Introduction
In 2020, esophageal cancer, a lethal type of cancer,
was responsible for ~5.5% of all cancer-associated fatalities
globally (1). Compared with other
regions globally, the incidence and mortality rates of esophageal
cancer are greater in Asian countries (2). According to the Taiwan Cancer Registry
Annual Report, esophageal cancer ranks ninth for cancer-related
mortality; squamous cell carcinoma (SCC) is the most frequent
histological subtype, accounting for up to 91.4% of all cases
(3). The prognosis is poor for
esophageal SCC (ESCC) due to its aggressive nature, which includes
early distant organ metastases and regional tracheal invasion
(4). Typically, metastasis affects
the liver, lung and lymph nodes (3,5). A
thorough understanding of the intricate processes underlying the
spread of ESCC and distant metastases is essential for future
advancements in early prevention and intervention.
Small non-coding RNA molecules known as microRNAs
(miRNAs/miRs) functionally control gene expression by degrading or
suppressing the translation of mRNA targets (6,7). These
compounds exert key regulatory effects on cellular functions such
as apoptosis, differentiation and cell cycle entry and progression
(8–10). miRNAs typically function as
oncogenes or tumor suppressors in different types of cancer. In
cancer, tumor development, aggressiveness and treatment evasion are
associated with dysregulation of miRNAs (6,11).
miRNAs primarily regulate intricate signaling pathways and networks
that regulate gene expression to regulate the growth and
progression of a tumor with metastases and treatment sensitivity
(12). Targeting oncogenic miRNAs
or boosting tumor suppressor miRNAs in cancer is considered to be a
unique form of cancer treatment (12).
Previous studies have demonstrated that obesity
enhances the risk of cancer progression and metastases,
particularly for malignancy of the kidney, prostate, endometrium,
breast, colon and esophagus (13,14).
Adipocytes secrete bioactive chemicals known as adipokines, which
are key in the advancement of cancer, metabolic disorder,
cardiovascular disease, inflammation and metastasis (15–17).
In the tumor microenvironment, adipokines may trigger the
epithelial-to-mesenchymal transition and enhance metastasis
(18). Visfatin was originally
revealed in visceral adipose tissue and is considered a
multifunctional adipokine and an extracellular nicotinamide
phosphoribosyltransferase enzyme (19). Patients with numerous types of
cancer have elevated serum levels of visfatin (20,21).
Visfatin is key for the invasion and metastasis of cancer (19,22).
In esophageal cancer, visfatin levels have been documented to be
upregulated compared with those of healthy controls and promote
VEGF-C-regulated lymphangiogenesis (3). However, the regulatory roles of
visfatin in miRNA synthesis and cell motility in esophageal cancer
remain unclear. The aim of the present study was to investigate the
regulatory role of visfatin in miRNA synthesis and to elucidate the
underlying mechanisms by which miRNA influences cell motility in
esophageal cancer.
Materials and methods
Materials
Vascular endothelial zinc finger 1 (VEZF1; cat. no
SC-365560; Santa Cruz Biotechnology, Inc.) β-actin (cat. no GT5512;
Genetex International Corporation), and versican (VCAN) antibody
(cat. no SAB1408906; MilliporeSigma) were used. Recombinant human
visfatin (cat. no. 130-09-25UG) was obtained from PeproTech, Inc.
The small interfering (si)RNAs targeting VEZF1 (cat no. sc-94046)
and VCAN (cat no. sc-41903) were purchased from Santa Cruz
Biotechnology, Inc. A non-targeting negative control siRNA (cat no.
D-001810-10-05) was purchased from Thermo Fisher Scientific, Inc.
PI3K (Ly294002) inhibitor (cat. no. ALX-270-038) was obtained from
Enzo Life Sciences, Inc. AKT (cat. no. A6730) and mTOR (rapamycin)
inhibitors (cat. no. R0395) were obtained from Sigma-Aldrich (Merck
KGaA). The miR-3613-5p mimic (5′-UGUUGUACUUUUUUUUUUGUUC-3′) and miR
mimic negative control (5′-UUGUACUACACAAAAGUACUG-3′) were purchased
from AllBio. All other reagents and chemicals were obtained from
MilliporeSigma, unless otherwise specified.
Cell culture
The invasive ESCC cell line KYSE-410 (cat no.
94072023) was obtained from the European Collection of Cell
Cultures and human ESCC cell line CE81T (cat. no. 60166) was
purchased from Bioresource Collection and Research Centre. Cells
were cultured in either RPMI-1640 or DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% heat-inactivated fetal calf
serum (Corning, Inc), 2 mM L-glutamine, 100 U/ml penicillin and 100
µg/ml streptomycin. All cells were maintained at 37°C in a
humidified atmosphere containing 5% CO2.
miRNA sequencing and bioinformatics
analysis
For miRNA sequencing (NEXTflex small RNA sequencing
kit v3, cat no. NOVA-5132-06, PerkinElmer) was utilized,
high-quality total RNA samples from the visfatin (30 ng/ml)-treated
for 24 h at 37°C and untreated control KYSE-410 cells were
utilized. The experimental workflow was performed by Azenta Life
Sciences. To prepare the library, 1 µg total RNA was used
quantified using both Qubit and qPCR methods. The qualified library
were sequenced pair end PE150 (150 pair-End) Small RNA sequencing
was conducted using the Illumina, Inc. HiSeq/Novaseq or MGI2000
platform. Total RNA was extracted using the TRIzol (Thermo Fisher
Scientific, Inc.) and RNA integrity was confirmed with an Agilent
2100 Bioanalyzer (RNA integrity number >7). To prevent
adaptor-dimer formation, an excess of 3′ SR Adaptor for Illumina
was hybridized with the SR RT Primer. Subsequently, the 5′ SR
Adaptor for Illumina was ligated to the small RNA using a 5′
Ligation Enzyme. First-strand cDNA synthesis was performed using
ProtoScript II Reverse Transcriptase (New England Biolabs, US).
Each sample was then amplified by PCR kit (Bioo Scientific,
PerkinElmer) using thermocycling conditions: initial denaturation
95°C for 2 min, 20–25 cycles of denaturation 95°C for 20 sec,
annealing 60°C for 30 sec, Extension 72°C for 15 sec, final
extension 95°C for 2 minusing P5 (Forward,
5′-GTTCAGAGTTCTACAGTCCGACGATC-3′) and P7 (Reverse,
5′-AGATCGGAAGAGCACACGTCT-3′) primers and the PCR product was
purified by DNA Clean Beads (Bioo Scientific, PerkinElmer). The
purified products of 140–160 bp were recovered and cleaned using
PAGE (6%) and validated using an Agilent 2100 Bioanalyzer. The raw
readings underwent quality control before processing, which
included removing adapter sequences and contaminants. To guarantee
data quality, analysis of the lengths and counts of the filtered
reads was performed by Trimmomatic (V0.30), Cutadapt (V1.3) and
FastQC (V0.10.1), along with an evaluation of the data volume
(9). DEseq2 (V1.6.3), DEseq
(V1.18.0), EdgeR (V3.4.6) and bowtie2 (V2.1.0), software was used
for Heatmap and volcano plot to analyzed differentially expressed
miRNA.
The levels of visfatin in patients with primary and
metastatic esophageal cancer were analyzed using a UALCAN, dataset
and cell motility genes associated with VEZF1 were obtained from
The Cancer Genome Atlas (TCGA) database (3,11).
Target genes were predicted using TargetScan
(targetscan.org/vert_80/), miRTarBase (https://mirtarbase.cuhk.edu.cn), miRDB (https://mirdb.org/) and ENCORI (https://rnasysu.com/encori/index.php) databases
(24). Spearman correlation was
used to analyze gene correlation. Gene expression levels in ESCC
patients were analyzed using the GEO dataset GSE161533.
Kaplan-Meier analysis was performed to assess the VCAN levels in
ESCC. Expression levels of Visfatin, VEZF1, and VCAN were evaluated
using GSE77861, while miR-3613-5p levels were examined using
GSE97051 in ESCC patient tissues.
Migration and invasion assay
Transwell inserts (Costar, Inc.; 8-µm pores) were
used in 24-well plates for the migration experiments and pre-coated
with a layer of Matrigel at 37°C for 30 min before the invasion
assay. KYSE410 and CE81T Cells were pretreated with or without (10
µM) of PI3K (Ly294002), AKT inhibitor (AKTi), mTOR (Rapamycin)
inhibitors. KYSE410 and CE81T cells were transfected using
(Lipofectamine 2000, Invitrogen, Thermo Fisher Scientific, Inc.)
with or without siVEZF1, siVCAN siRNAs and miRNA mimics (50 nM) and
incubated at 37°C for 24 h, immediately followed by migration and
invasion assay. The upper chamber contained 1×104
KYSE410 and CE81T cells in 200 µl serum-free medium (DMEM or RPMI),
whereas the lower chamber contained 300 µl 10% FBS and medium with
various concentrations (1,3,10, 30)
ng/ml of visfatin and incubated for 24 h at 37°C in 5%
CO2. Cotton-tipped swabs were used to remove the
Matrigel from the upper side of the filters, and PBS was used to
wash the filters (23,24). Cells were fixed in 3.7% formaldehyde
for 5 min at room temperature, incubated for 24 h at 37°C in 5%
CO2 and stained with 0.05% crystal violet in PBS for 20
min at room temperature. Stained cells were observed under an
Olympus CKX53 inverted light microscope, and quantified by ImageJ
(V1.52a; imagej.net/ij/) and GraphPad prism (V8.0;
graphpad.com/guides/prism/8/user-guide/index.htm).
Reverse transcription-quantitative
(RT-q)PCR
TRIzol (Thermo Fisher Scientific) cat no. 12183555)
was used to extract total RNA from the esophageal cancer KYSE410
cells. In summary, oligo-dT primers (Thermo Fisher Scientific,
Inc.), 10 mM dNTP (Cyrusbioscience), 5× standard buffer
(Invitrogen; Thermo Fisher Scientific) and 0.1 M DTT (Invitrogen;
Thermo Fisher Scientific) were used to reverse-transcribe 1 µg RNA
into cDNA in compliance with the manufacturer's instructions. The
KAPA SYBER FAST qPCR kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) was used to mix 100 ng cDNA sample with specific
primers. qPCR was performed using a Senso Quest Labcycler thermal
cycler. The thermocycling conditions were as follows: Initial
denaturation at 95°C for 6 min, followed by 40 cycles of
denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec and
extension at 72°C for 1 min, with a final extension at 72°C for 10
min. The Mir-XTM miRNA First Strand Synthesis kit (Takara Bio Inc.)
was used to create cDNA from 100 ng total RNA for the miRNA assay.
The endogenous control GAPDH was used to achieve relative
quantification of gene expression. The comparative Cq approach was
performed to calculate the relative expression (25,26).
The sequences of primers were as follows: miR-3613-5p,
5′-TGTTGTACTTTTTTTTTTGTTC-3′ (melting temperature (Tm), 43.7°C;
length, 22 bases); VEZF1: Forward, 5′-GGTTCTGCAGCATTTCACCC-3′ and
reverse, 5′-TGATGGGAAGCTTCATGGGC-3′ (Tm, 53.8°C; length, 20 bases
each); VCAN: Forward, 5′-GTAACCCATGCGCTACATAAAGT-3′ (Tm, 53.5°C;
length, 23 bases) and reverse, 5′-GGCAAAGTAGGCATCGTTGAAA-3′ (Tm,
53.0°C; length, 22 bases) and GAPDH: Forward
5′-ACCACAGTCCATGCCATCAC-3′ (Tm, 53.5°C, length, 20 bases) and
reverse, 5′-TCCACCACCCTGTTGCTGTA-3′ (Tm, 53.8, length, 20 bases).
Relative gene expression levels were calculated using the
2−ΔΔCq method (27).
Western blot analysis
Proteins from KYSE410 cells were extracted using
RIPA lysis buffer (cat. No. P0012, Beyotime Institute of
Biotechnology). The total protein concentration was determined
using a BCA Protein Assay Kit and 30 µg of protein per lane was
used for analysis. Proteins were separated by SDS-PAGE using a 10%
gel and transferred onto PVDF membranes (Merck KGaA). Membranes
were blocked with 5% non-fat milk at room temperature for 1 h and
incubated with primary antibodies VEZF1 (1:1,000), VCAN (1:1,000)
as target proteins and β-actin (1:3,000) for an entire night (18–20
h) at 4°C. Membranes were incubated for 1 h at room temperature
with horseradish peroxidase-conjugated secondary antibodies
(1:5,000) goat anti-mouse IgG (cat. no. sc-516102), Santa Cruz
Biotechnology, Inc. An ECL kit (MilliporeSigma) was used to detect
the expression of the target protein and an ImageQuant LAS 4000
biomolecular imager was used for visualization (28,29).
Luciferase reporter assay
A luciferase assay kit was utilized to track the
luciferase activity in order to measure the 3′-untranslated region
(UTR). After transfecting the cells with either the wild-type (wt)-
or mutant (mt)-VEZF1-3′-UTR luciferase plasmid (Stratagene; Agilent
Technologies, Inc.), using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific) the cells were transfected for 24 h at 37°C
using miR-3613-5p mimic (sequence: 5′-ACAAAAAAAAAAGUACAACAUU-3′;
(AllBio). Following 24 h transfection, cells were lysis and
instantly the luciferase activity was measured using the
Dual-Luciferase® Reporter Assay System (Promega
Corporation). Firefly luciferase activity was normalized to Renilla
luciferase activity to control for transfection efficiency.
Statistical analysis
Data were analyzed using ImageJ software (V1.52a)
(https://imagej.net/ij/) and GraphPad Prism
software (V8.0) (graphpad.com/guides/prism/8/user-guide/index.htm).
All data are presented as the mean ± SD from 3 independent
experiments. Statistical significance between two groups was
assessed using the unpaired Student's t-test. Comparisons involving
>2 groups with a single variable were performed using one-way
ANOVA followed by Bonferroni's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
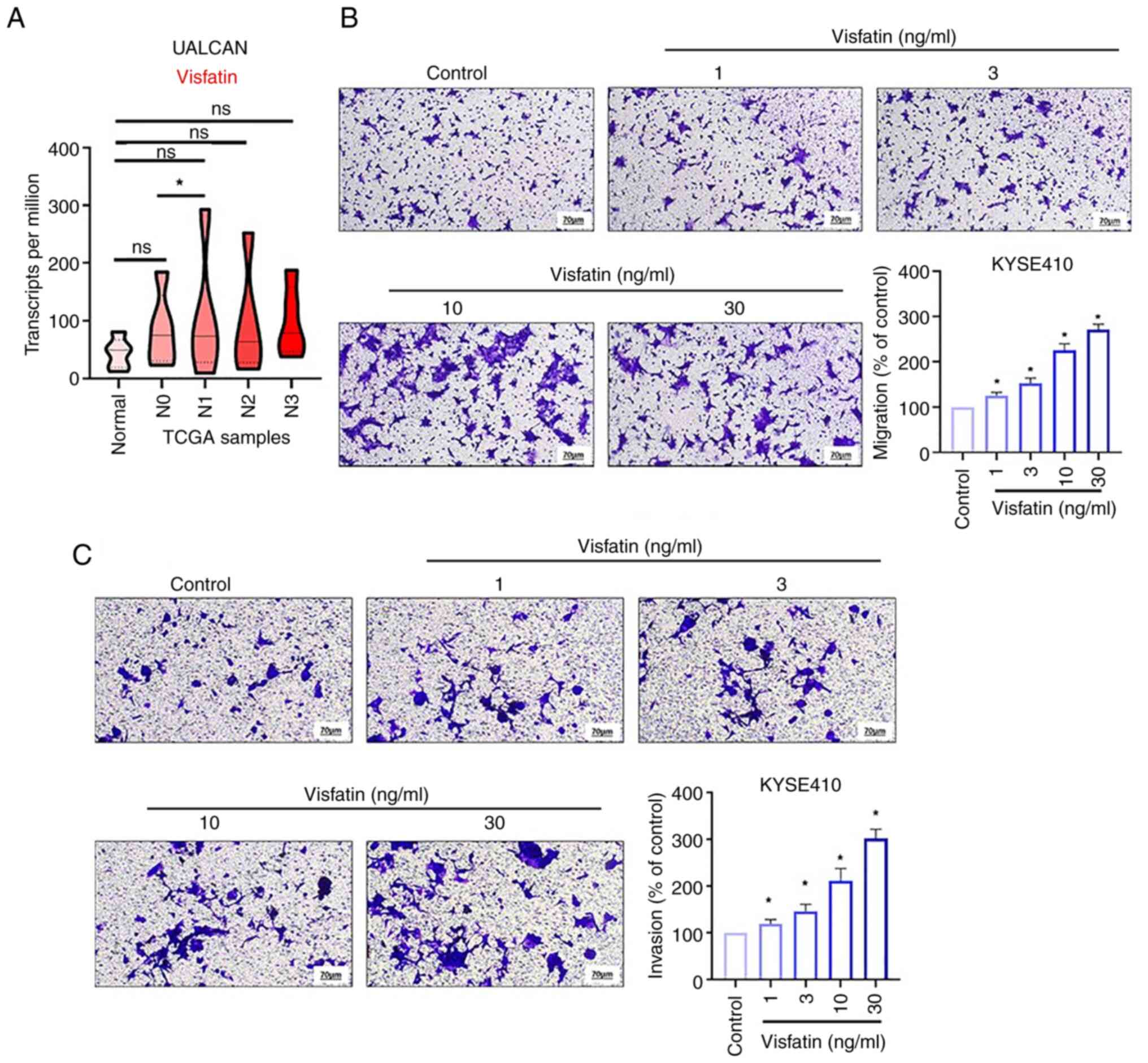
Visfatin promotes esophageal cancer
migration by inhibiting miR-3613-5p
Visfatin is key for the development of numerous
types of cancer (3,30). However, the mechanisms through which
visfatin affects esophageal cancer metastases remain unknown. The
UALCAN data revealed that the levels of visfatin were higher in
patients with metastatic esophageal cancer than in those with
primary esophageal cancer (Fig.
1A). Transwell migration assay was used to examine the effects
of visfatin on the migration of esophageal cancer cells. Visfatin
promoted the migration of KYSE-410 esophageal cancer cells in a
concentration-dependent manner (Fig.
1B). Additionally, visfatin enhanced the invasive ability of
esophageal cancer cells (Fig.
1C).
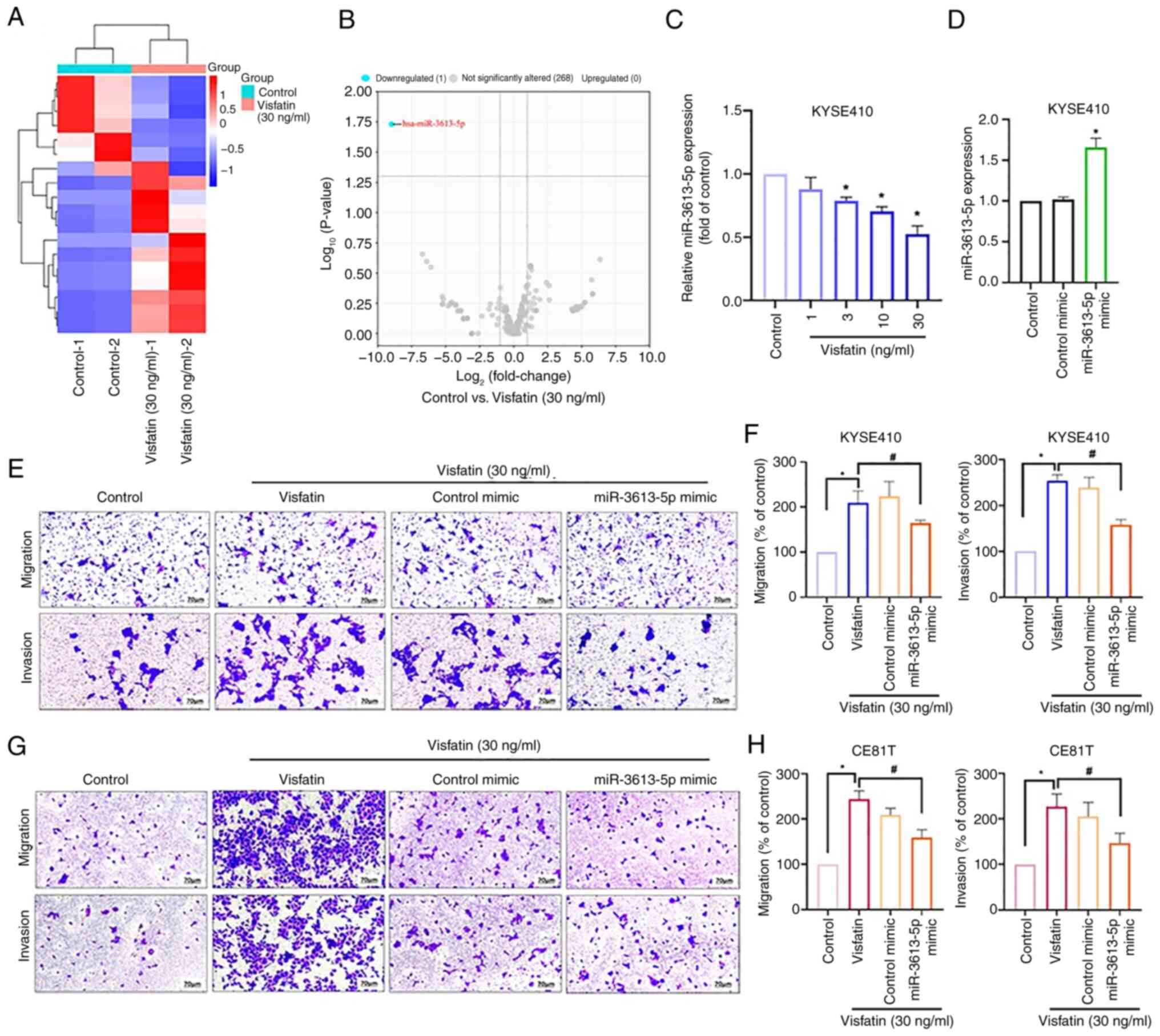
The metastasis of esophageal cancer is associated
with dysregulated miRNAs (31). To
investigate whether miRNAs mediate visfatin-induced esophageal
cancer cell migration, miRNA sequencing was performed to assess the
differential expression of miRNAs in KYSE-410 cells treated with
visfatin. The resulting heatmap and volcano plot illustrated
differentially expressed miRNAs (Fig.
2A and B); among these, miR-3613-5p was the most downregulated
(Fig. 2B). Visfatin (≥3 ng/ml)
inhibited miR-3613-5p synthesis in a concentration-dependent manner
(Fig. 2C). Transfection with a
miR-3613-5p mimic increased the miR-3613-5p expression compared
with control (Fig. 2D). miR-3613-5p
mimic antagonized the visfatin-induced promotion of cell migration
and invasion (Fig. 2E and F).
Consistent with the KYSE-410 results, miR-3613-5p mimic treatment
inhibited visfatin-induced migration and invasion in CE81T cells
(Fig. 2G and H). Thus, these
findings demonstrate that visfatin promoted esophageal cancer cell
migration by decreasing miR-3613-5p production.
miR-3613-5p inhibits the VEZF1/VCAN
axis and mediates visfatin-promoted cell migration
A total of four publicly available miRNA databases
(TargetScan, miRTarBase, miRDB and ENCORI) predicted that
miR-3613-5p targets seven potential candidates (Fig. 3A). Among these, VEZF1 was markedly
upregulated compared with ANP32B in patients with esophageal cancer
(Fig. 3B). Additionally, VEZF1 was
more highly upregulated in patients with metastatic than in those
with primary esophageal cancer, however this was not significant
(Fig. 3C). The direct application
of visfatin to esophageal cancer cells enhanced VEZF1 mRNA and
protein expression (Fig. 3D and E).
Transfection of the cells with VEZF1 siRNA, effectively suppressed
VEZF1 protein levels, as confirmed by western blot analysis
(Fig. 3F), and inhibited the
visfatin-induced promotion of cell migration and invasion (Fig. 3G-I). These findings were validated
in CE81T cells, where VEZF1 siRNA also reversed visfatin-induced
cell motility, confirming a consistent effect across ESCC cell
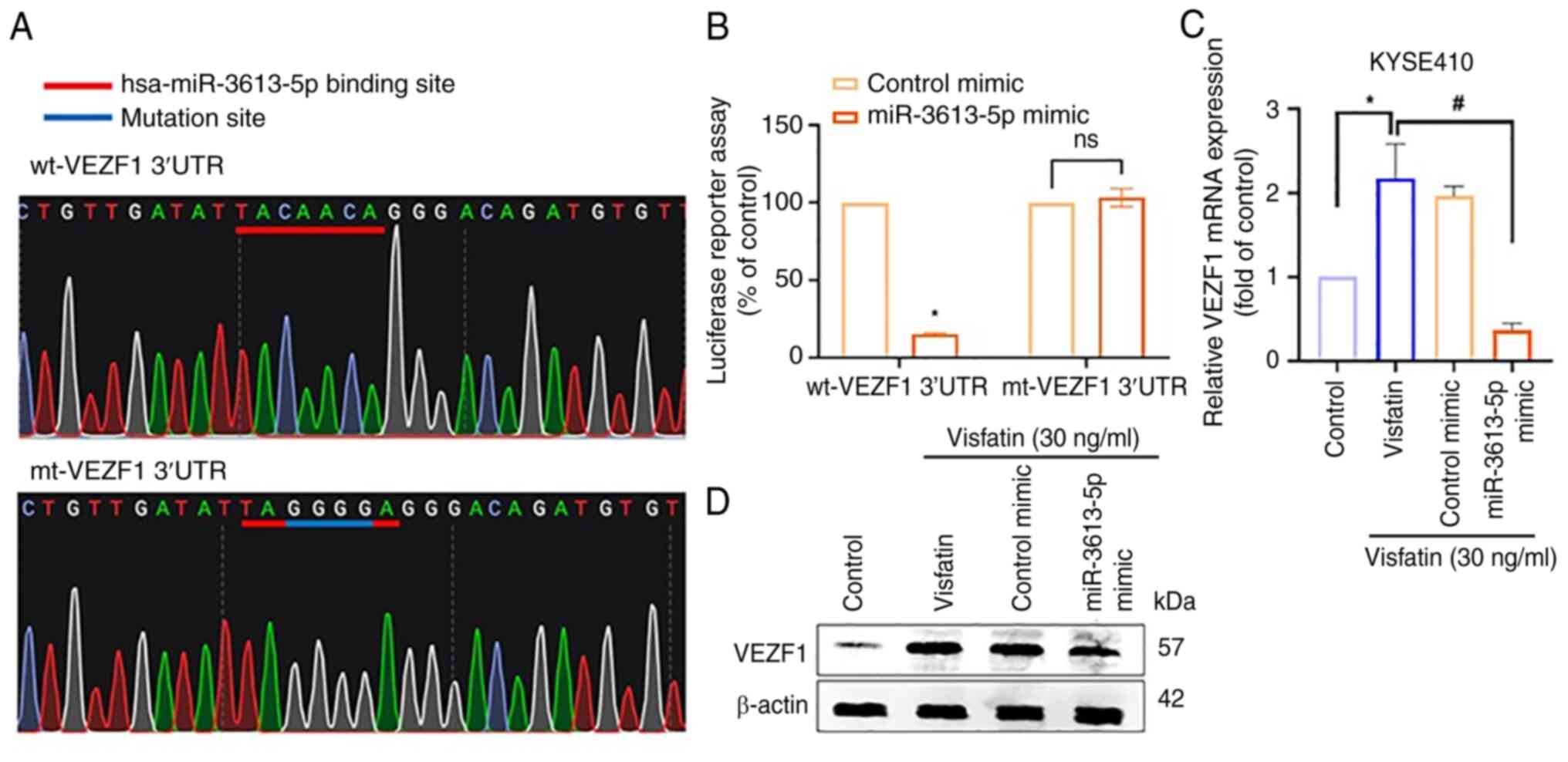
lines (Fig. 3J-L). Subsequently,
wt- and mt-VEZF1-3′-UTR luciferase plasmids were generated to
examine the direct binding effects of miR-3613-5p on VEZF1
(Fig. 4A). The miR-3613-5p mimic
decreased wt-VEZF1-3′-UTR, but not mt-VEZF1-3′-UTR luciferase
activity (Fig. 4B). Furthermore,
the miR-3613-5p mimic blocked visfatin-induced VEZF1 mRNA
expression (Fig. 4C) and protein
levels (Fig. 4D).
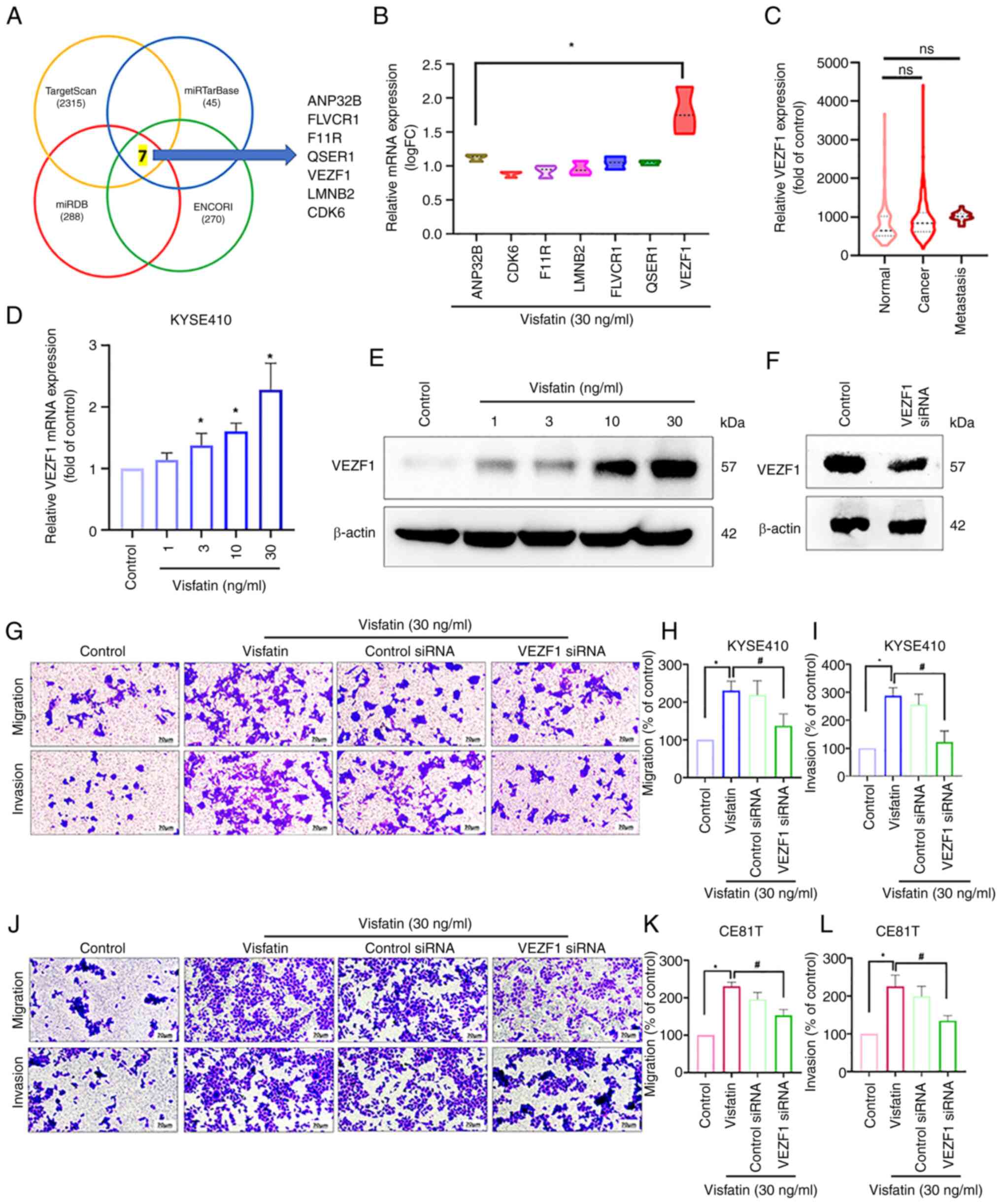 | Figure 3.VEZF1, regulated by miR-3613-5p, is
involved in visfatin-induced esophageal cancer cell migration. (A)
miR databases (TargetScan, miRTarBase, miRDB and ENCORI) predicted
that miR-3613-5p targets seven potential candidates. (B) Gene
levels in patients with esophageal cancer retrieved from TCGA. (C)
VEZF1 gene levels in normal tissue and tissues from patients with
primary and metastatic esophageal cancer retrieved from TCGA. (D)
KYSE 410 cells were stimulated with visfatin, and VEZF1 expression
was examined using reverse transcription-quantitative PCR and (E)
western blot analysis. (F) VEZF1 siRNA transfection efficiency
examined by western blot. (G) KYSE410 cells were transfected with
VEZF1 siRNA and treated with visfatin, then cell (H) migration and
(I) cell invasion was examined. (J) CE81T cells were transfected
with VEZF1 siRNA followed by visfatin treatment and then (K)
Migration and (L) invasion was examined. *P<0.05 vs. control;
#P<0.05 vs. visfatin. miR, microRNA; TCGA, The Cancer
Genome Atlas; VEZF1, Vascular endothelial Zinc Finger 1; si, small
interfering; FC, fold change; ANP32B, Acidic Leucine-Rich Nuclear
Phosphoprotein 32 Family Member B; CDK, Cyclin-dependent kinase;
F11R, F11 receptor; LMNB2, Lamin B2; FLVCR, Feline Leukemia Virus
subgroup C Receptor 1; QSER, Glutamine and Serine-rich protein 1;
ns, not significant. |
To investigate whether the visfatin-induced
expression of VEZF1 regulates cell motility genes, TCGA database
was searched for genes associated with VEZF1 (Fig. 5A). Among 300 genes exhibiting a
positive correlation, 13 genes were also associated with cell
adhesion functions (Fig. 5B). Data
from the GSE161533 database indicated that VCAN was the most
upregulated gene in patients with esophageal cancer (Fig. 5C). Kaplan-Meier analysis confirmed
that the VCAN levels were higher in patients with esophageal cancer
than in healthy controls (Fig. 5D).
Visfatin promoted the mRNA and protein expression of VCAN (Fig. 6A and B). Transfection with VCAN
siRNA effectively decreased VCAN protein levels, as confirmed by
western blot analysis (Fig. 6C),
and also attenuated visfatin-induced cell motility (Fig. 6D-F); this effect was validated in
CE81T cells (Fig. 6G-I).
miR-3613-5p mimic and VEZF1 siRNA also suppressed visfatin-induced
VCAN expression (Fig. 6J and K),
indicating that the inhibition of miR-3613-5p and the promotion of
VEZF1 occurred upstream of visfatin-induced VCAN expression and
cell motility. Mechanically, visfatin inhibits miR-1264 and
promotes platelet derived growth factor C (PDGF-C) synthesis
through activation of the PI3K/AKT/mTOR signaling pathway (19). To elucidate the molecular mechanism,
the present study examined the role of the PI3K/AKT/mTOR pathway in
visfatin-mediated ESCC migration and invasion Transwell assay
results indicate that treatment with PI3K (Ly294002), AKTi and mTOR
(rapamycin) pathway inhibitors effectively reversed the migration
and invasion effects induced by visfatin, confirming the
involvement of PI3K/AKT/mTOR signaling axis in visfatin-driven
migration and invasion (Fig. 6L-Q).
To explore the clinical relevance of visfatin-related genes,
GSE77861 dataset was analyzed to compare gene expression between
normal and esophageal cancer tissue. Visfatin (Fig. 6R), VEZF1 (Fig. 6T) and VCAN (Fig. 6U) were significantly upregulated in
esophageal cancer tissues compared with normal controls
(P<0.05). In addition, analysis of the GSE97051 dataset showed
that miR-3613-5p expression was slightly decreased in cancerous
tissue compared with normal samples (Fig. 6S). Together, these findings support
the potential involvement of the visfatin/miR-3613-5p/VEZF1/VCAN
axis in the progression of esophageal cancer.
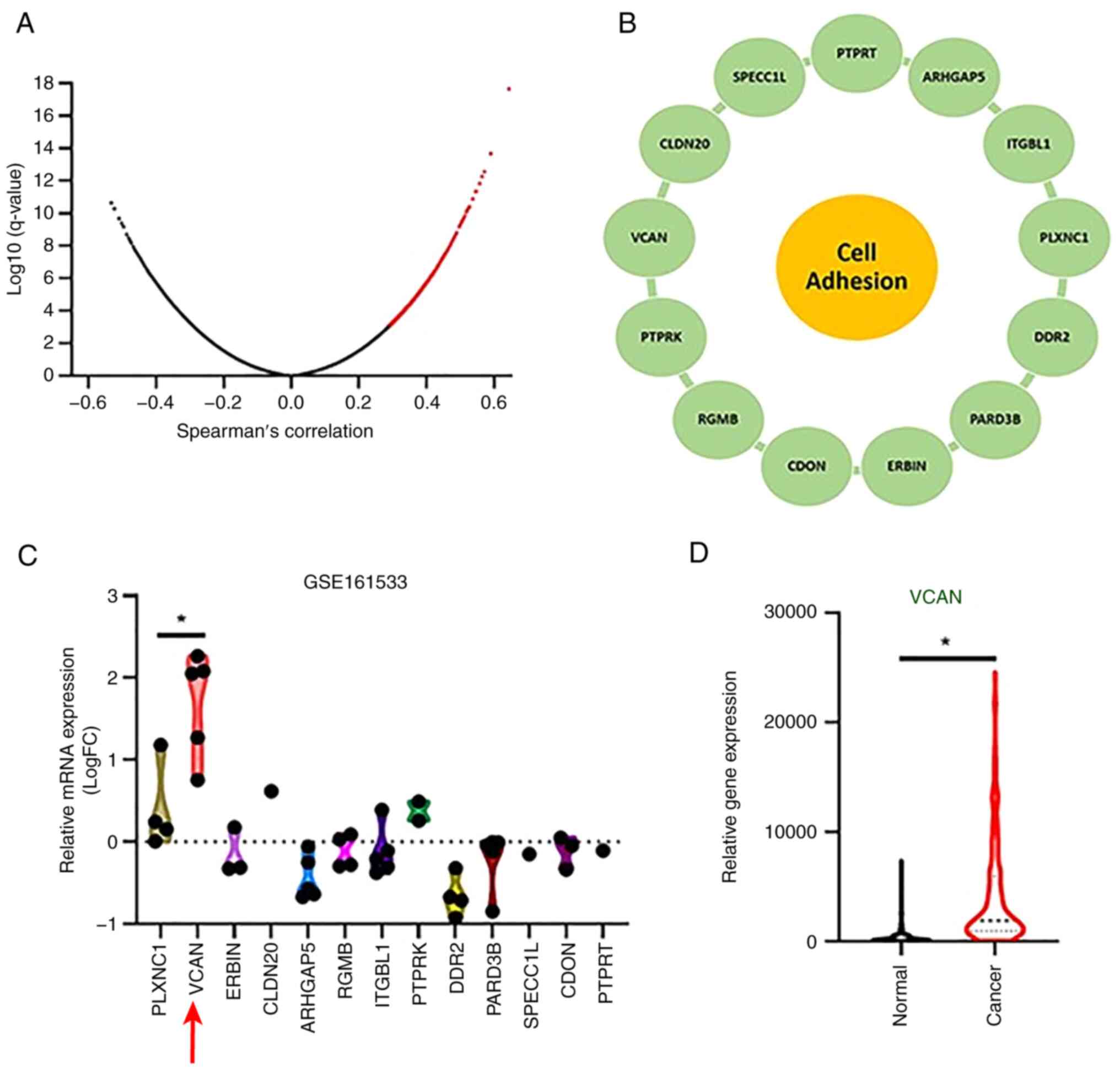 | Figure 5.VCAN is highly expressed in patients
with esophageal cancer. (A) Spearman correlation analysis from
TCGA. (B) A total of 13 genes correlated with VEZF1 and were
associated with cell adhesion functions. (C) Gene levels in
patients with esophageal cancer retrieved from the GSE161533 and
TCGA database. (D) Kaplan-Meier analysis of VCAN level in normal
and cancer stage in ESCC *P<0.05. VCAN, Versican; VEZF1,
Vascular Endothelial Zinc Finger 1; TCGA, The Cancer Genome Atlas;
FC, Fold Change; PLXNC1, Plexin C1; ERBIN, interacting protein;
CLDN, claudin; ARAHGAP, Rho GTPase-activating protein 5; RGMB,
Repulsive Guidance Molecule BMP Co-Receptor B; ITGBL, Integrin
subunit beta 1; PTPRK, Protein Tyrosine Phosphatase Receptor Type
Kappa; DDR, DNA Damage Response; PARD, Par-3 family cell polarity
regulator; SPECC1L, sperm antigen with calponin homology and
coiled-coil domains 1 like; CDON, Cell Adhesion Associated,
Oncogene Regulated; PTPRT, protein tyrosine phosphatase, receptor
type, T. |
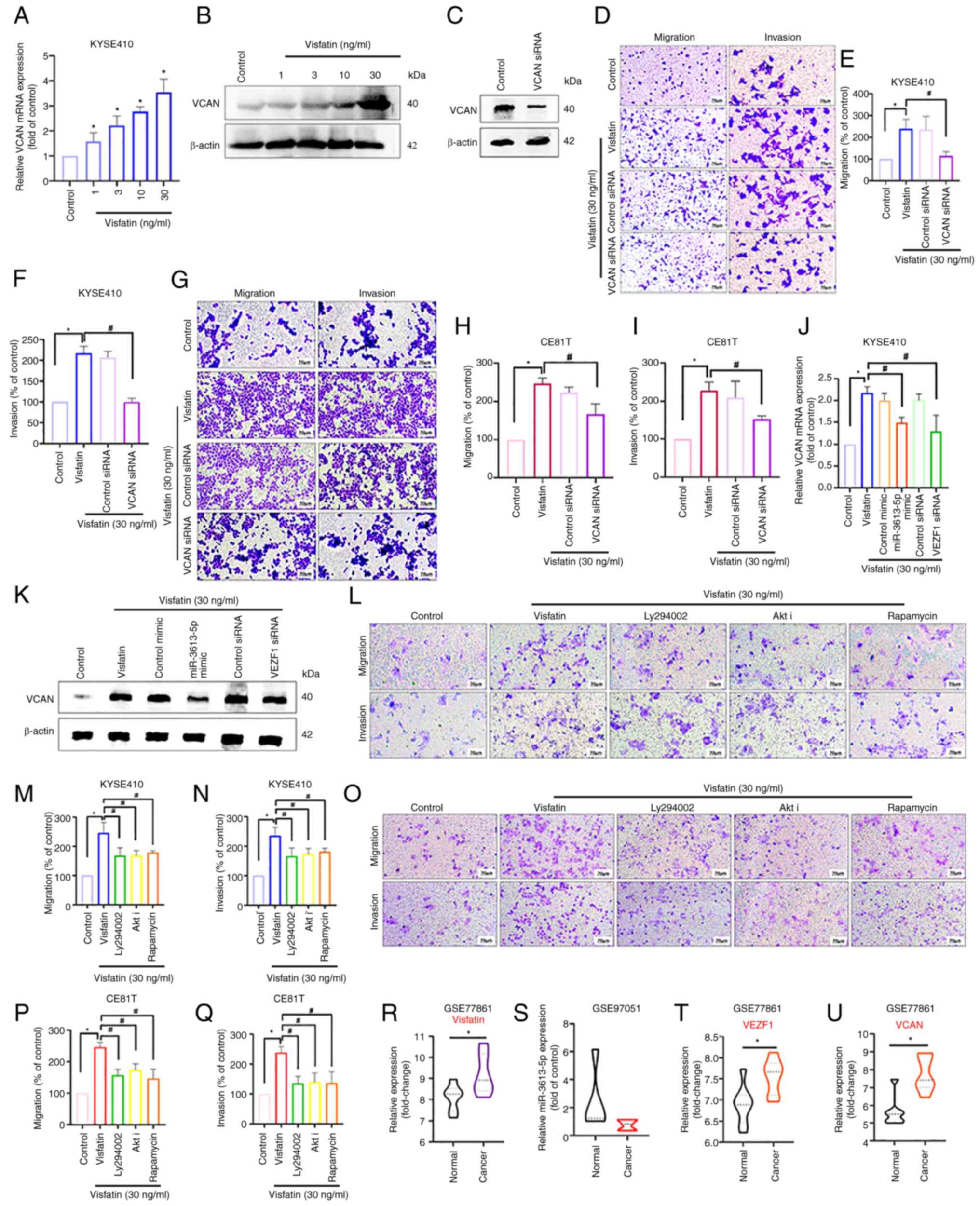 | Figure 6.VCAN is involved in visfatin-induced
esophageal cancer migration. Cells were stimulated with visfatin
and VCAN expression was examined using (A) RT-qPCR and (B) western
blot analysis. (C) VCAN siRNA transfection efficiency confirmed by
western blot analysis. (D) KYSE410 cells were transfected with or
without VCAN siRNA followed by visfatin treatment and cell (E)
migration and (F) invasion were examined. (G) CE81T cells were
transfected with VCAN siRNA and treated with visfatin; cell (H)
migration and (I) invasion were examined. Cells were transfected
with miR-3613-5p mimic or VEZF-1 siRNA and treated with visfatin;
VCAN expression was examined using (J) RT-qPCR and (K) western
blotting. (L) KYSE 410 cells treated with PI3K (Ly294002), AKT and
mTOR (rapamycin) inhibitors and visfatin treatment to assess
effects on (M) migration and (N) invasion. (O) CE81T cells treated
with PI3K (Ly294002), AKT and mTOR (rapamycin) inhibitors and
visfatin treatment were assayed for (P) cell migration and (Q)
invasion. Gene Expression Omnibus dataset GSE77861 shows
significantly increased mRNA expression of (R) NAMPT, (S)
miR-3613-5p, (T) VEZF1 and (U) VCAN in esophageal cancer compared
with normal tissue. *P<0.05 vs. control; #P<0.05
vs. visfatin. miR, microRNA; VCAN, versican; RT-q, Reverse
Transcriptase Quantitative; si, small interfering; VEZf-1, Vascular
Endothelial Zinc Finger 1; NAMPT, Nicotinamide
phosphoribosyltransferase; Akti, Akt inhibitor. |
Discussion
Esophageal cancer is a relatively prevalent
malignancy worldwide, marked by a poor prognosis and a strong
tendency for metastasis. It is the eighth most commonly diagnosed
cancer and the sixth leading cause of cancer-related deaths
globally. Notably, over 80% of all cases and fatalities are
reported in developing countries. In the United States alone, the
National Cancer Institute estimated approximately 18,000 new cases
and over 15,000 deaths due to esophageal cancer in 2013 (32). The ESCC subtype, which accounts for
almost 90% of esophageal malignancies in Asia, has a high mortality
rate and poor prognosis (33).
Despite progress in detection and treatment, the 5-year survival
rate of patients with esophageal cancer is relatively low (34). The high mortality rate from
esophageal cancer may be decreased with improved treatment
approaches (35). The present study
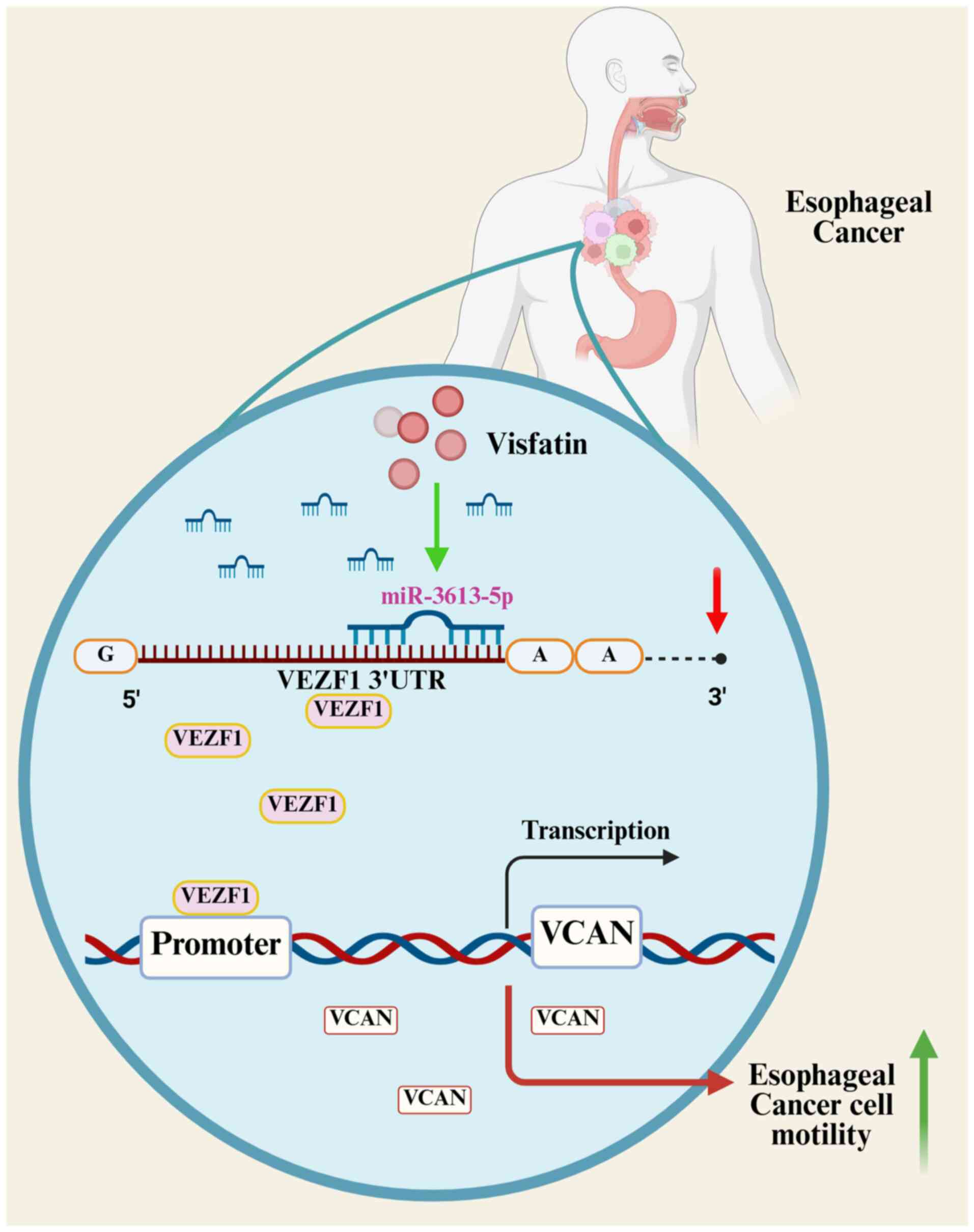
demonstrated that the levels of visfatin were associated with
metastasis in patients with esophageal cancer. The present study
showed that inhibition of miR-3613-5p and the promotion of the
VEZF1/VCAN axis mediated visfatin-facilitated esophageal cancer
cell motility (Fig. 7). The present
study also demonstrated the expression of visfatin, VEZF1, and VCAN
in normal and cancer samples from the GEO database. However, a
limitation is the lack of experimental validation using clinical
samples from patients with ESCC, underscoring the need for further
investigation.
Adipocytokines are associated with development,
spread, recurrence and metastasis of numerous types of malignancy
(36). Lower resistin mRNA levels
are found in ESCC samples and blood compared with normal esophageal
samples (37), Patients with ESCC
have lower adiponectin levels compared with controls (38). Additionally, a significant
association has been found between leptin levels and advanced tumor
stage in ESCC, as well as lymph node involvement (39). Specifically, visfatin serves a key
role in inflammation and cancer. Additionally, visfatin promotes
the metastasis of chondrosarcoma (40). Visfatin is associated with a higher
disease stage in ESCC tissue and promotes lymphangiogenesis. Our
previous study demonstrated that visfatin is highly expressed in
ESCC N1 and N2 stage samples compared with N0 and is associated
with lymph node metastasis (3). In
the present study, Transwell migration and Matrigel invasion assays
revealed that visfatin facilitated the migration and invasion of
esophageal cancer cells. To the best of our knowledge, the present
study is the first to demonstrate that visfatin promotes cell
motility in esophageal cancer.
At the post-transcriptional level, small, non-coding
miRNAs are key for regulating gene expression (41). This regulation controls
physiological and pathological processes, including cancer, by
destroying or inhibiting the translation of target mRNAs (42–44). A
promising treatment strategy to combat tumor metastasis is to alter
miRNA expression through pharmacological intervention, which may be
utilized to inhibit cancer cells from migrating (45,46).
In the present study, the miRNA sequencing analysis revealed that
miR-3613-5p was the most downregulated miRNA following the use of
visfatin. Subsequent experiments demonstrated that visfatin reduced
miR-3613-5p expression and introducing a miR-3613-5p mimic into
esophageal cancer cells reversed visfatin-induced cell motility.
These findings indicated that visfatin promoted esophageal cancer
cell migration and invasion by suppressing miR-3613-5p synthesis.
Additionally, visfatin-induced inhibition of miR-1264 promotes
PDGF-C synthesis via the PI3K/AKT/mTOR pathway (19). In the present study, Transwell
assays demonstrated that inhibition of the PI3K/AKT/mTOR pathway
effectively reversed the metastatic effects induced by visfatin. To
the best of our knowledge, however, there is no direct evidence
linking the PI3K/AKT/mTOR pathway to the regulation of miR-3613-5p.
Absence of direct evidence limits understanding of the upstream
regulatory network controlling miR-3613-5p expression in response
to visfatin stimulation. Whether the PI3K/AKT/mTOR pathway is
involved in visfatin-mediated regulation of miR-3613-5p expression
needs further investigation.
With its six-type zinc finger motifs, poly glutamine
domain and proline-rich region, VEZF1 is a potential zinc finger
transcription factor that is key for angiogenesis (47). Initially, VEZF1 expression was found
in both the embryo proper and the mesodermal components of the
extraembryonic mesoderm (48).
Subsequently, endothelial cells that emerge during angiogenesis
were found to express VEZF1 (48).
By targeting downstream genes, such as metallothionein 1 and
stathmin, VEZF1 controls different phases of angiogenesis (49). VEZF1 transcriptional activity also
controls the metastasis of hepatocellular carcinoma (50). According to four publicly accessible
miRNA databases, miR-3613-5p targets seven possible candidates, and
patients with esophageal cancer have significantly higher levels of
VEZF1. Visfatin-induced cell migration and invasion were reduced by
VEZF1 siRNA, suggesting that VEZF1 mediated the motility of
esophageal cancer. The present study also identified VCAN as a
downstream molecule of VEZF1. Therefore, the VEZF1/VCAN axis may
mediates visfatin-induced esophageal cancer cell migration.
In conclusion, the present study demonstrated that
visfatin facilitated the migration and invasion of esophageal
cancer cells. The inhibition of miR-3613-5p and the promotion of
the VEZF1/VCAN axis mediated visfatin-induced esophageal cancer
cell motility.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Science and
Technology Council (grant nos. 113-2320-B-039-049-MY3,
113-2320-B-371-002- and 112-2314-B-039-018-MY3), China Medical
University (grant no. CMU111-ASIA-05) and China Medical University
Hospital (grant nos. DMR-114-014, DMR-114-021, DMR-113-008 and
DMR-114-069).
Availability of data and materials
The data generated in the present study may be found
in the Gene Expression Omnibus under accession number GSE298998 or
at the following URL: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE298998).
Authors' contributions
CLH, CHT and PIL wrote the manuscript. SSG, JHG,
CLL, YHC and CLH performed experiments and analyzed data. HCT, PIL,
YHC, MYL and CHT analyzed data. HCT, SSG and CHT edited the
manuscript. All authors have read and approved the final
manuscript. CLH, MYL and CHT confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
Zhang HZ, Jin GF and Shen HB:
Epidemiologic differences in esophageal cancer between Asian and
Western populations. Chin J Cancer. 31:281–286. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang CL, Achudhan D, Liu PI, Lin YY, Liu
SC, Guo JH, Liu CL, Wu CY, Wang SW and Tang CH: Visfatin
upregulates VEGF-C expression and lymphangiogenesis in esophageal
cancer by activating MEK1/2-ERK and NF-κB signaling. Aging (Albany
NY). 15:4774–4793. 2023.PubMed/NCBI
|
|
4
|
Shai SE, Lai YL, Tang HW and Hung SC:
Treatment of trivial esophageal cancer with huge devastating airway
obstruction via the use of a modified emergency tracheostomy under
local anesthesia. Asian J Surg. 43:1182–1185. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Quint LE, Hepburn LM, Francis IR, Whyte RI
and Orringer MB: Incidence and distribution of distant metastases
from newly diagnosed esophageal carcinoma. Cancer. 76:1120–1125.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ and Hammond SM: A microRNA polycistron as a potential
human oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Achudhan D, Lai YL, Lin YY, Huang YL, Tsai
CH, Ho TL, Ko CY, Fong YC, Huang CC and Tang CH: CXCL13 promotes
TNF-α synthesis in rheumatoid arthritis through activating ERK/p38
pathway and inhibiting miR-330-3p generation. Biochem Pharmacol.
221:1160372024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chang TK, Ho TL, Lin YY, Thuong LHH, Lai
KY, Tsai CH, Liaw CC and Tang C: Ugonin P facilitates chondrogenic
properties in chondrocytes by inhibiting miR-3074-5p production:
Implications for the treatment of arthritic disorders. Int J Biol
Sci. 21:1378–1390. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tran NB, Chang TK, Chi NDP, Lai KY, Chen
HT, Fong YC, Liaw CC and Tang CH: Ugonin inhibits chondrosarcoma
metastasis through suppressing cathepsin V via promoting
miR-4799-5p expression. Int J Biol Sci. 21:1144–1157. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thuong LHH, Huang CL, Fong YC, Liu CL, Guo
JH, Wu CY, Liu PI and Tang CH: Bone sialoprotein facilitates
anoikis resistance in lung cancer by inhibiting miR-150-5p
expression. J Cell Mol Med. 28:e701552024. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin LW, Lin TH, Swain S, Fang JK, Guo JH,
Yang SF and Tang CH: Melatonin inhibits ET-1 production to break
crosstalk between prostate cancer and bone cells: Implication for
osteoblastic bone metastasis treatment. J Pineal Res.
76:e700002024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Annett S, Moore G and Robson T: Obesity
and cancer metastasis: Molecular and translational perspectives.
Cancers (Basel). 12:37982020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin CJ, Chang YC, Hsu HY, Tsai MC, Hsu LY,
Hwang LC, Chien KL and Yeh TL: Metabolically healthy
overweight/obesity and cancer risk: A representative cohort study
in Taiwan. Obes Res Clin Pract. 15:564–569. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ouchi N, Parker JL, Lugus JJ and Walsh K:
Adipokines in inflammation and metabolic disease. Nat Rev Immunol.
11:85–97. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rana MN and Neeland IJ: Adipose tissue
inflammation and cardiovascular disease: An update. Curr Diab Rep.
22:27–37. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
de Liyis BG, Nolan J and Maharjana MA:
Fibroblast growth factor receptor 1-bound extracellular vesicle as
novel therapy for osteoarthritis. Biomedicine (Taipei). 12:1–9.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Akrida I and Papadaki H: Adipokines and
epithelial-mesenchymal transition (EMT) in cancer. Mol Cell
Biochem. 478:2419–2433. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song CY, Chang SL, Lin CY, Tsai CH, Yang
SY, Fong YC, Huang YW, Wang SW, Chen WC and Tang CH:
Visfatin-Induced inhibition of miR-1264 facilitates PDGF-C
synthesis in chondrosarcoma cells and enhances endothelial
progenitor cell angiogenesis. Cells. 11:34702022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cymbaluk-Płoska A, Chudecka-Głaz A,
Pius-Sadowska E, Sompolska-Rzechuła A, Machaliński B and Menkiszak
J: Circulating serum level of visfatin in patients with endometrial
cancer. Biomed Res Int. 2018:85761792018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu SL, Liu SC, Lin CY, Fong YC, Wang SS,
Chen LC, Yang SF and Tang CH: Genetic associations of visfatin
polymorphisms with clinicopathologic characteristics of prostate
cancer in Taiwanese males. Int J Med Sci. 21:2494–2501. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin CY, Law YY, Yu CC, Wu YY, Hou SM, Chen
WL, Yang SY, Tsai CH, Lo YS, Fong YC and Tang CH: NAMPT enhances
LOX expression and promotes metastasis in human chondrosarcoma
cells by inhibiting miR-26b-5p synthesis. J Cell Physiol.
239:e313452024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chang AC, Chen PC, Lin YF, Su CM, Liu JF,
Lin TH, Chuang SM and Tang CH: Osteoblast-secreted WISP-1 promotes
adherence of prostate cancer cells to bone via the VCAM-1/integrin
α4β1 system. Cancer Lett. 426:47–56. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang YH, Huang YL, Tsai HC, Chang AC, Ko
CY, Fong YC and Tang CH: Chemokine ligand 2 promotes migration in
osteosarcoma by regulating the miR-3659/MMP-3 axis. Biomedicines.
11:27682023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee HP, Chen PC, Wang SW, Fong YC, Tsai
CH, Tsai FJ, Chung JG, Huang CY, Yang JS, Hsu YM, et al: Plumbagin
suppresses endothelial progenitor cell-related angiogenesis in
vitro and in vivo. J Funct Foods. 52:537–544. 2019. View Article : Google Scholar
|
|
26
|
Lee HP, Wang SW, Wu YC, Lin LW, Tsai FJ,
Yang JS, Li TM and Tang CH: Soya-cerebroside inhibits
VEGF-facilitated angiogenesis in endothelial progenitor cells. Food
and Agricultural Immunology. 31:193–204. 2020. View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee KT, Su CH, Liu SC, Chen BC, Chang JW,
Tsai CH, Huang WC, Hsu CJ, Chen WC, Wu YC and Tang C:
Cordycerebroside A inhibits ICAM-1-dependent M1 monocyte adhesion
to osteoarthritis synovial fibroblasts. J Food Biochem.
46:e141082022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu SC, Tsai CH, Wu TY, Tsai CH, Tsai FJ,
Chung JG, Huang CY, Yang JS, Hsu YM, Yin MC, et al:
Soya-cerebroside reduces IL-1β-induced MMP-1 production in
chondrocytes and inhibits cartilage degradation: Implications for
the treatment of osteoarthritis. Food and Agricultural Immunology.
30:620–632. 2019. View Article : Google Scholar
|
|
30
|
Song CY, Wu CY, Lin CY, Tsai CH, Chen HT,
Fong YC, Chen LC and Tang CH: The stimulation of exosome generation
by visfatin polarizes M2 macrophages and enhances the motility of
chondrosarcoma. Environ Toxicol. 39:3790–3798. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He Z, Ji Y, Yuan Y, Liang T, Liu C, Jiao
Y, Chen Y, Yang Y, Han L, Hu Y and Cong X: Uncovering the role of
microRNAs in esophageal cancer: From pathogenesis to clinical
applications. Front Pharmacol. 16:15325582025. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Napier KJ, Scheerer M and Misra S:
Esophageal cancer: A review of epidemiology, pathogenesis, staging
workup and treatment modalities. World J Gastrointest Oncol.
6:112–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li D, Zhang L, Liu Y, Sun H, Onwuka JU,
Zhao Z, Tian W, Xu J, Zhao Y and Xu H: Specific DNA methylation
markers in the diagnosis and prognosis of esophageal cancer. Aging
(Albany NY). 11:11640–11658. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang L, Han H, Wang Z, Shi L, Yang M and
Qin Y: Targeting the microenvironment in esophageal cancer. Front
Cell Dev Biol. 9:6849662021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
He S, Xu J, Liu X and Zhen Y: Advances and
challenges in the treatment of esophageal cancer. Acta Pharm Sin B.
11:3379–3392. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim JW, Kim JH and Lee YJ: The role of
adipokines in tumor progression and its association with obesity.
Biomedicines. 12:972024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hung AC, Wang YY, Lee KT, Chiang HH, Chen
YK, Du JK, Chen CM, Chen MY, Chen KJ, Hu SC and Yuan SF: Reduced
tissue and serum resistin expression as a clinical marker for
esophageal squamous cell carcinoma. Oncol Lett. 22:7742021.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dalamaga M, Diakopoulos KN and Mantzoros
CS: The role of adiponectin in cancer: A review of current
evidence. Endocr Rev. 33:547–594. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ray A and Cleary MP: The potential role of
leptin in tumor invasion and metastasis. Cytokine Growth Factor
Rev. 38:80–97. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hung SY, Lin CY, Yu CC, Chen HT, Lien MY,
Huang YW, Fong YC, Liu JF, Wang SW, Chen WC and Tang CH: Visfatin
promotes the metastatic potential of chondrosarcoma cells by
stimulating AP-1-dependent MMP-2 production in the MAPK pathway.
Int J Mol Sci. 22:86422021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tehrani SS, Zaboli E, Sadeghi F, Khafri S,
Karimian A, Rafie M and Parsian H: MicroRNA-26a-5p as a potential
predictive factor for determining the effectiveness of trastuzumab
therapy in HER-2 positive breast cancer patients. Biomedicine
(Taipei). 11:30–39. 2021.PubMed/NCBI
|
|
42
|
Kanwal R, Plaga AR, Liu X, Shukla GC and
Gupta S: MicroRNAs in prostate cancer: Functional role as
biomarkers. Cancer Lett. 407:9–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tsai HC, Lai YY, Hsu HC, Fong YC, Lien MY
and Tang CH: CCL4 stimulates cell migration in human osteosarcoma
via the mir-3927-3p/Integrin αvβ3 axis. Int J Mol Sci.
22:127372021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tzeng HE, Lin SL, Thadevoos LA, Lien MY,
Yang WH, Ko CY, Lin CY, Huang YW, Liu JF, Fong YC, et al: Nerve
growth factor promotes lysyl oxidase-dependent chondrosarcoma cell
metastasis by suppressing miR-149-5p synthesis. Cell Death Dis.
12:11012021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bruderer M, Alini M and Stoddart MJ: Role
of HOXA9 and VEZF1 in endothelial biology. J Vasc Res. 50:265–278.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xiong JW, Leahy A, Lee HH and Stuhlmann H:
Vezf1: A Zn finger transcription factor restricted to endothelial
cells and their precursors. Dev Biol. 206:123–141. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Miyashita H, Kanemura M, Yamazaki T, Abe M
and Sato Y: Vascular endothelial zinc finger 1 is involved in the
regulation of angiogenesis: Possible contribution of stathmin/OP18
as a downstream target gene. Arterioscler Thromb Vasc Biol.
24:878–884. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shi X, Zhao P and Zhao G: VEZF1,
destabilized by STUB1, affects cellular growth and metastasis of
hepatocellular carcinoma by transcriptionally regulating PAQR4.
Cancer Gene Ther. 30:256–266. 2023. View Article : Google Scholar : PubMed/NCBI
|