Introduction
p16 is a 148 amino acid-protein encoded by the
INK4a gene, which binds to cyclin-dependent kinases (CDKs)
and through the inactivation of CDKs, induces growth arrest
(1). p16 is the second
most common tumour suppressor gene, and the genetic inactivation
due to missense mutations or promoter methylation of p16
itself is frequently found in cancer (2), namely in approximately 26% of all
tumours (3). However, p16
mutations appear to be infrequent in prostate cancer (4); moreover, p16 overexpression
in cancer has not yet been fully clarified. Certain types of
tumour, such as melanoma, HPV-associated tumours, non-small cell
lung cancer, mesothelioma and lymphoma exhibit diminished p16
protein levels (5-7),
while in other tumours, including prostate cancer, the
overexpression of wild-type or mutant p16 has been found
(8,9). p16 overexpression is
associated with tumour recurrence (10), and with a poor clinical course in
patients with erythroblast transformation-specific-related
gene (ERG)-negative prostate cancer (11); therefore, although p16 acts
as a tumour suppressor gene, stress or oncogenic factors or
alternative molecular events may overcome the role of p16 as
a negative cell cycle regulator (12). Metabolic cellular reprogramming
represents a key event in cancer cells, and p16 seems to be
involved in the metabolic switch to glycolysis during
tumorigenesis, also regulating NADPH Oxidase 4 (NOX4)
expression in pancreatic ductal adenocarcinoma (13). Pyruvate Kinase type M2
(PKM2) also plays a central role in cancer, modulating
glucose metabolism to support malignant cell proliferation
(14). PKM2 expression has
been in fact shown to be associated with tumour progression in
several studies (15-19);
however, its involvement in prostate cancer remains to be fully
elucidated (20). Moreover, in
recent years, microRNAs (miRNAs or miRs) have been evaluated as
cancer regulators with a huge impact (21) on the management of several types
of tumour, including prostate cancer.
This study attempted to assess the interaction
between p16 and metabolic factors, such as NOX4 and
PKM2, with a putative involvement of the miRNA-mediated
regulation in prostate cancer. Thus, The Cancer Genome Atlas (TCGA)
prostate cancer data were accessed, with the hypothesis that
p16 plays a central role in prostate cancer progression,
with an interaction with metabolic factors, as well as with
miRNAs.
Taken together, p16, NOX4 and
PKM2 expression were found to be decreased, possibly due to
miR-625-5p, miR-23a-3p and miR-122-5p regulation, respectively, in
cancer tissues with a low Gleason score, suggesting a deeper
understanding of their interplay and of their regulation by miRNAs
for developing novel therapeutic strategies for prostate
tumours.
Materials and methods
TCGA database
IlluminaHiSeq expression data were extracted from
the TCGA data portal (http://tcga.cancer.gov/; accessed October, 2017);
expression data were downloaded along with the corresponding
clinicopathological characteristics of 243 patients with prostate
cancer.
Statistical analysis
One-way ANOVA and Tukey's test as a post hoc test
were used for multiple comparisons. The Student's t-test was
applied for comparisons of the mean expression values between 2
groups. Survival analyses were performed using the Kaplan-Meier
method with the log-rank test for statistical significance. Using
JMP10 software (SAS), a P-value <0.05 was considered to indicate
a statistically significant difference.
miRNA regulation prediction
analysis
Bioinformatic analysis was performed using the
online database for miRNA target prediction, TargetScan (http://www.targetscan.org/), in order to reveal
potential binding sites for miRNAs in the 3'UTR of the genes
analysed in this study.
Results
TCGA database
A cohort of 243 patients with prostate cancer was
extracted from the TCGA database. A lower Gleason score (6,7)
was observed in 144 out of the 243 TCGA cases, a Gleason score of 8
was observed in 27 cases, and a score of 9 in 71 cases; there was
only 1 patient with a Gleason score of 10. As regards tumour stage,
there were 9 cases with stage T2a, 3 with T2b, 81 with T2c, 75 with
T3a, 68 with T3b and 3 with T4 stage disease; in 4 cases, the T
stage was unknown. The surgical margin resection status was R0 in
147 cases (62.9%), R1 in 75 cases (32%), R2 in 4 cases (1.7%) and
RX in 8 cases (3.4%); in 9 cases, the surgical margin resection
status was unknown (Table I).
 | Table IClinicopathological characteristics
of the 243 prostate cancer patients extracted from the TCGA
database. |
Table I
Clinicopathological characteristics
of the 243 prostate cancer patients extracted from the TCGA
database.
|
Characteristics | No. of samples |
|---|
| Gleason score |
|
6-7 | 144 |
|
8 | 27 |
|
9-10 | 72 |
| T stage |
|
T2a | 9 |
|
T2b | 3 |
|
T2c | 81 |
|
T3a | 75 |
|
T3b | 68 |
|
T4 | 3 |
|
Unknown | 4 |
| Surgical margin
status |
|
R0 | 147 |
|
R1 | 75 |
|
R2 | 4 |
|
RX | 8 |
|
Unknown | 9 |
The disease free interval range was 0.76-165 months
(median value, 34.4 months); the overall survival range was
0.76-165 months (median value, 37.8 months).
p16 expression and clinicopathological
characteristics
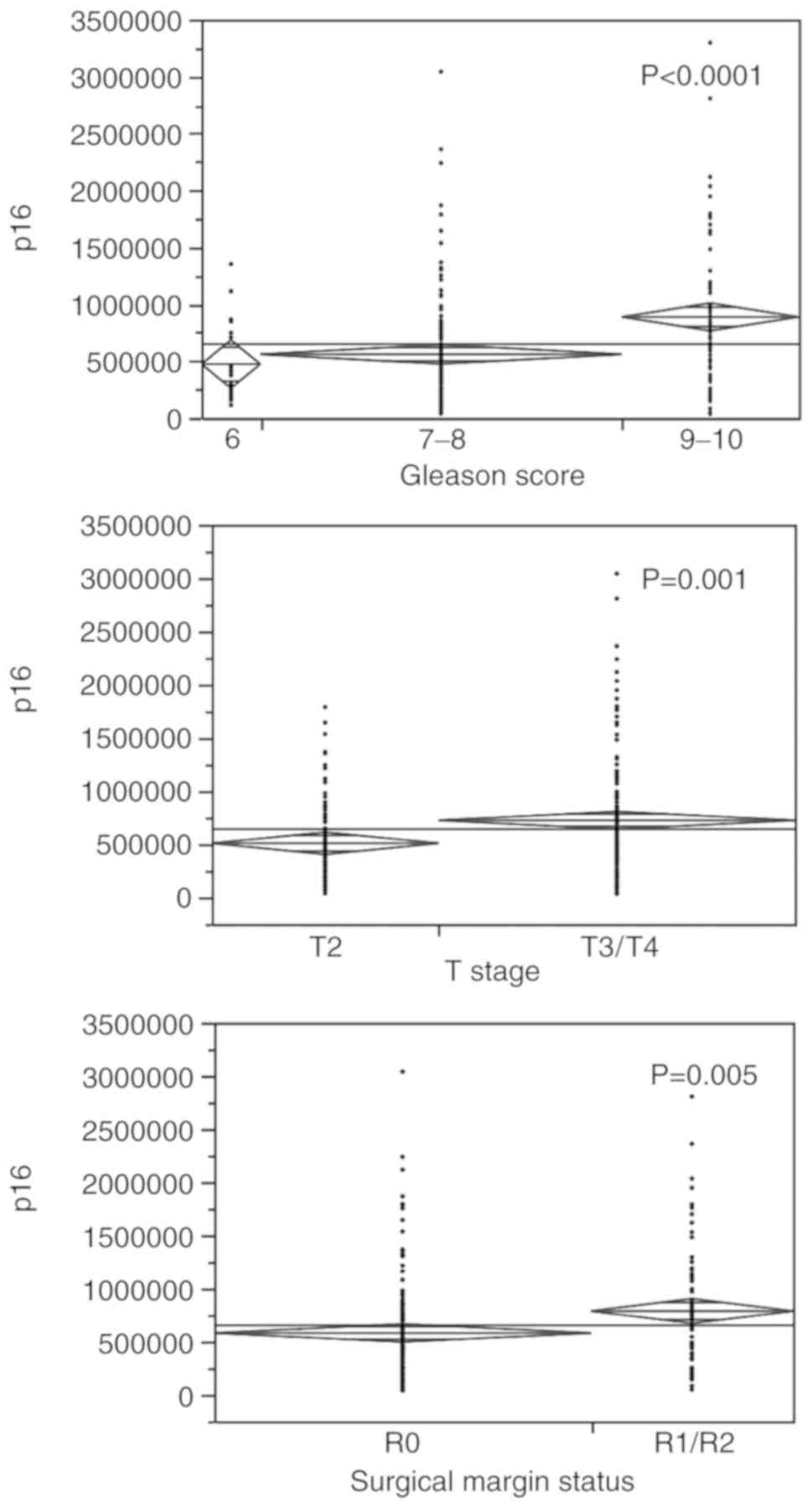
An elevated p16 expression level was
significantly associated with a high Gleason score, decreasing with
the score (P<0.0001); the mean value was 886,646 in cases with a
Gleason score of 9-10, 558,832 in those with a score of 7-8 and
472,610 in cases with a score of 6. When comparing tumour stage T2
versus T3/T4, it was found that high p16 levels were associated
with an advanced tumour stage (t-test; P=0.001), as well as with a
positive surgical resection margins status R1/R2 in comparison to
R0 (t-test; P=0.005) (Fig.
1).
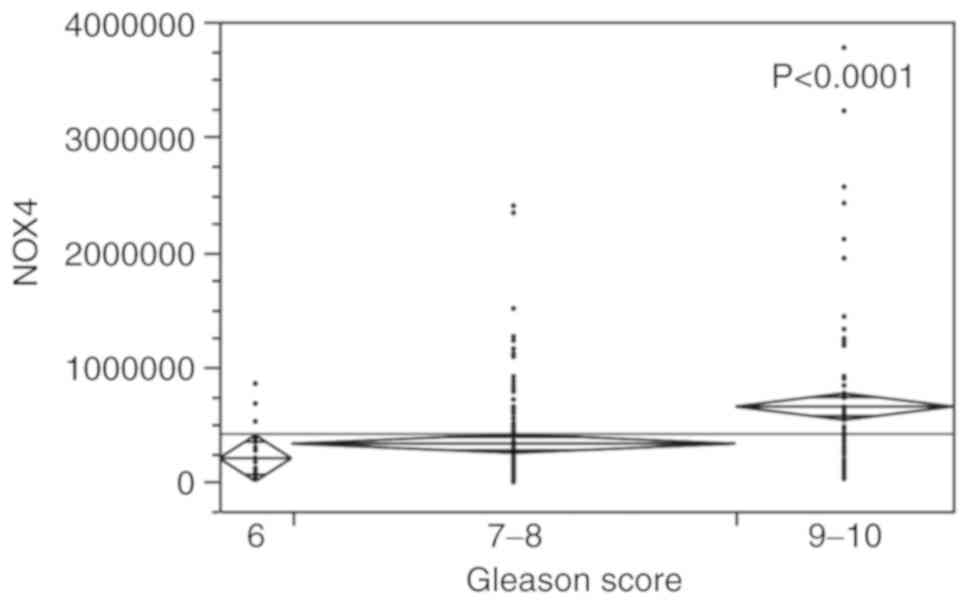
NOX4 expression
The samples with a Gleason of 9-10 exhibited a
higher value of NOX4 expression (664,737±59,395), those with
a score of 6 exhibited the lowest (20,7001±102,158), and those with
a score of 7-8 (333,929±41,275) exhibited intermediate and
increasing values (P<0.0001) (Fig.
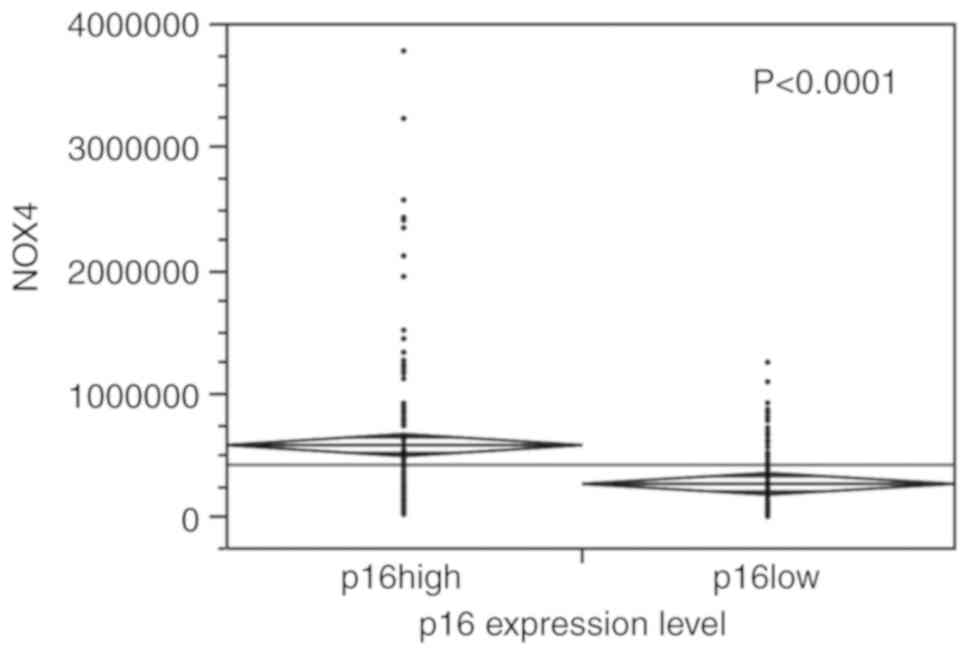
2). p16 and NOX4 expression were positively
associated (P<0.0001) (Fig.
3).
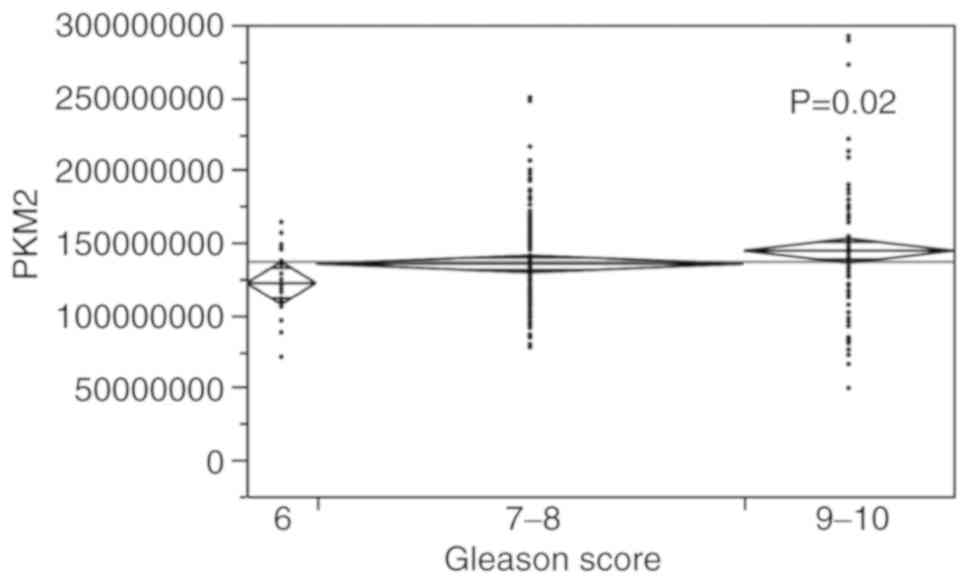
PKM2 expression
PKM2 expression exhibited a similar trend
(P=0.02), as that of p16; the samples with a Gleason score
of 9-10 exhibited a higher value of PKM2 expression, while a
lower PKM2 value was found in the prostate tumours with a
score of 6 (Fig. 4). A high
p16 expression was significantly associated with elevated
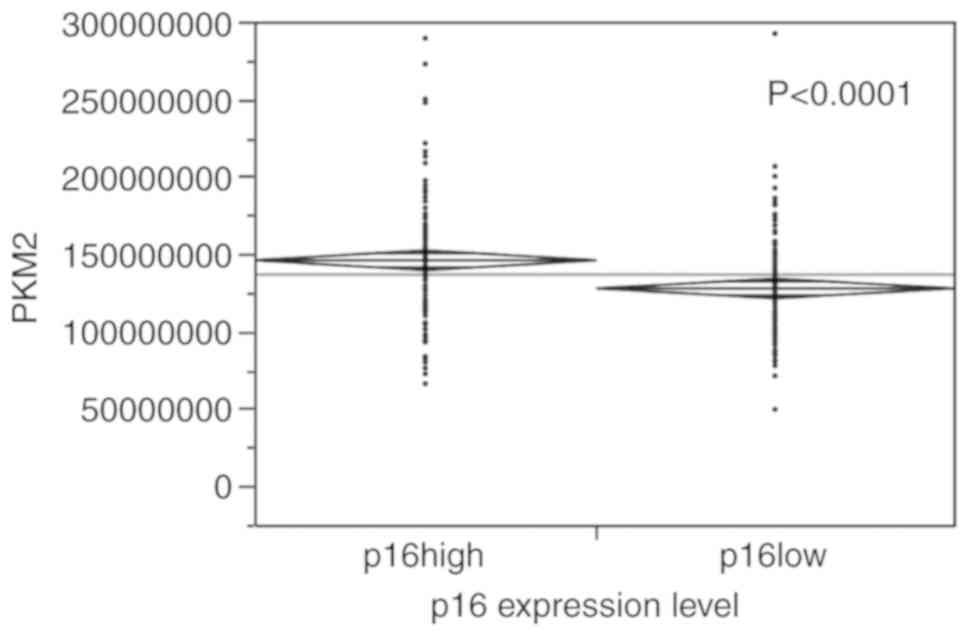
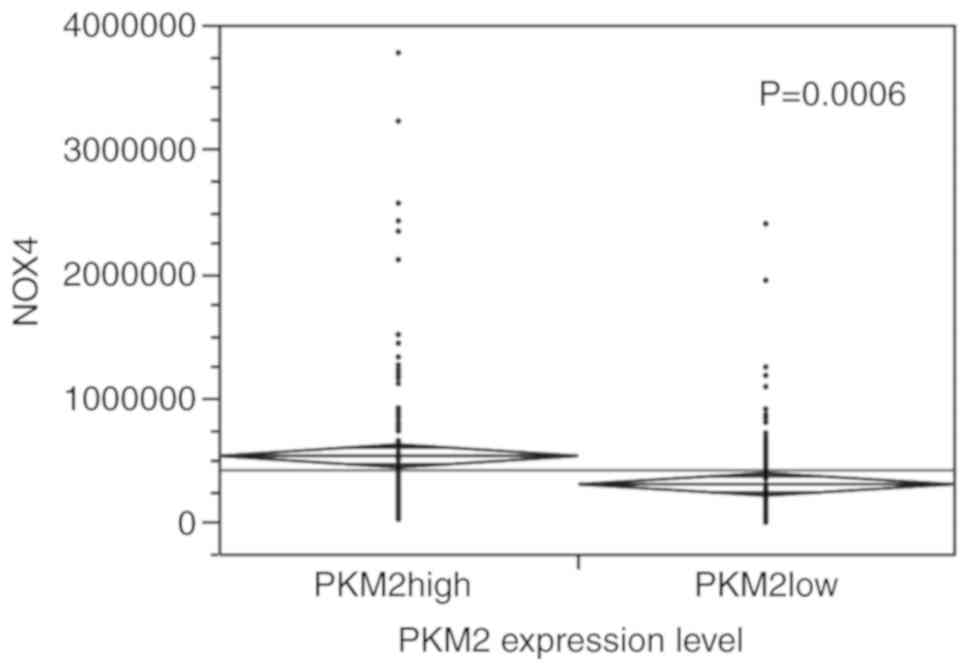
PKM2 levels (P<0.0001) (Fig. 5); moreover, as shown in Fig. 6, a positive association was
observed between PKM2 and NOX4 (P=0.0006).
miRNA regulation
Bioinformatics analysis using TargetScan revealed
that miR-625-5p could bind to the 3'UTR of p16. In addition,
a consequential pairing of the NOX4 and PKM2 target
region with miR-23a-3p and miR-122-5p, respectively was found. The
predicted pairing with miRNAs is illustrated in Fig. 7. Of note, the miR-625-5p levels
were inversely correlated with p16 expression, and
miR-23a-3p and miR-122-5p correlated with NOX4 and
PKM2, respectively (data not shown).
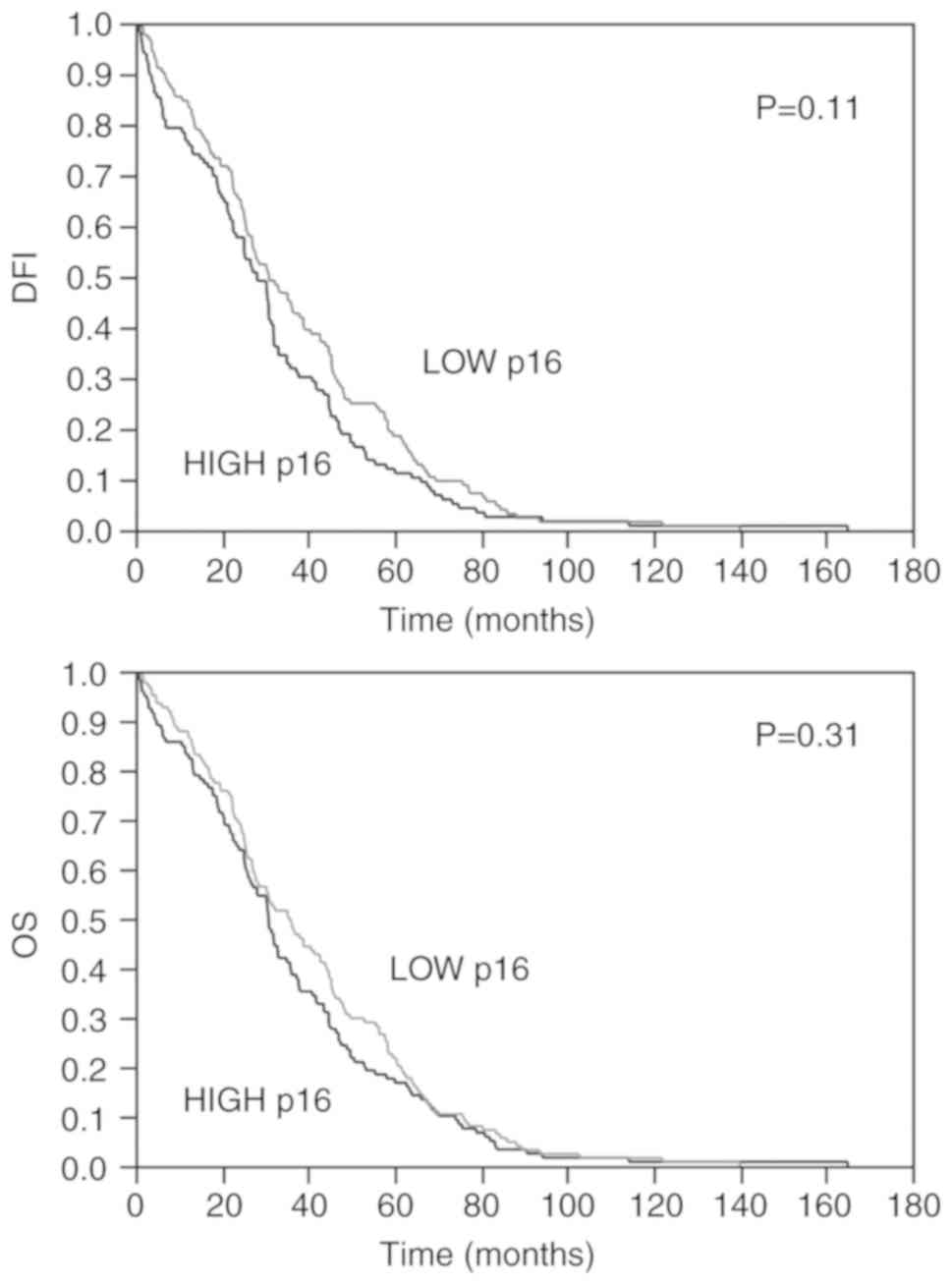
p16 expression and survival
As regards patient survival, as shown in Fig. 8, Kaplan-Meier analysis revealed a
shorter disease-free or overall survival in patients with a high
p16 expression (28.25±2.38 months and 30.85±2.44,
respectively) in comparison to those with low p16 levels
(30.98±2.33 and 36.01±2.36), although no statistically significant
differences were observed (P=0.11 and P=0.31, for disease-free
interval and overall survival, respectively); this may be due to
the absolute excellent survival rate in prostate cancer.
Discussion
The exact role of p16 in prostate cancer progression
has not yet been fully determined. p16 is one of the proteins which
has been most extensively studied over the past three decades, with
its high expression being associated with a more aggressive
clinical development in several neoplastic diseases, such as
melanoma, lymphoma and non-small cell lung cancer (5-7,22).
On the contrary, recently, patients with prostate cancer with
p16 overexpression were shown to have a more aggressive
behaviour (11). The
overexpression of wild-type or mutant p16 may involve an
effort to recover p16 functions in certain types of tumour;
however, the mechanisms responsible for the development and
progression of prostate cancer remain unknown. p16
dysregulation has been shown to be involved in metabolic
reprogramming in pancreatic cancer cells (13), inducing NOX4 activity, one of the
NADPH oxidases required to maintain glycolysis for tumoural cell
growth. Recently, NOX4 silencing has been suggested as a
theoretical target in prostate cancer, considering the effect of
repressed glycolysis, with decreased lactate and ATP production, on
cell proliferation (23). PKM2 is
another well-established regulator of glycolysis and energy
metabolism of cancer cells (24).
Apart from its role in aerobic glycolysis, PKM2 seems to be also
involved in non-metabolic functions, such as cell cycle
progression; however, the mechanisms underlying its regulation in
tumour progression are not yet clear, and this may be dependent on
the cancer cell type (25).
This study accessed the public TCGA database in
order to clarify the role of p16 in prostate cancer and its
putative interaction with NOX4 and PKM2; a possible
miRNA regulation was also analysed. It was found that the p16
expression level was increased in samples with a higher Gleason
score and in those with an advanced tumour stage. It was also
associated with positive surgical resection margins and with a
trend to a worse survival, confirming the involvement of p16
in the biological switch to advanced prostate cancer. p16
upregulation was linked to either a NOX4 or PKM2 high
expression level, suggesting a putative network linking p16
and metabolic genes in modulating growth control and prostate
cancer progression. The role of NOX4 and PKM2 in
reprogramming the p16-induced metabolic switch to glycolysis may
identify a novel therapeutic target for prostate tumours.
miRNAs are small, non-coding molecules involved in
repressing translation by binding the 3'UTR of their target genes
(26). In recent years, several
studies have demonstrated the role of miRNAs in diagnosis,
prognosis and as predictive biomarkers in different cancer types
(27-30).
In this study, bioinformatics analysis using TargetScan revealed
that either p16 or NOX4 and PKM2 3'UTR could
bind to miR-625-5p, miR-23a-3p and miR-122-5p, respectively, with
downregulation due to consequential pairing, suggesting the three
genes as physiologically targets of corresponding miRNAs.
p16 may be involved in overlapping pathways;
its contribution as a tumour suppressor gene has been
well-established, while the particular role of p16
deregulation to the development and progression of a specific
tumour remain to be further explored. A complex coordination of
p16 with other molecular events occurring in the same tumour
microenvironment may explain this intricate role of p16.
Taken together, the data of this study suggest
interplay of p16 with modulating metabolism markers, with a
regulation by miRNAs in prostate tumours; uncovering the putative
regulatory network of p16 may have a potential clinical
impact on the development of novel therapeutic strategies for
prostate cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data supporting the conclusions of this article
are available from the corresponding author on reasonable
request.
Authors' contributions
All the authors (LB, FM, CS, RB and PF) conceived
the study. LB and PF were involved in data collection, project
development and management, manuscript writing and editing. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Serrano M, Hannon GJ and Beach D: A new
regulatory motif in cell cycle control causing specific inhibition
of cyclinD-cdk4. Nature. 366:704–707. 1993.PubMed/NCBI View
Article : Google Scholar
|
|
2
|
Serrano M: The tumor suppressor protein
p16INK4a. Exp Cell Res. 237:7–13. 1997.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Liggett WH and Sidransky D: Role of the
p16 tumor suppressor gene in cancer. J Clin Oncol. 16:1197–1206.
1998.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Tamimi Y, Bringuier PP, Smit F, van
Bokhoven A, Debruyne FM and Schalken JA: p16 mutations/deletions
are not frequent events in prostate cancer. Brit J Cancer.
74:120–122. 1996.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Taga S, Osaki T, Ohgami A, Imoto H,
Yoshimatsu T, Yoshino I, Yano K, Nakanishi R, Ichiyoshi Y and
Yasumoto K: Prognostic value of the immunohistochemical detection
of p16INK4 expression in non small cell lung carcinoma. Cancer.
80:389–395. 1997.PubMed/NCBI View Article : Google Scholar
|
|
6
|
García-Sanz R, González M, Vargas M,
Chillón MC, Balanzategui A, Barbón M, Flores MT and San Miguel JF:
Deletions and rearrangements of cyclin-dependent kinase 4 inhibitor
gene p16 are associated with poor prognosis in B cell non-Hodgkin's
lymphomas. Leukemia. 11:1915–1920. 1997.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hwang HC, Sheffield BS, Rodriguez S,
Thompson K, Tse CH, Gown AM and Churg A: Utility of BAP1
immunohistochemistry and p16 (CDKN2A) FISH in the diagnosis of
malignant mesothelioma in effusion cytology specimens. Am J Surg
Pathol. 40:120–126. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Mahajan A: Practical issues in the
application of p16 immunohistochemistry in diagnostic pathology.
Hum Pathol. 51:64–74. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Serra S and Chetty R: p16. J Clin Pathol.
71:853–858. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lee CT, Capodieci P, Osman I, Fazzari M,
Ferrara J, Scher HI and Cordon-Cardo C: Overexpression of the
cyclin-dependent kinase inhibitor p16 is associated with tumor
recurrence in human prostate cancer. Clin Cancer Res. 5:977–983.
1999.PubMed/NCBI
|
|
11
|
Burdelski C, Dieckmann T, Heumann A,
Hube-Magg C, Kluth M, Beyer B, Steuber T, Pompe R, Graefen M, Simon
R, et al: p16 upregulation is linked to poor prognosis in ERG
negative prostate cancer. Tumour Biol. 37:12655–12663.
2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li J, Poi MJ and Tsai MD: Regulatory
mechanisms of tumor suppressor P16(INK4A) and their relevance to
cancer. Biochemistry. 50:5566–5582. 2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ju HQ, Ying H, Tian T, Ling J, Fu J, Lu Y,
Wu M, Yang L, Achreja A, Chen G, et al: Mutant Kras- and
p16-regulated NOX4 activation overcomes metabolic checkpoints in
development of pancreatic ductal adenocarcinoma. Nat Commun.
8(14437)2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhu H, Luo H and Zhu X, Hu X, Zheng L and
Zhu X: Pyruvate kinase M2 (PKM2) expression correlates with
prognosis in solid cancers: A meta-analysis. Oncotarget.
8:1628–1640. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Goldberg MS and Sharp PA: Pyruvate kinase
M2-specific siRNA induces apoptosis and tumor regression. J Exp
Med. 209:217–224. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Hsu MC, Hung WC, Yamaguchi H, Lim SO, Liao
HW, Tsai CH and Hung MC: Extracellular PKM2 induces cancer
proliferation by activating the EGFR signalling pathway. Am J
Cancer Res. 6:628–638. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Singh H, Longo DL and Chabner BA:
Improving prospects for targeting RAS. J Clin Oncol. 33:3650–3659.
2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sun Q, Chen X, Ma J, Peng H, Wang F, Zha
X, Wang Y, Jing Y, Yang H, Chen R, et al: Mammalian target of
rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is
critical for aerobic glycolysis and tumor growth. Proc Natl Acad
Sci USA. 108:4129–4134. 2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yang W, Xia Y, Ji H, Zheng Y, Liang J,
Huang W, Gao X, Aldape K and Lu Z: Nuclear PKM2 regulates
beta-catenin transactivation upon EGFR activation. Nature.
480:118–122. 2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wong N, Yan J, Ojo D, De Melo J, Cutz JC
and Tang D: Changes in PKM2 associate with prostate cancer
progression. Cancer Invest. 32:330–338. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Straume O and Akslen L: Alterations and
prognostic significance of p16 and p53 protein expression in
subgroups of cutaneous melanoma. Int J Cancer. 74:535–539.
1997.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wu QQ, Zheng B, Weng GB, Yang HM, Ren Y,
Weng XJ, Zhang SW and Zhu WZ: Downregulated NOX4 underlies a novel
inhibitory role of microRNA-137 in prostate cancer. J Cell Biochem.
120:10215–10227. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yoshida GJ: Metabolic reprogramming: The
emerging concept and associated therapeutic strategies. J Exp Clin
Cancer Res. 34(111)2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Dey P, Kundu A, Sachan R, Park JH, Ahn MY,
Yoon K, Lee J, Kim ND, Kim IS, Lee BM and Kim HS: PKM2 knockdown
induces autophagic cell death via AKT/mTOR pathway in human
prostate cancer cells. Cell Physiol Biochem. 52:1535–1552.
2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Etheridge A, Lee I, Hood L, Galas D and
Wang K: Extracellular microRNA: A new source of biomarkers. Mutat
Res. 717:85–90. 2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Volinia S and Croce CM: Prognostic
microRNA/mRNA signature from the integrated analysis of patients
with invasive breast cancer. Proc Natl Acad Sci USA. 110:7413–7417.
2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Feber A, Xi L, Pennathur A, Gooding WE,
Bandia S, Wu M, Luketich JD, Godfrey TE and Litle VR: MicroRNA
prognostic signature for nodal metastases and survival in
esophageal adenocarcinoma. Ann Thorac Surg. 91:1523–1530.
2011.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Beer DG, Kardia SL, Huang C, Giordano TJ,
Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al:
Gene-expression profiles predict survival of patients with lung
adenocarcinoma. Nat Med. 8:816–824. 2002.PubMed/NCBI View
Article : Google Scholar
|
|
30
|
Shukla S, Evans JR, Malik R, Feng FY,
Dhanasekaran SM, Cao X, Chen G, Beer DG, Jiang H and Chinnaiyan AM:
Development of a RNA-Seq based prognostic signature in lung
adenocarcinoma. J Natl Cancer Inst. 109:2016.PubMed/NCBI View Article : Google Scholar
|