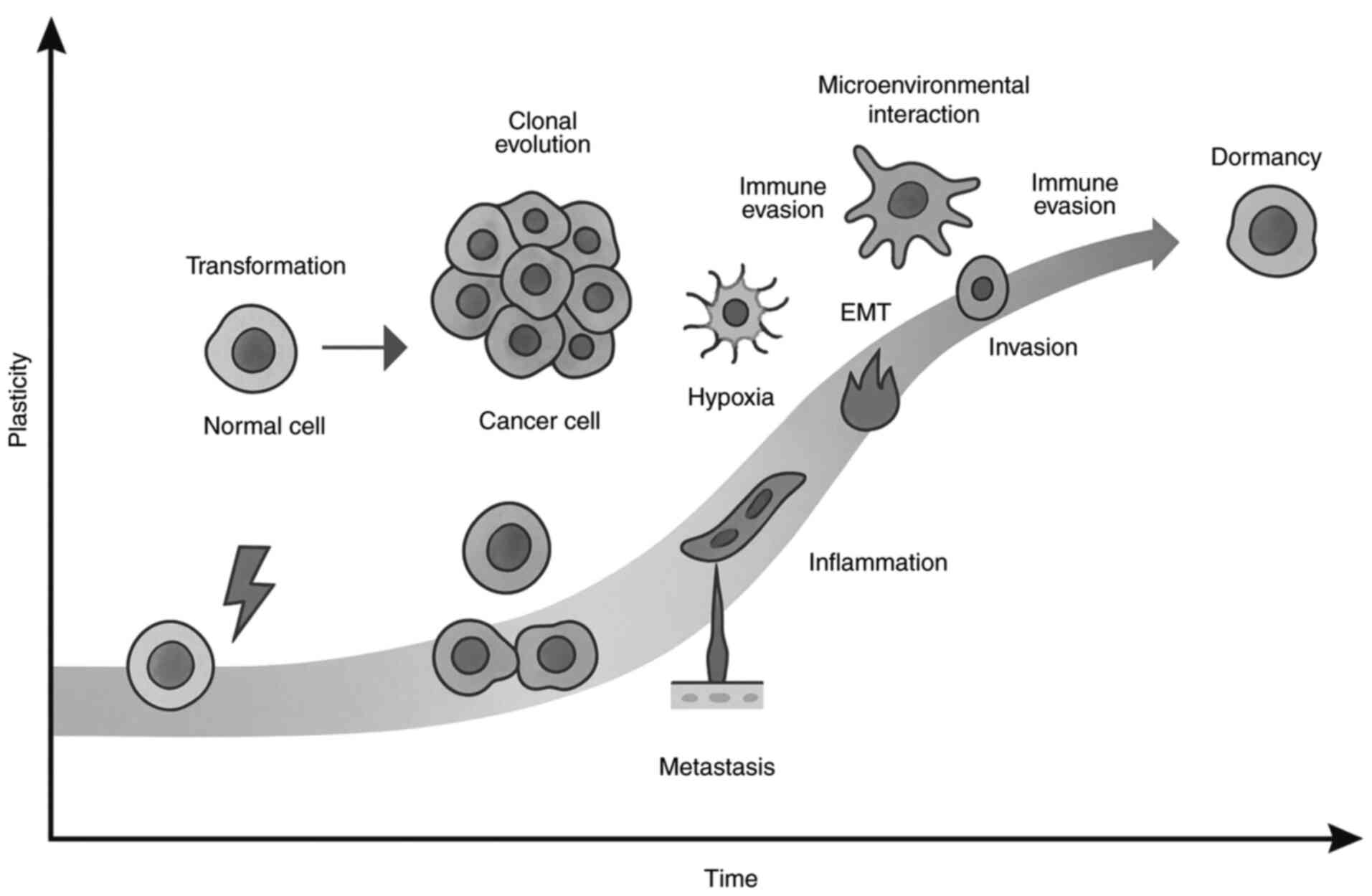
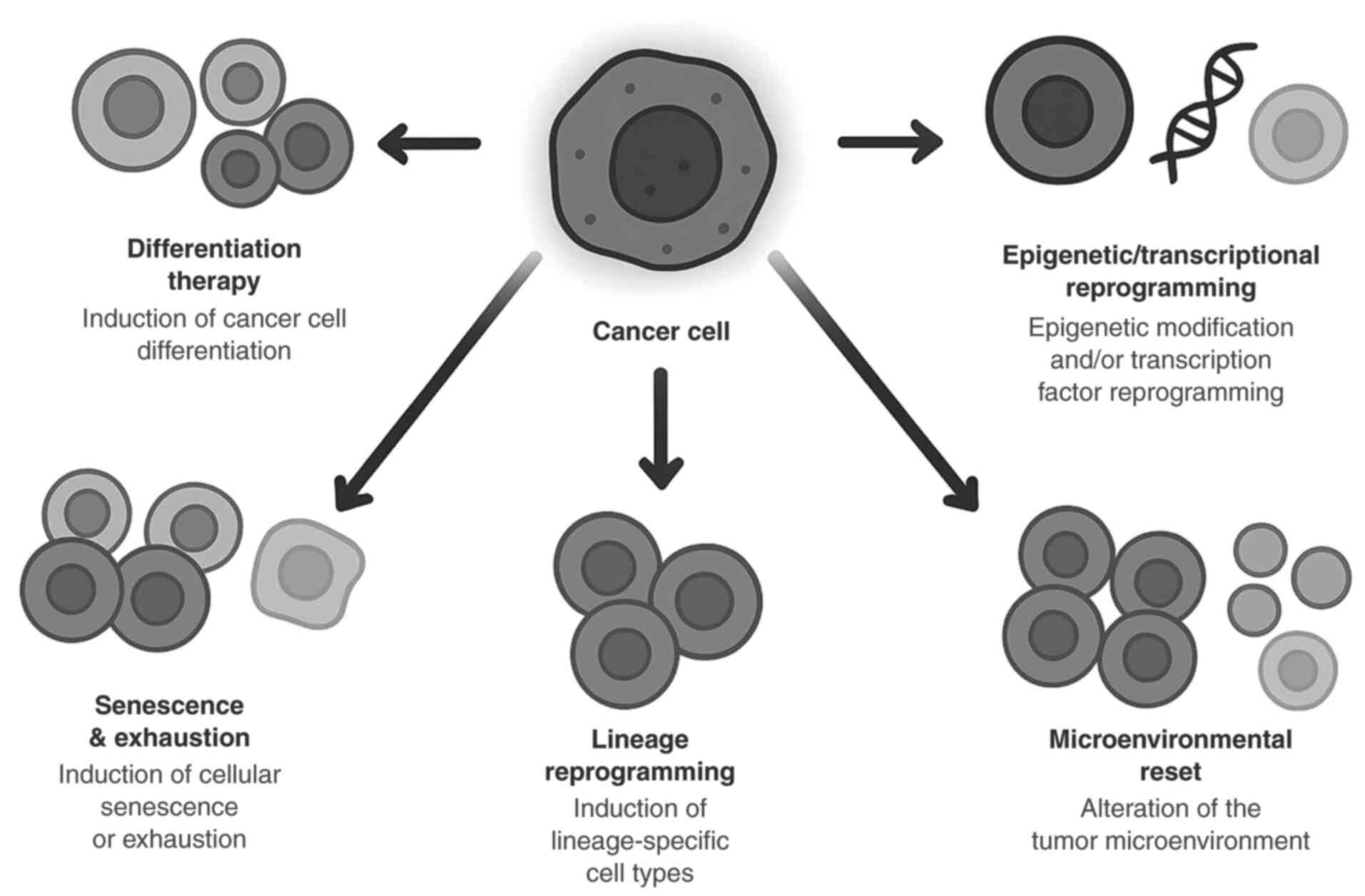
|
1
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Greaves M and Maley CC: Clonal evolution
in cancer. Nature. 481:306–313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gerlinger M, Rowan AJ, Horswell S, Math M,
Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N,
Stewart A, et al: Intratumor heterogeneity and branched evolution
revealed by multiregion sequencing. N Engl J Med. 366:883–892.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marusyk A, Almendro V and Polyak K:
Intra-tumour heterogeneity: A looking glass for cancer? Nat Rev
Cancer. 12:323–334. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nieto MA, Huang RY, Jackson RA and Thiery
JP: EMT: 2016. Cell. 166:21–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yan M and Liu Q: Differentiation therapy:
a promising strategy for cancer treatment. Chin J Cancer. 35:32016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khoshchehreh R, Totonchi M, Carlos Ramirez
J, Torres R, Baharvand H, Aicher A, Ebrahimi M and Heeschen C:
Epigenetic reprogramming of primary pancreatic cancer cells
counteracts their in vivo tumourigenicity. Oncogene. 38:6226–6239.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gong JR, Lee CK, Kim HM, Kim J, Jeon J,
Park S and Cho KH: Control of cellular differentiation trajectories
for cancer reversion. Adv Sci (Weinh). 12:e24021322025. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim DJ: The role of the DNA
methyltransferase family and the therapeutic potential of DNMT
inhibitors in tumor treatment. Curr Oncol. 32:882025. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Alexandrov LB, Kim J, Haradhvala NJ, Huang
MN, Tian Ng AW, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom
EN, et al: The repertoire of mutational signatures in human cancer.
Nature. 578:94–101. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Weiler P, Lange M, Klein M, Pe'er D and
Theis F: CellRank 2: Unified fate mapping in multiview single-cell
data. Nat Methods. 21:1196–1205. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qiu X, Zhang Y, Martin-Rufino JD, Weng C,
Hosseinzadeh S, Yang D, Pogson AN, Hein MY, Hoi Joseph Min K, Wang
L, et al: Mapping transcriptomic vector fields of single cells.
Cell. 185:690–711.e45. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Quinn JJ, Jones MG, Okimoto RA, Nanjo S,
Chan MM, Yosef N, Bivona TG and Weissman JS: Single-cell lineages
reveal the rates, routes, and drivers of metastasis in cancer
xenografts. Science. 371:eabc19442021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Aiello NM, Maddipati R, Norgard RJ, Balli
D, Li J, Yuan S, Yamazoe T, Black T, Sahmoud A, Furth EE, et al:
EMT subtype influences epithelial plasticity and mode of cell
migration. Dev Cell. 45:681–695.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pastushenko I, Brisebarre A, Sifrim A,
Fioramonti M, Revenco T, Boumahdi S, Van Keymeulen A, Brown D,
Moers V, Lemaire S, et al: Identification of the tumour transition
states occurring during EMT. Nature. 556:463–468. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Quail DF and Joyce JA: Microenvironmental
regulation of tumor progression and metastasis. Nat Med.
19:1423–1437. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Joyce JA and Fearon DT: T cell exclusion,
immune privilege, and the tumor microenvironment. Science.
348:74–80. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Semenza GL: Tumor metabolism: Cancer cells
give and take lactate. J Clin Invest. 118:3835–3837.
2008.PubMed/NCBI
|
|
21
|
Erdogan B and Webb DJ: Cancer-associated
fibroblasts modulate growth factor signaling and extracellular
matrix remodeling to regulate tumor metastasis. Biochem Soc Trans.
45:229–236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu F, Yang J, Liu J, Wang Y, Mu J, Zeng Q,
Deng S and Zhou H: Signaling pathways in cancer-associated
fibroblasts and targeted therapy for cancer. Signal Transduct
Target Ther. 6:2182021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kalluri R: The biology and function of
fibroblasts in cancer. Nat Rev Cancer. 16:582–598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu T, Han C, Wang S, Fang P, Ma Z, Xu L
and Yin R: Cancer-associated fibroblasts: An emerging target of
anti-cancer immunotherapy. J Hematol Oncol. 12:862019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sibai M, Cervilla S, Grases D, Musulen E,
Lazcano R, Mo CK, Davalos V, Fortian A, Bernat A, Romeo M, et al:
The spatial landscape of cancer hallmarks reveals patterns of tumor
ecological dynamics and drug sensitivity. Cell Rep. 44:1152292025.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu W, Puri A, Fu D, Chen L, Wang C,
Kellis M and Yang J: Dissecting the tumor microenvironment in
response to immune checkpoint inhibitors via single-cell and
spatial transcriptomics. Clin Exp Metastasis. 41:313–332. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lambert AW, Pattabiraman DR and Weinberg
RA: Emerging biological principles of metastasis. Cell.
168:670–691. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aguirre-Ghiso JA: Models, mechanisms and
clinical evidence for cancer dormancy. Nat Rev Cancer. 7:834–846.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sosa MS, Bragado P and Aguirre-Ghiso JA:
Mechanisms of disseminated cancer cell dormancy: an awakening
field. Nat Rev Cancer. 14:611–622. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ghajar CM: Metastasis prevention by
targeting the dormant niche. Nat Rev Cancer. 15:238–247. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shay JW and Wright WE: Role of telomeres
and telomerase in cancer. Semin Cancer Biol. 21:349–353. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Barthel FP, Wei W, Tang M,
Martinez-Ledesma E, Hu X, Amin SB, Akdemir KC, Seth S, Song X, Wang
Q, et al: Systematic analysis of telomere length and somatic
alterations in 31 cancer types. Nat Genet. 49:349–357. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Negrini S, Gorgoulis VG and Halazonetis
TD: Genomic instability-an evolving hallmark of cancer. Nat Rev Mol
Cell Biol. 11:220–228. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Flynn RL, Cox KE, Jeitany M, Wakimoto H,
Bryll AR, Ganem NJ, Bersani F, Pineda JR, Suvà ML, Benes CH, et al:
Alternative lengthening of telomeres renders cancer cells
hypersensitive to ATR inhibitors. Science. 347:273–277. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Behan FM, Iorio F, Picco G, Gonçalves E,
Beaver CM, Migliardi G, Santos R, Rao Y, Sassi F, Pinnelli M, et
al: Prioritization of cancer therapeutic targets using CRISPR-Cas9
screens. Nature. 568:511–516. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Koschmann C, Calinescu AA, Nunez FJ,
Mackay A, Fazal-Salom J, Thomas D, Mendez F, Kamran N, Dzaman M,
Mulpuri L, et al: ATRX loss promotes tumor growth and impairs
nonhomologous end joining DNA repair in glioma. Sci Transl Med.
8:328ra282016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Plotnikov A, Kozer N, Cohen G, Carvalho S,
Duberstein S, Almog O, Solmesky LJ, Shurrush KA, Babaev I, Benjamin
S, et al: PRMT1 inhibition induces differentiation of colon cancer
cells. Sci Rep. 10:200302020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Campos B, Wan F, Farhadi M, Ernst A,
Zeppernick F, Tagscherer KE, Ahmadi R, Lohr J, Dictus C, Gdynia G,
et al: Differentiation therapy exerts antitumor effects on
stem-like glioma cells. Clin Cancer Res. 16:2715–2728. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu K, Xia Y and Tian X, He Y, Zhou J, Han
R, Guo H, Song T, Chen L and Tian X: Characterization and
therapeutic perspectives of differentiation-inducing therapy in
malignant tumors. Front Genet. 14:12713812023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jin X, Jin X and Kim H: Cancer stem cells
and differentiation therapy. Tumour Biol. 39:10104283177299332017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun X, Zhang J, Zhao Q, Chen X, Zhu W, Yan
G and Zhou T: Stochastic modeling suggests that noise reduces
differentiation efficiency by inducing a heterogeneous drug
response in glioma differentiation therapy. BMC Syst Biol.
10:732016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Geng Y, Amante JJ, Goel HL, Zhang X,
Walker MR, Luther DC, Mercurio AM and Rotello VM: Differentiation
of cancer stem cells through nanoparticle surface engineering. ACS
Nano. 14:15276–15285. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shen S, Xu X, Lin S, Zhang Y, Liu H, Zhang
C and Mo R: A nanotherapeutic strategy to overcome chemotherapeutic
resistance of cancer stem-like cells. Nat Nanotechnol. 16:104–113.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
He C, Jiang Y, Guo Y and Wu Z: Amplified
ferroptosis and apoptosis facilitated by differentiation therapy
efficiently suppress the progression of osteosarcoma. Small.
19:23025752023. View Article : Google Scholar
|
|
45
|
Wang Y, Wang H, Lv X, Liu C, Qi L, Song X
and Yu A: Enhancement of all-trans retinoic acid-induced
differentiation by pH-sensitive nanoparticles for solid tumor
cells. Macromol Biosci. 14:369–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Guinea-Viniegra J, Zenz R, Scheuch H,
Jiménez M, Bakiri L, Petzelbauer P and Wagner EF:
Differentiation-induced skin cancer suppression by FOS, p53, and
TACE/ADAM17. J Clin Invest. 122:2898–2910. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yan Y, Li Z, Xu X, Chen C, Wei W, Fan M,
Chen X, Li JJ, Wang Y and Huang J: All-trans retinoic acids induce
differentiation and sensitize a radioresistant breast cancer cells
to chemotherapy. BMC Complement Altern Med. 16:1132016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lotan R, Francis GE, Freeman CS and Waxman
S: Differentiation therapy. Cancer Res. 50:3453–3464.
1990.PubMed/NCBI
|
|
49
|
Xie J, Wang Z, Fan W, Liu Y, Liu F, Wan X,
Liu M, Wang X, Zeng D, Wang Y, et al: Targeting cancer cell
plasticity by HDAC inhibition to reverse EBV-induced
dedifferentiation in nasopharyngeal carcinoma. Signal Transduct
Target Ther. 6:3332021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jamshidi F, Zhang J, Harrison JS, Wang X
and Studzinski GP: Induction of differentiation of human leukemia
cells by combinations of COX inhibitors and 1,25-dihydroxyvitamin
D3 involves Raf1 but not Erk 1/2 signaling. Cell Cycle. 7:917–924.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yao J, Li G, Cui Z, Chen P, Wang J, Hu Z,
Zhang L and Wei L: The histone deacetylase inhibitor I1 induces
differentiation of acute leukemia cells with MLL gene
rearrangements via epigenetic modification. Front Pharmacol.
13:8760762022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sherif ZA, Ogunwobi OO and Ressom HW:
Mechanisms and technologies in cancer epigenetics. Front Oncol.
14:15136542025. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Akar RO, Selvı S, Ulukaya E and Aztopal N:
Key actors in cancer therapy: Epigenetic modifiers. Turk J Biol.
43:155–170. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Geissler F, Nesic K, Kondrashova O,
Dobrovic A, Swisher EM, Scott CL and J Wakefield M: The role of
aberrant DNA methylation in cancer initiation and clinical impacts.
Ther Adv Med Oncol. 16:175883592312205112024. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hassell KN: Histone deacetylases and their
inhibitors in cancer epigenetics. Diseases. 7:572019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bojang P Jr and Ramos KS: The promise and
failures of epigenetic therapies for cancer treatment. Cancer Treat
Rev. 40:153–169. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhao H, Wu L, Yan G, Chen Y, Zhou M, Wu Y
and Li Y: Inflammation and tumor progression: Signaling pathways
and targeted intervention. Signal Transduct Target Ther. 6:2632021.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zheng S, Bian H, Li J, Shen Y, Yang Y and
Hu W: Differentiation therapy: Unlocking phenotypic plasticity of
hepatocellular carcinoma. Crit Rev Oncol Hematol. 180:1038542022.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chao J, Zhao S and Sun H:
Dedifferentiation of hepatocellular carcinoma: Molecular mechanisms
and therapeutic implications. Am J Transl Res. 12:2099–2109.
2020.PubMed/NCBI
|
|
60
|
Gong L, Yan Q, Zhang Y, Fang X, Liu B and
Guan X: Cancer cell reprogramming: A promising therapy converting
malignancy to benignity. Cancer Commun (Lond). 39:482019.PubMed/NCBI
|
|
61
|
Uppaluri KR, Challa HJ, Gaur A, Jain R,
Krishna Vardhani K, Geddam A, Natya K, Aswini K, Palasamudram K and
K SM: Unlocking the potential of non-coding RNAs in cancer research
and therapy. Transl Oncol. 35:1017302023. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Xu X, Peng Q, Jiang X, Tan S, Yang Y, Yang
W, Han Y, Chen Y, Oyang L, Lin J, et al: Metabolic reprogramming
and epigenetic modifications in cancer: from the impacts and
mechanisms to the treatment potential. Exp Mol Med. 55:1357–1370.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Nardella C, Clohessy JG, Alimonti A and
Pandolfi PP: Pro-senescence therapy for cancer treatment. Nat Rev
Cancer. 11:503–511. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Romesser PB and Lowe SW: The potent and
paradoxical biology of cellular senescence in cancer. Annu Rev
Cancer Biol. 7:207–228. 2023. View Article : Google Scholar
|
|
65
|
Wang L, Lankhorst L and Bernards R:
Exploiting senescence for the treatment of cancer. Nat Rev Cancer.
22:340–355. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Maggiorani D, Le O, Lisi V, Landais S,
Moquin-Beaudry G, Lavallée VP, Decaluwe H and Beauséjour C:
Senescence drives immunotherapy resistance by inducing an
immunosuppressive tumor microenvironment. Nat Commun. 15:24352024.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Jackson J: Abstract SoA2-02: Implications
of therapy induced senescence in breast cancer. Cancer Res.
84:SoA2–02. 2024. View Article : Google Scholar
|
|
68
|
Ji P, Wang C, Liu Y, Guo X, Liang Y, Wei
J, Liu Z, Gong L, Yang G and Ji G: Targeted clearance of senescent
cells via engineered extracellular vesicles reprograms tumor
immunosuppressive microenvironment. Adv Healthc Mater.
13:e24009452024. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Beauséjour CM, Krtolica A, Galimi F,
Narita M, Lowe SW, Yaswen P and Campisi J: Reversal of human
cellular senescence: Roles of the p53 and p16 pathways. EMBO J.
22:4212–4222. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhao Y, Shao Q and Peng G: Exhaustion and
senescence: two crucial dysfunctional states of T cells in the
tumor microenvironment. Cell Mol Immunol. 17:27–35. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liu X, Si F, Bagley D, Ma F, Zhang Y, Tao
Y, Shaw E and Peng G: Blockades of effector T cell senescence and
exhaustion synergistically enhance antitumor immunity and
immunotherapy. J Immunother Cancer. 10:e0050202022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Triana-Martínez F, Loza MI and Domínguez
E: Beyond tumor suppression: Senescence in cancer stemness and
tumor dormancy. Cells. 9:3462020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Chakradeo S, Elmore LW and Gewirtz DA: Is
senescence reversible? Curr Drug Targets. 17:460–466. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Demirci D, Dayanc B, Mazi FA and Senturk
S: The jekyll and hyde of cellular senescence in cancer. Cells.
10:2082021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ashrafizadeh M, Paskeh MDA, Mirzaei S,
Gholami MH, Zarrabi A, Hashemi F, Hushmandi K, Hashemi M, Nabavi N,
Crea F, et al: Targeting autophagy in prostate cancer: Preclinical
and clinical evidence for therapeutic response. J Exp Clin Cancer
Res. 41:1052022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yu H, Liu R, Ma B, Li X, Yen HY, Zhou Y,
Krasnoperov V, Xia Z, Zhang X, Bove AM, et al: Axl receptor
tyrosine kinase is a potential therapeutic target in renal cell
carcinoma. Br J Cancer. 113:616–625. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yin JJ, Selander K, Chirgwin JM, Dallas M,
Grubbs BG, Wieser R, Massagué J, Mundy GR and Guise TA: TGF-beta
signaling blockade inhibits PTHrP secretion by breast cancer cells
and bone metastases development. J Clin Invest. 103:197–206. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Liu Y, Lomeli I and Kron SJ:
Therapy-induced cellular senescence: potentiating tumor elimination
or driving cancer resistance and recurrence? Cells. 13:12812024.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Shin D, Gong JR, Jeong SD, Cho Y, Kim HP,
Kim TY and Cho KH: Attractor landscape analysis reveals a reversion
switch in the transition of colorectal tumorigenesis. Adv Sci
(Weinh). 12:24125032025. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Oh J, Kim Y, Baek D and Ha Y: Malignant
gliomas can be converted to non-proliferating glial cells by
treatment with a combination of small molecules. Oncol Rep.
41:361–368. 2019.PubMed/NCBI
|
|
81
|
Knappe N, Novak D, Weina K, Bernhardt M,
Reith M, Larribere L, Hölzel M, Tüting T, Gebhardt C, Umansky V and
Utikal J: Directed dedifferentiation using partial reprogramming
induces invasive phenotype in melanoma cells. Stem Cells.
34:832–846. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Yi Y, Che W, Xu P, Mao C, Li Z, Wang Q,
Lyu J and Wang X: Conversion of glioma cells into neuron-like cells
by small molecules. iScience. 27:1110912024. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Câmara DAD, Porcacchia AS, Costa AS,
Azevedo RA and Kerkis I: Murine melanoma cells incomplete
reprogramming using non-viral vector. Cell Prolif. 50:e123522017.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Hollebecque A, Salvagni S, Plummer R,
Niccoli P, Capdevila J, Curigliano G, Moreno V, de Braud F, de
Villambrosia SG, Martin-Romano P, et al: Clinical activity of
CC-90011, an oral, potent, and reversible LSD1 inhibitor, in
advanced malignancies. Cancer. 128:3185–3195. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Hiatt JB, Sandborg H, Garrison SM, Arnold
HU, Liao SY, Norton JP, Friesen TJ, Wu F, Sutherland KD, Rienhoff
HY, et al: Inhibition of LSD1 with bomedemstat sensitizes small
cell lung cancer to immune checkpoint blockade and T cell killing.
Clin Cancer Res. 28:4551–4564. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Khan S, Zhang X, Lv D, Zhang Q, He Y,
Zhang P, Liu X, Thummuri D, Yuan Y, Wiegand JS, et al: A selective
BCL-XL PROTAC degrader achieves safe and potent antitumor activity.
Nat Med. 25:1938–1947. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
de Visser KE and Joyce JA: The evolving
tumor microenvironment: From cancer initiation to metastatic
outgrowth. Cancer Cell. 41:374–403. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Yum S, Li M, Fang Y and Chen ZJ: TBK1
recruitment to STING activates both IRF3 and NF-κB that mediate
immune defense against tumors and viral infections. Proc Natl Acad
Sci USA. 118:e21002251182021. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yang H, Wang H, Ren U, Chen Q and Chena
ZJ: cGAS is essential for cellular senescence. Proc Natl Acad Sci
USA. 114:E4612–E4620. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li Q, Wu P, Du Q, Hanif U, Hu H and Li K:
cGAS-STING, an important signaling pathway in diseases and their
therapy. MedComm (2020). 5:e5112024. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Beernaert B and Parkes EE: cGAS-STING
signalling in cancer: Striking a balance with chromosomal
instability. Biochem Soc Trans. 51:539–555. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Shen M, Jiang X, Peng Q, Oyang L, Ren Z,
Wang J, Peng M, Zhou Y, Deng X and Liao Q: Correction: The
cGAS-STING pathway in cancer immunity: Mechanisms, challenges, and
therapeutic implications. J Hematol Oncol. 18:492025. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ying X, Chen Q, Yang Y, Wu Z, Zeng W, Miao
C, Huang Q and Ai K: Nanomedicines harnessing cGAS-STING pathway:
sparking immune revitalization to transform ‘cold’ tumors into
‘hot’ tumors. Mol Cancer. 23:2772024. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Mahin J, Xu X, Li L and Zhang C:
cGAS/STING in skin melanoma: From molecular mechanisms to
therapeutics. Cell Commun Signal. 22:5532024. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Murayama T, Mahadevan NR, Meador CB,
Ivanova EV, Pan Y, Knelson EH, Tani T, Nakayama J, Ma X, Thai TC,
et al: Targeting TREX1 induces innate immune response in
drug-resistant small-cell lung cancer. Cancer Res Commun.
4:2399–2414. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Giustarini G, Pavesi A and Adriani G:
Nanoparticle-based therapies for turning cold tumors hot: How to
treat an immunosuppressive tumor microenvironment. Front Bioeng
Biotechnol. 9:6892452021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Saha S, Xiong X, Chakraborty PK, Shameer
K, Arvizo RR, Kudgus RA, Dwivedi SK, Hossen MN, Gillies EM,
Robertson JD, et al: Gold nanoparticle reprograms pancreatic tumor
microenvironment and inhibits tumor growth. ACS Nano.
10:10636–10651. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Campbell CJ and Booth BW: The influence of
the normal mammary microenvironment on breast cancer cells. Cancers
(Basel). 15:5762023. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Cai Q and Xu Y: The microenvironment
reprograms circuits in tumor cells. Mol Cell Oncol. 2:e9696342015.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Cheng YQ, Wang SB, Liu JH, Jin L, Liu Y,
Li CY, Su YR, Liu YR, Sang X, Wan Q, et al: Modifying the tumour
microenvironment and reverting tumour cells: New strategies for
treating malignant tumours. Cell Prolif. 53:e128652020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Xu Y, Yang Y, Wang Z, Sjöström M, Jiang Y,
Tang Y, Cheng S, Deng S, Wang C, Gonzalez J, et al: ZNF397
deficiency triggers TET2-driven lineage plasticity and AR-targeted
therapy resistance in prostate cancer. Cancer Discov. 14:1496–1521.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Cai W, Xiao C, Fan T, Deng Z, Wang D, Liu
Y, Li C and He J: Targeting LSD1 in cancer: Molecular elucidation
and recent advances. Cancer Lett. 598:2170932024. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Xu Z, Jiang G and Dai J: Tumor
therapeutics in the era of ‘RECIST’: Past, current insights, and
future prospects. Oncol Rev. 18:14359222024. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Gong D, Arbesfeld-Qiu JM, Perrault E, Bae
JW and Hwang WL: Spatial oncology: Translating contextual biology
to the clinic. Cancer Cell. 42:1653–1675. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Bartolomucci A, Nobrega M, Ferrier T,
Dickinson K, Kaorey N, Nadeau A, Castillo A and Burnier JV:
Circulating tumor DNA to monitor treatment response in solid tumors
and advance precision oncology. NPJ Precis Oncol. 9:842025.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Yue B, Gao W, Lovell JF, Jin H and Huang
J: The cGAS-STING pathway in cancer immunity: Dual roles,
therapeutic strategies, and clinical challenges. Essays Biochem.
69:EBC202530062025. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Andrews HS, Zariffa N, Nishimura KK, Deng
Y, Eisele M, Ensor J, Espenschied C, Fabrizio D, Goren EM, Haddad
V, et al: Molecular response cutoffs and ctDNA collection
timepoints influence on interpretation of associations between
early changes in ctDNA and overall survival in patients treated
with anti-PD(L)1 and/or chemotherapy. J Immunother Cancer.
13:e0124542025. View Article : Google Scholar : PubMed/NCBI
|