 interacts with
PTEN, decreases the phosphorylation of PTEN and promotes the
activation of the PI3K/AKT pathway. Therefore, our results suggest
that PTP4A1 is a promising therapeutic target for ICC.
Materials and methods
Patients and specimens
ICC tissues and matched adjacent non-tumor tissues
were obtained from 60 patients (37 men and 23 women; median age, 64
years; age range, 41–83 years) who underwent radical resection for
ICC at the Hunan Provincial People's Hospital (Changsha, China)
between January 2021 and January 2023. The diagnoses were
pathologically confirmed by two independent pathologists. None of
the patients received any anticancer therapy prior to surgery. A
total of 40 pairs of fresh ICC specimens and matched adjacent
non-tumor tissues were stored in liquid nitrogen immediately after
resection and subsequently stored at −80°C for reverse
transcription-quantitative PCR (RT-qPCR) and WB. A total of 20
pairs of ICC specimens and matched adjacent non-tumor tissues were
fixed in 4% paraformaldehyde (PFA) at room temperature for 12–24 h
and subsequently subjected to paraffin embedding and
immunohistochemical (IHC) analysis. All experiments in our research
were approved (approval no. 2023–151) by the Ethics Committee of
the Hunan Provincial People's Hospital (Changsha, China) and
carried out in accordance with the Declaration of Helsinki. Written
informed consent was obtained from all participants.
Sample size selection and power
calculation
In more than 75% of normal lung tissue samples, the
percentage of PTP4A1-positive cells among the total cells is
<50%, which is considered as low expression of PTP4A1 (9). In the present study, it was assumed
that the probability of PTP4A1 overexpression (OE) in ICC adjacent
tissues (P1) was 0.3, and that in the ICC tissue (P2) was 0.7.
Using the T-test method, with a significance level (ɑ) set at 0.05
and a statistical power (β) set at 0.8, the calculated sample size
based on the sample size formula
{n=(Zɑ/2+Zβ)2 × [p1(1-p1) + p2
(1-p2)]/(p1-p2)2]} is 21. Using G*Power, it was found
that a sample size of 21 per group achieves a power of ~0.8 (80%).
In the present study, IHC results are subjective to some extent and
may be affected by technical variability. Therefore, a relatively
small sample size is needed to balance resource consumption and the
reliability of the results. A sample size of 20 pairs was selected
to avoid excessive experimental costs and workload. Western
blotting (WB) and RT-qPCR are quantitative detection techniques
that can accurately measure the expression levels of PTP4A1, with
high sensitivity and specificity. A total of 10 pairs of ICC tumor
tissues and adjacent tissues were analyzed by to determine the
expression of PTP4A1. For RT-qPCR assay, a sample size of 40 pairs
can provide sufficient statistical power to identify potential
association between PTP4A1 expression and ICC clinical
features.
Cell culture and transfection
The ICC cell lines HCCC-9810 and RBE were purchased
from the Cell Bank of the Chinese Academy of Sciences (Shanghai,
China). The cells were cultured in DMEM supplemented with 10% fetal
bovine serum (FBS) in a 5% CO2, 37°C incubator.
Lentiviruses containing specific short hairpin RNA (shRNA)
targeting PTP4A1 (sh-PTP4A1: ATCCAACCAATGCGACCTT) or shRNA negative
control (sh-NC: TTCTCCGAACGTGTCACGT) were purchased from Shanghai
GenePharma Co., Ltd. The PTP4A1 coding sequence was synthesized,
cloned and inserted into a lentiviral GV492 vector. The empty GV492
vector was considered an empty vector (EV). A total of
5×104 HCCC-9810 and RBE cells were seeded into 6-well
plates for 24 h. When the cells confluence was 30%, they were
infected with the appropriate lentiviruses. The optimal infection
conditions were 10 µl lentivirus (108 TU/ml) + 40 µl 25
× HiTransG lentivirus infection reagent (GeneChem, Inc.) + 950 µl
complete medium. After 16 h of infection, the medium was replaced
with complete medium, and the cells were cultured for 72 h. Then,
these cells were selected with 2 µg/ml of puromycin (cat. no.
ab141453; Abcam) for three days and harvested for further
studies.
Plasmids
A Flag-tagged PTEN wild-type (WT) plasmid
[pLV3-CMV-PTEN (human)-3X FLAG-Puro; cat. no. P64887] was purchased
from Wuhan MiaoLing Biotech Science Co., Ltd. PTEN S380A mutants
were generated by Hieff Mut™ Site-Directed Mutagenesis Kit (cat.
no. 11003ES10; Shanghai Yeasen Biotechnology Co., Ltd.). The
primers used were as follows: forward,
5′-TATAGATATGCTGACACCACTGACTCTGATCCAGAGA-3′ and reverse,
5′-TGGTGTCAGCATATCTATAATGATCAGGTTCATTGTCA-3′. Transfections were
conducted using Lipofectamine™ 3000 (cat. no. L3000015; Invitrogen'
Thermo Fisher scientific, Inc.).
Proliferation, colony formation and
5-ethynyl-2′-deoxyuridine (EdU) assays
For the proliferation assay, RBE and HCCC-9810 cells
transfected with the PTP4A1-knockdown (KD) or OE lentiviral vector
(3,000 per well) were seeded into 96-well plates and treated with
Cell Counting Kit-8 (CCK-8) reagent (10 µl/well; Beyotime Institute
of Biotechnology) at 37°C for 2 h. Subsequently, cell viability at
24, 48 or 72 h was evaluated using a microplate reader (Thermo
Fisher Scientific, Inc.) at 450 nm. Experiments were performed in
triplicate.
For the colony formation assay, RBE and HCCC-9810
cells transfected with the PTP4A1-KD or OE lentiviral vector (500
per well) were seed into 6-well plates and cultured in DMEM plus
10% FBS in a 5% CO2 incubator at 37°C for 14 days and
then fixed with 4% PFA for 1 h at room temperature. The cells were
then stained with crystal violet (0.1%) at room temperature for 30
min. Images of the colonies were subsequently captured with an
inverted light microscope with camera functionality at ×10
magnification (3 images per sample). Experiments were performed in
triplicate.
For the EdU assay, 3×105 RBE and
HCCC-9810 cells transfected with the PTP4A1-KD were inoculated into
24-well plates and treated with 2 µM PTEN inhibitor SF1670 (cat.
no. S7310; Selleck Chemicals) or 10 µM GSK3 inhibitor SB216763
(cat. no. S1075; Selleck Chemicals) for 24 h. Then, the cells were
incubated with 20 µM EdU (Guangzhou RiboBio, Co., Ltd.) at 37°C for
2 h. Subsequently, the cells were fixed and permeated with 4% PFA
for 30 min and 0.5% Triton X-100 for 10 min at room temperature.
After being washed with PBS, the cells were reacted with 300 µl of
Apollo reaction cocktail (Guangzhou RiboBio, Co., Ltd.) for 30 min,
and the cell nuclei were stained with 300 ml of 1X Hoechst 33342
(Guangzhou RiboBio, Co., Ltd.) for 30 min. Finally, the cells were
analyzed by fluorescence microscopy (magnification, ×200; Nikon
Corporation). Experiments were performed in triplicate.
Tumorigenesis in nude mice
A total of 10 male BALB/c nude mice (5 weeks-old;
weighing 20 g) purchased from Hunan SJA Laboratory Animal Co.,
Ltd., were used to establish a xenograft model and were maintained
in an environment at 24±2°C and 50±5% humidity with a 12/12-h
light/dark cycle. The animals were fed an autoclaved laboratory
rodent diet. RBE/HCCC-9810 cell lines stably transfected with
PTP4A1-KD and NC lentiviral vectors (5×106 cells/mouse)
in 0.1 ml PBS were subcutaneously injected into the axillae of the
anterior left limbs of the mice. Tumor volume was measured every
four days for 4 weeks. The tumor volume was calculated as follows:
Volume=0.5 × long diameter × short diameter2. A tumor
diameter exceeding 17 mm, weight loss >20% of body weight, the
animal exhibiting cachexia or wasting syndrome or the size of the
solid tumor being >10% of body weight were considered as humane
endpoint. The animals were sacrificed if any humane endpoints were
reached. In the present study, the maximum tumor volume and
diameter measured in vivo were 408.35 mm3 and 12
mm respectively. Notably, none of the mice succumbed to humane
endpoints during the experimental process. The mice were euthanized
by cervical dislocation, in accordance with the recommendations of
the American Veterinary Medical Association Guidelines for the
Euthanasia of Animals. Tumors were harvested for imaging and
monitored using Vernier calipers. All animal experiments described
in the present study were reviewed and approved by Hunan Provincial
People's Hospital (The First Affiliated Hospital of Hunan Normal
University) (approval no. 2024-156). All animal protocols complied
with the ARRIVE guidelines.
Wound healing assays
A total of 3×105 RBE and HCCC-9810 cells
transfected with PTP4A1-KD or OE lentiviral vectors were cultured
in 6-well plates for 24 h, after which an artificial scratch wound
was generated with a sterile pipette tip (200 µl). The floating
cells were then gently removed. The cells were subsequently
cultured with serum-free medium at 37°C for 24 h. The width of each
scratch was determined using an inverted microscope with camera
functionality at 0 and 24 h after scratching, and the width of each
wound was detected using ImageJ software (version 1.8.0; National
Institutes of Health). Experiments were performed in
triplicate.
Migration and invasion assays
Cell invasion and migration abilities were analyzed
in Transwell assays using Transwell chambers (pore size: 8 mm,
24-well; Corning, Inc.) with or without Matrigel (BD Biosciences),
respectively. Matrigel basement membrane matrix was diluted with
serum-free DMEM (1:1). Matrigel matrix (100 µl/well) was added into
the upper chamber and then solidified in an incubator at 37°C for 2
h. A total of 5×104 or 1×105 RBE or HCCC-9810
cells were suspended in 200 µl of serum-free medium and seeded into
the upper chamber for migration and invasion, respectively. A total
of 600 µl of medium plus 10% FBS was added to the lower chamber.
Then the cells were treated with PTEN inhibitor SF1670 or GSK3
inhibitor SB216763. After incubation at 37°C with 5% CO2
for 24 h, the cells were fixed with 4% PFA at room temperature for
30 min and stained with 0.5% crystal violet at room temperature for
15 min. Images of the migratory and invasive cells were captured
using an inverted light microscope with camera functionality.
Experiments were performed in triplicate.
WB
The cells and tissues were lysed on ice in
radioimmunoprecipitation assay (RIPA) lysis buffer (Shanghai Yeasen
Biotechnology Co., Ltd.) supplemented with protease inhibitor
cocktail tablets. Cell lysates were clarified by centrifugation (20
min at 15,000 × g at 4°C) and protein concentration determined
using the BCA protein assay (Biosharp Life Sciences). Equal amounts
of protein (40 µg) were subjected to 8–12% SDS-PAGE, transferred
onto polyvinylidene difluoride (PVDF) membranes, blocked in 10%
milk at room temperature for 1 h and probed with anti-PTP4A1
(1:1,000; cat. no. 67584-1-Ig; Proteintech Group, Inc.), anti-AKT
(1:1,000; cat. no. 9272S; Cell Signaling Technology, Inc.),
anti-phosphorylated (p)-AKT (1:1,000; cat. no. 4060S; Cell
Signaling Technology, Inc.), anti-PI3K (1:1,000; cat. no. R22768;
Zen-bio, Inc.), anti-p-PI3K (1:1,000; cat. no. 341468; Zen-bio,
Inc.), anti-PTEN (1:1,000; cat. no. R381415; Zen-bio, Inc.) and
anti-p-PTEN (1:1,000; cat. no. 9551; Cell Signaling Technology,
Inc.) antibodies at 4°C overnight. The membranes were subsequently
incubated with anti-mouse HRP-conjugated antibodies (1:2,000; cat.
no. SA00001-1; Proteintech Group, Inc.) or anti-rabbit
HRP-conjugated antibodies (1:2,000; cat. no. SA00001-2; Proteintech
Group, Inc.) for 1 h at room temperature. An enhanced
chemiluminescence (ECL; cat. no. AC13895; Acmec Biochemical)
detection system was used to visualize the protein bands.
Densitometric analysis of western blots was performed using Image
Lab Software (version 6.1; Bio-Rad Laboratories, Inc.). Experiments
were performed in triplicate.
RNA extraction and RT-qPCR
Total RNA was extracted from 1×106 cells
or 0.5 g clinical samples using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, reverse
transcription was performed using a PrimeScript RT Reagent Kit
(cat. no. BL1018A; Biosharp Life Sciences) according to the
manufacturer's protocols. The synthesized cDNA was used as a
template for PCR, which was performed using an ABI 7000
quantitative PCR instrument (Applied Biosystems; Thermo Fisher
Scientific, Inc.) with a SYBR Green PCR kit (Vazyme Biotech Co.,
Ltd.). The following thermocycling conditions were used for the
qPCR: Initial denaturation at 95°C for 30 sec, followed by 40
cycles of 95°C for 5 sec and annealing at 60°C for 30 sec, followed
by 1 cycle of 95°C for 15 sec, 60°C for 30 sec and 95°C for 15 sec.
Each reaction was performed in triplicate, and the expression
values were normalized to those of the internal control GAPDH. mRNA
expression was analyzed based on the 2−ΔΔCq relative
quantification method (21). The
primers used for amplification were as follows: PTP4A1 forward,
5′-ATTGAAGGTGGAATGAAATACGAAG-3′ and reverse,
5′-TACTTCTCCAAATACAGAAGTTGCT-3′; and GAPDH forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG'. Experiments were performed in
triplicate.
IHC
ICC tumor tissues, matched adjacent non-tumor
tissues tumor xenografts derived from HCCC-9810 and RBE cell lines
were fixed in 4% PFA at room temperature for 24 h, paraffin
embedded and sectioned to a thickness of ~5 µm. The embedded
tissues were dewaxed with xylene (10 min × 3 times), followed by
rehydration with gradient ethanol (anhydrous ethanol, 95% ethanol,
90% ethanol, 80% ethanol, 70% ethanol, 5 min each), and antigen
retrieval using microwave for 15 min. The primary antibody
anti-PTP4A1 (1:200), anti-Ki67 (1:200; cat. no. R381101; Zen-bio,
Inc.) and its corresponding secondary antibody (1:1,000) were
subsequently applied, and the samples were incubated with the
tissue slides in a wet box at 37°C for 2 h. Nuclei were
counterstained with hematoxylin. The cells were imaged using a
light microscope with camera functionality. ImageJ software
(version 1.8.0; National Institutes of Health) was then used to
calculate the average value and immunoreactivity score (IRS). The
percentages of positively stained cells were scored from 1–4 as
follows: 0, 0–5; 1, 6–25; 2, 26–50; 3, 51–75; and 4, 76–100%. The
staining intensity score was scored from 0–3 as follows: 0, no
staining signals; 1, weak staining; 2, moderate staining; and 3,
strong staining. The IRS was the comprehensive score obtained by
multiplying the staining cell score by the staining intensity score
(0–12).
Immunofluorescence (IF)
RBE cells transfected with the PTP4A OE vector were
seeded on coverslips (1×103 cells/cm2). After
48 h, the cells were fixed with 4% PFA at 4°C overnight. Then, the
cells were permeabilized with 0.1% Triton-X 100 (Sigma-Aldrich;
Merck KGaA) for 30 min, blocked with 5% BSA (Beijing Solarbio
Science & Technology Co., Ltd.) in PBS at room temperature for
2 h, and incubated with p-PTEN and PTP4A1 antibodies (1:100) at 4°C
overnight. The coverslips were subsequently incubated with the Goat
Anti-Rabbit IgG/Alexa Fluor 647 (1:1,000; cat. no. HY-P80952;
MedChemExpress) and Goat Anti-Mouse IgG (H+L) Cy3 (1:500; cat. no.
S0012; Affinity Biosciences) for 1 h at room temperature. Finally,
the slides were incubated with 1 µg/ml DAPI at room temperature for
30 min, and the cells were observed under a confocal microscope.
Images from the basal plane of the cells were captured and stored
as digital images. Each group was treated in triplicate.
Co-immunoprecipitation (Co-IP)
RBE cells were lysed on ice in RIPA lysis buffer
supplemented with protease inhibitor cocktail tablets. A total of 1
µg of PTP4A1 antibody was added to the cell lysate, which was
subsequently incubated at 4°C overnight with slow shaking.
Subsequently, 10 µl of protein A agarose beads was added to the
cell lysates, which were subsequently incubated overnight with the
PTP4A1 antibody and then for 4 h at 4°C with slow shaking. 1 µg
normal rabbit IgG (cat. no. 2729S; Cell Signaling Technology, Inc.)
were included as a NC. The agarose beads were subsequently
centrifuged at at 600 × g for 3 min at 4°C. The supernatant was
carefully aspirated. Then, 20 µl of 2X SDS gel loading buffer was
added to the beads, which were boiled for 6 min for WB. Experiments
were performed in triplicate.
Statistical analysis
Data analysis was performed using SPSS 20.2 (IBM
Corp.). The values represent the mean ± SD from 3 independent
experiments. Unpaired Student's t-tests were performed to compare
the differences between two groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
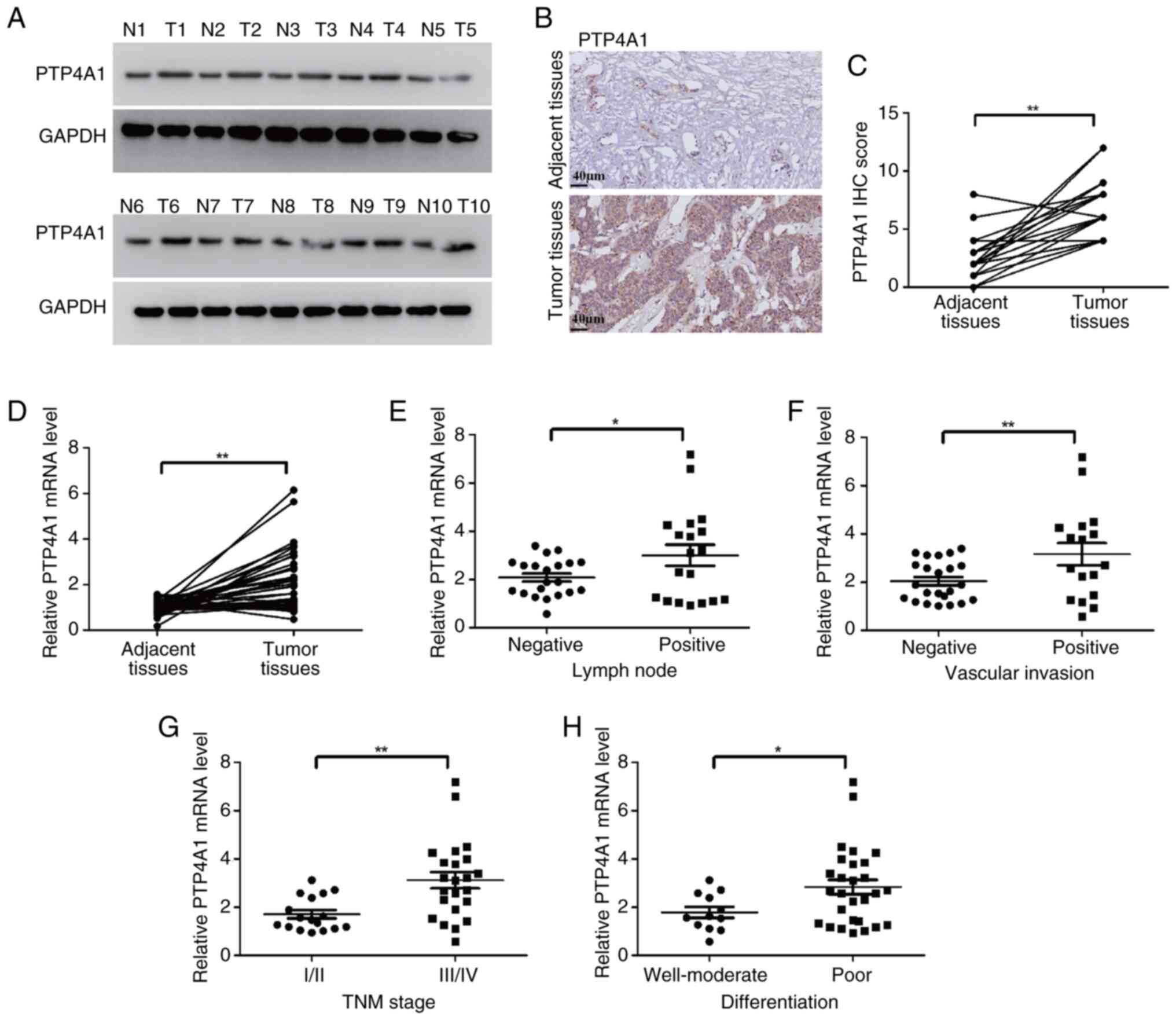
PTP4A1 is upregulated and associated
with invasive pathological features in ICC
To investigate the function of PTP4A1 in ICC, PTP4A1
expression was first detected in ICC tissues using WB. The results
revealed that PTP4A1 is overexpressed in tumor tissues compared
with adjacent normal tissues (Fig.
1A). To further verify PTP4A1 expression in ICC, the expression
levels of PTP4A1 in 20 pairs of ICC tissues and matched adjacent
normal tissues were analyzed using IHC. The results revealed that
PTP4A1 was highly expressed in ICC tissues (Fig. 1B and C). RT-qPCR was subsequently
performed to evaluate the mRNA levels of PTP4A1 in 40 pairs of ICC
tumor tissues and matched adjacent normal tissues. PTP4A1 mRNA was
frequently upregulated in ICC (Fig.
1D). Furthermore, the relationships between PTP4A1 expression
and the clinicopathological characteristics of patients with ICC
were explored. Compared with that in patients with ICC with
negative lymph node metastasis, PTP4A1 mRNA expression was higher
in patients with ICC with positive lymph node metastasis (Fig. 1E). In addition, the PTP4A1 mRNA was
overexpressed in patients with ICC with vascular invasion (Fig. 1F). Compared with patients with I/II
TNM stage ICC, PTP4A1 was highly expressed in patients with III/IV
TNM stage ICC (Fig. 1G). Moreover,
the expression of PTP4A1 was related to the differentiation of ICC.
PTP4A1 mRNA expression in poorly differentiated ICC tissues was
higher than the levels in well-differentiated and moderately
differentiated ICC tissues (Fig.
1H). Collectively, these results indicated that PTP4A1 is
overexpressed and associated with aggressive pathological
characteristics in ICC.
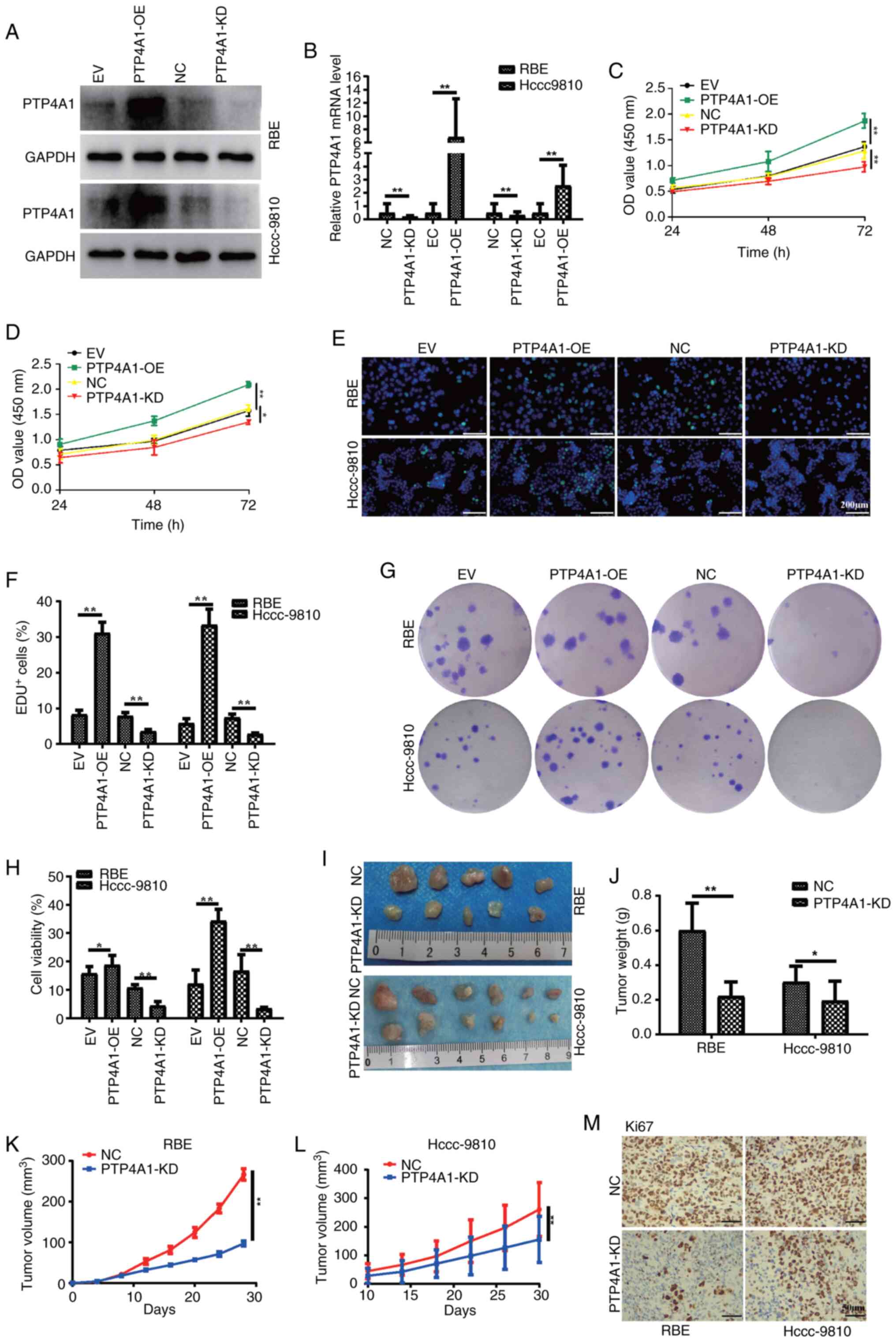
PTP4A1 promotes ICC cell
proliferation, migration and invasion in vitro and in vivo
To investigate the potential biological function of
PTP4A1 in ICC, RBE and HCCC-9810 cells were stably transfected with
PTP4A1 KD and OE lentiviruses. The expression levels of PTP4A1 in
the ICC cell lines RBE and HCCC-9810 were analyzed using WB and
RT-qPCR. As demonstrated in Fig. 2A and
B, the expression of PTP4A1 was significantly decreased in RBE
and HCCC-9810 cells transfected with PTP4A1 KD lentivirus and
significantly increased in REB and HCCC-9810 cells transfected with
PTP4A1-OE lentivirus compared with that in ICC cells transfected
with NC or EV lentivirus. CCK-8, EdU and colony formation assays
were subsequently performed to evaluate the role of PTP4A1 in the
proliferation of ICC cells. The results of the CCK-8 assays
suggested that PTP4A1 promoted the proliferation of RBE and
HCCC-9810 cells (Fig. 2C and D).
The results of the EdU assays demonstrated that the viabilities of
the RBE and HCCC-9810 cells transfected with the PTP4A1-KD
lentivirus were lower than those of the corresponding NC cells
(Fig. 2E and F). Consistent with
these findings, the viabilities of RBE and HCCC-9810 cells
transfected with the PTP4A1-OE lentivirus were greater than those
of the corresponding EV-transfected cells (Fig. 2E and F). Additionally, the results
of the colony formation assay revealed that PTP4A1 increased the
survival ability of RBE and HCCC-9810 cells (Fig. 2G and H). Furthermore, a subcutaneous
tumor formation experiment was conducted in nude mice in which RBE
and HCCC-9810 cells were transfected with PTP4A1-KD lentivirus or
NC lentivirus. All the mice successfully formed tumors. ICC tumor
tissue images revealed that the tumors that originated from RBE and
HCCC-9810 cells transfected with the PTP4A1-KD lentivirus were
smaller than those in the NC groups (Fig. 2I). The tumor growth curves revealed
that the tumors in RBE and HCCC-9810 cells transfected with
PTP4A1-KD lentivirus grew slower than those in the NC groups did at
the same time points (Fig. 2J). The
volume of tumor tissues derived from RBE and HCCC-9810 cells
transfected with the PTP4A1-KD lentivirus was significantly smaller
than that derived from the NC cells (Fig. 2K and L). The IHC results revealed
that the expression of the proliferation marker Ki67 was decreased
in tumor tissues derived from RBE and HCCC-9810 cells transfected
with the PTP4A1-KD lentivirus (Fig.
2N). Collectively, these results indicated that PTP4A1 promotes
ICC proliferation in vitro and in vivo.
To further explore the function of PTP4A1 in ICC
metastasis, wound healing assays were performed, and the results
revealed that PTP4A1-OE increased the migration of RBE and
HCCC-9810 cells. However, the migration abilities of
PTP4A1-downregulated RBE and HCCC-9810 cells were reduced (Fig. 3A and B). Similarly, the results of
migration assays demonstrated that PTP4A1 promoted the migration of
RBE and HCCC-9810 cells (Fig. 3C and
D). Invasion assays suggested that PTP4A1 increased invasion in
RBE and HCCC-9810 cells (Fig. 3E and
F). In addition, PTP4A1-OE promoted epithelial-mesenchymal
transition (EMT), and PTP4A1-KD inhibited EMT in RBE and HCCC-9810
cells (Fig. 3G-N). Furthermore, the
results showed that PTP4A1-OE significantly increases the
expression levels of MMP2 and MMP9, which are key enzymes involved
in tumor invasion and metastasis. Conversely, PTP4A1-KD leads to a
decrease in MMP2 and MMP9 expression (Fig. 3G-N). Expression of the epithelial
cell marker E-cadherin was decreased and that of the mesenchymal
cell marker N-cadherin was increased in tumorigenic tissues derived
from RBE and HCCC-9810 cells transfected with PTP4A1-KD lentivirus
(Fig. 3M). These results
demonstrated that PTP4A1 promoted migration, invasion and EMT in
ICC.
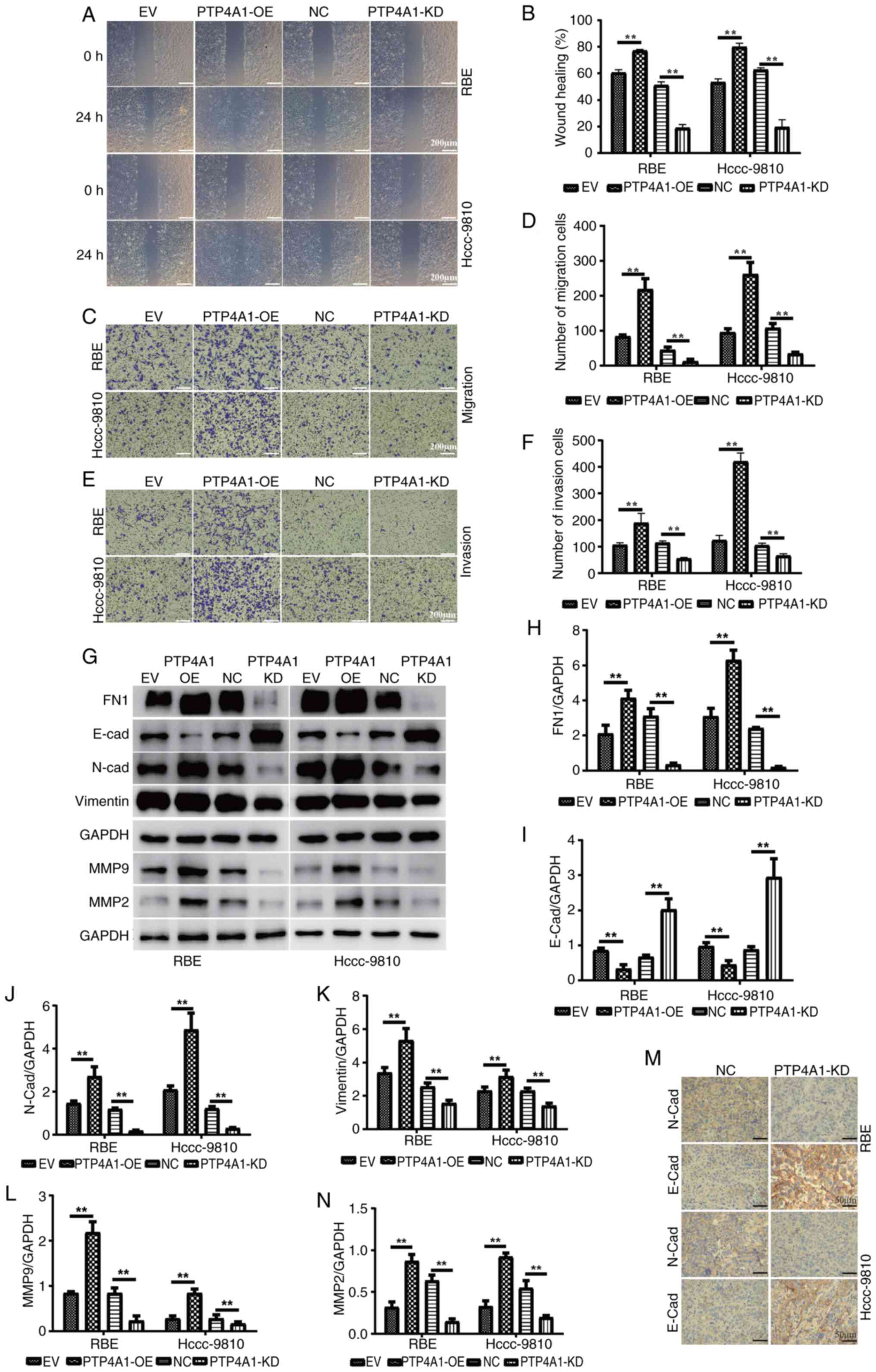 | Figure 3.PTP4A1 promotes migration, invasion
and EMT in intrahepatic cholangiocarcinoma. (A and B) Wound healing
and (C and D) migration assays were used to detect the effect of
PTP4A1 on the migration of RBE and HCCC-9810 cells. Invasion assays
(E and F) were used to evaluate the invasion abilities of RBE and
HCCC-9810 cells stably transfected with PTP4A1-overexpressing and
PTP4A1-knockdown lentiviruses. (G) EMT marker and MMP2/MMP9
expression in RBE and HCCC-9810 cells stably transfected with
PTP4A1-OE and PTP4A1-KD lentiviruses. (H-N) Quantitative analysis
of (H) FN1, (I) E-cadherin, (J) N-cadherin, (K) Vimetin, (L) MMP9
and (N) MMP2 expression levels. (M) E-cadherin and N-cadherin
expression levels in tumorigenic tissues were measured by
immunohistochemistry. **P<0.01. PTP4A1, protein tyrosine
phosphatase 4A1; EMT, epithelial-mesenchymal transition; OE,
overexpression; KD, knockdown; NC, negative control; EV, empty
vector. |
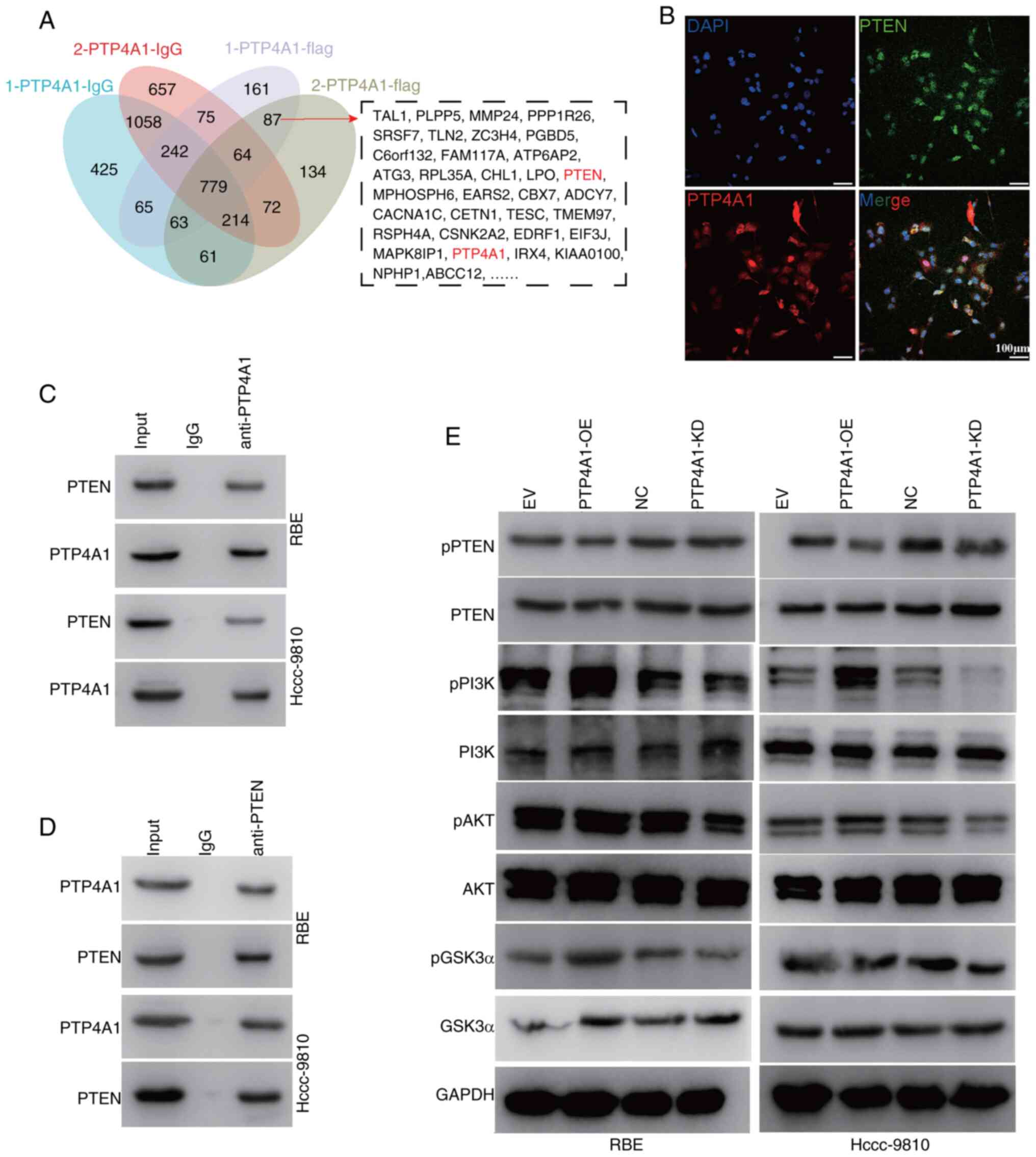
PTP4A1 interacts with PTEN and is
involved in the activation of the PI3K/AKT/GSK3α pathway
To further clarify the mechanism by which PTP4A1
promotes the progression of ICC, Co-IP-MS was performed to screen
for proteins that interact with PTP4A1. A Venn diagram revealed
that 87 proteins may interact with PTP4A1 (Fig. 4A). Among them, focus was addressed
on PTEN. To further verify the interaction between PTP4A1 and PTEN,
IF staining was performed. The results revealed that PTP4A1
colocalized with PTEN in RBE cells (Fig. 4B). Co-IP analysis verified that
PTP4A1 interacted with PTEN in RBE cells transfected with PTP4A1-OE
lentivirus (Fig. 4C and D). PTP4A1
belongs to the PTP family and may play a role in the
dephosphorylation of its interacting proteins. Next, the effect of
PTP4A1 on PTEN phosphorylation was evaluated. The results revealed
that PTP4A1-OE decreased PTEN phosphorylation and that KD of PTP4A1
increased the phosphorylation level of PTEN in RBE and HCCC-9810
cells, indicating that PTP4A1 inhibited the activation of PTEN
(Fig. 4E). PTEN is a negative
regulator of the PI3K/AKT/GSk3α signaling pathway. The activation
of PI3K/AKT/GSk3α signaling in PTP4A1-OE and PTP4A1-KD RBE and
HCCC-9810 cells was subsequently detected (Fig. 4E). These results indicated that
PTP4A1 contributes to the activation of the PI3K/AKT/GSk3α
signaling pathway by interacting with and regulating PTEN.
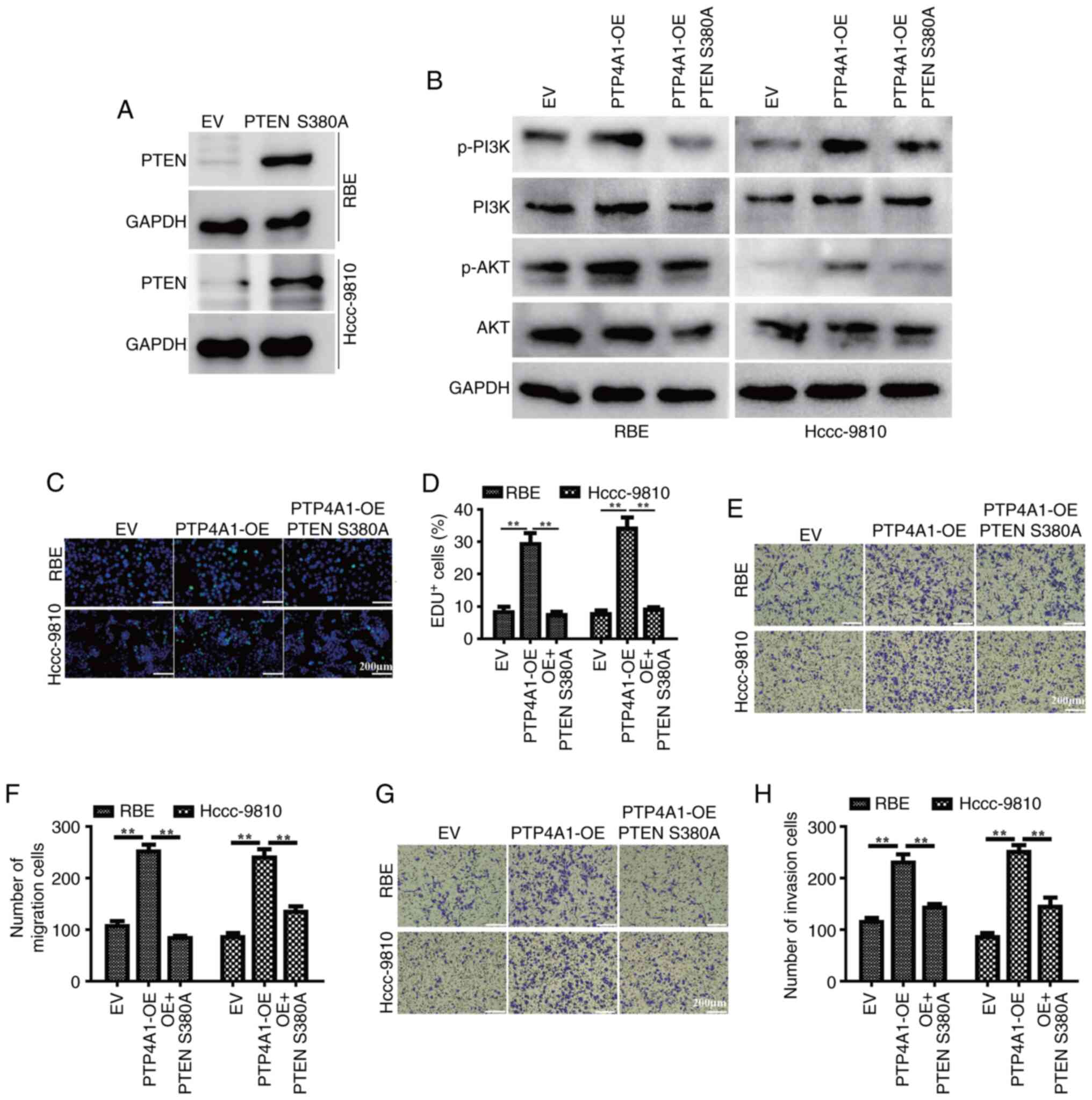
To further validate the inhibitory role of PTP4A1 in
PTEN phosphorylation, a phospho-mimetic PTEN mutant plasmid (PTEN
S380A) was first constructed and transfected into RBE and HCCC-9810
cells. The results of WB showed the successful OE of PTEN S380A
(Fig. 5A). Subsequently, PTEN S380A
plasmid was overexpressed in RBE and HCCC-9810 cells with stable
PTP4A1-OE. WB revealed that the phospho-mimetic PTEN S380 mutant
markedly diminished the phosphorylation levels of PI3K and AKT
(Fig. 5B). Additionally, mimetic
phosphorylation of PTEN S380 attenuated the PTP4A1-driven promotion
of cell proliferation (Fig. 5C and
D), invasion (Fig. 5E and F)
and metastasis (Fig. 5G and H) in
RBE and HCCC-9810 cells. These findings suggest that PTP4A1 may
exert its oncogenic effects by modulating the PTEN/PI3K/AKT
signaling pathway.
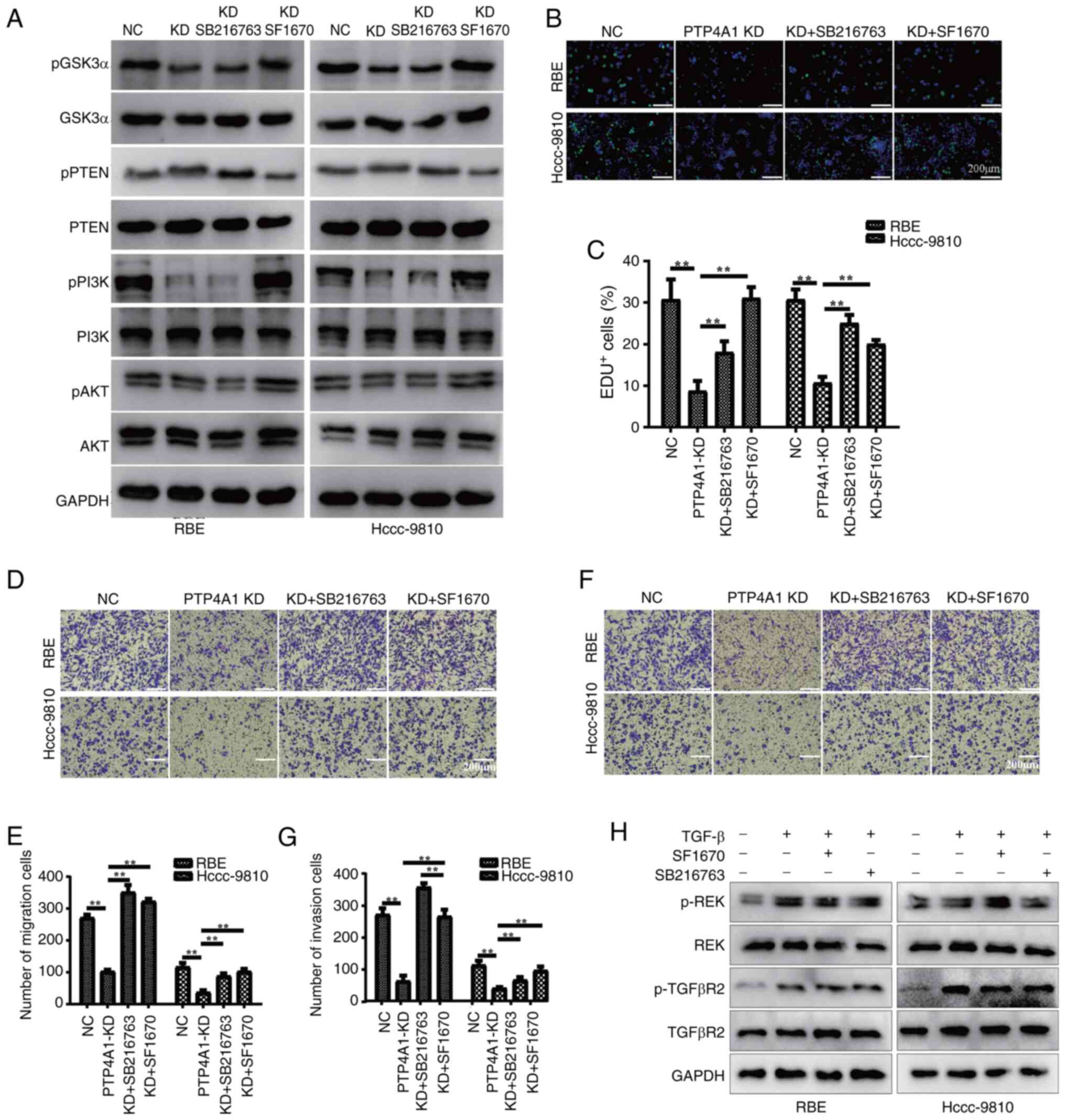
PTP4A1 promotes ICC progression
through regulating the PTEN/PI3K/AKT/GSK3α signaling pathway
PTP4A1 promoted cell proliferation, migration and
invasion; interacted with PTEN; and contributed to the activation
of the PI3K/AKT/GSk3α signaling pathway. In addition, the
PI3K/AKT/GSk3α pathway is involved in tumor progression. Thus, it
was hypothesized that the promotion of cell proliferation, invasion
and metastasis by PTP4A1 is dependent on the regulation of the
PTEN/PI3K/AKT/GSk3α pathway. To test this hypothesis, RBE and
HCCC-9810 cells were stably transfected with PTP4A1-KD lentivirus
with the PTEN inhibitor SF1670 or the GSK3α inhibitor SB216763. The
WB results revealed that the KD of PTP4A1 increased PTEN
phosphorylation and decreased the activation of the PI3K/AKT/GSk3α
pathway in RBE and HCCC-9810 cells (Fig. 6A). Additionally, the PTEN inhibitor
SF1670 significantly inhibited PTEN phosphorylation and activated
the PI3K/AKT/GSk3α pathway in RBE and HCCC-9810 cells with PTP4A1
KD (Fig. 6A). As a downstream
molecule of PI3K/AKT, the GSK3α inhibitor SB216763 did not affect
the phosphorylation of PTEN, PI3K or AKT (Fig. 6A). EdU assays revealed that the PTEN
inhibitor SF1670 and the GSK3α inhibitor SB216763 significantly
attenuated the inhibitory effect of PTP4A1 KD on the proliferation
of RBE and HCCC-9810 cells (Fig. 6B and
C). Furthermore, the results of migration (Fig. 6D and E) and invasion assays
(Fig. 6F and G) revealed that the
PTEN inhibitor SF1670 and the GSK3α inhibitor SB216763 reversed the
inhibitory effects of PTP4A1 KD on the migration and invasion of
RBE and HCCC-9810 cells. To validate the off-target effects of the
PTEN inhibitor SF1670 and GSK3α inhibitor SB216763 on other
pathways (ERK and TGF-β), the ERK and TGF-β signaling pathways were
evaluated in RBE and HCCC-9810 cells co-treated with SF1670 or
SB216763 and TGF-β. The results demonstrated that SF1670 and
SB216763 did not significantly affect ERK phosphorylation levels or
the activation status of the TGF-β signaling pathway in these
cells, suggesting that these inhibitors did not exhibit off-target
effects (Fig. 6H). Collectively,
these results indicate that PTP4A1 promotes the progression of ICC
by regulating the PTEN/PI3K/AKT/GSk3α pathway.
Discussion
ICC is a highly aggressive tumor with an advanced
clinical stage, limited therapeutic options, and a high
postoperative recurrence rate (22,23).
Moreover, the prognosis of ICC is extremely poor, with a 5-year
survival rate of only 25–40% (5);
moreover, the median overall survival (OS) time of patients with
advanced ICC is as low as ~22 months (24,25).
The molecular pathogenesis of ICC is complex and not fully
understood. A deeper understanding of the molecular mechanisms of
ICC may contribute to the development of novel therapeutic
approaches to improve ICC prognosis. In the present study, it was
revealed that PTP4A1 was overexpressed and associated with
aggressive clinicopathological characteristics in ICC. Furthermore,
it was demonstrated that PTP4A1 promoted cell proliferation,
migration and invasion by interacting with PTEN and activating the
PI3K/AKT/GSK3α pathway.
Accumulating data suggest that PTP4A1 functions as
an oncogene and is involved in tumor progression (26). An increasing number of studies,
including the authors' previous study (20), have revealed that the OE of PTP4A1
is associated with poor OS in various types of tumors, such as
NSCLC (9), oral squamous cell
carcinoma (OSCC) (18), cervical
cancer (27) and HCC (16,19,28).
Similarly, it was revealed that PTP4A1 was highly expressed in ICC
tissues. In addition, the present study demonstrated that PTP4A1 is
associated with lymph node metastasis, vascular invasion, advanced
TNM stage and poor differentiation of ICC. Consistent with the
present findings, overexpressed PTP4A1 was significantly correlated
with advanced TNM stage in patients with HCC (16,19)
and with lymphatic metastasis in patients with NSCLC (9). Based on the aforementioned results, it
was hypothesized that PTP4A1 plays an essential role in tumor
progression.
Furthermore, the findings of the present study
showed that PTP4A1 KD decreased ICC cell proliferation, migration
and invasion in vivo and in vitro. Similarly, PTP4A1
OE increased these processes, which is consistent with the findings
of previous studies in colorectal cancer (17,29),
OSCC (18), esophageal squamous
cell carcinoma (ESCC) (30) and HCC
(16,19). Mechanistically, a previous study
indicated that PTP4A1 regulated proliferation and apoptosis by
downregulating the protein level of P53 (17) and promoted EMT via the
ERK/GSK3β/β-catenin pathway in colon cancer (29). In cervical cancer, PTP4A1 aggravated
malignant progression by activating the ERK pathway (13,31).
Additionally, PTP4A1 contributes to metastasis through altering
mitochondrial metabolic reprogramming mediated by PMK2 and ACO2 in
OSCC (18). These results indicate
that PTP4A1 exerts oncogenic effects through diverse mechanisms
across distinct tumor types. However, it has been reported that
PTP4A1 promotes migration and invasion by activating the PI3K/AKT
pathway in HCC (16,19) and ESCC (30). ICC, a liver malignancy with a
relatively low incidence rate, shares similarities with HCC in
terms of oncogenesis.
PTP4A1 has been reported to suppress E-cadherin and
increase migration and invasion via the PI3K/AKT and ERK pathways
in HCC (16,19), promote TGF-β signaling in human
fibroblasts through interacting with SRC and activating the ERK
pathway (15).
The present study demonstrated that PTP4A1 promoted
cell viability, migration and invasion by activating the PI3K/AKT
pathway in ICC. Nevertheless, the molecular mechanism by which
PTP4A1 activates the PI3K/AKT pathway remains unclear. PTEN is a
classical tumor suppressor that antagonizes PI3K/AKT signaling
(32,33). In the present study, it was found
that PTP4A1 interacted with PTEN. PTP4A1 is a protein tyrosine
phosphatase, and its OE decreased the phosphorylation of PTEN and
increased the phosphorylation of PI3K, AKT and its downstream
protein GSK3α. By contrast, PTP4A1 KD increased PTEN
phosphorylation and inactivated PI3K/AKT/GSK3α, indicating that
PTP4A1 activated the PI3K/AKT pathway by suppressing the activation
of PTEN. In addition, the PTEN inhibitor SF1670 and the GSK3α
inhibitor SB216763 significantly attenuated the inhibitory effects
of PTP4A1 KD on the proliferation, migration and invasion of ICC
cells.
In conclusion, the present study demonstrated that
PTP4A1 was overexpressed and associated with aggressive
pathological characteristics in ICC. As an oncogene, PTP4A1
promotes cell proliferation, migration and invasion in a manner
dependent on the regulation of the PTEN/PI3K/AKT/GSk3α pathway,
indicating that PTP4A1 may serve as a valuable therapeutic target
for ICC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Fund of Science and
Technology Department of Hunan (grant no. 2018SK50726).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
OL was responsible for project development, data
collection and manuscript writing. YP collected and analyzed data.
JC collected data. YL contributed to the study conception and
design, revised and edited the manuscript. All authors read and
approved the final version of the manuscript. OL and YP confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
The experimental protocols were approved (approval
no. 2023-151) by the Ethics Committee of Hunan Provincial People's
Hospital (The First Affiliated Hospital of Hunan Normal University;
Changsha, China). Written informed consent was obtained from all
participants. All animal experiments described in the present study
were reviewed and approved by Hunan Provincial People's Hospital
(The First Affiliated Hospital of Hunan Normal University)
(approval no. 2024-156). All animal protocols complied with the
ARRIVE guidelines. Euthanasia was performed by means of cervical
dislocation, in accordance with the recommendations of the American
Veterinary Medical Association Guidelines for the Euthanasia of
Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
Moris D, Palta M, Kim C, Allen PJ, Morse
MA and Lidsky ME: Advances in the treatment of intrahepatic
cholangiocarcinoma: An overview of the current and future
therapeutic landscape for clinicians. CA Cancer J Clin. 73:198–222.
2023.PubMed/NCBI
|
|
3
|
Banales JM, Marin JJG, Lamarca A,
Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen
JB, Braconi C, et al: Cholangiocarcinoma 2020: The next horizon in
mechanisms and management. Nat Rev Gastroenterol Hepatol.
17:557–588. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Greten TF, Schwabe R, Bardeesy N, Ma L,
Goyal L, Kelley RK and Wang XW: Immunology and immunotherapy of
cholangiocarcinoma. Nat Rev Gastroenterol Hepatol. 20:349–365.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mazzaferro V, Gorgen A, Roayaie S, Droz
Dit Busset M and Sapisochin G: Liver resection and transplantation
for intrahepatic cholangiocarcinoma. J Hepatol. 72:364–377. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kelley RK, Bridgewater J, Gores GJ and Zhu
AX: Systemic therapies for intrahepatic cholangiocarcinoma. J
Hepatol. 72:353–363. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brindley PJ, Bachini M, Ilyas SI, Khan SA,
Loukas A, Sirica AE, The BT, Wongkham S and Gores GJ:
Cholangiocarcinoma. Nat Rev Dis Primers. 7:652021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vogel A, Segatto O, Stenzinger A and
Saborowski A: FGFR2 inhibition in cholangiocarcinoma. Annu Rev Med.
74:293–306. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang T, Shi X, Wang Z, Liu X, Zhang G, Zhu
Q, Mi L and Wang R: Overexpression of PTP4A1 is associated with
poor overall survival in non-small cell lung cancer. Int J Clin Exp
Pathol. 11:3583–3590. 2018.PubMed/NCBI
|
|
10
|
Sun JP, Luo Y, Yu X, Wang WQ, Zhou B,
Liang F and Zhang ZY: Phosphatase activity, trimerization, and the
C-terminal polybasic region are all required for PRL1-mediated cell
growth and migration. J Biol Chem. 282:29043–29051. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bai Y, Luo Y, Liu S, Zhang L, Shen K, Dong
Y, Walls CD, Quilliam LA, Wells CD, Cao Y and Zhang ZY: PRL-1
protein promotes ERK1/2 and RhoA protein activation through a
non-canonical interaction with the Src homology 3 domain of p115
Rho GTPase-activating protein. J Biol Chem. 286:42316–42324. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu LZ, He YZ, Dong PP, Ma LJ, Wang ZC,
Liu XY, Duan M, Yang LX, Shi JY, Zhou J, et al: Protein tyrosine
phosphatase PTP4A1 promotes proliferation and
epithelial-mesenchymal transition in intrahepatic
cholangiocarcinoma via the PI3K/AKT pathway. Oncotarget.
7:75210–75220. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li X, Ma N, Zhang Y, Wei H, Zhang H, Pang
X, Li X, Wu D, Wang D, Yang Z and Zhang S: Circular RNA circNRIP1
promotes migration and invasion in cervical cancer by sponging
miR-629-3p and regulating the PTP4A1/ERK1/2 pathway. Cell Death
Dis. 11:3992020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo Y, Liang F and Zhang ZY: PRL1 promotes
cell migration and invasion by increasing MMP2 and MMP9 expression
through Src and ERK1/2 pathways. Biochemistry. 48:1838–1846. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sacchetti C, Bai Y, Stanford SM, Di
Benedetto P, Cipriani P, Santelli E, Piera-Velazquez S, Chernitskiy
V, Kiosses WB, Ceponis A, et al: PTP4A1 promotes TGFβ signaling and
fibrosis in systemic sclerosis. Nat Commun. 8:10602017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jin S, Wang K, Xu K, Xu J, Sun J, Chu Z,
Lin D, Koeffler PH, Wang J and Yin D: Oncogenic function and
prognostic significance of protein tyrosine phosphatase PRL-1 in
hepatocellular carcinoma. Oncotarget. 5:3685–3696. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu H, Ye L and Liu Z: GINS2 regulates the
proliferation and apoptosis of colon cancer cells through PTP4A1.
Mol Med Rep. 25:1172022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu B, Si W, Wei B, Zhang X and Chen P:
PTP4A1 promotes oral squamous cell carcinoma (OSCC) metastasis
through altered mitochondrial metabolic reprogramming. Cell Death
Discov. 9:3602023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu JW, Chang JG, Yeh KT, Chen RM, Tsai JJ,
Su WW and Hu RM: Increased expression of PRL-1 protein correlates
with shortened patient survival in human hepatocellular carcinoma.
Clin Transl Oncol. 14:287–293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li O, Jiang B, Yi WM, Zhang Y, Yang PZ,
Guo C, Sun ZP and Peng C: LncRNA NEAT1 promotes cell proliferation,
migration, and invasion via the miR-186-5p/PTP4A1 axis in
cholangiocarcinoma. Kaohsiung J Med Sci. 37:379–391. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bekki Y, Von Ahrens D, Takahashi H,
Schwartz M and Gunasekaran G: Recurrent intrahepatic
cholangiocarcinoma-review. Front Oncol. 21:7768632021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sapisochin G, Ivanics T and Heimbach J:
Liver transplantation for intrahepatic cholangiocarcinoma: Ready
for prime time? Hepatology. 75:455–472. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Edeline J, Touchefeu Y, Guiu B, Farge O,
Tougeron D, Baumgaertner I, Ayav A, Campillo-Gimenez B, Beuzit L,
Pracht M, et al: Radioembolization plus chemotherapy for first-line
treatment of locally advanced intrahepatic cholangiocarcinoma: A
phase 2 clinical trial. JAMA Oncol. 6:51–59. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shi GM, Huang XY, Wu D, Sun HC, Liang F,
Ji Y, Chen Y, Yang GH, Lu JC, Meng XL, et al: Toripalimab combined
with lenvatinib and GEMOX is a promising regimen as first-line
treatment for advanced intrahepatic cholangiocarcinoma: A
single-center, single-arm, phase 2 study. Signal Transduct Target
Ther. 8:1062023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Giménez-Mascarell P, Oyenarte I, Hardy S,
Breiderhoff T, Stuiver M, Kostantin E, Diercks T, Pey AL,
Ereño-Orbea J, Martínez-Chantar ML, et al: Structural basis of the
oncogenic interaction of phosphatase PRL-1 with the magnesium
transporter CNNM2. J Biol Chem. 292:786–801. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Reich R, Hadar S and Davidson B:
Expression and clinical role of protein of regenerating liver (PRL)
phosphatases in ovarian carcinoma. Int J Mol Sci. 12:1133–1145.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fu Y, Li B, Huang R, Ji X and Bai WK: Long
noncoding RNA DLEU2 promotes growth and invasion of hepatocellular
carcinoma by regulating miR-30a-5p/PTP4A1 axis. Pathol Res Pract.
238:1540782022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang JX, Mai SJ, Huang XX, Wang FW, Liao
YJ, Lin MC, Kung HF, Zeng YX and Xie D: MiR-29c mediates
epithelial-to-mesenchymal transition in human colorectal carcinoma
metastasis via PTP4A and GNA13 regulation of β-catenin signaling.
Ann Oncol. 25:2196–2204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zou J, Ma Q, Gao C, Yang M, Wen J, Xu L,
Guo X, Zhong X and Duan Y: WTAP promotes proliferation of
esophageal squamous cell carcinoma via m6A-dependent epigenetic
promoting of PTP4A1. Cancer Sci. 115:2254–2268. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen M, Chi Y, Chen H and Zhao L: Long
non-coding RNA USP30-AS1 aggravates the malignant progression of
cervical cancer by sequestering microRNA-299-3p and thereby
overexpressing PTP4A1. Oncol Lett. 22:5052021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Glaviano A, Foo ASC, Lam HY, Yap KCH,
Jacot W, Jones RH, Eng H, Nair MG, Makvandi P, Geoerger B, et al:
PI3K/AKT/mTOR signaling transduction pathway and targeted therapies
in cancer. Mol Cancer. 22:1382023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Parsons R: Discovery of the PTEN tumor
suppressor and its connection to the PI3K and AKT oncogenes. Cold
Spring Harb Perspect Med. 10:a0361292020. View Article : Google Scholar : PubMed/NCBI
|