1. Introduction
A large-scale novel type of pneumonia, caused by
coronavirus disease 2019 (COVID-19), began in Wuhan, China at the
end of 2019, and it has affected numerous countries and regions
worldwide as of early June, 2020. The outbreak of COVID-19 was
caused by a coronavirus infection, similar to the prior infectious
disease outbreaks Severe acute respiratory syndrome (SARS) and
Middle East respiratory syndrome (MERS) (1,2).
According to the current epidemiological analysis, COVID-19 may be
far more infectious compared with SARS-CoV, while it has been
reported that this virus is transmitted by humans, albeit with
weaker pathogenicity than SARS-CoV (3,4). To
this end, governments worldwide have adopted strict measures for
the isolation and control of the disease in order to prevent its
further large-scale spread, including the lockdown of cities, the
isolation of suspected populations and the establishment of new
COVID-19 treatment hospitals (5).
However, this virus still poses a serious threat to human health.
Currently, in addition to numerous epidemiological studies
gradually revealing the possible origin, transmission routes and
characteristics of COVID-19, several preliminary studies have
investigated the histopathological changes, potential pathogenesis
and treatment approaches for COVID-19. The results of these studies
may provide an important theoretical basis for the understanding
and prevention of COVID-19 infection. Furthermore, an in-depth
study of COVID-19 by scientists of different specialties worldwide
may gradually lead to the elucidation of the pathophysiology of the
disease.
2. Transmission and origin
The World Health Organization (WHO) officially named
the novel coronavirus pneumonia as COVID-19(6). On December 30, 2019, the Wuhan Health
Commission of China issued an emergency notice stating that
patients with pneumonia of unknown origin were admitted to various
hospitals in the country, which immediately attracted the attention
of the Chinese government, which in turn appointed relevant experts
to investigate and analyze this newly identified type of pneumonia.
It was finally confirmed that this type of pneumonia was caused by
a virus. Based on information obtained from patients, it was
initially considered that this virus could have originated from the
Wuhan HuaNan Seafood Market. Chinese scholars gradually realized
that the virus was rapidly spreading among humans (3). The number of infected individuals
significantly increased within a short period of time, and
pneumonia cases were first reported outside China in mid-January,
2020. At the end of January, 2020, the Chinese government imposed
stricter measures in order to prevent the epidemic from spreading
further, and organized the country's resources in the fight against
this novel coronavirus-associated pneumonia. In mid-to-late
February, 2020, the number of new pneumonia cases was gradually
declining in China, while the number of patients discharged from
hospital was gradually increasing. In early March, with the
exception of Wuhan, the number of suspicious infections in China
stabilized, and the epidemic was considered to be effectively
controlled. However, the COVID-19 outbreak is significantly more
severe compared with the previous SARS outbreak. By August, 2020,
according to the report of the Health Commission (7), the total number of COVID-19 infection
cases in China exceeded 90,000, with >4,700 deaths. Although
this disease has had a relatively serious negative impact on the
Chinese population, this was markedly restricted due to the
successful strict measures imposed by the Chinese government.
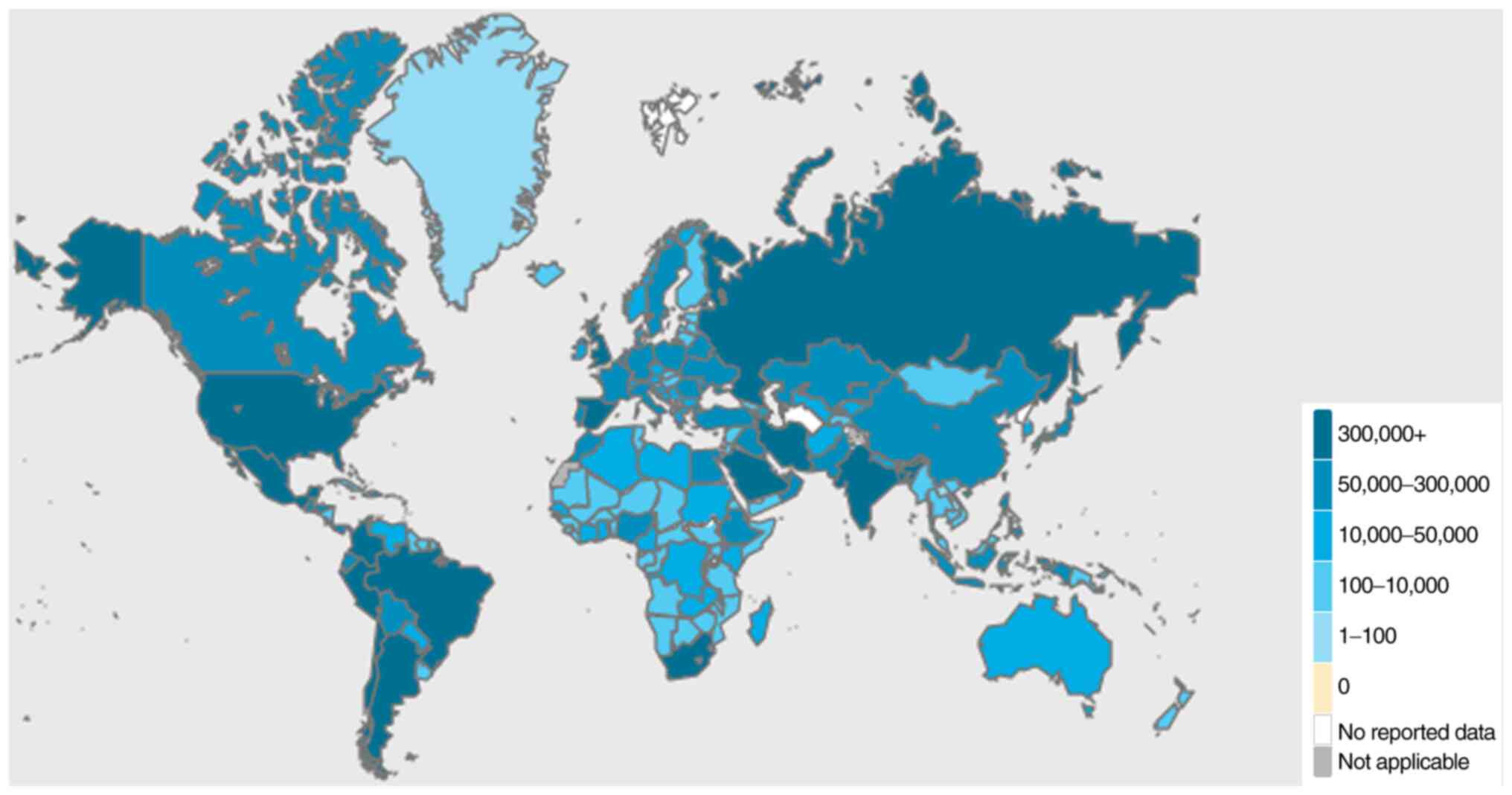
As regards countries other than China, Thailand
confirmed the first imported case of COVID-19 as early as January,
2020(8). By February, 2020, 24
countries globally had reported confirmed cases of
COVID-19(9). Due to the initial
lack of awareness regarding the novel coronavirus-mediated
pneumonia, the number of cases in several countries rapidly
increased. As of August, 2020, the United States of America was the
country with the highest total number of confirmed cases (>6
million cases), while Brazil was the country with highest number of
confirmed cases per day (>50,000 cases). As regards Asia, India
had >3 million individuals diagnosed with COVID-19, while ~1
million cases were confirmed in Russia. Ιn the African continent, a
total of 57 countries reported >1 million confirmed cases. At
the time of the writing of the present review article, 13 countries
in Africa had reported >10,000 confirmed cases of COVID-19,
among which South Africa, Egypt and Nigeria were shown to have a
markedly increased incidence of the disease (Fig. 1).
The similarity comparison and mutation site analysis
of the COVID-19 genome, obtained from the 2019 novel coronavirus
resource library released by the National Genomics Scientific Data
Center, revealed that the sequence similarity between SARS-CoV-2
(COVID-19) and SARS-CoV, responsible for the 2003 outbreak, was
80%, whereas it was ~50% for MERS-CoV. In addition, SARS-CoV-2 was
clustered with SARS-CoVs in the phylogenetic tree of SARS-related
coronaviruses (10-12).
SARS-CoV-2 belongs to the ‘Coronavirus’ family, the ‘β-coronavirus’
genus, and the ‘severe acute respiratory syndrome-associated
coronavirus’ species, which also includes SARS-CoV (13). Based on currently available
analyses, the COVID-19 appears to be more infectious than SARS-CoV
(3). In addition, it has been
reported that COVID-19 exhibits the highest similarity (88%) with
the genomic sequence of bat-isolated SARS-like coronavirus
(bat-SL-CoVZC45). Bat-SL-CoVZC45 was first isolated from domestic
bats in February, 2017 (14,15).
Furthermore, SARS-CoV-2 is also closely associated with a type of
coronavirus isolated from bats, namely RaGT13-CoV, with a
nucleotide identity of 96%, thus indicating that SARS-CoV-2 may
have also originated from bats. However, whether SARS-CoV-2 was
transmitted directly from bats to humans or through intermediate
hosts remains unclear (16). Some
studies have suggested that pangolins may also be the host of
SARS-CoV-2. Notably, SARS-CoV-2 encompasses a unique peptide (PRRA)
insertion; however, this element is lacking from the
pangolin-carried coronavirus (17). Therefore, further in-depth research
into the identification of the virus hosts may provide the
necessary knowledge for preventing these diseases.
3. Diagnosis
In the context of the current SARS-CoV-2 pandemic,
accurate and rapid diagnostic tests are crucial for the detection
of COVID-19 infection. With the identification of the COVID-19
virus sequence, reverse transcription-quantitative PCR (RT-qPCR)
analysis and gene sequencing have been recommended for the
diagnosis of COVID-19(18).
However, due to the urgency of the epidemic and since gene
sequencing is a time-consuming method, this approach is less
frequently adopted. Currently, the most commonly used clinical
diagnostic tests for COVID-19 are divided into 3 categories:
Molecular testing for the detection of viral RNA (RT-qPCR);
serological testing for the detection of anti-SARS-CoV-2
immunoglobulins; and computed tomography (CT). Several
manufacturers have developed and produced different diagnostic kits
for COVID-19, and have gradually optimized the detection efficiency
and detection time. The virus is mainly detected in throat swabs,
sputum, alveolar lavage fluid, serum and plasma (19).
RT-qPCR, used to detect viral RNA, is considered as
the ‘standard diagnostic tool’. A disadvantage of RT-qPCR testing
is the risk of false-negative and false-positive results. Emerging
evidence has suggested that SARS-CoV-2 is characterized by genetic
diversity and rapid evolution. Therefore, the results of RT-qPCR
using primers for different genes may be affected by changes in the
viral RNA sequence (20,21). Although RT-qPCR assays are designed
to be as accurate as possible based on the conserved regions of the
viral genome, genome variability may cause mismatches between the
primer/probes and the target sequences may lead to decreased
accuracy and potential false-negative results (22). The kinetics of the viral load may
also lead to false-negative results to a large extent. Currently,
rapid and efficient nasal and throat swabs are recommended for
sample collection (23). The
Coronavirus Standards Working Group (https://jimb.stanford.edu/covid-19-standards) led by
the Joint Biometrics Project has developed a set of guidelines to
ensure the accuracy of the diagnostic test results (24).
Compared with tests using nucleic acid, serological
testing requires less technical knowledge and equipment, and is
considered as an easy method to perform. The majority of
serological assays are based on SARS-CoV-2 nucleocapsid protein
(N), transmembrane spike protein (S) or S receptor-binding domain
(RBD), due to their high antigenicity (25,26).
The spike (S) protein on the surface of the virus mediates the
adhesion of the virus to human respiratory cells through its RBD,
which, in turn, promotes the fusion of the virus with the cell
membrane. A previous study demonstrated that individuals who were
not exposed to SARS-CoV-2 had no spike protein at all, and their
serum samples showed little or no response in ELISA (27). In another study, the seroconversion
of IgM and IgG occurred simultaneously or sequentially, and the
median interval for seroconversion for both immunoglobulins was 13
days following the onset of COVID-19, and 19 days report the onset
of symptoms (28). Additionally,
the IgG seroconversion rate reached 100%, and the sensitivity test
range for IgM and IgG was between 72.7 and 100%, while the
specificity range was between 98.7 and 100%, respectively. When
IgM/IgG ELISA was combined with PCR, the detection rate was
significantly improved compared with PCR alone (98.6 vs. 51.9%)
(29,30). However, serological tests are
affected by several factors, such as specimens, reagents and
operations; therefore, false-negative results may occur, affecting
the clinical treatment and prevention of the infection.
CT examination is also an important method (31,32),
which may help clarify the false-negative results in the detection
of COVID-19 obtained by PCR and serological examinations. During or
outside the COVID-19 incubation period, a CT scan of infected
patients may reveal some characteristic lesions in their lungs that
are indicative of infection. The most common chest CT manifestation
is bilateral ground-glass opacity (GGO) (33). This is defined as a hazy area in
the lung that exhibits a slight increase in density, without
blurring of the edges of the bronchi and blood vessels, which is
caused by the partial displacement of air promoted by the partial
filling of the alveolar cavity or the thickening of the gap
(34). The presence of GGO usually
indicates pulmonary edema or hyaline membrane formation (33). In addition, the lung CT examination
of patients with COVID-19 may also reveal a reticular pattern,
crazy paving pattern and the air bubble sign (35-37).
The pathological fluid covers the edges of the underlying blood
vessels and airway walls and increases the density of the lung
parenchyma, which is referred to as consolidation (38). In patients with COVID-19,
consolidation may be associated with cellulose and mucus exudates
in alveolar cells (39). It was
recently reported that consolidation is considered a sign of
disease progression, while GGO in a lung CT scan is considered as
an early finding of SARS-CoV-2 infection (40). GGO may evolve into consolidation
after 1-3 weeks (33). In patients
with severe COVID-19 infection, multifocal or large-area sheet
consolidation may appear in the lungs, which is characterized by
large white areas on lung CT images, also referred to as ‘white
lungs’ (41). All the
aforementioned detection techniques are associated with
shortcomings; therefore, in order to improve the detection rate of
COVID-19, clinicians may opt to use a combination of these methods.
Patients with ‘white lungs’ are usually infected within their
familiar environment and, although they may not have been tested or
tested negative for COVID-19 by RT-qPCR analysis, some researchers
refer to such patients as ‘clinical diagnostic cases’. Although
these patients are not really confirmed cases, they are recorded as
confirmed or suspected cases, requiring high caution and immediate
isolation (42).
4. Pathology and mechanisms
Understanding the pathological changes of the
disease is a prerequisite for its treatment. A pathological
examination of the lungs of patients with COVID-19 revealed that
the lung tissue was grossly affected by diffuse congestion and
partial necrosis, while the bronchi were lined with a large amount
of mucus and exudate (43-47).
Microscopic analysis also revealed extensive hemorrhage,
infarction, pulmonary interstitial fibrosis, extensive type I
alveolar epithelial cell damage and atypical proliferation of type
II alveolar cells, hyaline membrane formation, exudation, pulmonary
edema and consolidation. Additionally, a large number of
inflammatory cells, including lymphocytes and plasma cells,
infiltrated the lung tissue, while increased vascular proliferation
was also observed. Several studies have also reported other
characteristics of the lungs of patients with COVID-19, such as the
loss and squamous metaplasia of alveolar epithelial cells and a
large amount of cellulose-like exudate in the alveoli. Other
pathological analyses demonstrated that patients with COVID-19
exhibited structural damage to the lung tissue and a large amount
of mucus and exudate blocking the airway lumen and alveoli, thus
leading to severe respiratory failure and insufficient spontaneous
breathing. Therefore, for patients with severe COVID-19, even the
use of extracorporeal assisted ventilation is ineffective. The
primary function of the lungs is to rely on normal physiological
movements to maintain the body's normal oxygen supply based on the
inhalation and discharge of oxygen or carbon dioxide (43-47).
Varga et al reported for the first time that patients with
COVID-19 exhibited endothelial cell damage in multiple organs,
including the heart, kidney, lung and small intestine, thus
indicating that SARS-CoV-2 infection may facilitate the induction
of endotheliitis, apoptosis and pyroptosis in several organs, as a
direct consequence of the host's inflammatory response (48). Necrotic lymphocyte infiltration,
interstitial edema and fibrosis have also been found in the
gastrointestinal tract (49),
kidneys (50) and liver (51) of patients, while axonal damage has
been observed in brain nerve cells (52). Furthermore, in several patients,
COVID-19 has been found to be accompanied by viral rashes and
vascular endodermatitis of the skin and blood vessels, respectively
(48,53). The aforementioned reports indicate
that lung injury is the most common lesion of COVID-19; however,
the damage is not limited to the lungs, suggesting that COVID-19 is
a systemic disease characterized by multiorgan injury.
It has been reported that the cytokine storm
(54) and angiotensin-converting
enzyme 2 (ACE-2) may be the potential mechanisms underlying the
aforementioned pathological changes (5,55).
The overactive immune responses of the host against the
SARS-CoV-2-infection may lead to excessive and aggressive
inflammatory responses, eventually leading to the release of a
large amount of pro-inflammatory cytokines, a process referred to
as cytokine storm (56). As
regards the innate immune responses following virus infection, the
pattern recognition receptor (PRR) recognizes the conserved
molecular structures of the invading virus, namely the
pathogen-associated molecular pattern (PAMP). The combination of
PAMP and PRR triggers the activation of a variety of signaling
pathways and transcription factors, which in turn induce the
expression of several genes associated with the immune responses
against viral infections, such as pro-inflammatory cytokines
(56,57). Macrophages, dendritic cells,
endothelial cells, natural killer cells and T and B lymphocytes are
the main immune cell subtypes responsible for the secretion of
cytokines. In addition, interleukin (IL)-1, IL-6, chemokines and
tumor necrosis factor-α (TNF-α) are mainly involved in the
inflammatory responses (58). The
normal and adequate release of cytokines is necessary for the human
body to be able to resist against various pathogens. In patients
with COVID-19, the balance of the secreted cytokines is disrupted.
Therefore, the increased secretion of cytokines within a short
period of time may cause damage to tissue endothelial cells, blood
vessels and alveoli and, may eventually, lead to organ failure
(15,56).
ACE-2 is expressed on bronchial and alveolar
epithelial cells, and it is also widely distributed in the
gastrointestinal tract, brain, heart and other organs (59). Recent studies have demonstrated
that both SARS-CoV-2 and SARS-CoV have ACE-2 cell invasion
receptors (5,11); therefore, it is likely to cause
acute lung injury by binding to the ACE-2 like SARS-CoV. The
S-protein of the coronavirus contains two functional units, namely
S1 and S2. S1 contains the RBD, which directly binds to the
coronavirus host receptor ACE-2. S2 is responsible for the fusion
of the virus with the host cell membrane. When S1 binds to the host
ACE-2 receptor, the cleavage site of S2 is exposed, producing lytic
host protease (60). The
expression of ACE-2 is downregulated in SARS-CoV-2-infected cells
(59). ACE-2 is a known peptidase
that regulates the renin-angiotensin-aldosterone system, thereby
controlling blood pressure. It has been reported that hypertension
and cardiovascular diseases are the most common complications of
COVID-19(61).
5. Clinical symptoms and treatment
According to the current epidemiological data,
SARS-CoV-2 is mainly transmitted through respiratory droplets and
close human contact. Increased amounts of aerosols and human
excreta are also considered as potential routes of transmission.
The incubation period of COVID-19 is estimated to be 1-14 days,
with an average of 3-7 days (15).
Furthermore, infected patients are primarily characterized by
symptoms of the upper respiratory tract, such as fever (highest
incidence), cough and runny nose. In addition, diarrhea and nervous
system abnormalities are often observed (62,63).
Therefore, individuals with the aforementioned symptoms residing in
areas with a high incidence of viral pneumonia are considered as
high-risk individuals. The majority of adults or children infected
with SARS-CoV-2 develop mild flu-like symptoms, while a small
number of patients, particularly those with cardiovascular diseases
and diabetes, are prone to rapid acute respiratory distress
syndrome, respiratory failure, multiple organ failure, or even
death (64).
COVID-19 is a severe infectious disease, and active
isolation measures are a prerequisite for all types of treatment.
Therefore, the interruption of the transmission route, the
protection of susceptible individuals and the active isolation of
the virus carriers are crucial. At present, numerous countries
worldwide, including China as well as countries first affected by
the pandemic, are adopting strict quarantine measures, including
the imposition of short-term lockdowns. Patients diagnosed with
COVID-19 are treated individually or collectively. Patients with
COVID-19 are most commonly treated with the timely inhalation of
effective hydrogen-oxygen mixture and antiviral therapy (65). Body temperature and oxygen
saturation should be recorded regularly. In patients with severe
COVID-19 infection, invasive mechanical ventilation or
extracorporeal membrane oxygenation may also be applied (66). In addition, treatment with plasma
isolated from recovered patients is also considered an effective
treatment approach (67). Among
conventional drugs, antiviral agents, such as oseltamivir and
acyclovir, and systemic glucocorticoids, such as
methylprednisolone, have been also used in the treatment of
patients with COVID-19. However, their effectiveness is
questionable (68).
Chloroquine/hydroxychloroquine, two antimalarial agents, can alter
the pH of cells and are stored in lysosomes in a protonated form.
It has been reported that these compounds weaken the ability of the
virus to release its genetic material into the cell and replicate
(69,70). Studies have demonstrated that the
combination of chloroquine/hydroxychloroquine, remdesivir and
azithromycin has shown great promise in the treatment of COVID-19
(71,72). Another study revealed that the
viral load was significantly reduced in patients treated with
lopinavir and ritonavir (73).
Based on the mechanism through which ACE2 mediates the invasion of
the human body by SARS-CoV-2, captopril is considered an inhibitor
of ACE2, which acts by attenuating the inflammatory reactions in
severely ill patients (72). The
immunopathology of COVID-19 is characterized by lymphopenia and
lymphocyte dysfunction (74). In
response to the immune characteristics of COVID-19, some
researchers have proposed several immunotherapeutic strategies,
such as enhancing the activity of lymphocytes or inhibiting
inflammation. NK cell-based therapies and immunomodulators are used
to enhance the activity of lymphocytes. To suppress inflammation,
mesenchymal stem cell-based therapies, regulatory T-cell-based
therapies and other strategies may be used (75).
The S protein on the surface of the coronavirus
binds to the target protein that invades the surface of the
receptor, thereby mediating the release of the viral genome into
the host cell for replication (60). Therefore, S protein has been used
as an immunogen for the development of antibodies (76). Compared with small-molecule drugs,
monoclonal antibodies are relatively costly and more difficult to
produce. However, they differ from other drugs, since they can
engage the host immune system through the binding of their constant
domains to the Fc gamma receptors on host immune cells. Antibodies
are key components of most vaccines and will likely prove crucial
for the development of an effective vaccine against SARS-CoV-2. It
has been reported that vaccines that selectively induce the
production of antibodies targeting the RBD of SARS-CoV-2 may be
particularly effective (25).
Currently, >100 vaccines against COVID-19 are under research,
including traditional live attenuated viral vaccines, inactivated
viral vaccines, vector-based vaccines, as well as a new generation
of safer recombinant-protein vaccines (77). Unfortunately, none of these has
been approved for large-scale clinical application.
An important feature in the landscape of vaccine
research and development for SARS-COV-2 is represented by the
varied range of evaluated technological platforms, including
nucleic acids (DNA and RNA), virus-like particles, peptides, viral
vector (replicative and non-replicative), recombinant proteins,
live attenuated viruses and inactivated viruses (78), potential vaccines must also pass
the same clinical trial phase. This is especially important when it
comes to safety issues, even during a pandemic.
6. Summary
SARS that erupted 17 years ago posed a threat to
human health, and the current spread of COVID-19 has again brought
feelings of fear. With the continuous research of scholars
worldwide, a basic understanding of the transmission mode,
pathological characteristics and potential pathogenesis of COVID-19
has now been acquired. Methods have also been developed to rapidly
identify the virus, and various treatment measures have been
systematically evaluated. This has strengthened the confidence of
researchers and has provided hope that COVID-19 will be defeated.
At present, some countries, including China, the United States, and
the United Kingdom have increased the speed of vaccine research
through open green channels, and some vaccines that have undergone
clinical trials are beginning to be administered. However, judging
by the current global pandemic trend, the threat of SARS-CoV-2
infection may continue for a long time to come.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
JL designed the theme of the present review. YS, XS,
HH and YL retrieved the relevant literature. XS wrote and reviewed
the article. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Meo SA, Alhowikan AM, Al-Khlaiwi T, Meo
IM, Halepoto DM, Iqbal M, Usmani AM, Hajjar W and Ahmed N: Novel
coronavirus 2019-nCoV: Prevalence, biological and clinical
characteristics comparison with SARS-CoV and MERS-CoV. Eur Rev Med
Pharmacol Sci. 24:2012–2019. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hui DS, Memish ZA and Zumla A: Severe
acute respiratory syndrome vs. the Middle East respiratory
syndrome. Curr Opin Pulm Med. 20:233–241. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chen J: Pathogenicity and transmissibility
of 2019-nCoV-A quick overview and comparison with other emerging
viruses. Microbes Infect. 22:69–71. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zhu N, Zhang D, Wang W, Li X, Yang B, Song
J, Zhao X, Huang B, Shi W, Lu R, et al: A novel coronavirus from
patients with pneumonia in China, 2019. N Engl J Med. 382:727–733.
2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kannan S, Shaik Syed Ali P, Sheeza A and
Hemalatha K: COVID-19 (Novel Coronavirus 2019)-recent trends. Eur
Rev Med Pharmacol Sci. 24:2006–2011. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wu YC, Chen CS and Chan YJ: The outbreak
of COVID-19: An overview. J Chin Med Assoc. 83:217–220.
2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Xu W, Wu J and Cao L: COVID-19 pandemic in
China: Context, experience and lessons. Health Policy Technol.
9:639–648. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sookaromdee P and Wiwanitkit V: Imported
cases of 2019-novel coronavirus (2019-nCoV) infections in Thailand:
Mathematical modelling of the outbreak. Asian Pac J Trop Med.
13(139)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Harapan H, Itoh N, Yufika A, Winardi W,
Keam S, Te H, Megawati D, Hayati Z, Wagner AL and Mudatsir M:
Coronavirus disease 2019 (COVID-19): A literature review. J Infect
Public Health. 13:667–673. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Benvenuto D, Giovanetti M, Salemi M,
Prosperi M, De Flora C, Junior Alcantara LC, Angeletti S and
Ciccozzi M: The global spread of 2019-nCoV: A molecular
evolutionary analysis. Pathog Glob Health. 114:64–67.
2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhou P, Yang XL, Wang XG, Hu B, Zhang L,
Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al: A pneumonia outbreak
associated with a new coronavirus of probable bat origin. Nature.
579:270–273. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Coronaviridae Study Group of the
International Committee on Taxonomy of Viruses. The species Severe
acute respiratory syndrome-related coronavirus: Classifying
2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 5:536–544.
2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Schoeman D and Fielding BC: Coronavirus
envelope protein: Current knowledge. Virol J. 16(69)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H,
Wang W, Song H, Huang B, Zhu N, et al: Genomic characterisation and
epidemiology of 2019 novel coronavirus: Implications for virus
origins and receptor binding. Lancet. 395:565–574. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Lai CC, Shih TP, Ko WC, Tang HJ and Hsueh
PR: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
and coronavirus disease-2019 (COVID-19): The epidemic and the
challenges. Int J Antimicrob Agents. 55(105924)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
da Silva SJR, Silva CTAD, Guarines KM,
Mendes RPG, Pardee K, Kohl A and Pena L: Clinical and laboratory
diagnosis of SARS-CoV-2, the virus causing COVID-19. ACS Infect
Dis. 6:2319–2336. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li X, Zai J, Zhao Q, Nie Q, Li Y, Foley BT
and Chaillon A: Evolutionary history, potential intermediate animal
host, and cross-species analyses of SARS-CoV-2. J Med Virol.
92:602–611. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chan JF, Yip CC, To KK, Tang TH, Wong SC,
Leung KH, Fung AY, Ng AC, Zou Z, Tsoi HW, et al: Improved molecular
diagnosis of COVID-19 by the novel, highly sensitive and specific
COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain
reaction assay validated in vitro and with clinical specimens. J
Clin Microbiol. 58:e00310–20. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ling Y, Xu SB, Lin YX, Tian D, Zhu ZQ, Dai
FH, Wu F, Song ZG, Huang W, Chen J, et al: Persistence and
clearance of viral RNA in 2019 novel coronavirus disease
rehabilitation patients. Chin Med J (Engl). 133:1039–1043.
2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Phan T: Genetic diversity and evolution of
SARS-CoV-2. Infect Genet Evol. 81(104260)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang
L, Zhou Z, Yang J, Zhong J, Yang D, et al: Genomic diversity of
severe acute respiratory syndrome-coronavirus 2 in patients with
coronavirus disease 2019. Clin Infect Dis. 71:713–720.
2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Tahamtan A and Ardebili A: Real-time
RT-PCR in COVID-19 detection: Issues affecting the results. Expert
Rev Mol Diagn. 20:453–454. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kim JY, Ko JH, Kim Y, Kim YJ, Kim JM,
Chung YS, Kim HM, Han MG, Kim SY and Chin BS: Viral load kinetics
of SARS-CoV-2 infection in first two patients in Korea. J Korean
Med Sci. 35(e86)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bustin SA and Nolan T: RT-qPCR testing of
SARS-CoV-2: A primer. Int J Mol Sci. 21(3004)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Robbiani DF, Gaebler C, Muecksch F,
Lorenzi JCC, Wang Z, Cho A, Agudelo M, Barnes CO, Gazumyan A,
Finkin S, et al: Convergent antibody responses to SARS-CoV-2 in
convalescent individuals. Nature. 584:437–442. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Stadlbauer D, Amanat F, Chromikova V,
Jiang K, Strohmeier S, Arunkumar GA, Tan J, Bhavsar D, Capuano C,
Kirkpatrick E, et al: SARS-CoV-2 seroconversion in humans: A
detailed protocol for a serological assay, antigen production, and
test setup. Curr Protoc Microbiol. 57(e100)2020.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Amanat F, Stadlbauer D, Strohmeier S,
Nguyen THO, Chromikova V, McMahon M, Jiang K, Arunkumar GA,
Jurczyszak D, Polanco J, et al: A serological assay to detect
SARS-CoV-2 seroconversion in humans. Nat Med. 26:1033–1036.
2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Van Elslande J, Houben E, Depypere M,
Brackenier A, Desmet S, André E, Van Ranst M, Lagrou K and
Vermeersch P: Diagnostic performance of seven rapid IgG/IgM
antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19
patients. Clin Microbiol Infect. 26:1082–1087. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zainol Rashid Z, Othman SN, Abdul Samat
MN, Ali UK and Wong KK: Diagnostic performance of COVID-19 serology
assays. Malays J Pathol. 42:13–21. 2020.PubMed/NCBI
|
|
30
|
Guo L, Ren L, Yang S, Xiao M, Chang D,
Yang F, Dela Cruz CS, Wang Y, Wu C, Xiao Y, et al: Profiling early
humoral response to diagnose novel coronavirus disease (COVID-19).
Clin Infect Dis. 71:778–785. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhu Y, Liu YL, Li ZP, Kuang JY, Li XM,
Yang YY and Feng ST: Clinical and CT imaging features of 2019 novel
coronavirus disease (COVID-19). J Infect: Mar 3, 2020 doi:
10.1016/j.jinf.2020.02.022 (Epub ahead of print).
|
|
32
|
Xie C, Jiang L, Huang G, Pu H, Gong B, Lin
H, Ma S, Chen X, Long B, Si G, et al: Comparison of different
samples for 2019 novel coronavirus detection by nucleic acid
amplification tests. Int J Infect Dis. 93:264–267. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ye Z, Zhang Y, Wang Y, Huang Z and Song B:
Chest CT manifestations of new coronavirus disease 2019 (COVID-19):
A pictorial review. Eur Radiol. 30:4381–4389. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Franquet T: Imaging of pulmonary viral
pneumonia. Radiology. 260:18–39. 2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Song F, Shi N, Shan F, Zhang Z, Shen J, Lu
H, Ling Y, Jiang Y and Shi Y: Emerging 2019 novel coronavirus
(2019-nCoV) pneumonia. Radiology. 295:210–217. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Pan F, Ye T, Sun P, Gui S, Liang B, Li L,
Zheng D, Wang J, Hesketh RL, Yang L and Zheng C: Time course of
lung changes at chest CT during recovery from coronavirus disease
2019 (COVID-19). Radiology. 295:715–721. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yoon SH, Lee KH, Kim JY, Lee YK, Ko H, Kim
KH, Park CM and Kim YH: Chest radiographic and CT findings of the
2019 novel coronavirus disease (COVID-19): Analysis of nine
patients treated in Korea. Korean J Radiol. 21:494–500.
2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Hansell DM, Bankier AA, MacMahon H, McLoud
TC, Müller NL and Remy J: Fleischner Society: Glossary of terms for
thoracic imaging. Radiology. 246:697–722. 2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Xu Z, Shi L, Wang Y, Zhang J, Huang L,
Zhang C, Liu S, Zhao P, Liu H, Zhu L, et al: Pathological findings
of COVID-19 associated with acute respiratory distress syndrome.
Lancet Respir Med. 8:420–422. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu
GM and Zhang LJ: Coronavirus disease 2019 (COVID-19): A perspective
from China. Radiology. 296:E15–E25. 2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Pan Y and Guan H: Imaging changes in
patients with 2019-nCov. Eur Radiol. 30:3612–3613. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Xu YH, Dong JH, An WM, Lv XY, Yin XP,
Zhang JZ, Dong L, Ma X, Zhang HJ and Gao BL: Clinical and computed
tomographic imaging features of novel coronavirus pneumonia caused
by SARS-CoV-2. J Infect. 80:394–400. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Liu J, Zheng X, Tong Q, Li W, Wang B,
Sutter K, Trilling M, Lu M, Dittmer U and Yang D: Overlapping and
discrete aspects of the pathology and pathogenesis of the emerging
human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J
Med Virol. 92:491–494. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Tian S, Hu W, Niu L, Liu H, Xu H and Xiao
SY: Pulmonary pathology of early phase 2019 novel coronavirus
(COVID-19) pneumonia in two patients with lung cancer. J Thorac
Oncol. 15:700–704. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Yao XH, Li TY, He ZC, Ping YF, Liu HW, Yu
SC, Mou HM, Wang LH, Zhang HR, Fu WJ, et al: A pathological report
of three COVID-19 cases by minimal invasive autopsies. Zhonghua
Bing Li Xue Za Zhi. 49:411–417. 2020.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
46
|
Wang C, Xie J, Zhao L, Fei X, Zhang H, Tan
Y, Nie X, Zhou L, Liu Z, Ren Y, et al: Alveolar macrophage
dysfunction and cytokine storm in the pathogenesis of two severe
COVID-19 patients. EBioMedicine. 57(102833)2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Calabrese F, Pezzuto F, Fortarezza F,
Hofman P, Kern I, Panizo A, von der Thüsen J, Timofeev S,
Gorkiewicz G and Lunardi F: Pulmonary pathology and COVID-19:
Lessons from autopsy. The experience of European Pulmonary
Pathologists. Virchows Arch. 477:359–372. 2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Varga Z, Flammer AJ, Steiger P, Haberecker
M, Andermatt R, Zinkernagel AS, Mehra MR, Schuepbach RA, Ruschitzka
F and Moch H: Endothelial cell infection and endotheliitis in
COVID-19. Lancet. 395:1417–1418. 2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Xiao F, Tang M, Zheng X, Liu Y, Li X and
Shan H: Evidence for gastrointestinal infection of SARS-CoV-2.
Gastroenterology. 158:1831–1833.e3. 2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Su H, Yang M, Wan C, Yi LX, Tang F, Zhu
HY, Yi F, Yang HC, Fogo AB, Nie X and Zhang C: Renal
histopathological analysis of 26 postmortem findings of patients
with COVID-19 in China. Kidney Int. 98:219–227. 2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Tabary M, Khanmohammadi S, Araghi F,
Dadkhahfar S and Tavangar SM: Pathologic features of COVID-19: A
concise review. Pathol Res Pract. 216(153097)2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Briguglio M, Bona A, Porta M, Dell'Osso B,
Pregliasco FE and Banfi G: Disentangling the hypothesis of host
dysosmia and SARS-CoV-2: The bait symptom that hides neglected
neurophysiological routes. Front Physiol. 11(671)2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Suchonwanit P, Leerunyakul K and
Kositkuljorn C: Cutaneous manifestations in COVID-19: Lessons
learned from current evidence. J Am Acad Dermatol. 83:e57–e60.
2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Chen C, Zhang XR, Ju ZY and He WF:
Advances in the research of cytokine storm mechanism induced by
Corona Virus Disease 2019 and the corresponding immunotherapies.
Zhonghua Shao Shang Za Zhi. 36(E005)2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Nitulescu GM, Paunescu H, Moschos SA,
Petrakis D, Nitulescu G, Ion GND, Spandidos DA, Nikolouzakis TK,
Drakoulis N and Tsatsakis A: Comprehensive analysis of drugs to
treat SARS-CoV-2 infection: Mechanistic insights into current
COVID-19 therapies (Review). Int J Mol Med. 46:467–488.
2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Ragab D, Salah Eldin H, Taeimah M, Khattab
R and Salem R: The COVID-19 Cytokine Storm; What We Know So Far.
Front Immunol. 11(1446)2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Thompson MR, Kaminski JJ, Kurt-Jones EA
and Fitzgerald KA: Pattern recognition receptors and the innate
immune response to viral infection. Viruses. 3:920–940.
2011.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Coperchini F, Chiovato L, Croce L, Magri F
and Rotondi M: The cytokine storm in COVID-19: An overview of the
involvement of the chemokine/chemokine-receptor system. Cytokine
Growth Factor Rev. 53:25–32. 2020.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Zhang X, Li S and Niu S: ACE2 and COVID-19
and the resulting ARDS. Postgrad Med J. 96:403–407. 2020.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Tortorici MA and Veesler D: Structural
insights into coronavirus entry. Adv Virus Res. 105:93–116.
2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Devaux CA, Rolain JM and Raoult D: ACE2
receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension,
multi-organ failure, and COVID-19 disease outcome. J Microbiol
Immunol Infect. 53:425–435. 2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Gu J, Han B and Wang J: COVID-19:
Gastrointestinal manifestations and potential fecal-oral
transmission. Gastroenterology. 158:1518–1519. 2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Li YC, Bai WZ and Hashikawa T: The
neuroinvasive potential of SARS-CoV2 may be at least partially
responsible for the respiratory failure of COVID-19 patients. J Med
Virol. 92:552–555. 2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu
Y, Zhang L, Fan G, Xu J, Gu X, et al: Clinical features of patients
infected with 2019 novel coronavirus in Wuhan, China. Lancet.
395:497–506. 2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Li H, Wang YM, Xu JY and Cao B: Potential
antiviral therapeutics for 2019 Novel Coronavirus. Zhonghua Jie He
He Hu Xi Za Zhi. 43(E002)2020.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
66
|
MacLaren G, Fisher D and Brodie D:
Preparing for the most critically ill patients with COVID-19: The
potential role of extracorporeal membrane oxygenation. JAMA.
323:1245–1246. 2020.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Chen L, Xiong J, Bao L and Shi Y:
Convalescent plasma as a potential therapy for COVID-19. Lancet
Infect Dis. 20:398–400. 2020.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD,
Jin HJ, Tan KS, Wang DY and Yan Y: The origin, transmission and
clinical therapies on coronavirus disease 2019 (COVID-19)
outbreak-an update on the status. Mil Med Res. 7(11)2020.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Becker RC: Covid-19 treatment update:
Follow the scientific evidence. J Thromb Thrombolysis. 50:43–53.
2020.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Dehelean CA, Lazureanu V, Coricovac D,
Mioc M, Oancea R, Marcovici I, Pinzaru I, Soica C, Tsatsakis AM and
Cretu O: SARS-CoV-2: Repurposed drugs and novel therapeutic
approaches-insights into chemical structure-biological activity and
toxicological screening. J Clin Med. 9(2084)2020.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Gautret P, Lagier JC, Parola P, Hoang VT,
Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Vieira VE,
et al: Hydroxychloroquine and azithromycin as a treatment of
COVID-19: Results of an open-label non-randomized clinical trial.
Int J Antimicrob Agents. 56(105949)2020.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Serafin MB, Bottega A, Foletto VS, da Rosa
TF, Hörner A and Hörner R: Drug repositioning is an alternative for
the treatment of coronavirus COVID-19. Int J Antimicrob Agents.
55(105969)2020.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Lim J, Jeon S, Shin HY, Kim MJ, Seong YM,
Lee WJ, Choe KW, Kang YM, Lee B and Park SJ: Case of the index
patient who caused tertiary transmission of COVID-19 Infection in
Korea: The application of lopinavir/ritonavir for the treatment of
COVID-19 infected pneumonia monitored by quantitative RT-PCR. J
Korean Med Sci. 35(e79)2020.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Lippi G and Plebani M: Laboratory
abnormalities in patients with COVID-2019 infection. Clin Chem Lab
Med. 58:1131–1134. 2020.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Yang L, Liu S, Liu J, Zhang Z, Wan X,
Huang B, Chen Y and Zhang Y: COVID-19: Immunopathogenesis and
Immunotherapeutics. Signal Transduct Target Ther.
5(128)2020.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Casadevall A and Pirofski LA: The Ebola
epidemic crystallizes the potential of passive antibody therapy for
infectious diseases. PLoS Pathogens. 11(e1004717)2015.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Wang J, Peng Y, Xu H, Cui Z and Williams
RO III: The COVID-19 vaccine race: Challenges and opportunities in
vaccine formulation. AAPS PharmSciTech. 21(225)2020.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Calina D, Docea AO, Petrakis D, Egorov AM,
Ishmukhametov AA, Gabibov AG, Shtilman MI, Kostoff R, Carvalho F,
Vinceti M, et al: Towards effective COVID-19 vaccines: Updates,
perspectives and challenges (Review). Int J Mol Med. 46:3–16.
2020.PubMed/NCBI View Article : Google Scholar
|