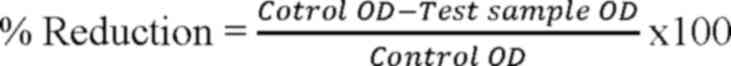Introduction
The most frequent growth pattern used by bacteria,
the biofilm, is now understood to have significant therapeutic
implications (1). Bacteria which
are in groups and that have created a self-made matrix and are
attached to a surface, as well as to one another are known as
bacterial biofilms. The biofilm matrix is composed of proteins
(such as fibrin), polysaccharides (such as alginate) and
extracellular DNA. Biofilms formed by bacteria can use a range of
ways of survival mechanisms that enable them to bypass the host's
defensive mechanisms in addition to the refuge provided by the
matrix (2,3). Treatment is made more difficult by
the development of bacterial biofilms as the illness progresses.
Enclosed cells inside the biofilm have distinct traits that lead to
an increase in antibiotic resistance, which is greater to that of
the planktonic condition, by 10 to 1,000-fold (4).
Antibiotic resistance is known as a serious health
issue and is mostly caused by the improper and excessive usage of
antibacterial substances (5).
Humans naturally acquire typhoidal and non-typhoidal
Salmonella from the environment or diet. Salmonella
can cause diarrheal disease, and can also create biofilms in the
intestines after being consumed by the host; Salmonella
Typhi can move into the gallbladder and develop biofilms on
cholesterol gallstones after obtaining access to the liver
systemically. These biofilms in humans enable both chronic
Salmonella infection and ongoing Salmonella host
shedding (6).
Antimicrobial proteins (AMPs), plant-derived
antimicrobial compounds, probiotics and bacteriophages as
biological alternatives to antibiotics are increasingly being
employed for the prevention and treatment of bacterial pathogenic
illnesses (7). Gram-negative
bacteria, including Escherichia coli (E. coli),
Klebsiella and Pseudomonas spp. produce AMPs known as
bacteriocins, which have narrow-spectrum action against
Gram-negative infections. Colicins, colicin-like bacteriocins,
microcins and phage tail-like bacteriocins are the four categories
into which they fall. Gram-negative bacteria that make bacteriocins
comparable to E. coli colicins in structure and function
include P. aeruginosa (e.g., S-type pyocins) and
Klebsiella spp. (8,9). Numerous types of bacteria create
bacteriocins, which are water-soluble protein toxins, in response
to nutritional deprivation and intra- and interspecies competition
(10). The bacteriocins produced
by Klebsiella are known as klebocins or klebicins (11). Klebocin is released from
Klebsiella pneumoniae (K. pneumoniae), and it appears
to exert effects against a variety of bacteria belonging to the
Enterobacteriaceae family (12).
Salmonella, Pseudomonas and E. coli are
examples of Gram-negative organisms that can be controlled by
bacteriocins, a class of non-antibiotic antibacterial proteins
(9). Klebocins are harmful for
Klebsiella species with a klebocinogenic plasmid that
carries the genetic components for klebocin synthesis, immunity and
release. Additionally, it was discovered that klebocins are
chromosomally encoded. The genetic analysis of the antibiotic
system of klebocin has demonstrated that it comprises proteins,
since it is expressed by distinct regulatory genes (13). The specific class of bacteriocins
known as klebocins exclusively exhibits homologous action, or
activity against bacteria that are closely related to one another
(14). By contrast, it has been
indicated that the antibacterial spectrum of klebocins from K.
pneumoniae is broad and unrestricted by the confines of the
genus or family (15). Thus, the
present study aimed to detect klebicin in K. pneumoniae
isolates and to evaluate their antimicrobial activity on biofilm
formation by other pathogenic bacteria, such as Salmonella
and Enterobacter.
Materials and methods
Isolation and identification of K.
pneumonia and Enterobacter
The protocol for the present study was approved by
the Ethics Committee at the Department of Biology, University of
Baghdad and the Iraqi Ministry of Health (Reference:
CSEC/0323/0056). Written informed consent was obtained from all the
patients. The study was carried out in accordance with the code of
Ethics of the World Medical Association (Declaration of Helsinki).
Different wound samples (surgical wounds and burn wounds; n=120; 67
males and 53 females) Wound samples (n=120) were collected from
Baghdad hospitals in Iraq (Al-Yarmouk Teaching Hospital and Baghdad
Teaching Hospital). Bacterial isolates were identified using the
traditional biochemical and morphological tests. They were cultured
on MacConkey agar (HiMedia Laboratories Private Limited), and
examined morphologically bacterial shape, size and arrangement
using an optical microscope. (Olympus Corporation) A motility test
was performed by stabbing in a semisolid medium, and the suspected
isolates were confirmed using the VITEK® 2 Compact
system (bioMérieux France).
Isolation and identification of
Salmonella
A total of 140 stool samples from patients (83 males
and 57 females) were collected at attended hospitals in Baghdad
(Al-Yarmouk Teaching Hospital and Baghdad Teaching Hospital) and
cultured on selenite broth for enrichment and selectivity, then
cultured on Salmonella-Shigella agar (HiMedia Laboratories Private
Limited), a selective and differential medium.
Antibiotic susceptibility testing for
Salmonella and Enterobacter
The Kirby-Bauer disc diffusion method (16), was used to conduct the antibiotic
sensitivity test, and according to the Clinical and Laboratory
Standards Institute (CLSI) (17),
results were obtained for six different antibiotics (gentamycin,
ciprofloxacin, azithromycin, tetracycline, cefotaxime and
chloramphenicol) as follows: In sterile plates, Mueller-Hinton agar
(HiMedia Laboratories Private Limited) was prepared and added. To
create a moderate turbidity of bacterial suspension compared to the
typical turbidity solution (McFarland standard 0.5), 3-5 colonies
of bacteria were moved into a tube containing 5 ml normal saline,
which roughly equates to 1.5x108 CFU/ml. The plates were
labelled. A cotton swab (sterile) was dipped into the inoculums and
used to apply the bacterial suspension to Mueller-Hinton agar
medium. By gently pressing and rubbing the swab on the tube's side
above the liquid level, the remaining material was eliminated. The
surface of the medium was rubbed with the swab three times, with
the plate turning at a 60˚ angle. The swab was then wound around
the border of the agar surface. The cover was closed and the
inoculums were allowed to dry for 5-10 min at room temperature.
With sterile forceps, the antibiotic disc was picked out, placed on
the inoculation plate (each plate contains four discs), and then
gently pressed on the agar to ensure it came into contact with the
agar. After 30 min, the plates were turned upside down and
incubated for 18 to 24 h at 37˚C.
Detection of bacteriocin (klebicin) in
K. pneumoniae isolates
The presence of the klebicin gene in 32 isolates of
K. pneumoniae was detected. DNA extraction was carried out
for 32 K. pneumoniae isolates using the OneTaq®
2X Master Mix kit (New England Biolabs, Inc.). From the isolated
DNA, the klebicin gene cluster was amplified using the following
primers: Forward, 5'-CATTAGCGTCCGCAGAACAAG-3' and reverse,
5'-GCCGACAGAGTAAAACCTCCA-3' (designed for the present study; the
primers were designed using Geneious prime 2023.1.1 software
depending on a reference sequence from GenBank with the accession
no. CP026155). The 16SrRNA gene was also amplified using the
following primers: Forward, 5'-GGACGGGTGAGTAATGTC-3' and reverse,
5'-TCTCAGACCAGCTAGGGATCG-3' (18).
The reaction mixture of PCR contained 2 µl of each forward and
reverse primer (10 mM), 3 µl template DNA, 12.5 µl Green master
mix, and 25 µl free nuclease water. The conditions for PCR were as
follows: 10 min at 94˚C (the initial denaturation temperature); 32
cycles with 1 min at 94˚C (the denaturation temperature), 40 sec at
54˚C (the annealing temperature), and 2 min at 72˚C (the extension
temperature); and 5 min at 72˚C (the final extension temperature).
Electrophoresis with 1% agarose gels stained with RedSafe™ Nucleic
Acid (Promega Corporation) was used to resolve the amplified DNA
products. The ability of 32 isolates to produce bacteriocin
(klebicin) was examined to detect the presence of the klebicin
gene.
Extraction of klebicin from producing
isolates
Klebicin was extracted from the Klebsiella
isolates and the crude extract of Klebicin was obtained as follows
(19): The bacterial isolates of
Klebsiella which were cultured overnight in 2.5 ml of LB
(HiMedia Laboratories Private Limited) was used to inoculate 100 ml
sterile Luria Bertani broth (HiMedia Laboratories Private Limited)
accompanied by 5% glycerol in a shaker incubator. Using a cooling
centrifuge set at 5,000 x g for 30 min (temperature, 4˚C), the
supernatant was separated. For the klebicin antibacterial activity
test and protein analysis, the supernatant was used. The Bradford
technique was used to estimate the protein content in the crude
extract of klebicin (20) and the
calculation of the protein concentration was by assessed using the
bovine serum albumin (BSA) (CDH Fine Chemical) standard curve.
Determination of the klebicin minimal
inhibitory (MIC) concentration
The resazurin-based turbidimetric assay was employed
for estimating the MIC (21) by
preparing (50, 25, 12.5, 6.25, 3.12, 1.56, 0.78, 0.39, 0.19 and
0.09%) from the stock 100% of klebicin extract. On a 96-well
microtiter plate, an aliquot of 100 µl double-strength
Muller-Hinton broth (HiMedia Laboratories Private Limited) was
added from the first to the 12th well in each row. Each first well
of the microtiter plate received 100 µl of the klebicin extract,
which was then pipetted in and mixed with the broth. The mixture
was then moved 100 µl from the first well to the second well and
carefully stirred. Up until the eleventh well, dilution was
ongoing. Subsequently, 100 µl were removed and discarded from the
eleventh well. The 12th well of each row served as a positive
control (a control well devoid of klebicin extract). To all but the
11th well, 20 µl of an overnight diluted bacterial suspension that
had its turbidity corrected to the 0.5 MacFarland standard was
added and thoroughly mixed. The 11th well hence acted as a negative
control. A total of 5 µl resazurin (Abcam) (6.75 mg/ml) were added
to each well followed by incubation at 37˚C for 18-24 h. Blue to
pink color shifts were observed and noted. Prior to the color
change, the lowest concentration was found to be the MIC.
Biofilm formation assay
A colorimetric microtiter plate technique was used
to determine biofilm development quantitatively (22): The isolates were inoculated in
brain heart infusion broth (HiMedia Laboratories Private Limited)
and then incubated for 24 h at 37˚C. Subsequently, 100 µl bacterial
growth and 2 ml ordinary saline were added to a tube, and the
turbidity was adjusted to the McFarland standard of 0.5. A 180 µl
of 1% glucose-containing brain heart infusion broth was added to
sterile, 96-well polystyrene microtiter plates with flat bottoms.
20 µl of an adjusted turbidity bacterial suspension was placed in
three wells of sterile flat-bottomed 96-well polystyrene microtiter
plates. In total, six wells of bacterial-free brain heart infusion
broth served as the negative control. The plates were not shaken
during the 24 h that they were incubated at 37˚C under their
covers. The plate was dried after three rounds of distilled water
washing following incubation. Following incubation for 15 min at
room temperature and the addition of 200 µl absolute methanol
(Alpha Chemika) to each well, the biofilms were fixed by washing
and air-drying the wells. The plates were stained for 15 min at
room temperature using 200 µl of a 0.5% crystal violet solution
(CDH Fine Chemical), washed three times in water, and dried for 30
min at 37˚C. A total of 200 µl glacial acetic acid (HiMedia
Laboratories Private Limited) and 100% ethanol [Thomas Baker
(Chemicals) Pvt. Ltd.] (1:1) were used to resolubilize the dye for
10 min. At 630 nm, the optical density (OD) of each well was
determined using a microtiter plate reader (BioTek Instruments,
Inc.; serial no. 130131A). It was found that the cut-off OD, or
ODc, was three standard deviations higher than the mean OD of the
negative control. All isolates were sorted into four categories
based on the ODc value as follows: Non-producers, weak biofilm,
moderate biofilm and strong biofilm, as shown in Table I.
 | Table IBacterial adhesion categorization on
microtiter plates (23). |
Table I
Bacterial adhesion categorization on
microtiter plates (23).
| Mean OD630 | Biofilm
intensity |
|---|
| OD ≤ ODc | Non-adherent |
| 2ODc > OD >
ODc | Weak |
| 4 ODc > OD >
2ODc | Moderate |
| OD > 4 ODc | Strong |
Detection of the antibiofilm activity
of the klebicin crude extract
Estimation of the antibiofilm activity of klebocin
was achieved by testing it on 32 isolates of Salmonella and
one Enterobacter (indicator isolates). The same procedure
designated above in the biofilm formation assay section was
followed for biofilm production, although 100 µl klebicin crude
extract subMIC were added to each well. The plate was then
incubated for 24 h at 37˚C. Following the incubation period, each
well was rinsed with water and stained, and the absorbance at 490
nm was then assessed using an ELISA reader (BioTek Instruments,
Inc.; serial no. 130131A).
Estimation of biofilm inhibition
The activity of klebicin crude extract as an
antibiofilm was tested on 32 isolates of Salmonella and one
Enterobacter, using a 96-well microtiter plate as previously
described (24). The formation of
biofilm was accomplished by the addition 100 µl of bacterial
suspension (108 cells/ml), followed by a 24-h incubation
period at 37˚C. The plate was incubated at 37˚C for 24 h with a
growth-free medium as a control before the antibiofilm substance
was added. The decrease in biofilm growth was calculated using the
following formula (25):
Results
Isolation and identification of
bacterial isolates
i) K. pneumonia: A total of 2 g negative
isolates of bacteria were obtained from 120 specimens of wounds
which were suspected to be K. pneumoniae after culturing on
MacConkey's agar medium. ii) Enterobacter: One bacterial
isolate was obtained from wound specimens; iii) Salmonella:
A total of 32 bacterial isolates were obtained from 140 stool
samples.
Antibiotic susceptibility testing for
Salmonella and Enterobacter
A total of six different antibiotics (gentamycin,
ciprofloxacin, azithromycin, tetracycline, cefotaxime and
chloramphenicol) were used to assess the susceptibility of
Salmonella and Enterobacter isolates towards them.
The results revealed that the Salmonella isolates were
resistant to gentamycin, tetracycline and cefotaxime with a
resistance of 68.75, 53.12 and 75% respectively, while they were
sensitive to ciprofloxacin and chloramphenicol with a sensitivity
of 68.75 and 65.6%, respectively. Azithromycin affected 50% of the
isolates, while 50% of them were resistant (Table II). Enterobacter exhibited
resistance to all the antibiotics tested, while it was sensitive to
ciprofloxacin only; 100% of the Enterobacter isolates were
resistant to gentamycin, tetracycline, cefotaxime, azithromycin and
chloramphenicol, and 100% of the isolates were sensitive to
ciprofloxacin.
 | Table IISusceptibility of Salmonella
isolates to antibiotics. |
Table II
Susceptibility of Salmonella
isolates to antibiotics.
| Antibiotic | Sensitive isolates
of Salmonella (%) | Resistant isolates
of Salmonella (%) |
|---|
| Gentamycin | 31.25 | 68.75 |
| Tetracycline | 46.87 | 53.12 |
| Ciprofloxacin | 68.75 | 31.25 |
| Cefotaxime | 25 | 75 |
| Azithromycin | 50 | 50 |
|
Chloramphenicol | 65.6 | 34.3 |
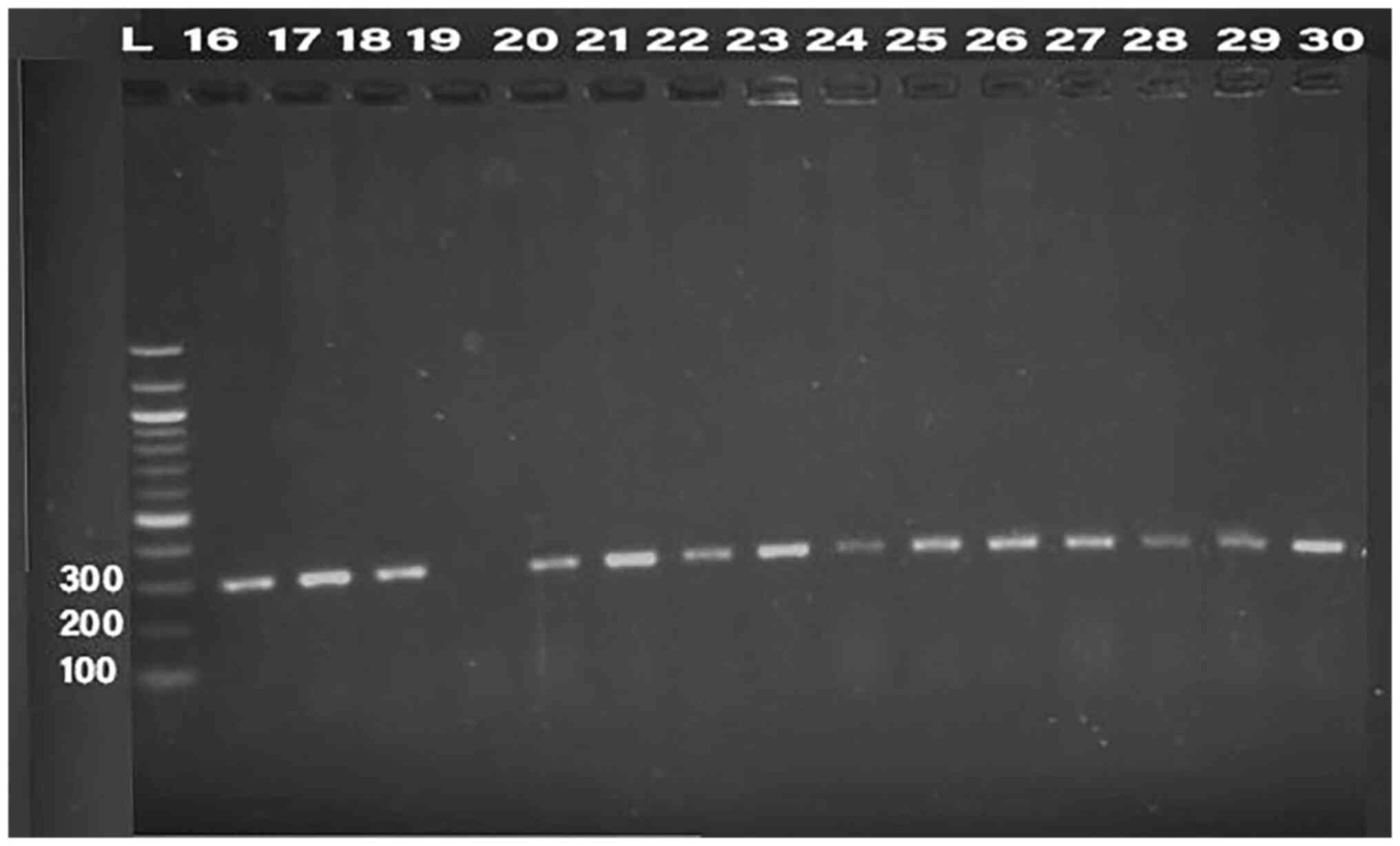
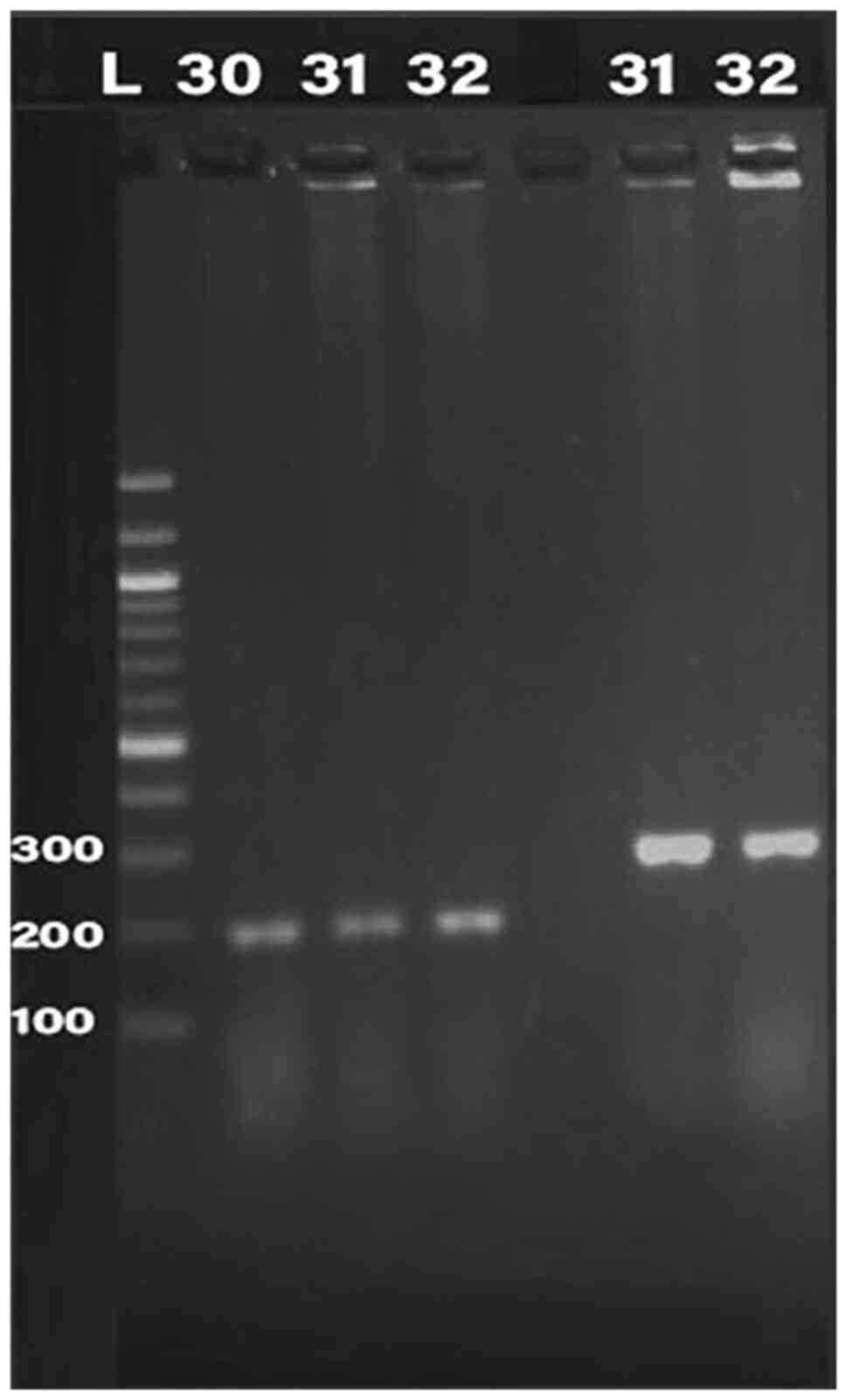
Detection of bacteriocin (klebicin) in
K. pneumoniae isolates
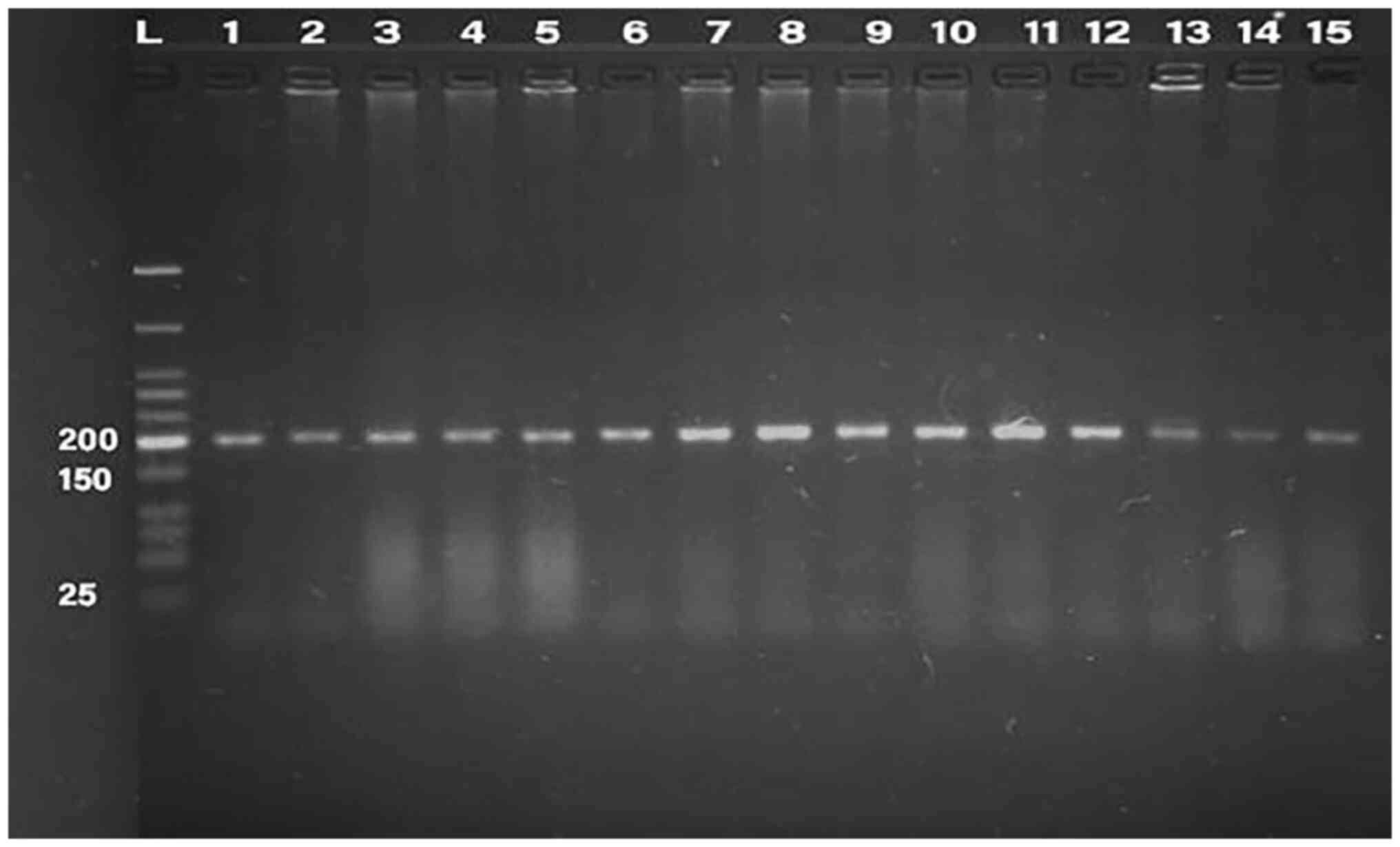
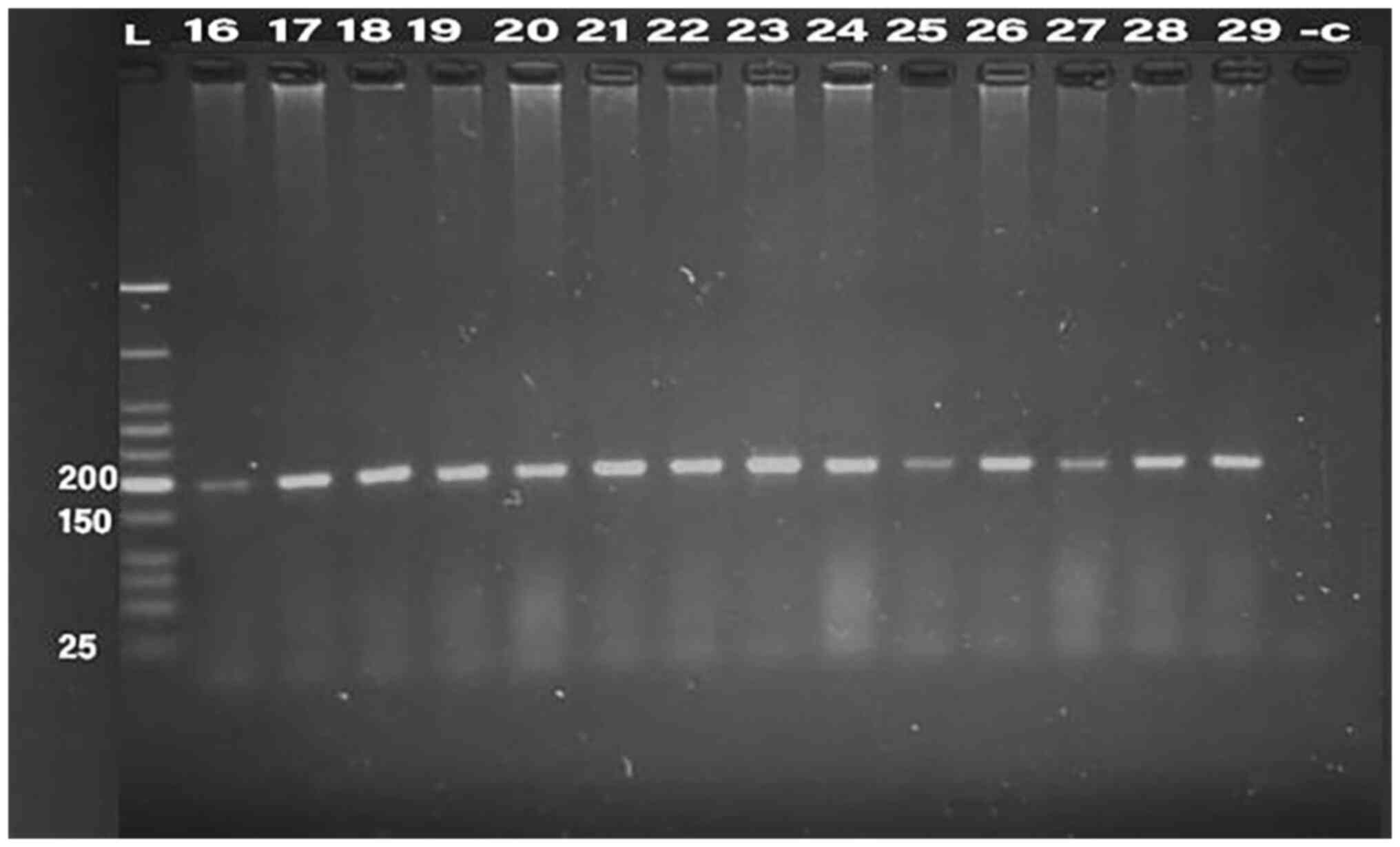
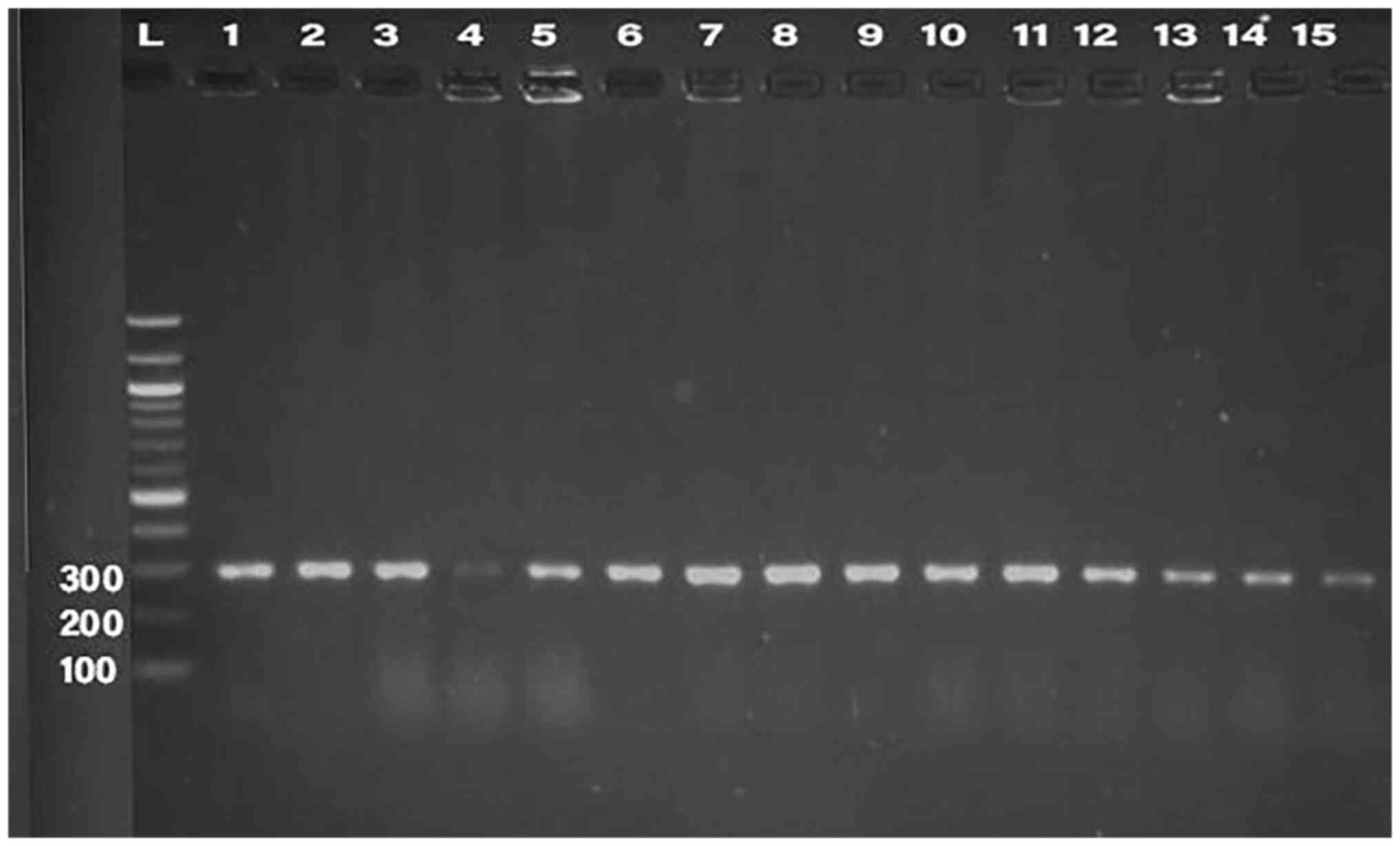
PCR was conducted for 32 isolates, using primer of
the 16SrRNA gene (198 bp) and klebicin gene (294 bp) for
amplification; gel electrophoresis was used to confirm the bands,
as presented in Fig. 1, Fig. 2, Fig.
3, Fig. 4 and Fig. 5. The result revealed that the
klebicin gene was detected in 31 (96.87%) of the K.
pneumoniae isolates and it was not detected only in isolate
19.
Protein concentration in the klebicin
crude extract
The protein concentration was calculated and the
highest protein concentration was detected in the extract of K5,
K10 and K17, as shown in Table
III. The K17 isolate, which had the highest protein
concentration in its extract, was used for further experiments in
the present study.
 | Table IIIProtein concentrations in the
klebicin crude extract. |
Table III
Protein concentrations in the
klebicin crude extract.
| Klebsiella
isolate | Protein
concentration (µg/ml) | Klebsiella
isolate | Protein
concentration (µg/ml) |
|---|
| K1 | 140.455 | K17 | 169.2953 |
| K2 | 135.3679 | K18 | 118.5249 |
| K3 | 98.1706 | K20 | 128.52927 |
| K5 | 157.96899 | K21 | 89.448 |
| K6 | 102.5634 | K22 | 131.6285 |
| K7 | 168.745 | K23 | 115.2993 |
| K8 | 96.486 | K24 | 99.318 |
| K9 | 136.423 | K25 | 124.4231 |
| K10 | 165.4775 | K26 | 135.6142 |
| K11 | 115.5543 | K27 | 130.6183 |
| K12 | 138.467 | K28 | 114.5521 |
| K13 | 122.532 | K29 | 122.7396 |
| K14 | 117.341 | K30 | 95.3185 |
| K15 | 101.138 | K31 | 140.4185 |
| K16 | 132.3853 | K32 | 138.817 |
Determination of the klebicin MIC
against Salmonella isolates
The MIC of klebicin was assessed, and the results
revealed that the Enterobacter isolate was inhibited by 50%
of the klebicin extract, while the majority of the
Salmonella isolates were inhibited by 50% of the klebicin
extract, and only (S3, S10, S16, S20, S25, S28 and S32) were
inhibited by 25% of the klebicin extract.
Antibiofilm activity of the klebicin
extract against Salmonella and Enterobacter and their dual biofilm
formation
Biofilm production was quantified using a
colorimetric microtiter plate approach. The results revealed that
prior to treatment with the klebicin extract, the
Enterobacter isolate was a strong biofilm producer; however,
following treatment, biofilm production became moderate. The
majority of the Salmonella isolates were strong biofilm
formers, while following treatment, most of them were moderate and
weak biofilm producers, as shown in Table IV.
 | Table IVBiofilm forming capacity of
Salmonella isolates. |
Table IV
Biofilm forming capacity of
Salmonella isolates.
| Isolate no. | Mean value (before
treatment) | Type of
thickness | Mean value (after
treatment) | Type of
thickness |
|---|
| 1 | 0.195 | Very strong | 0.1077 | Weak |
| 2 | 0.367 | Strong | 0.2369 | Moderate |
| 3 | 0.361 | Strong | 0.1534 | Moderate |
| 4 | 0.293 | Strong | 0.2194 | Moderate |
| 5 | 0.297 | Strong | 0.1898 | Weak |
| 6 | 0.302 | Strong | 0.2114 | Moderate |
| 7 | 0.377 | Very strong | 0.2763 | Strong |
| 8 | 0.378 | Very strong | 0.2456 | Strong |
| 9 | 0.243 | Strong | 0.2175 | Moderate |
| 10 | 0.534 | Strong | 0.1903 | Moderate |
| 11 | 0.537 | Very strong | 0.2923 | Strong |
| 12 | 0.394 | Strong | 0.2014 | Moderate |
| 13 | 0.361 | Strong | 0.1977 | Moderate |
| 14 | 0.590 | Strong | 0.2006 | Moderate |
| 15 | 0.284 | Strong | 0.2367 | Moderate |
| 16 | 0.401 | Very strong | 0.2449 | Strong |
| 17 | 0.185 | Moderate | 0.1742 | Moderate |
| 18 | 0.495 | Strong | 0.1155 | Weak |
| 19 | 0.372 | Very strong | 0.2771 | Strong |
| 20 | 0.502 | Very strong | 0.2425 | Strong |
| 21 | 0.225 | Moderate | 0.1742 | Moderate |
| 22 | 0.406 | Strong | 0.1162 | Weak |
| 23 | 0.403 | Strong | 0.2104 | Moderate |
| 24 | 0.503 | Strong | 0.1807 | Moderate |
| 25 | 0.453 | Strong | 0.2131 | Moderate |
| 26 | 0.527 | Very strong | 0.2776 | Strong |
| 27 | 0.283 | Strong | 0.2377 | Moderate |
| 28 | 0.351 | Very strong | 0.2755 | Strong |
| 29 | 0.286 | Strong | 0.2028 | Moderate |
| 30 | 0.496 | Strong | 0.184 | Moderate |
| 31 | 0.277 | Very strong | 0.2831 | Strong |
| 32 | 0.294 | Strong | 0.174 | Moderate |
The results of dual biofilm formation by bacterial
isolates of (Salmonella and Enterobacter) revealed
that all of them were strong biofilm producers prior to treatment
with the klebicin extract, while they became weak biofilm producers
following treatment, as shown in Table
V.
 | Table VBiofilm forming capacity of dual
bacterial isolates (Salmonella and Enterobacter). |
Table V
Biofilm forming capacity of dual
bacterial isolates (Salmonella and Enterobacter).
| Isolate no. | Mean value (before
treatment) | Type of
thickness | Mean value (after
treatment) | Type of
thickness |
|---|
| 1 | 1.054 | Strong | 0.073 | Weak |
| 2 | 1.065 | Strong | 0.079 | Weak |
| 3 | 1.057 | Strong | 0.075 | Weak |
| 4 | 1.054 | Strong | 0.331 | Weak |
| 5 | 1.068 | Strong | 0.080 | Weak |
| 6 | 1.079 | Strong | 0.076 | Weak |
| 7 | 1.060 | Strong | 0.076 | Weak |
| 8 | 1.068 | Strong | 0.074 | Weak |
| 9 | 1.057 | Strong | 0.081 | Weak |
| 10 | 1.071 | Strong | 0.077 | Weak |
| 11 | 1.067 | Strong | 0.077 | Weak |
| 12 | 1.072 | Strong | 0.084 | Weak |
| 13 | 1.065 | Strong | 0.072 | Weak |
| 14 | 1.068 | Strong | 0.089 | Weak |
| 15 | 1.061 | Strong | 0.078 | Weak |
| 16 | 1.086 | Strong | 0.088 | Weak |
| 17 | 1.062 | Strong | 0.071 | Weak |
| 18 | 1.080 | Strong | 0.088 | Weak |
| 19 | 1.063 | Strong | 0.070 | Weak |
| 20 | 1.070 | Strong | 0.075 | Weak |
| 21 | 1.035 | Strong | 0.072 | Weak |
| 22 | 1.074 | Strong | 0.082 | Weak |
| 23 | 1.078 | Strong | 0.080 | Weak |
| 24 | 2.970 | Strong | 1.973 | Weak |
| 25 | 1.042 | Strong | 0.068 | Weak |
| 26 | 1.088 | Strong | 0.077 | Weak |
| 27 | 1.075 | Strong | 0.084 | Weak |
| 28 | 1.066 | Strong | 0.077 | Weak |
| 29 | 1.085 | Strong | 0.089 | Weak |
| 30 | 1.079 | Strong | 0.087 | Weak |
| 31 | 1.075 | Strong | 0.084 | Weak |
| 32 | 2.405 | Very strong | 1.514 | Strong |
Estimation of biofilm inhibition
The use of the klebicin extract was obviously
effective against biofilm reduction. In particular, it was more
effective against dual biofilm formation by both bacteria,
Salmonella and Enterobacter, compared to each
bacteria alone Table VI.
 | Table VIBiofilm reduction of the
Salmonella and the combination of the Salmonella and
Enterobacter isolates following treatment with klebicin
extract. |
Table VI
Biofilm reduction of the
Salmonella and the combination of the Salmonella and
Enterobacter isolates following treatment with klebicin
extract.
| Salmonella
isolates | Biofilm reduction
following treatment with klebicin extract (%) | Salmonella
and Enterobacter isolates | Biofilm reduction
following treatment with klebicin extract extract (%) |
|---|
| S1 | 44.7 | S1 + E | 93 |
| S2 | 35.4 | S2 + E | 92.5 |
| S3 | 57.5 | S3 + E | 92.9 |
| S4 | 25.1 | S4 + E | 68.5 |
| S5 | 36 | S5 + E | 92.5 |
| S6 | 30 | S6 + E | 92.9 |
| S7 | 26.7 | S7 + E | 92.8 |
| S8 | 35 | S8 + E | 93 |
| S9 | 10.4 | S + 9E | 92.3 |
| S10 | 64.3 | S10 + E | 92.8 |
| S11 | 45.5 | S11 + E | 92.7 |
| S12 | 48.8 | S12 + E | 92.1 |
| S13 | 45.2 | S13 + E | 93.2 |
| S14 | 66 | S14 + E | 91.6 |
| S15 | 16.6 | S15 + E | 92.6 |
| S16 | 38.9 | S16 + E | 91.8 |
| S17 | 5.8 | S17 + E | 93.3 |
| S18 | 76.6 | S18 + E | 91.8 |
| S19 | 25.5 | S19 + E | 93.4 |
| S20 | 51.6 | S20 + E | 92.9 |
| S21 | 22.5 | S21 + E | 93 |
| S22 | 71.3 | S22 + E | 92.3 |
| S23 | 47.7 | S23 + E | 92.5 |
| S24 | 64 | S24 + E | 33.5 |
| S25 | 52.9 | S25 + E | 93.4 |
| S26 | 47.3 | S26 + E | 92.9 |
| S27 | 16 | S27 + E | 92.1 |
| S28 | 21.5 | S28 + E | 92.7 |
| S29 | 29 | S29 + E | 91.7 |
| S30 | 62.9 | S30 + E | 91.9 |
| S31 | 5.3 | S31 + E | 92.1 |
| S32 | 40.8 | S32 + E | 37.1 |
Discussion
In order to preferentially separate Gram-negative
and enteric (often found in the digestive system) bacilli and
identify them based on fermenting lactose, MacConkey agar, a
differential and selective growth medium is used, and Gram-positive
organisms are prevented from growing by crystal violet and bile
salts (26). In the present study,
this medium was used to isolate K. pneumoniae and
Enterobacter. Isolates which were suspected to be K.
pneumoniae were confirmed using VITEK-2 compact system.
Following culture on MacConkey's agar medium, Enterobacter
colonies were lactose fermented and motile compared with K.
pneumoniae confirmed by VITEK- 2 compact system.
Salmonella isolates were obtained following culture on
selenite broth and transferred to Salmonella-Shigella agar that
appears as pale colonies with a black center. Selenite broth, as a
selective and enrichment medium, is used for the cultivation of
Salmonella spp., that may be present in small numbers in the
intestine and competing with its flora (27). The identification of isolates was
confirmed using the VITEK-2 compact system.
Antibiotic susceptibility was performed to exhibit
the high resistance rate in bacterial isolates, and resistance to
more than one antibiotic was observed; thus, this leads to the
necessity for alternative mechanisms to combat antibiotic
resistance. A previous study reported that Salmonella
isolates in high proportions were resistant to tetracycline
(n=53.9%) and ciprofloxacin (n=47.2%) (28). Another study demonstrated that 98%
of Salmonella isolates were non-susceptible to ciprofloxacin
(29). Patil and Mule (30) reported that all isolates (100%) of
Salmonella were sensitive to cefixime, ceftriaxone and
azithromycin, and 94.4% (237/251) of the bacterial isolates were
significantly sensitive to chloramphenicol.
In the present study, PCR was conducted for 32
isolates of K. pneumoniae, and the klebicin gene was found
in the majority of these isolates, apart from one. In their study,
Kareem et al (31) reported
PCR amplification results. PCR products corresponding to the
klebicin gene appeared in 15 isolates (48.39%) (31).
Herein, the protein concentration was determined in
the klebicin crude extract of all K. pneumoniae isolates and
the isolate K17 with the highest protein concentration extract was
used to perform the ensuing analyses. Bacteriocins are ribosomal
proteins or peptides. When bacteriocin-producing bacteria release
bacteriocin, this can interact with the appropriate receptor on the
surface of the vulnerable bacteria to kill it (32).
Klebicin crude extract of the K17 isolate had an
inhibitory effect on Salmonella isolates with two MICs. The
sensitive bacteria are killed by klebicin via a number of
mechanisms. Klebicin binds to certain receptors, which are outer
membrane proteins that are used for the entrance of various
nutrients. Subsequently, either the Tol or TonB systems transport
klebicin over the periplasm and through the outer membrane
(33). The action of klebicins
would include either creating a channel (voltage-dependent) into
the inner membrane or by using their activity of endonuclease on
DNA, rRNA, or tRNA to reach their target (34,35).
Klebicins are proteins that are encoded by both chromosomes and
plasmids (14).
Klebicin crude extract markedly affected the biofilm
formation of Salmonella and Enterobacter isolates and
reduced it. High percentages of inhibition were observed in the
dual biofilm formation with two types of bacteria. Khalaf and
Hussein (36) reported that
klebicin crude extract affected the formation of biofilm in some
bacterial isolates, such as Klebsiella, Proteus and
E. coli.
To date, only a limited number of studies have
examined the effects of klebicin on bacteria and their biofilm
development compared to other types of bacteriocins produced by
other bacteria, such as colicin from E. coli and pyocin from
Pseudomonas. Colicins, which are derived from E.
coli, and other bacteriocins that are similar to colicins,
which are derived from a variety of Gram-negative bacteria, are
poisonous to bacterium closely similar to the strain that produces
it. Of note, >90% of Pseudomonas aeruginosa strains
generate pyocins, and each strain is capable of producing several
pyocins. The pyocin genes are found on the chromosome of
Pseudomonas aeruginosa (37). S2-pyocin and antibiotics were
tested against P. aeruginosa biofilms in vitro in the
study by Smith et al (38).
S2-pyocin was shown to be the most effective against Pseudomonas
aeruginosa biofilms, resulting in a 4 log decrease in
Pseudomonas aeruginosa survival (38). Another study revealed that K.
pneumoniae clinical isolates were capable of producing
bacteriocin (klebocin) which affected pathogenic isolates, such as
other Klebsiella, E. coli and Proteus
(14). Klebocin had a strong
antibacterial and antibiofilm action. Therefore, it was concluded
that these outcomes may be a potential source of antimicrobial
agents (14). The heterologous
action of klebocins on numerous pathogenic species of Gram-negative
and some Gram-positive bacteria, those isolated from individuals
with persistent otitis media and pyelonephritis, in particular, may
suggest that these klebocins can be used as an alternative to
broad-spectrum antibiotics (39).
In the present study, the effects of klebicin crude
extract on Salmonella and Enterobacter isolates was
notable according to the data obtained, particularly on biofilm
formation reduction, since the current low efficacy of antibiotics
towards bacterial infections caused by biofilms can be largely
attributed to the fact that the biofilm mode of development is
characterized by prolonged infection by resistant pathogens;
therefore, antibiotic alternatives should be researched. The data
obtained herein suggest the possibility of the use of bacteriocin
(klebicin) as an antibacterial and antibiofilm agent. Further
studies need to be conducted to purify and characterize klebicin,
examine its effects on biofilm formation and examine it under an
electron microscope.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NSA and HKT contributed to the conception and design
of the study. All authors (NSA, HKT and AHO) were involved in the
study methodology. NSA and HKT contributed to the data collection
and analysis. NSA was involved in the writing of the manuscript.
NSH and HKT confirmed the authenticity of all the raw data. All
authors have read and agreed to the published version of the final
manuscript.
Ethics approval and consent to
participate
The protocol for the present study was approved by
the Ethics Committee at the Department of Biology (University of
Baghdad) and the Iraqi Ministry of Health (Reference:
CSEC/0323/0056). Written informed consent was obtained from all the
patients. The study was carried out in accordance with the code of
Ethics of the World Medical Association (Declaration of
Helsinki).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hoiby N, Ciofu O, Johansen HK, Song ZJ,
Moser C, Jensen PO, Molin S, Givskov M, Tolker-Nielsen T and
Bjarnsholt T: The clinical impact of bacterial biofilms. Int J Oral
Sci. 3:55–65. 2011.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Stoodley P and Hall-Stoodley L: Evolving
concepts in biofilm infections. Cell Microbiol. 11:1034–1043.
2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Donlan RM and Costerton JW: Biofilms:
Survival mechanisms of clinically relevant microorganisms. Clin
Microbiol Rev. 15:167–193. 2002.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Mah TFC and O'Toole GA: Mechanisms of
biofilm resistance to antimicrobial agents. Trends Microbiol.
9:34–39. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Davies J: Origins and evolution of
antibiotic resistance. Microbiologia. 12:9–16. 2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Harrell JE, Hahn MM, D'Souza SJ, Vasicek
EM, Sandala JL, Gunn JS and McLachlan JB: Salmonella Biofilm
formation, chronic infection, and immunity within the intestine and
Hepatobiliary Tract. Front Cell Infect Microbiol.
10(624622)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lojewska E and Sakowicz T: An alternative
to antibiotics: Selected methods to combat zoonotic foodborne
bacterial infections. Curr Microbiol. 78:4037–4049. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Parret AH and De Mot R: Bacteria killing
their own kind: Novel bacteriocins of pseudomonas and other
gamma-proteobacteria. Trends Microbiol. 10:107–112. 2002.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Denkovskiene E, Paškevičius S, Misiūnas A,
Stočkūnaitė B, Starkevič U, Vitkauskiene A, Hahn-Löbmann S, Schulz
S, Giritch A, Gleba Y and Ražanskienė A: Broad and efficient
control of klebsiella pathogens by peptidoglycan-degrading and
pore-forming bacteriocins klebicins. Sci Rep.
9(15422)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Grinter R, Milner J and Walker D:
Ferredoxin containing bacteriocins suggest a novel mechanism of
iron uptake in Pectobacterium spp. PLoS One.
7(e33033)2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hardgree M and Tu A (eds): Bacterial
Toxins: Handbook of Natural Toxins. CRC Press, Boca Raton, FL,
p488, 1988.
|
|
12
|
Thomas X, Destoumieux-Garzon D, Peduzzi J,
Afonso C, Blond A, Birlirakis N, Goulard C, Dubost L, Thai R, Tabet
JC and Rebuffat S: Siderophore Peptide, a new type of
post-translationally modified antibacterial peptide with potent
activity. J Biol Chem. 279:28233–28242. 2004.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cascales E, Buchanan S, Duché D,
Kleanthous C, Lloubès R, Postle K, Riley M, Slatin S and Cavard D:
Colicin biology. Microbiol Mol Biol Rev. 71:158–229.
2007.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Maresz-Babczyszyn J, Durlakowa I,
Lachowicz Z and Hamon Y: Characteristics of bacteriocins produced
by Klebsiella bacilli. Arch Immunol Ther Exp (Warsz). 15:530–539.
1967.PubMed/NCBI
|
|
15
|
Albesa I, Finola MS, Moyano S, Frigerio CI
and Eraso AJ: Klebocin activity of Klebsiella strains and its
heterologous effect on Staphylococcus sp. Microbiologica. 12:35–41.
1989.PubMed/NCBI
|
|
16
|
Vandeppitte J, Verhaegen J, Engbaek K,
Rohner P, Piot P and Heuck C: Basic Laboratory Procedures in
Clinical Bacteriology (2nd edition). World Health Organization,
Geneva, 2003.
|
|
17
|
Clinical and Laboratory Standards
Institute (CLSI). Performance Standards for Antimicrobial
Susceptibility Testing. 33rd edition. CLSI document
M100, 2023.
|
|
18
|
Girlich D, Poirel L and Nordmann P: CTX-M
expression and selection of ertapenem resistance in Klebsiella
pneumoniae and Escherichia coli. Antimicrob Agents Chemother.
53:832–834. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Herschman HR and Helinski DR: Purification
and characterization of colicin E2 and colicin E3. J Biol Chem.
212:5360–5368. 1967.PubMed/NCBI
|
|
20
|
Bradford MM: A dye binding assay for
protein. Anal Biochem. 72:248–254. 1976.
|
|
21
|
The CH, Nazni WA, Nurulhusna AH, Norazah A
and Lee HL: Determination of antibacterial activity and minimum
inhibitory concentration of larval extract of fly via
resazurinbased turbidometric assay. BMC Microbiol. 17:1–8.
2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Jaffar N, Miyazaki T and Maeda T: Biofilm
formation of periodontal pathogens on hydroxyapatite surfaces:
Implications for periodontium damage. J Biomed Mater Res A.
104:2873–2880. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhao T and Liu Y: N-acetylcysteine inhibit
biofilms produced by Pseudomonas aeruginosa. BMC Microbiol.
10(140)2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Selim SA, Adam ME, Hassan SM and Albalawi
AR: Chemical composition, antimicrobial and antibiofilm activity of
the essential oil and methanol extract of the Mediterranean cypress
(Cupressus sempervirens L.). BMC Complement Altern Med.
14(179)2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Mathur S, Gutte M, Paul D and Udgire M:
Study the effect of essential oils on microbial biofilm formation
by Klebsiella pneumoniae. Sch Acad J Biosci. 1:76–79. 2013.
|
|
26
|
Anderson C, Johnson TR, Case CL,
Cappuccino JG and Sherman N: Great Adventures in the Microbiology
Laboratory. 7th edition. pp175-176, 2013.
|
|
27
|
Orden B and Franco A: Wellcolex colour
salmonella test and Selenite-F broth. J Clin Microbiol.
31:2249–2250. 1993.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Peruzy MF, Capuano F, Proroga YTR,
Cristiano D, Carullo MR and Murru N: Antimicrobial susceptibility
testing for salmonella serovars isolated from food samples:
Five-year monitoring (2015-2019). Antibiotics (Basel).
9(365)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Veeraraghavan B, Pragasam AK, Ray P, Kapil
A, Nagaraj S, Perumal SP, Saigal K, Thomas M, Gupta M,
Rongsen-Chandola T, et al: Evaluation of antimicrobial
susceptibility profile in salmonella typhi and salmonella paratyphi
A: Presenting the current scenario in india and strategy for future
management. J Infect Dis. 224:S502–S516. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Patil N and Mule P: Sensitivity pattern Of
Salmonella typhi And Paratyphi A isolates to chloramphenicol and
other anti-typhoid drugs: An in vitro study. Infect Drug Resist.
12:3217–3225. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kareem AA, Al-Salmani TS and AlSalmani MS:
Detection of klebicin gene cluster from klebsiella pneumoniae
isolated from sputum at baghdad city. Indian J Public Health Res
Develop. 10:4970–4973. 2019.
|
|
32
|
Yang SC, Lin CH, Sung CT and Fang JY:
Antibacterial activities of bacteriocins: application in foods and
pharmaceuticals. Front Microbiol. 5(241)2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lloubès R, Cascales E, Walburger A,
Bouveret E, Lazdunski C, Bernadac A and Journet L: The Tol-Pal
proteins of Escherichia coli cell envelope: An energized system
required for outer membrane integrity. Res Microbiol. 152:523–529.
2001.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chavan M, Rafi H, Wertz J, Goldstone C and
Riley MA: Phage associated bacteriocins reveal a novel mechanism
for bacteriocin diversification in klebsiella. J Mol Evol.
60:546–556. 2005.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Riley M, Pinou T, Wertz J, Tan Y and
Valletta C: Molecular characterization of the klebicin B plasmid of
Klebsiella pneumoniae. Plasmid. 45:209–221. 2001.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Khalaf ZZ and Hussein AR: Antibiofilm
activity of klebocin crude extract against Some Species of
Enterobacteriaceae. Iraj J Sci. 59:1826–1835. 2018.
|
|
37
|
Michel-Briand Y and Baysse C: The Pyocins
of Pseudomonas aeruginosa. Biochimie. 84:499–510. 2002.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Smith K, Martin L, Rinaldi A, Rajendran R,
Ramage G and Walker D: Activity of Pyocin S2 against Pseudomonas
aeruginosa Biofilms. Antimicrob Agents Chemother. 56:1599–1601.
2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Al-Charrakh AH, Yousif SY and AlJanabi HS:
Antimicrobial spectrum of the action of bacteriocins from
Klebsiella isolates from Hilla/Iraq. Iran J Microbiol.
2(1)2011.
|