Introduction
Lycium is a species of plant commonly known
as box-thorn, and is a genus of flowering plants of the Solanaceae
family. There are numerous species of Lycium, such as
Lycium barbarum, Lycium chinense, Lycium
europeaum [also known as Gouji (pinyin: gǒuqǐ), or Goji
(1). Carl Linnaeus, a naturalist,
provided the genus name Lycium in the year 1753, and gave
the species name barbarum, while Philip Miller, a botanist,
described Lycium chinense after 15 years (2). The Lycium fruit, known as
‘Gou qi zi’, ‘Gojizi’, ‘Wolfberry’, or ‘Goji berry’, is the red
berry obtained from two closely related species, Lycium
chinense Miller and Lycium barbarum L. of the box-thorn
in the family, Solanaceae, originating from Northwestern
China, mainly in Ningxia, Qinghai, Gansu and Inner Mongolia
(3-5).
The Lycium plant is extensively cultivated in most parts of
China, particularly in the Ningxia Hui Autonomous Region; however,
it is also cultivated in many other parts of China and worldwide
(1,5,6).
Lycium barbarum is regarded as Ningxia Goji, and its
products have a number of medicinal properties; thus, they are a
main part of Traditional Chinese Medicine (TCM).
The root bark (Digupi) and the fruit (Goji) of the
Lycium plant have long been used in TCM. This therapeutic
approach including Goji was first promoted by Jingyue Zhang,
indicated in his book ‘Jingyue Quanshu’ (1640 A.D.); he advised
using gentle heating and using ‘thick’ tonic herbs for nurturing
the internal organs. During the Ming Dynasty (1368-1644 A.D.), the
book ‘Bencao Gangmu’, written by Shizhen Li reported that the
regular consumption of Goji berries generates vital energy,
strengthens one's physique and increases longevity. It is well
known that the Goji berry has valuable properties in nourishing the
blood, enriching the ‘Yin’ (opposite to the ‘Yang’ in TCM), and is
beneficial to the kidneys, liver and lungs (4-6).
It is used in the treatment of consumptive diseases that are
associated with symptoms, such as thirst, dizziness, hypoplasia and
chronic cough. Furthermore, recent pharmaceutical investigations
have focused on proteoglycans, such as ‘Lycium barbarum
polysaccharide (LBP)’, which has antioxidant properties and may be
effective against age-related diseases (3,7).
Currently, based on folk remedies and research studies, Goji berry
or LBP is most well-known for the treatment of poor vision
(1,8), anemia (3), diabetes mellitus, memory enhancement
and liver disease (6,8-11),
as well as Alzheimer's disease (12), atherosclerosis (13) and other diseases (14-17).
The juice products of Goji, known as Himalayan Goji Juice, are
popular in the new food markets in developed countries, such as the
USA and UK. In first decade of the 21st century, UK, USA, Canada
and some other countries began cultivating Goji commercially to
meet potential markets for fresh fruits, juice and other products
(2-6).
Random amplified polymorphic DNA (RAPD), as one of
the important molecular marker techniques, was first reported in
1990 by Williams et al (18). Subsequently, RAPD, alone or in
combination with other molecular marker techniques, for example,
inter-simple sequence repeat (ISSR), simple sequence repeat (SSR)
or variable number tandem repeat (VNTR), oligonucleotide
polymorphism (OP), sequence-characterized amplified region (SCAR),
single nucleotide polymorphism (SNP) and amplified fragment length
polymorphism (AFLP) has been widely utilized in the analysis of
genetic or molecular diversity in various organisms, germplasm
characterization, genotype identification and fingerprinting,
estimating distances between species or offspring, and molecular
marker-assisted breeding (19-25).
Although RAPD is popular due to its numerous
advantages, it also possesses some drawbacks, such as poor
reproducibility and a lower production rate. By using a technique
of improved RAPD-PCR or random amplified microsatellite
polymorphism (RAMP)-PCR, its production and resolution can be
markedly increased, and its ramp time can be prolonged from 0 min
(as in regular PCR) to 2-3 min at annealing to the extension stage
in the PCR machine (termed RAMP-PCR) (26,27).
There are countless Lycium species or
cultivars, and confusion arises regarding the naming system and
identification process. For example, the same cultivars grown in
different soils or climates may produce very dissimilar fruits.
Therefore, the medicinal or/and nutritional values of Lycium
fruits can be significantly variable. Zhongning County in Ningxia,
China is the largest producer (approximately 20%) of Goji berries
worldwide, where Lycium barbarum plantations range between
40 and 400 hectares (100-1,000 acres). As regards the importance of
Lycium species and the roles of LBP in the new food market
and modern medicine, research into the genetic authentication and
characterization of TCM, particularly with DNA-based molecular
techniques is necessary. In the present study, varieties of their
samples were collected from different localities, and they were
characterized genetically by RAMP-PCR, and the results were also
verified by ISSR markers. Thus, the present study may provide
valuable insight into genetic information and biological diversity
of these medicinal plants.
Materials and methods
Experimentals
RAPD primers (2.5 µmol/l) are listed in
Table I and ISSR primers (2.5
µmol/l) are listed in Table
II. Taq Mastermix (2X PCR; TianGen Biotech Co. Ltd.) and the
DNA molecular weight marker, DL2000 (Takara Biotechnology Co. Ltd.)
were applied for PCR amplification. Other reagents that were used
were all of analytical grade and have been previously described
(26,27).
 | Table IRAPD primer sequences. |
Table I
RAPD primer sequences.
| Primer | 5'-3' sequence | Primer | 5'-3' sequence |
|---|
| SBS-A1 | CAGGCCCTTC | SBS-A7 | GAAACGGGTG |
| SBS-A11 | CAATCGCCGT | SBS-A12 | TCGGCGATAG |
| SBS-A16 | ACCTGGACAC | SBS-I8 | TTTGCCCGGT |
| SBS-I10 | ACAACGCGAG | SBS-I18 | TGCCCAGCCT |
| SBS-Q3 | ACCTCAGCTC | SBS-Q4 | AGTGCGCTGA |
| SBS-Q12 | AGTAGGGCAC | SBS-Q16 | AAGCGACCTG |
| SBS-Q18 | AGGCTGGGTG | | |
 | Table IIISSR primer sequences. |
Table II
ISSR primer sequences.
| Primer | 5'-3' sequence | Primer | 5'-3' sequence |
|---|
| UBC807 |
AGAGAGAGAGAGAGAGT | UBC811 |
GAGAGAGAGAGAGAGAC |
| UBC825 | ACACACACACACACA
CT | UBC847 |
CACACACACACACACARC |
| UBC851 |
GTGTGTGTGTGTGTGTYG | UBC876 |
GACAGACAGACAGACA |
| UBC879 |
CTTCACTTCACTTCA | UBC880 | GGAGAG GAG AGG
AGA |
| UBC885 | ACACACACACACACA
CYT | UBC886 |
ACACACACACACACACYA |
Plant sample collection
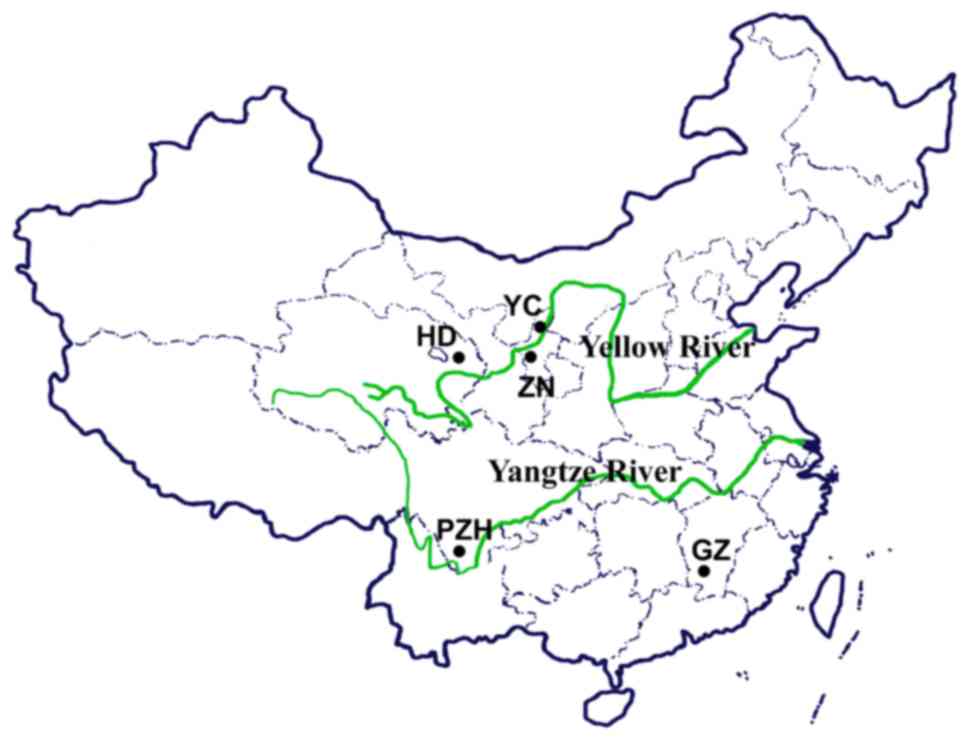
A total of 16 Lycium species or cultivars
were collected from 6 different regions of China and the USA: One
from Zhongning, Lingxia (ZN), one from Gongzhou, Jiangxi (GZ), one
from Panzhihua, Sichuan (PZH), 3 from Haidong, Qinghai (HD), 9 from
the Ningxia Academy of Agriculture and Forestry Sciences (YC) and
the last one from Houston, TX, USA (TX) (Fig. 1 and Table III).
 | Table IIILycium species sources for
RAPD-ISSR analysis. |
Table III
Lycium species sources for
RAPD-ISSR analysis.
| No. | Species or
cultivars | Sources | Abbreviation |
|---|
| 1 | Lycium
chinense Miller | Zhongning,
Lingxia | ZN |
| 2 | Lycium
chinense Miller | Gongzhou,
Jiangxi | GZ |
| 3 | Lycium
chinense Miller | Panzhihua,
Sichuan | PZH |
| 4 | Lycium
chinense Miller (207) | Haidong,
Qinghai | HD |
| 5 | Lycium
chinense Miller | Haidong,
Qinghai | HD |
| 6 | Lycium
barbarum ‘Ningqi-1’ | Haidong,
Qinghai | HD |
| 7 | Lycium
barbarum ‘Ningqi-1’ | NAAFS, Yinchuan,
Lingxia | YC |
| 8 | Lycium
barbarum ‘Ningqi-2’ | NAAFS, Yinchuan,
Lingxia | YC |
| 9 | Lycium
barbarum ‘Ningqi-3’ | NAAFS, Yinchuan,
Lingxia | YC |
| 10 | Lycium
barbarum ‘Ningqi-4’ | NAAFS, Yinchuan,
Lingxia | YC |
| 11 | Lycium
barbarum ‘Ningqi-5’ | NAAFS, Yinchuan,
Lingxia | YC |
| 12 | Lycium
barbarum ‘Ningqi-6’ | NAAFS, Yinchuan,
Lingxia | YC |
| 13 | Lycium
barbarum L.cv. ‘Ningqi-7’ | NAAFS, Yinchuan,
Lingxia | YC |
| 14 | Lycium
chinense Mill. ‘Cai-1’ | NAAFS, Yinchuan,
Lingxia | YC |
| 15 | Lycium
barbarum ‘Ningqi-9’ | NAAFS, Yinchuan,
Lingxia | YC |
| 16 | Lycium
chinense Miller | Houston, TX,
USA | TX |
DNA isolation
The plant genomic DNA was isolated from fresh or
dried leaves using the cetyltrimethylammonium bromide (CTAB) method
as previously described (27-29).
DNA quality and concentration were detected by agarose gel
electrophoresis and spectrophotometry (NonoDrop 2000
spectrophotometer, Thermo Fisher Scientific Inc.) (28). DNA samples were adjusted to 10
ng/µl as a final concentration and used for the next step of
DNA amplification.
RAMP-PCR marker amplification
In total, 15 different SBS primers were first used
to evaluate polymorphic detection using RAMP-PCR marker
amplification. Among these, 13 primers amplified products with
valuable polymorphic bands for the following data analysis (listed
in Table II). Thermal cycling of
PCR (10 µl in total) was carried out with the following
reaction conditions: 1 µl of primers, 1 µl of DNA templates
for Lycium cultivars or species (10 ng in total), 5
µl of Master Mix buffer and 3 µl of double-distilled
water. The PCR conditions were as follows: i) Pre-denaturation at
95˚C for 1 min and 30 sec; ii) 40 PCR cycles of 40 sec
at 94˚C, 1 min at 36˚C, 1 min and 30 sec at 72˚C; and iii)
extension for 5 min at 72˚C. PCR amplification was performed in a
thermal cycler (Applied Biosystems Veriti® 96-Well
Thermal Cycler, Life Technologies; Thermo Fisher Scientific, Inc.).
The ramp rate (the stage from annealing to extension) was set from
5% (0.125˚C/sec) to 10% (0.25˚C/sec) or to 40% (1˚C/sec) or to 100%
(2.5˚C/sec), respectively for samples using RAMP-PCR to compare the
reproducibility in this study. A ramp rate set to 0.125˚C/sec (5%
ramp rate) was used to execute in triplicates for each of the 16
samples (27).
ISSR marker amplification
ISSR amplification reactions were also executed in
10 µl reaction volumes containing 1 µl of ISSR primers, 1
µl of templates for DNA of Lycium cultivars or
species, 5 µl of Master Mix, and 3 µl of
double-distilled water. The steps of the PCR reaction were as
follows: i) Denaturation at 95˚C for 1 min and 30 sec; ii) 35
cycles of 40 sec at 94˚C, 30 sec at 50˚C, 1 min and 30 sec at 72˚C;
and iii) final extension for 5 min at 72˚C (24). The PCR reaction was performed in
the aforementioned PCR machine. In total, 17 primers were used
initially, among which 10 primers (listed in Table II) amplified products well with
high number of polymorphic bands.
Agarose gel analysis and data
analysis
The PCR-amplified products were then tested on a
1.8% agarose gel electrophoresis. Ethidium bromide (EtBr) staining
was used for visualizing the gels and the images were captured on a
Chemi Doc XRS system (Bio-Rad Laboratories, Inc.). Bands visualized
by ethidium bromide were selected on scoring for data analysis. ‘1’
was used to mark the presence of a clear band in the gel, and ‘0’
was used to denote that the corresponding band was absent in other
sample(s). The similarity index (SI) and similarity matrix (SM)
were calculated using the SM coefficient. Based on the unweighted
pair group method with arithmetic averages (UPGMA), the sequential,
agglomerative, hierarchical, and nested clustering (SAHN) module
was used to produce the dendrograms (30).
Results
Technical comparison between regular
RAPD amplification and RAMP-PCR
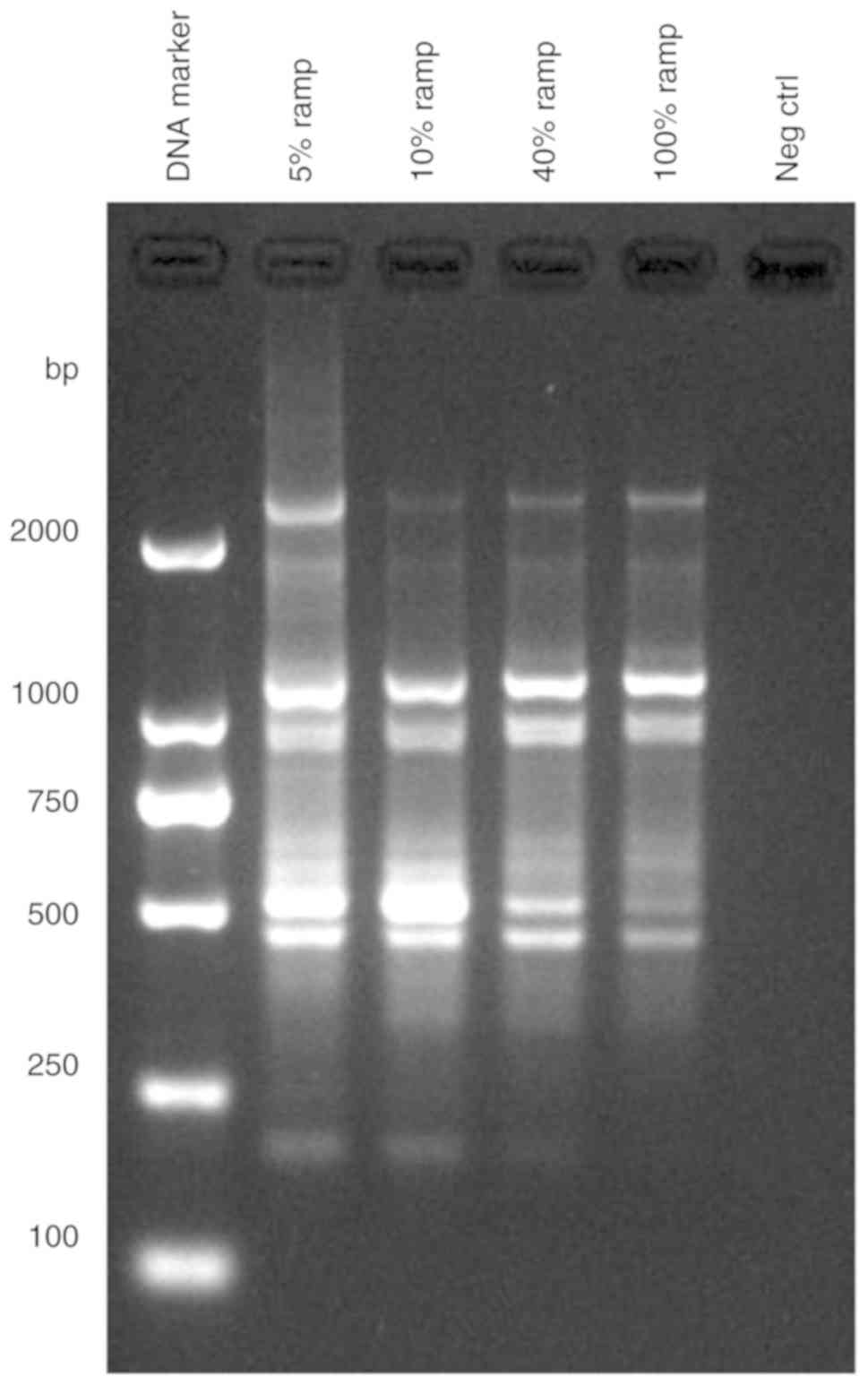
To obtain more specific bands from RAMP-PCR
amplification, at first, the primer SBS-I4 was used for
amplification with a ramp rate (from annealing to extension) of 5,
10, 40 and 100% (2.5˚C/sec), respectively, from the Lycium
barbarum ‘Ningqi-1’ sample (no. 7) (listed in Table III) in a PCR thermocycler. As
shown in Fig. 2, the PCR bands
were quantitatively increased with ramp rates from 100 to 5%. The
5% ramp rate had the most bands with stronger signals. As a
negative control, without a template, no bands were visible in the
5% ramp rate. Specifically, the band numbers were 4 by regular PCR
(with 100% ramp rate), and this then increased to 7 in RAMP-PCR
(with a 5% ramp rate), and signals that had at least 2 PCR bands
with the 5% ramp rate were much stronger than those with the 100%
ramp rate (Fig. 2). This finding
indicates that the decreased ramp rate (for example, 5%), markedly
increases the band numbers and production. Therefore, the 5% ramp
rate has better RAPD amplification, and this optimized RAMP-PCR
condition was then used to complete the amplification of all 16
samples of the Lycium species or cultivars using RAPD
primers.
Amplification of Lycium species or
cultivars with RAMP-PCR
To compare whether other samples can also get more
and specific bands in this study, either regular RAPD or RAMP-PCR
was applied to amplify the DNA samples by setting the ramp time
with a ramp rate of 5 and 100% using Lycium samples (shown
in Table III). The PCR product
amounts and the bands numbers were obviously increased by RAMP-PCR
when the ramp rate was set from 100 to 5% (data not shown).
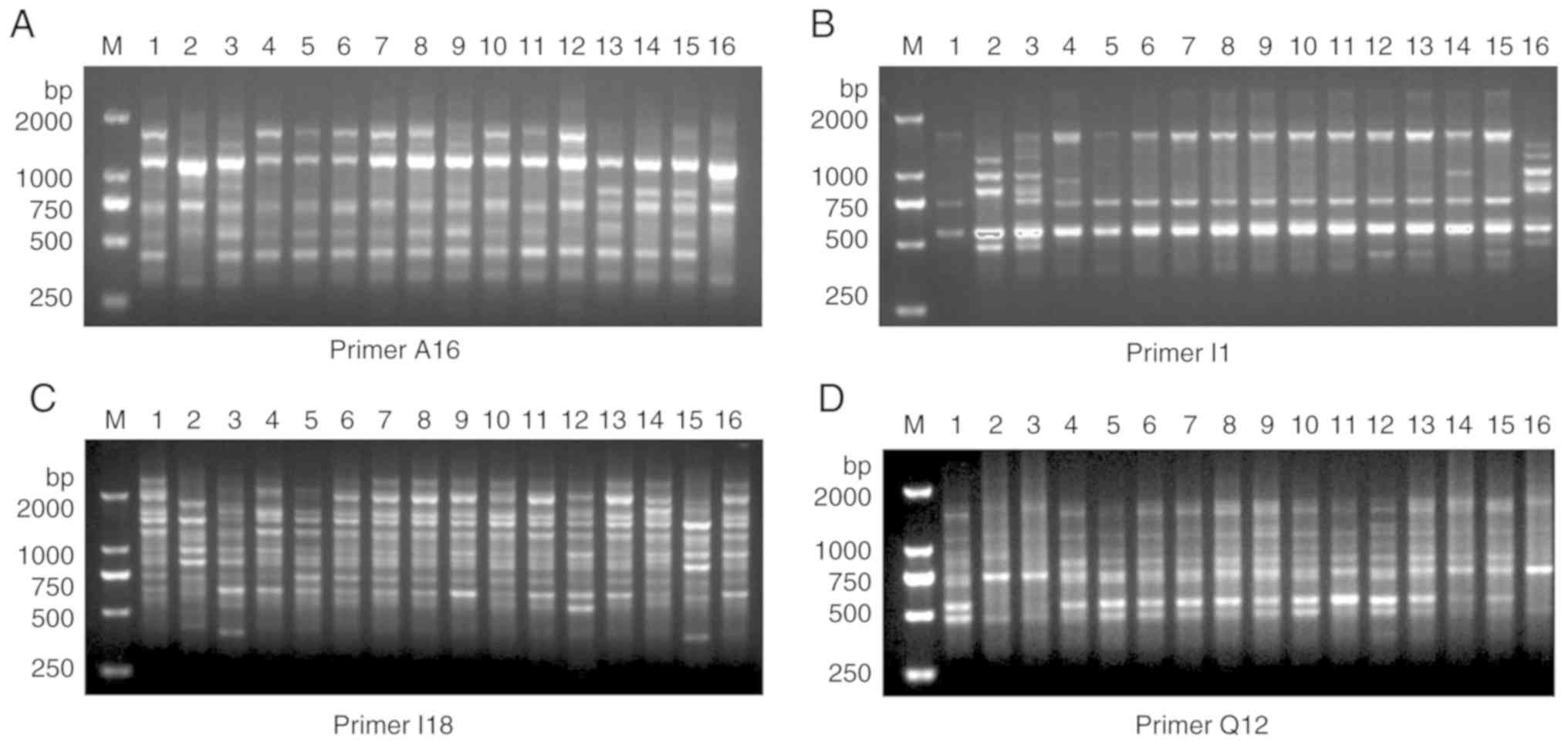
For the estimation of polymorphisms, 15 RAPD primers
were applied for RAMP-PCR analysis and 13 of these (shown in
Table I) produced polymorphic
amplification bands, which were highly reproducible. In Fig. 3, 4 representative primers
(SBS-A16, SBS-I1, SBS-I18 and SBS-Q12) from 16 samples are
presented. From these 13 primers, a total of 1,249 bands were
gained in total, where each primer exhibited 2-9 valuable bands
with an average of 6 per primer. The band size was in the range of
200-2,200 bp, and 89.05% of these were polymorphic in the 16
samples.
Genetic distance analysis based on
RAMP-PCR results
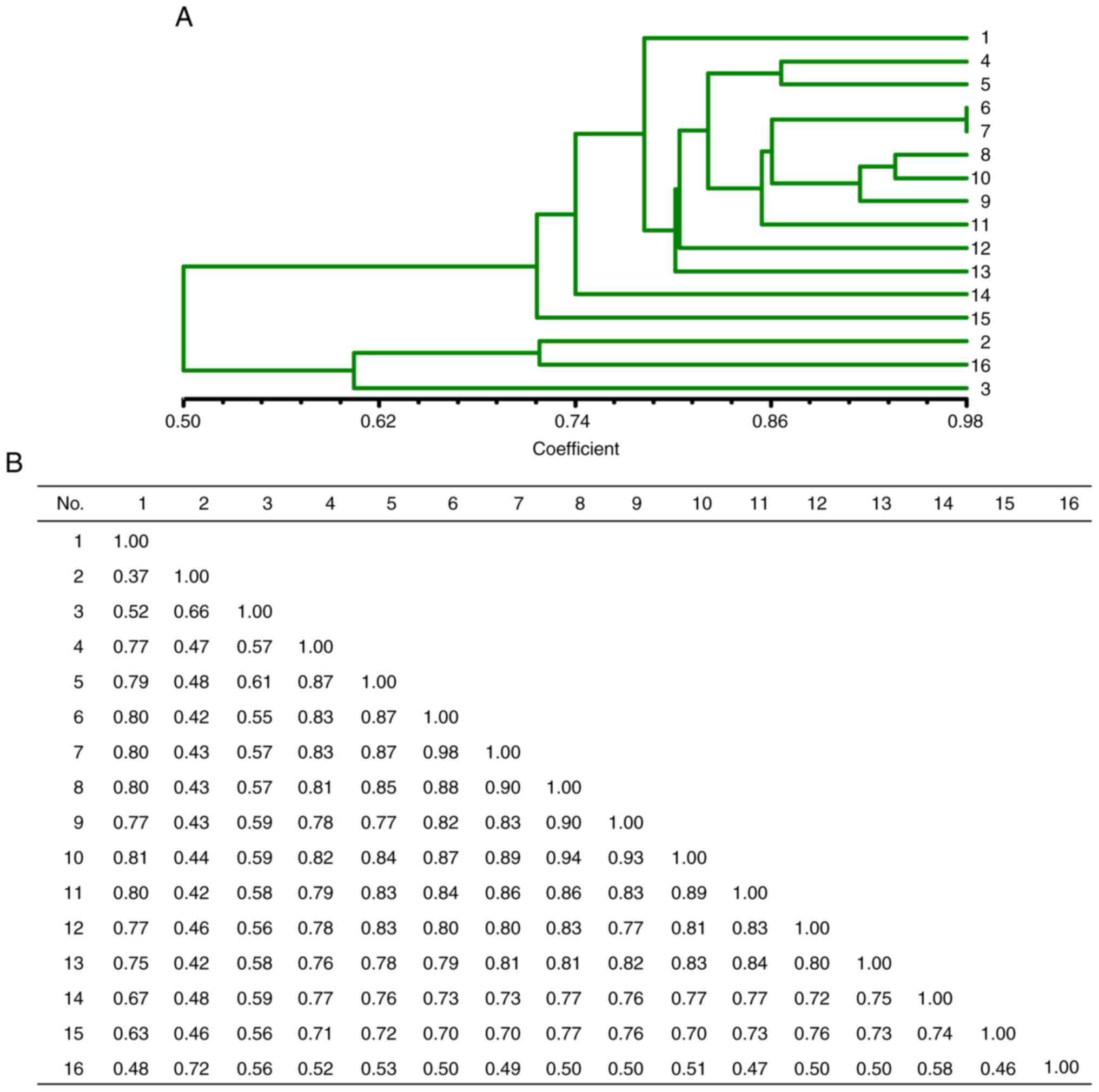
The cluster dendrogram based on the RAMP-PCR
amplified bands is presented in Fig.
4. The dendrogram results presented a similarity coefficient
(SC) index ranging from 0.37 to 0.98. The SC index between sample 1
(Lycium chinense Miller from Zhongning, Linxia) and sample 2
(from Gongzhou, Jiangxi) was the lowest (0.37), while that between
samples 6 and 7 (Lycium barbarum ‘Ningqi-1’ from Haidong,
Qinghai and form the Ningxia Academy of Agriculture and Forestry
Sciences) was the highest (0.98) (Fig. 4B). Samples 6 and 7, where were
obtained from different localities, were identified with the highes
SC index 0.98, indicating that they indeed originated from the same
cultivar.
Amplification of Lycium species or
cultivars DNAs by ISSR
In ISSR-PCR, 10 primers amplified well, which
generated 956 reproducible bands from 16 samples. Each reproducible
primer amplified 1-10 bands and revealed an average number of 5.9
bands per ISSR primer. The band size ranged between 200-2,000 bp,
and 88.28% of the bands were revealed as polymorphic in these 16
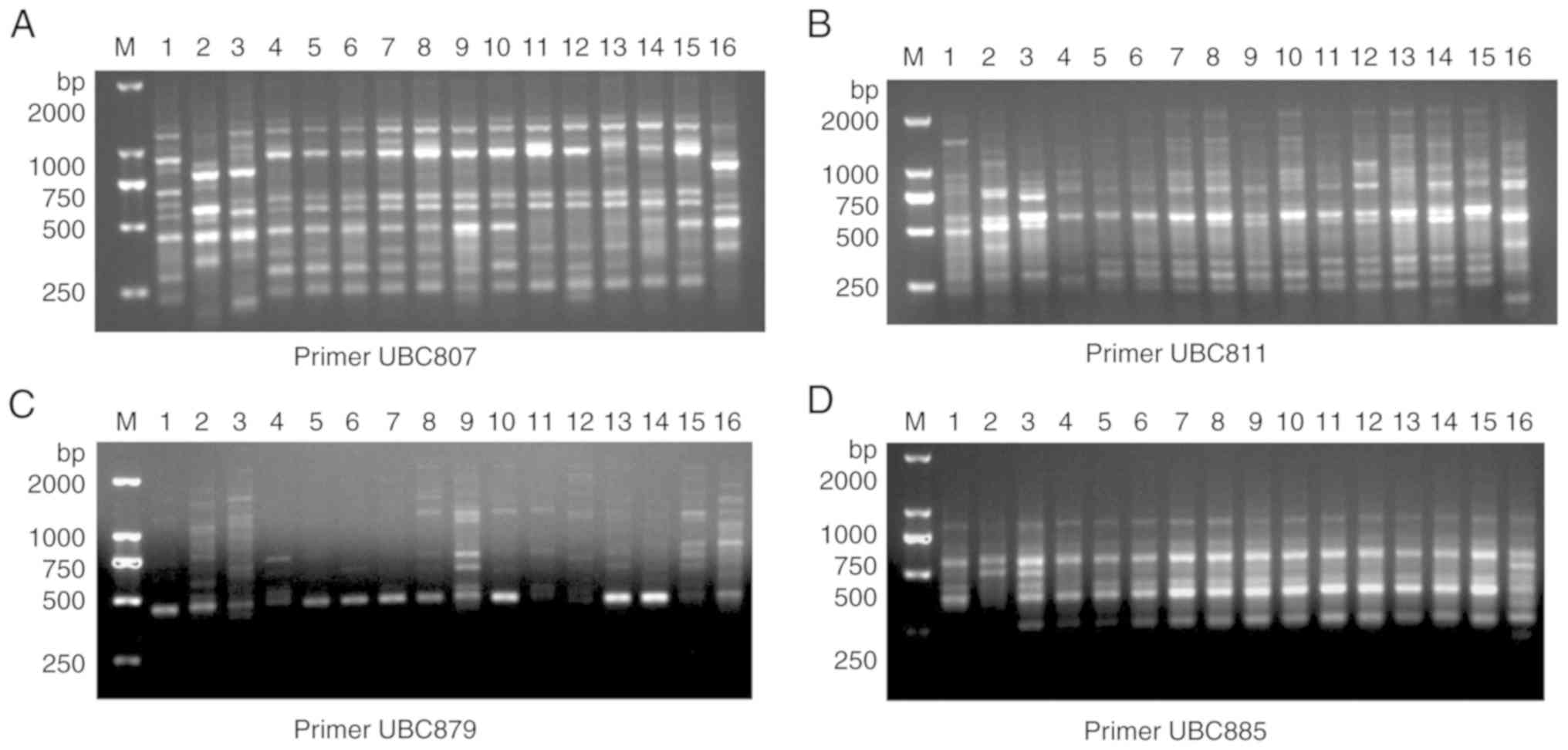
samples. The representative results of the primers UBC807, UBC811,
UBC879 and UBC885 are presented in Fig. 5. These findings provided a very
clear detection of DNA polymorphisms in Lycium species or
cultivars.
Results of genetic distance and
cluster analysis in ISSR amplification
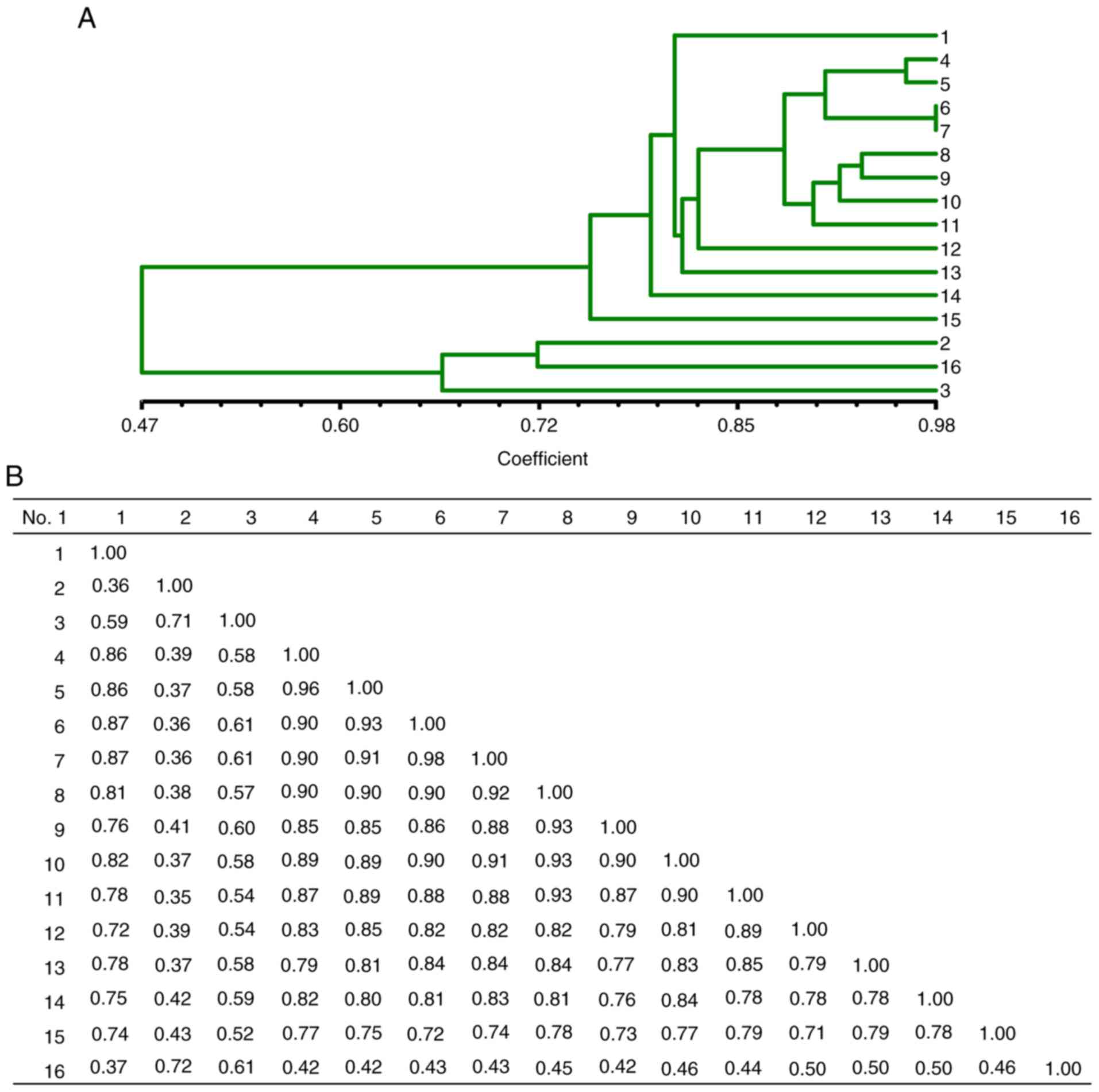
A cluster dendrogram was also obtained from ISSR
amplification profiles. It revealed similar results to those
produced by RAMP-PCR as regards the SC index among Lycium
samples (Fig. 6). The dendrogram
illustrated that the SC index among the samples ranged from 0.36 to
0.98. The SC index between sample 1 and sample 2 (Lycium
chinense Miller from Zhongning, Linxia and from Gongzhou,
Jiangxi) was the lowest (0.36), and that between samples 6 and 7 of
Lycium barbarum (‘Ningqi-1’ from Haidong, Qinghai and from
the Ningxia Academy of Agriculture and Forestry Sciences), was the
highest (0.98) (Fig. 6B),
indicating that they are the same cultivar. All these data are
consistent with the RAMP-PCR results and genetic analysis.
Discussion
The present study presented the first overview, to
the best of our knowledge, of the genetic variability of Lycium
barbarum cultivars or species, using both developed RAMP-PCR
and ISSR molecular markers. Notably, the Standardization
Administration of China (SAC), implemented in July 1, 2015, has
approved ‘Rules for agricultural seed testing-Verification of
genuineness and cultivar’ (GB/T3543.5-1995; https://www.codeofchina.com/standard/GBT3543.5-1995.html)
as the national standard, no. 1 modification item, in which the
regulation for variety authenticity or identity allows DNA
molecular detection methods, including SSR and SNP molecular
markers, and provides a strong basis for combating various illegal
acts of counterfeit rapidly and accurately. The classic markers
used by genetic ecologists are DNA sequencing which has also been
required to develop SNPs, another codominant and highly polymorphic
marker. Based on direct DNA sequencing, such as next generation
sequencing (NGS), short tandem repeat (STR) and SNP markers can
provide large genome coverage, and exhibit a high level of
variability, and thus can be used for phylogenetic studies
(31). STR analysis has been
successfully and widely used to evaluate genotypes in humans in
judicial authentication and forensic sciences (32). However, due to plant genomic
complexity, the genomic information remains largely unknown; STR
and SNP markers cannot be used for the genetic authentication of
plants or herbs.
Given the advantages and disadvantages of different
molecular genetic marker techniques, it is important to design the
most effective method in order to address particular ecological
questions. In RAMP-PCR, where the ramp time is prolonged at the
stage from annealing to extension, the resolution and production
are greatly increased compared to regular RAPD (25,26,33). The present study, using RAMP-PCR
and ISSR marker techniques, successfully characterized and
authenticated Lycium varieties from different geographic
regions indicating that samples 6 and 7 were the same cultivar.
Moreover, the similarity coefficient index between samples 2
(Lycium chinense Miller from Gongzhou, Jiangxi) and 16
(Lycium chinense Miller from Houston, TX, USA) was found to
be 0.72 by RAMP-PCR and 0.72 by ISSR, which clustered together,
indicating that these two cultivars have a good genetic
association, although a diverse geographic distance. The results of
RAMP-PCR and ISSR were both mutually consistent. RAMP is a
PCR-based technique, which combines ISSR and RAPD analysis, which
can generate useful molecular markers, investigate DNA
polymorphisms, and can be used to elucidate the genetic
associations among accessions, including Lycium varieties
(34).
In conclusion, the present study combined RAMP-PCR
and ISSR to analyze the genetic association and distance of
Lycium varieties comprehensively. To the best of our
knowledge, the present study is the first to genetically
characterize the molecular diversity of Lycium varieties by
combining these two methods, and this molecular characterization
may prove to be useful for the conservation and preservation of DNA
diversity of Lycium species.
Acknowledgements
The authors would like to thank Miss S. Fu from the
University of Houston for providing critical comments and advice on
the manuscript.
Funding
The present study was funded in part by the National
Natural Science Foundation of China (grant no. 81672887) and the
Joint Research Foundation of Luzhou City and Southwest Medical
University (grant no. 2018LZXNYD-YL01).
Availability of data and materials
The data and material that support the findings of
this study are available from the corresponding author upon
reasonable request.
Authors' contributions
XL, JD, CW and ZM were involved in the study
methodology and in performing the experiments. JC was involved in
data analysis. JF was involved in the conception of the study, and
in the writing and preparation of the original draft. MK and JF
were involved in the conception of the study, and in the editing
and revising of the manuscript. HC, TH and JF designed and
supervised the study. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dong JZ, Yang JJ and Wang Y: Resources of
Lycium species and related research progress. Zhongguo Zhong Yao Za
Zhi. 33:2020–2027. 2008.PubMed/NCBI(In Chinese).
|
|
2
|
Ram VJ: Herbal preparations as a source of
hepatoprotective agents. Drug News Perspect. 14:353–363.
2001.PubMed/NCBI
|
|
3
|
Potterat O: Goji (Lycium barbarum
and L. chinense): Phytochemistry, pharmacology and safety in
the perspective of traditional uses and recent popularity. Planta
Med. 76:7–19. 2010.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ulbricht C, Bryan JK, Costa D, Culwell S,
Giese N, Isaac R, Nummy K, Pham T, Rapp C, Rusie E, et al: An
evidence-based systematic review of Goji (Lycium spp.) by
the natural standard research collaboration. J Diet Suppl.
12:184–240. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Jin M, Huang Q, Zhao K and Shang P:
Biological activities and potential health benefit effects of
polysaccharides isolated from Lycium barbarum L. Int J Biol
Macromol. 54:16–23. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chang RC and So KF: Use of anti-aging
herbal medicine, Lycium barbarum, against aging-associated
diseases. What do we know so far? Cell Mol Neurobiol. 28:643–652.
2008.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ma ZF, Zhang H, The SS, Wang CW, Zhang Y,
Hayford F, Wang L, Ma T, Dong Z, Zhang Y and Zhu Y: Goji berries as
a potential natural antioxidant medicine: An insight into their
molecular mechanisms of action. Oxid Med Cell Longev.
2019(2437397)2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liu L, Lao W, Ji QS, Yang ZH, Yu GC and
Zhong JX: Lycium barbarum polysaccharides protected human
retinal pigment epithelial cells against oxidative stress-induced
apoptosis. Int J Ophthalmol. 8:11–16. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lam CS, Tipoe GL, So KF and Fung ML:
Neuroprotective mechanism of Lycium barbarum polysaccharides
against hippocampal-dependent spatial memory deficits in a rat
model of obstructive sleep apnea. PLoS One.
10(e0117990)2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yu H, Wark L, Ji H, Willard L, Jaing Y,
Han J, He H, Ortiz E and Zhang Y: Medeiros DM and Lin D: Dietary
wolfberry upregulates carotenoid metabolic genes and enhances
mitochondrial biogenesis in the retina of db/db diabetic mice. Mol
Nutr Food Res. 57:1158–1169. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lin J, Zhang Y, Wang X and Wang W:
Lycium ruthenicum extract alleviates high-fat diet-induced
nonalcoholic fatty liver disease via enhancing the AMPK signaling
pathway. Mol Med Rep. 12:3835–3840. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ye M, Moon J, Yang J, Hwa Lim H, Bin Hong
S, Shim I and Bae H: The standardized Lycium chinense fruit
extract protects against Alzheimer's disease in 3xTg-AD mice. J
Ethnopharmacol. 172:85–90. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lin L, Li J, Lv H, Ma Y and Qian Y: Effect
of Lycium ruthenicum anthocyanins on atherosclerosis in
mice. Zhongguo Zhong Yao Za Zhi. 37:1460–1466. 2012.PubMed/NCBI(In Chinese).
|
|
14
|
Zhao R, Cai Y, Shao X and Ma B: Improving
the activity of Lycium barbarum polysaccharide on sub-health
mice. Food Funct. 6:2033–2040. 2015.
|
|
15
|
Dursun R, Zengin Y, Gündüz E, İçer M,
Durgun HM, Dağgulli M, Kaplan İ, Alabalık U and Güloğlu C: The
protective effect of goji berry extract in ischemic reperfusion in
testis torsion. Int J Clin Exp Med. 8:2727–2733. 2015.PubMed/NCBI
|
|
16
|
Telang N, Li G, Sepkovic D, Bradlow HL and
Wong GY: Comparative efficacy of extracts from Lycium
barbarum bark and fruit on estrogen receptor positive human
mammary carcinoma MCF-7 cells. Nutr Cancer. 66:278–284.
2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chao JC, Chiang SW, Wang CC, Tsai YH and
Wu MS: Hot water-extracted Lycium barbarum and Rehmannia
glutinosa inhibit proliferation and induce apoptosis of
hepatocellular carcinoma cells. World J Gastroenterol.
12:4478–4484. 2006.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Williams JG, Kubelik AR, Livak KJ,
Rafalski JA and Tingey SV: DNA polymorphisms amplified by arbitrary
primers are useful as genetic markers. Nucleic Acids Res.
18:6531–6535. 1990.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Hess J, Kadereit JW and Vargas P: The
colonization history of Olea europaea L. In Macaronesia based on
internal transcribed spacer 1 (ITS-1) sequences, randomly amplified
polymorphic DNAs (RAPD), and inter simple sequence repeats (ISSR).
Mol Ecol. 9:857–868. 2000.
|
|
20
|
Agarwal M, Shrivastava N and Padh H:
Advances in molecular marker techniques and their applications in
plant sciences. Plant Cell Rep. 27:617–631. 2008.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Varela ES, Lima JP, Galdino AS, Pinto Lda
S, Bezerra WM, Nunes EP, Alves MA and Grangeiro TB: Relationships
in subtribe Diocleinae (Leguminosae; Papilionoideae) inferred from
internal transcribed spacer sequences from nuclear ribosomal DNA.
Phytochemistry. 65:59–69. 2004.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Taški-Ajduković K, Nagl N, Ćurčić Z and
Zorić M: Estimation of genetic diversity and relationship in sugar
beet pollinators based on SSR markers. Electronic J Biotechnol.
27:1–7. 2017.
|
|
23
|
Pachuau L, Atom AD and Thangjam R: Genome
classification of musa cultivars from northeast india as revealed
by ITS and IRAP markers. Appl Biochem Biotechnol. 172:3939–3948.
2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Mei Z, Zhang X, Khan MA, Imani S, Liu X,
Zou H, Wei C and Fu J: Genetic analysis of Penthorum
chinense Pursh. By improved-RAPD and ISSR in China. Electron J
Biotechnol. 30:6–11. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Fu S, Cheng J, Wei C, Yang L, Xiao X,
Zhang D, Stewart MD and Fu J: Development of diagnostic SCAR
markers for genomic DNA amplifications in breast carcinoma by DNA
cloning of high-GC RAMP-PCR fragments. Oncotarget. 8:43866–43877.
2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Fu JJ, Li LY, Xu X, Wang Z, Tang G, Yin C
and Lu G: An improved method for increasing the efficiency of the
technique of random amplified polymorphic DNA (RAPD). Hereditas.
22:251–252. 1999.
|
|
27
|
Fu J, Yang L, Khan MA and Mei Z: Genetic
characterization and authentication of Lonicera japonica Thunb. By
using improved RAPD analysis. Mol Biol Rep. 40:5993–5999.
2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Fu J: Short protocols in medical molecular
biology. China Medical Science Press, 2012.
|
|
29
|
Mei Z, Khan MA, Zhang X and Fu J: Rapid
and accurate genetic authentication of Penthorum chinense by
improved RAPD-derived species-specific SCAR markers. Biodiversitas.
18:1243–1249. 2017.
|
|
30
|
Rohlf FJ: NTSYS-pc: Numerical Taxonomy and
Multivariate Analysis System, version 2.1. Exeter Publishing,
Setauket, NY, 2000.
|
|
31
|
Robertson M and Richards C: Opportunities
and challenges of next-generation sequencing applications in
ecological epigenetics. Mol Ecol. 24:3799–37801. 2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Fu J, Cheng J, Liu X, Li J, Wei C, Zheng
X, He T and Fu J: Evaluation genotypes of cancer cell lines HCC1954
and SiHa by short tandem repeat (STR) analysis and DNA sequencing.
Mol Biol Rep. 45:2689–2695. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Mei Z, Zhang X, Liu X, Imani S and Fu J:
Genetic analysis of Canarium album in the different areas of China
by improved-RAPD and ISSR. CR Biol. 340:558–564. 2017.
|
|
34
|
Liang HY, Liu XJ and Yang MS: Analysis of
Lycium chinense variety revealed by RAMP-PCR method. Chinese
Agric Sci Bull. 26:61–64. 2011.
|