1. Introduction
Over the years, research on the gut microbiota and
its influence on respiratory health has evolved into the concept of
the gut-lung axis. Initially regarded as a simple connection,
studies in the 1990s and early 2000s began exploring the role of
probiotics in supporting immune function and reducing respiratory
infections (1-7).
The discoveries laid the foundation for understanding the intricate
association between the microbiome and the immune system in
maintaining lung health. A deeper understanding of the role of the
microbiome in both healthy and diseased individuals has
significantly contributed to identifying biomarker patterns, aiding
in early disease detection and personalized treatment strategies.
The integration of microbiome research into precision medicine has
been particularly impactful, as specific beneficial bacterial
strains have shown resilience to physiological stress, providing
promising therapeutic potential. Patients with a well-defined
microbial niche could benefit from next-generation treatments
tailored to their microbiome composition (8).
Microbial communities within the human body play
essential metabolic and immunological roles. The lung microbiome,
primarily composed of Pseudomonas, Streptococcus, Prevotella,
Fusobacterium, Haemophilus, Veillonella and
Porphyromonas (9),
contributes to respiratory health through the production of
small-chain fatty acids (SCFAs), which help regulate immune
responses. While precision medicine was initially centered on
genetic and phenotypic variations, there is currently a paradigm
shift towards the understanding the microbiome as a key factor
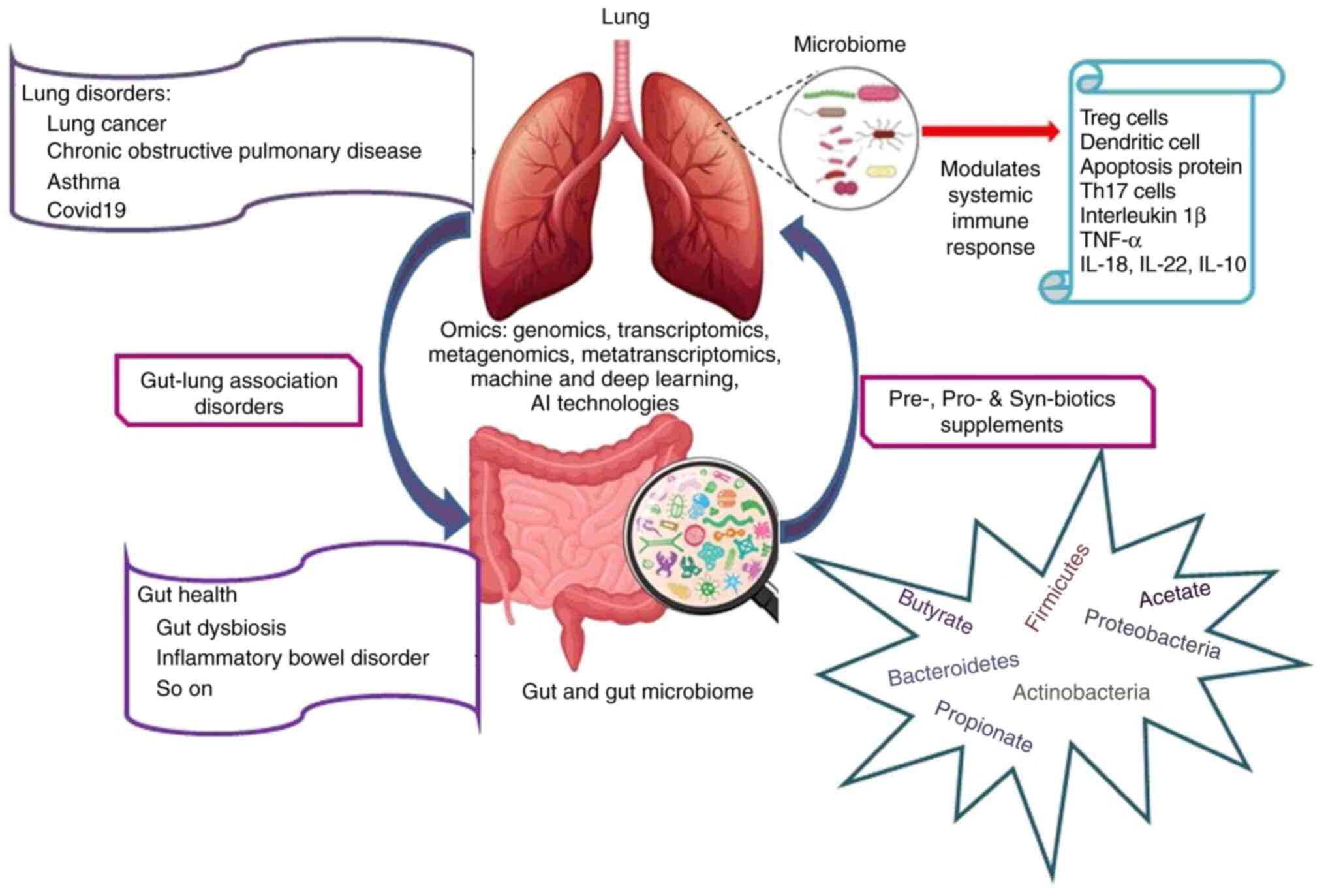
influencing health and disease (10). Investigating the crosstalk between
gut and lung microbiomes in various diseases holds promise for the
development of novel diagnostic markers and advancing personalized
medicine (Fig. 1). This evolving
perspective opens new avenues for integrating microbiome-targeted
therapies into clinical practice.
2. Precision medicine and the human
microbiome
Precision medicine is based on the deep
characterization of the genome, proteome, metabolome and microbiome
of an individual. Rapidity and technical advancements in genome
studies (DNA-Seq, RNA-Seq, Exosome-Seq, etc.) decreased the cost of
sequencing and genotyping that typically shifted the paradigm of
precision medicine from academic exercise to clinical applications
(11). The milestone projects,
such as complete human genome sequencing, HapMap (Haplotype Map)
project, Phase 1- NIH human microbiome project, and the phase 1-
European Union MetaHit (Metagenomics of the Human Intestinal Tract)
Project contributed to the understanding of the molecular basis of
diseases and to the development of initiatives to propose new
prototypes of personalized medicine (12,13).
The most recent advancement in understanding human disease is to
derive its association with the human microbiota. The human
microbiome is referred to as the second genome, as trillions of
microbial inhabitants are integrated ecologically and symbiotically
with their host. The analyzed omics profile of an individual aids
in the prevention, diagnosis and treatment of diseases. Knowledge
of human genomic variants and phenotypic changes is critical for
understanding the molecular basis of the disease (14). The human body colonizes diverse
communities of microbes, and also resists the colonization of
pathogenic organisms, such as Clostridium difficile
(15), which clearly suggests the
existence of symbiotic associations between the human body and its
microbiota. A previous study confirmed the presence of bacterial
peptides in blood serum and plasma of approximate range/of 0.1 nM
to 1 µM (16). That study utilized
the human microbiota protein sequences from NIH Human Microbiome
Project which as useful for determining the microbiota composition
in the intestinal regions. Well-structured databases are available
to provide the genotypic and phenotypic related variants and
related traits [ClinGen (https://clinicalgenome.org/), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/),
dbVar (https://www.ncbi.nlm.nih.gov/dbvar/), The Cancer
Genome Atlas (TCGA) (https://www.cancer.gov/ccg/research/genome-sequencing/tcga),
dbGENVOC (https://research.nibmg.ac.in/dbcares/dbgenvoc/home.php),
HGMD (https://www.hgmd.cf.ac.uk/ac/index.php), OMIM
(https://omim.org/), etc.]. The concept of ‘One dose
fits all’ is ideologically replaced with personalized medicine to
ensure a tailored treatment with effective and safe drug usage. For
an accelerated research, the accessibility to Electronic Health
Records (Electronic Medical Records and Genomics-Pharmacogenomics
(eMERGE-PGx) project, GANI_MED project, SCAN-B initiative and
Cancer 2015) provides an in-depth analysis of the medical history
of an individual against various infections and higher predictive
nature to derive the susceptibility of the patient to other
comorbid diseases. Other than this, knowledge-based approaches
derived from documented literature utilize sophisticated
text-mining and natural language processing for literature curation
and annotation to extract the vast amount of information buried
about the genetic variants and their functional phenotypic
associations (17).
3. Role of the gut microbiome in precision
medicine: Gut-lung axis microbiome
In recent days, ‘Gut Microbiota’ is a buzz word and
often termed as a forgotten organ/metabolic organ that plays a
fine-tuned symbiotic association with the host. Hundreds of
trillions of diverse microbial communities that include protozoa,
fungi, archaea, viruses, protists and bacteria reside within the
gastrointestinal tract (GIT) compared to the regions of skin, eye,
urogenital and epithelial layers of the respiratory system. In
general, the microbiome of an individual is more complex than the
human genome and it imparts a specific immunity to the individual
that characterizes the need to have personalized medicine.
According to the studies of the Human Microbiome project (18), it was revealed that the ratio of
commensal microbial genes to the total number of human genes was
high and the adult human gut can harbor trillions of microbes;
therefore, it exceeds the total number of somatic and germ cells by
10-fold (19).
The composition and pattern of the microbiota of an
individual is very specific and hence, it enables the
identification of disease-specific microbial signatures, providing
personalized predictive biomarkers for various diseases (20,21).
The insights from the gut-lung axis and the identification of
microbial patterns aids in the early identification and diagnostics
based on various immunological responses. With respect to
conventional therapies, this also triggers microbiome-targeted
interventions, such as probiotics, prebiotics and fecal microbiota
transplantation, which are more promising and safe strategies
(22,23). Given the complexity of microbiome
data, analyzing it is a challenge. which is strategically now being
done using various machine learning, deep learning and more
recently quantum methods (24).
The integration of microbiome-based diagnostics and therapies into
clinical frameworks has the potential to optimize respiratory
illness care, while maintaining microbial equilibrium, as precision
medicine moves beyond genetic and phenotypic differences.
The reasons for studying the gut
microbiome instead of the lung microbiome for addressing lung
disorders
The lung was originally described to be a sterile
organ; however, with technical advancements, the lung microbiome
was then defined, which is composed of a complex and diverse
bacterial community, with a low biomass. There is substantial
variation between upper and lower respiratory tract microbiomes due
to higher flux exhibited by microbes (25). Different environmental niches are
created as a result of the differential availability of pH and
nutritional factors, which plays a critical role during
inflammation or through structural changes in chronic respiratory
diseases (CRDs) (26). The
respiratory microbiome is also combined with the upper respiratory
tract and gut microbiota, which renders the assessment of its role
difficult (27,28).
The gut-lung axis refers to the communication
between the gut, lung and vice versa, where the gut microbial
metabolites enter the lung via the blood stream and influence the
pulmonary microbiome. Emerging scientific reports demonstrate the
modulation of systemic immune responses through altering the diet
and antibiotic treatment. In addition, an altered lung microbiome
in various chronic lung disorders indicates that the gut-lung axis
is bidirectional (29).
Immunological crosstalk between the respiratory and gut microbiome
is reflected by the fact that patients with chronic respiratory
disease are prone to GIT-related issues and vice-versa, where
patients with inflammatory bowel disease display pulmonary
involvement (30).
The gut and lung microbial communities overlap in
their phylum, but differ in local compositions and total microbial
biomass (31). This overlap may be
responsible for microbial interference in different disease
associations. The gut microbiota modulates immune responses
differently in healthy individuals and in patients with lung
disorders, which is dependent on diet and food intake.
Diet-Microbiota-Immunity link supplemented by high fiber diet is
well explained in different studies (30). The fiber intake influences the gut
microbiota by increasing the production of bacterial metabolites,
such as SCFAs (e.g., propionate, acetate and butyrate) (32); this in turn modulates different
aspects of defense lines, such as promoting goblet cell
differentiation and mucus production, stimulating the proliferation
of T-regulatory cells which produces anti-inflammatory cytokine
(e.g. IL-10), as well as the production of intestinal IgA by
affecting plasma B-cell metabolism and modulating tight junction
permeability to enhance intestinal epithelial barrier function in
the gut of the host (33).
The so-called exposome that includes socio-economic
factors, nutrition, stress, environmental factors (pollutants,
antibiotics, etc.) and the individual's genetics, sex and age
composes the microbial diversity of the GIT (34). Metabolites produced by the gut
microbiota are capable of eliciting or activating the downstream
cascades of reactions. The continuous interplay between the gut
microbiota and intestinal epithelium sends signals for the
regulation of immune responses to infectious diseases and
disorders. In the case of infectious diseases, the microbial
colonization of commensals and pathogenic organisms undergo
crosstalk with host immune responses (35). The consumption of a healthy diet
could modulate the local and systemic host physiology, metabolism,
immune function and nutrition state.
In the GIT, the bacterial cell-cell communication
takes place through the exchange of chemical messages using quorum
sensing mechanisms that are capable of triggering changes in the
gene expression. Extensive studies on quorum sensing between
bacterial communities in a local niche are being carried out; the
GIT is a complex system where bacteria respond to a milieu of
chemical signals to synchronize bacterial behavior and convert them
into reliable messages to coordinate the relative species, as well
as with the host immune system. Hence, in the GIT, multidirectional
communication pathways of quorum sensing occur and are referred
generally as interspecies communications and interkingdom signaling
(36). Gram-negative bacteria
produce N-acyl-homoserine lactones (AHLs) that bind to LuxR
receptors to communicate with each other and their accumulation to
a threshold concentration activates the gene expression for toxin
production and biofilm formation. Certainly, AHLs influence the
host immune system through the secretion of IL-10, where their
specific functions are to limit the host immune response towards
pathogens; in addition, due to the lipid solubility nature of AHLs,
they diffuse into the mammalian cells and induces apoptosis through
cascades of reactions (37). A
more systematic detail of the crosstalk of metabolites through
quorum sensing that interrelates with the gut microbiota and host
immune system has been previously discussed (36).
4. Microbial niches and their metabolites in
the gut homeostasis
Gut microbes mostly constitute the phyla of
Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria, and
occasionally, Verrucomicrobia and Fusobacteria (38-40).
Of these, Bateroidetes and Firmicutes constitute ~90% of intestinal
microbiota. Firmicutes are abundant in populations consuming
animal-based diets, whereas Bacteroidetes are largely found in
populations consuming plant-based diets (41). The genes for the hydrolysis of
plant-based polysaccharides are reported in the bacterial strains
of Prevotella and Xylanibacter; therefore, the GIT
possesses higher counts of these organisms in populations depending
on plant-based food varieties. Among the Indian population, the
genera of Prevotella, Lactobacillus and
Carnobacterium are abundant, owing to vegetarian diets of
the majority of the population (42). SCFAs are the most abundant form of
saturated aliphatic organic acids where 10% of SCFAs are
constituted by the daily diet and 70% are generated as a product
synthesized through the microbial niche (43), found within the GIT (Table I). The concentration of SCFA plays
a crucial role at local (colon) and systemic (blood) levels for
immune regulations (44,45). SCFAs supplied from the diet are
utilized by the host for its other metabolic process, whereas
microbial-synthesized SCFAs are employed as energy sources for
epithelial and endothelial cells of the colon (colonocytes). In a
normal human gut, the ratio of the SCFA concentration comprises 60%
of acetate, 25% of propionate and 15% of butyrate (46). Alterations in the microbiome may
influence the susceptibility of an individual to infectious
diseases. For example, the rate of ethanol-induced liver damage is
reduced with the supplementation of butyrate than acetate (47). SCFAs regulate immune functions via
several receptors and pathways; specifically, G-protein coupled
receptors (GPCRs) namely GPCR43 and GPCR120 enhance reactive oxygen
species-mediated killing, phagocytosis and cytokine production,
leading to apoptosis (48).
 | Table IFormation of SCFA products in the
GIT. |
Table I
Formation of SCFA products in the
GIT.
| Name of SCFA |
Formation/pathways | Microbes utilizes
the pathway | Function | (Refs.) |
|---|
| Acetate | 1. Decarboxylation
of pyruvate followed by Acetyl-CoA is hydrolyzed to acetate by an
acetyl- CoA hydrolase 2. Wood-Ljungdahl pathway | Prevotella
spp., Ruminococcus spp., Bifidobacterium spp.,
Bacteroides spp., Clostridium spp.,
Streptococcus spp., Akkermansia muciniphila,
Blautia hydrogenotrophica | 1. Insulin level
maintenance 2. Body weight loss 3. Energy production for other
cells of host 4. Lipid synthesis 5. Protein acetylation | (49) |
| Propionate | 1. Succinate
pathway 2. Acrylate pathway 3. Propanediol pathway | Bacteroidetes and
several Firmicutes, Coprococcus catus, Salmonella
enterica serovar Typhimurium, Roseburia
inulinivorans, Akkermansia municiphilla | Gluconeogenesis in
the liver Inhibits histone deacetylase | (50) |
| Butyrate | 1. Classical
Pathway: phosphotransbutyrylase and butyrate kinase enzymes are
involved 2. Butyryl-CoA: acetate CoA-transferase converts
butyryl-CoA into butyrate and acetyl-CoA | Ruminococcus
bromii, Coprococcus species, Eubacterium rectale,
Eubacterium rectale/Roseburia spp., Faecalibacterium
prausnitzii | Intestinal and
extra-intestinal functions | (51) |
5. Significance of pre- and probiotics
Prebiotics are non-digestible food ingredients that
have a positive impact on the composition of the gut microbiome and
their metabolic function at the level of the small intestine and
colon. Alternating the nutrient supplements selectively stimulates
the growth and/or activity of one or a bacterial community within
the colon. Prebiotics and dietary fibers promote the growth of
beneficial bacteria in the gut by acting as a substrate for
fermentation (49-51).
Prebiotic supplements maintain the homeostasis barrier and elevate
mineral absorption, thereby modulating energy metabolism and
satiety. The relevance of microbial diversity and its association
with metabolizing prebiotics and dietary fiber in the upper and
lower GIT, as well as the colon has been previously discussed
(52,53).
Probiotics are supplements with live bacterial feed;
when supplied in adequate quantity, they provide health benefits to
the host. The genera Lactococcus and Bifidobacterium
strains are predominant in the gut. These microbes are considered
significant for use as probiotics due to their exclusive
properties, such as acid and bile tolerance, adhesion to mucosal
and epithelial surfaces, antimicrobial activity against pathogenic
bacteria and bile salt hydrolase activity. The rationale behind the
supply of probiotics is to reshape and strengthen the commensal
microbes to compete for nutritional and functional resources with
the pathogens and to produce antimicrobial substances (54). However, factors such as safety,
product compatibility, viability of the microorganism, pH,
packaging, storage conditions and food processing should be
considered in the case of probiotics supplementation. Therefore,
probiotics should be improved for their viability and stability to
achieve the 100% establishment of microbes in the GIT. Soft cheeses
have an added advantage over yogurts and fermented products for
their delivery system of viable products to the GIT. The selection
of probiotics strains should meet up with the regulations according
to the World Health Organization (WHO), the European Food Safety
Authority (EFSA) and Food and Drug Administration (FDA) for their
safety and functional criteria (55). Technologies, including
microencapsulation, straw delivery system and viable spores of
spore forming organisms are being improvised to overcome the
delivery of live microorganisms into the GIT; however, advanced
techniques to supply other beneficial microbes as probiotics need
to be identified (56).
Dietary supplements can be a combination of
prebiotics and probiotics, that are together represented as either
conbiotics and as synbiotics. The use of synbiotics improves the
survival rate of beneficial microbes and supplies the respective
food or feed required for their proliferation. The challenge in
synbiotics supplementation is to identify the right formulations of
prebiotics and probiotics that are capable of modulating the
intestinal microbiota (55).
6. Human microbiome and its association with
diseases
Microbiome disease associations are very complex and
various omics approaches are integrated to understand them. A major
concern is the ongoing evolution of the microbiome and its direct
effect on metabolite production and its susceptibility towards any
disease, since it has the ability to modulate the immune system.
The human gut is inhabited symbiotically by a complex and
metabolically active microbial ecosystem, which systematically
controls the metabolic activity of humans. In healthy individuals,
the microbiota bacteria present in the lungs are
Pseudomonas, Streptococcus, Prevotella,
Fusobacterium, Haemophilus, Veillonella and
Porphyromonas (57). The
microbial metabolic products are of great use for maintaining gut
homeostasis and their varied taxonomic associations provide
detailed information on the diagnostic features of the individual
for personalized medicine. Nutritional supplements mostly include
dietary fibers, prebiotics and probiotics that play a critical role
in the gut homeostasis, and have also been reported to ameliorate
chronic disorders (58). The
association between food intake, microbiome health and the
occurrence of various diseases has been studied. CRDs includes a
range of lung disorders and are major concerns among the global
health burdens. CRDs affect the airways and lungs; among these,
chronic obstructive pulmonary disease (COPD) is the third leading
cause of mortality worldwide (59). Frequent exposure to polluted air,
hazardous chemical substances such as carcinogens, poor lifestyle
habits (smoking and alcohol consumption) are key factors for the
increased prevalence of asthma and lung cancer (60,61).
COVID-19, although an acute respiratory syndrome, increased risk of
mortality of patients infected with COPD and asthma (62).
The microbial composition in diseased lower airways
exhibits a decreased abundance of the phylum Bacteroidetes and
increased Gammaproteobacteria in contrast to the healthy
population. These Gram-negative pathogens of
Gammaproteobacteria are capable of utilizing the
inflammatory byproducts for their survival under anaerobic and
lower oxygen conditions (63).
Cancer
The association between diet and cancer (colorectal,
post-menopausal breast cancer) was recognized by the Dietary
Guideline Advisory Committee (DGAC) in 2015(64). It has been observed that the
consumption of a high-fat ketogenic diet (HFD) affects the
intestinal stem cell functions through the metabolite
β-hydroxybutyrate-mediated Notch signaling through the expression
of the gene, 3-hydroxy-3-methylglutaryl-CoA synthetase 2. A HFD
along with glucose inhibits the 3-hydroxy-3-methylglutaryl-CoA
synthase 2, an enzyme involved in the production of beneficial
ketone bodies (65). Switching
from glycogenic to ketogenic energy consumption benefits the growth
of normal cells instead of cancer cells by limiting the glucose
production (66). At present, the
University of Iowa is investigating the potential effect of a
ketogenic diet along with chemoradiation therapy for non-small cell
lung cancer (NCT01419587).
Butyrate has pleiotropic functions and plays a role
in the apoptosis of colon cancer cells, intestinal gluconeogenesis
and in controlling gut dysbiosis. Butyrate indirectly regulates the
health of the lungs through systemic inflammatory processes;
treatment with butyrate attenuates inflammation and promotes mucus
production in the lungs (67).
Mucin production is promoted by the butyrate-producing bacteria,
Roseburia spp. Although the mechanisms connecting the
gut-lung axis have not yet been elucidated, there is some
scientific evidence indicating that variant associations among
species and their relative abundance can be used as markers for
lung cancer prediction (68). The
enhanced activation of glutamate metabolism requires the
consumption of the majority of butyrate in the cells of lung
cancer; therefore, the shortage of butyrate for mucin production
leads to its destruction and causes inflammation (69). Consistently, dysbiosis has been
found to occur in the gut microbiota among patients with lung
cancer and the decrease in butyrate-producing bacteria, such as
Faecalibacterium prausnitzii, Clostridium leptum,
Clostridium cluster I, Ruminococcus spp.,
Clostridium cluster XIVa and Roseburia spp. are
observed in NSCLC. Therefore, the mentioned butyrate-producing
bacterial strains along with Clostridium cluster IV and
Eubacterium rectalis can be considered as gut probiotics. In
addition, these butyrate-producing bacteria have attracted
attention due to their ability to maintain the gut homeostasis
(70). The three proposed theories
for microbial dysbiosis for the onset of carcinogenesis include the
disruption of the immune equilibrium, the initiation of chronic
inflammation and the activation of cancer-causing pathways
(71).
In lung cancer, cell proliferation is associated
with various microbial genera. The microbiome number is altered in
the normal and diseased condition. The most dominant phyla in the
lungs of healthy individuals are Actinobacteria,
Bacteroides, Firmicutes and Proteobacteria
(9). Variations in these
microorganisms are observed in different types of cancer. For
example, in the patients affected with squamous cell carcinoma of
the lung and adenocarcinoma, the genera Capnocytophaga,
Selenomonas and Veillonella are abundant when
compared with the genus Neisseria (72). These microbiota thrive due to their
expression and their role in the activation of signaling and cell
proliferation pathways. For example, the β-catenin signaling
pathway, which promotes cell proliferation, is activated by
Fusobacterium nucleatum. Estimating the occurrence of levels
bacteria such as Fusobacterium nucleatum may be useful for
predicting the progression of cancer. It has also been observed
that patients with lung cancer, when compared with those with
benign disease, have an abundant population of Veillonella
and Megasphaera. Genetic variations in individuals are also
helpful for developing biomarkers. Patients suffering from squamous
cell carcinoma of the lung, lung cancer or benign lung disease with
the TP53 mutation have been shown to have abundant levels of the
genus Acidovorax. Microbiota dysbiosis can contribute to the
pathogenesis of lung cancer, as it leads to the upregulation of
IL6/8, IL17A, MAPK and inflammasome pathways (72).
In a previous study, when probiotic supplements with
aqueous extracts of Bifidobacterium species were to NSCLC
cell lines (A549, H1299 and HCC827) particularly reduced cell
proliferation with the increased expression of cleaved caspase-3
and cleaved poly(ADP-ribose) polymerase (PARP) associated with the
apoptotic pathway (73). In
another study, the additional supplementation of probiotics
containing Bifidobacterium and Clostridium butyricum
in patients with NSCLC receiving anti-programmed cell death-1
therapy (nivolumab, or pembrolizumab monotherapy) increased their
survival state (74).
Bifidobacterium activates dendritic cell function and
directs the T-cell mediated antitumor efficiency, whereas
Clostridium butyricum arrests cancer cell invasion and
migration by reducing Th17 cells (75). It has been shown that bacterial
supplements with Akkermansia muciniphila along with butyrate
inhibit tumor cell proliferation and migration by upregulating the
expression of miRNA (interference RNA) (76). Treatment of NSCLC with propionate
regulates survivin, an inhibitor of the apoptosis protein family
and suppresses p21 expression and the cell cycle (77). To date, the full regulatory
mechanisms of the gut microbiome in systemic inflammation and lung
diseases are not yet completely known; thus, it is necessary to
take a multidisciplinary approach to elucidate these
mechanisms.
COPD and asthma
COPD is a fatal lung disorder and an irreversible
obstruction in the respiratory tract potentially accompanied with
progressive decline in the function of lungs. The pathologies
include chronic bronchitis, airway remodeling and emphysema. One of
the major causes of COPD is smoking. There is a visible difference
between the gut microbiota of healthy smokers and non-smokers
(78). The intestinal levels of
Firmicutes are decreased and the count of Bacteroidetes increases
in smokers, whereas when an individual quits smoking, the effects
are vice versa. The production of SCFAs is highly variable between
smokers, non-smokers and in those who have quit smoking, and this
has direct response to the immune response exerted in the lungs.
The core microbiome of the lungs is mainly composed of
Pseudomonas, Streptococcus, Prevotella, Fusobacterium,
Haemophilus, Veillonella and Porphyromonas; these also
play a protective role by producing SCFAs. For example, butyrate
secreted by the gut microbiota can reduce the lung damage caused by
cigarette smoke-induced-COPD, by inhibiting the mevalonate pathway
in the gut, lungs and liver. However, the inflammation/emphysema
pathological condition can be reduced by supplementing with a high
fiber diet and probiotic intake of Lactobacillus rhamnosus
and Bifidobacterium (79,80).
The lung microbiota of patients with COPD exhibits an increase in
Gammaproteobacteria, whereas healthy lungs exhibit
Bacteroidetes (81).
The lung microbiome is frequently assessed through
samples, such as serum, sputum, bronchoalveolar lavage fluid (BALF)
and lung tissue, whereas for the gut microbiome, fecal samples are
used. In a number of respiratory disease conditions, the spatial
heterogeneity in the composition of the lung microbiome is highly
variable; however, within an individual, the microbial spatial
variation is less among healthy populations and either the segment
of lingula or BALF from the right middle lobe are used for
microbial assessment (82). Of
note, in COPD the microbial diversity varies with the sampling from
upper and lower bronchial regions of the lung (83). Exacerbations of COPD, are often
caused by respiratory viruses or bacteria; these exacerbations are
associated with markedly increased morbidity and mortality rates
(84). Studies on the dynamics of
the microbiome in different exacerbation phenotypes have indicated
that diverse patterns of microbiota existence are associated with
the types of exacerbation events (types 1, 2 and 3) (85,86).
Therefore, for the understanding of disease etiology, associated
changes in the microbial community and cytokine profiles are key
features to be explored towards the development of more effective
therapeutic strategies. The microbial niches and their associated
mediators of inflammation for Haemophilus are IL-1β and
tumor necrosis factor-α in sputum, as well as neutrophilic
inflammation and Moraxella, which are related to Th1
pathways such as interferon signaling (87). The evaluation of the gut microbiome
does not indicate the severity of COPD; however, the varied
prevalence of microbiota at different stages of COPD has been
observed (88). Diet plays a major
role in maintaining microbial diversity; mortality rates due to
COPD have been found to increase with western diets and may be
reduced among populations consuming diets rich in antioxidants and
fiber (88).
Medications for COPD include antibiotics, steroids
and the use of bronchodilators, which exert effects on microbial
composition. Recent day challenges, such as the emergence of
multidrug resistance for several antibiotics and metal-based drugs
with the capability of modulating the microbiome in COPD have been
previously discussed (71). A
comprehensive elucidation of the existing evidence, challenges and
the future concerns regarding the possible use of the lung
microbiome either as a potential biomarker or therapeutic target
for COPD has been previously provided (89). The microbial distribution of the
lung and gut in COPD has also bene previously detailed (83).
COPD and asthma are the two most frequently
diagnosed chronic respiratory diseases and it has been suggested
that the oral, lung and gut microbiota are associated with these
conditions. Asthma is a multifactorial and heterogeneous disease
characterized by the presence of airflow limitation associated with
chronic bronchitis or emphysema. Microbiota composition in the
respiratory tract mainly consists of Actinobacteria, Firmicutes,
Proteobacteria, Bacteroidetes, etc. and it is suggested that the
development of allergic diseases such as asthma is dependent on
these, as they influence inflammation via the gut-lung axis
(90). As regards gut microbial
dysbiosis, the lower abundance of Firmicutes increases the risk of
asthma and as with COPD lung microbial composition,
Proteobacteria such as Haemophilus and
Moraxella are the predominant among the lung microbiome
(91).
Delivering probiotics orally has its own efficiency
in immune cell regulation in the respiratory system; the most
common probiotics include lactic acid-producing bacterial species
such as Lactobacillus, Streptococcus, Bifidobacterium and
Enterococcus, as well as the non-pathogenic yeast,
Saccharomyces boulardii. The systemic immune modulating
capability of Lactobacilli has been found to affect the cytokine
profile, T-cell proliferation, and the phagocytic activity of
mononuclear cells and natural killer (NK) cells (92,93).
Research has demonstrated the involvement of immune cells like Th17
(T-helper) and Treg (regulatory T-cells), which indicates the role
of CD4+ T-cells in the production of pro-inflammatory
cytokines, as well as the transcription factor FoxP3 + CD4 cells in
the immune homeostasis. The administration of Lactobacillus
reuteri, Bifidobacterium lactis Bb12, Lacticaseibacillus
rhamnosus strain GG (LGG) and Lactobacillus casei
increase the levels of these immune cells (94,95).
In asthmatic conditions, the reduction of airway
hyperresponsiveness and the number of inflammatory cells in BALF
and in lung tissue have been reported with the supplementation of
Lactobacillus reuteri, LGG and Bifidobacterium breve
(96). Furthermore, probiotic
supplements with the heat-killed Mycobacterium vaccae confer
protection against airway allergic inflammation (97). Intaking Enterococcus
faecalis FK-23 attenuates TH17 cell development as an asthmatic
response, whereas NK cell impairment in lungs due to cigarette
smoking indicates an increase in NK cell activity with the
supplementation of Lactobacillus casei (98). The markers of systemic inflammation
and oxidative stress, such as plasma C-reactive proteins and
8-isoprostane are significantly reduced by probiotic treatment.
Moreover, multistrain probiotics stabilize the neuromuscular
junction by improvising muscle strength and its functional
performance by reducing the intestinal permeability (a randomized
controlled clinical trial no. GMC-CREC-00263) (99,100). The therapeutic application of the
gut bacteriome for the treatment of COPD and asthma has been
previously discussed (101).
The bacterium Parabacteroides goldsteinii
ameliorates COPD by reducing intestinal inflammation and enhancing
cellular mitochondrial activities, restoring the abnormal amino
acids metabolism through ribosomal activities. The derived
lipopolysaccharides of P. glodsteinii are efficient to
interact with Toll-like receptor (TLR)4 signaling pathways, thereby
acting as an anti-inflammatory agent (102). Similarly, the derived saponins of
the plant American ginseng have been reported to ameliorate COPD
(103). Butyrate has pleiotropic
functionality and has several beneficial effects on the maintenance
of homeostasis of the gut and lung microbiota. Despite the short
half-life of butyrate, rendering it a poor oral supplement for lung
disorders, a pre-clinical evaluation of the administration of
butyrate and metformin through inhalation was performed, to
directly target the lung (103).
COVID-19
COVID-19 is the recent pandemic caused by a virus
mainly affecting the respiratory tract. Recent research has
indicated the association between gut microbiome alterations and
COVID-19 severity, providing evidence that the translocation of
bacteria into blood is associated with gut microbiome dysbiosis
(104). This can lead to the
occurrence of secondary infections. In this recent study,
alterations in Paneth cells, goblet cells and markers of
permeability barriers in association with alterations in the gut
microbiome were observed. The gut-lung crosstalk is bidirectional,
where the metabolites produced by the altered gut microbes
transfuse through and exert affect the lungs, and lung inflammation
affects the gut microbiota, and vice versa (104). Angiotensin converting enzyme
(ACE2) is the enzyme that is located on the outer surface and
functions as the gateway for viral entry and it regulates
intestinal inflammation (105).
The components of the bacterial envelope (lipopolysaccharides and
peptidoglycans) facilitate the interaction of viral proteins, which
enhances thermostability in Polio and other viruses (106). This indicates the importance of
commensals in the increasing/decreasing of viral infections.
The gut microbiota of patients with COVID-19 are
abundant with opportunistic pathogens, such as Acinetobacter
baumannii and Candida spp. Furthermore, the functional
loss of ACE2 results in gut microbiome imbalance and a leaky gut.
These changes affect the host immune responses to SARS-CoV-2
infection (107). The pro- and
pre-biotic treatment for other viral influenza confirms their
potency to combat COVID-19. The flagellin of commensal bacteria are
capable of activating pattern-recognition receptors (PRRs) of the
pathogen and stimulate the release of IL-18 and IL-22, which are
cytokine derivatives of the intestinal epithelium and significantly
contribute to host defense mechanisms against inflammation and
intestinal infection (108).
Treatment with retinoic acid mediates the production of interferons
through the bacterium Lactobacillus sp. and also increases
the antiviral activity of vitamin A (109,110). Gut commensal Clostridium
orbiscindens promotes host immune protection against influenza
infection by enhancing the type I interferon signaling (111). The antiviral role of TLRs are
scientifically proven mechanisms of host immune responses and the
gut commensal microbiomes are expected to activate the TLRs and the
release of antiviral protein cathelicidin from mast cells (112). Through TLR ligand stimulation,
the death rate due to viral infections is significantly reduced
with the restoring responses of interferon gamma and
CD4+ T-cell responses (113). The bacterial species
Lactobacillus paracasei and Lactobacillus plantarum
activate the release of the cytokine IL-10 and suppress the
inflammatory responses in the lungs (114,115).
Apart from bacterial balances within the gut, the
intra-kingdom crosstalk between fungi, virus and bacteria has a
marked influence in maintaining the gut homeostasis (116). A significant attenuation of
COVID-19 infection and a reduced risk of respiratory failure was
observed when patients were provided with standard medication and
additional supplements with bacterial strains (Bifidobacterium
longum, Bifidobacterium animalis subsp. Lactis and Lactobacillus
rhamnosus, in addition vitamin D, zinc and selenium); this
clinical trial was registered with the number NCT04666116(117).
Dietary fibers, such as fructans and
galactooligosaccharides increase the prevalence of
Bifidobacteria and Lactobacilli (118). The intestinal commensals,
Bifidobacteria and Lactobacilli, which are essential
for inhibiting the colonization of pathogenic organisms in the gut
epithelial cells. Adhesin production by commensals and probiotic
supplements regenerates the intestinal mucosa, thereby suppressing
viral adhesion and replication, and reducing the risk of
respiratory tract infections. Peptides produced by
Lactobacillus and Paenibacillus block the binding of
SARS-CoV-2 with ACE2(119). The
lesions in the lung affects the gas exchange capability among
COVID-19-infected patients, where the probiotic formulation SLAB51
combats the breathing difficulty by reducing nitric oxide synthesis
within the gut (120).
Polyphenols of beverages and other foods favor the growth of
beneficial gut bacteria and provide intestinal barrier function
(120). The active clinical
trials for lung cancer, COPD, asthma and COVID-19 are presented in
Table II.
 | Table IIActive clinical trials utilizing pre-
and probiotics for lung cancer, COPD, asthma and COVID-19
(https://clinicaltrials.gov/). |
Table II
Active clinical trials utilizing pre-
and probiotics for lung cancer, COPD, asthma and COVID-19
(https://clinicaltrials.gov/).
| Clinical Trial Gov.
identifier | Subject or
title |
|---|
| NCT04699721 | Clinical study of
neoadjuvant chemotherapy and immunotherapy combined with probiotics
in patients with potential/resectable NSCLC |
| NCT05094167 |
Lactobacillus Bifidobacterium
V9(Kex02) improving the efficacy of carilizumab combined with
platinum in non-small cell lung cancer |
| NCT03642548 | Probiotics combined
with chemotherapy for patients with advanced-stage NSCLC |
| NCT03068663 | Microbiota and the
lung cancer |
| NCT04871412 | The thoracic
peri-operative integrative surgical care evaluation trial- Stage II
(POISE) |
| NCT05037825 | The gut microbiome
and immune checkpoint inhibitor therapy in solid tumors
(PARADIGM) |
| NCT05303493 | Camu-Camu prebiotic
and immune checkpoint inhibition in patients with non-small cell
lung cancer and melanoma |
| NCT05492448 | Probiotic on type 2
diabetes and chronic obstruction pulmonary disease |
| NCT05126654 | Partially
hydrolyzed Guar Gum (PHGG) for amelioration of chronic obstructive
pulmonary disease (COPD) |
| NCT05523180 | A study to evaluate
the effect of probiotic supplement on quality of life |
| NCT04366089 | Oxygen-ozone as
adjuvant treatment in early control of COVID-19 progression and
modulation of the gut microbial flora (PROBIOZOVID) |
| NCT04368351 | Bacteriotherapy in
the treatment of COVID-19 (BACT-ovid) |
| NCT00298337 | Use of probiotic
bacteria in prevention of allergic disease in children
1999-2008 |
| NCT01419587 | Ketogenic diet with
chemoradiation for lung cancer (KETOLUNG) |
Bacterial pneumonia and its impact on
the gut microbiota
Bacterial pneumonia remains a leading cause of
respiratory illness and mortality worldwide, with treatment relying
heavily on antimicrobial agents (121). According to recent research, the
human gut microbiota plays a critical role in protecting the body
from infections due to antimicrobial resistance. The body is
protected by a balanced microbiota through defense mechanisms, such
as nutritional competition, niche exclusion, colonization
resistance by secreting antibacterial substances, restoring the
mucosal barrier, suppressing inflammation and immune modulation
etc., rendering it a vital ally in the battle against antimicrobial
resistance. However, this protection is compromised by gut
dysbiosis, which makes it easier for resistant infections to
persist, colonize and spread (122). While antibiotics are essential
for managing the infection, they can significantly disrupt the gut
microbiota, leading to dysbiosis and potential long-term health
consequences (123). Antibiotic
resistance genes are derived from the gut microbiome, which has
developed into a major part of the resistome. Antibiotics alter the
natural microbiota of the host by favoring resistant microorganisms
that may cause opportunistic illnesses (124). The dysbiosis in the upper
respiratory tract microbiome has been shown to be a major cause of
bacterial pneumonia in the elderly and in young adults (125). Gut dysbiosis may be linked to
subtherapeutic antibiotic treatment, the consumption of low-dosage
antibiotics from food and horizontal gene transfer, leading to gene
acquisition or loss, bacterial biofilms, quorum sensing and other
environment factors (125).
Commonly prescribed antibiotics, including β-lactams
(such as penicillins and cephalosporins), macrolides (such as
azithromycin and clarithromycin), fluoroquinolones (such as
levofloxacin and ciprofloxacin), tetracyclines and aminoglycosides,
have been shown to reduce microbial diversity and deplete
beneficial gut bacteria, such as Bifidobacterium and
Lactobacillus. This disruption weakens immune function,
promotes the growth of opportunistic pathogens, and increases the
risk of developing antibiotic-resistant infections, such as
Clostridium difficile. Over time, these alterations can
contribute to metabolic disorders and chronic health issues
(126). Restoring the gut balance
through probiotics, prebiotics, fecal microbiota transplantation
and dietary interventions may help mitigate these effects and
support overall gut and immune health.
7. OMICS techniques: Metagenomics,
metatranscripto-mics, metaproteomics and metabolomics
The human intestine is colonized by >1,000
microbial species. The gut microbial community plays a critical
role in the protection of the host against pathogenic microbes,
modulating immunity and regulating metabolic processes (127,128). Since the beginning of microscopy,
several human gut microbial communities have been identified.
Recent technological advances in genomics and microbial genetics
have led to the identification of human microbiota associated with
various diseases.
Molecular biological technology plays a vital role
in the intestinal microbiome analysis; in particular, metagenomic
sequencing using the next-generation sequencing (NGS) techniques
has led to notable developments in the study of the human gut
microbiome. Metagenomics is widely used in the study of human gut
microbiome diversity and to gain the knowledge of its insight with
relation to various diseases, as well as its association with
health and disease (129,130). This aids in the identification of
taxonomic identification, and in the detection and characterization
of microbial species (131).
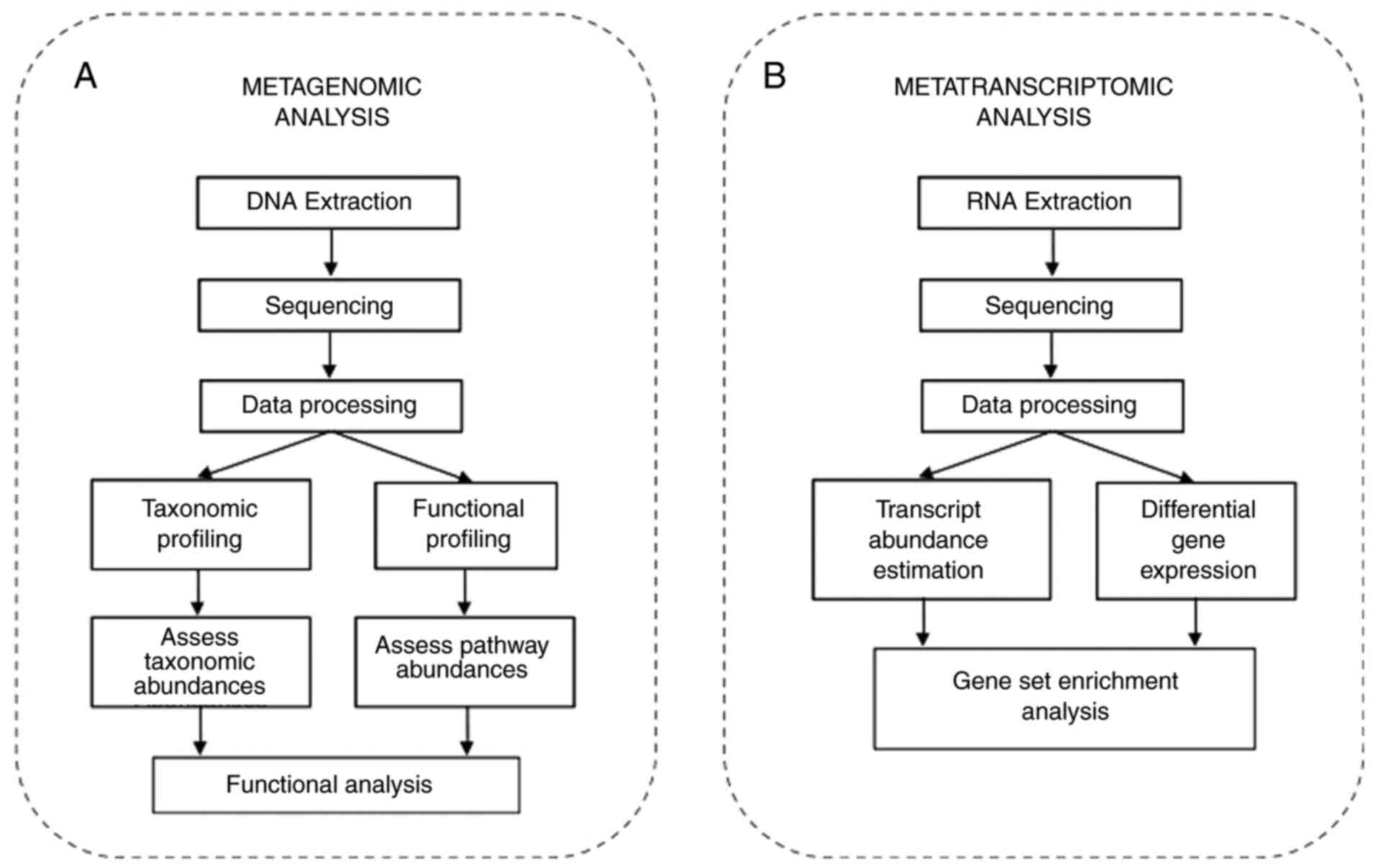
Metagenomic analysis involves the extraction of genomic DNA and
sequencing. The next step involves annotation after assembly and
the mapping of sequences to the reference database (Fig. 2A). Further, computational
annotation is performed using bioinformatics tools, such BLAST, COG
and KEGG, and statistical analysis can be performed using R and
Bioconductor (132,133).
The application of metagenomics involves the
identification of novel functional genes, microbial pathways,
antibiotic resistance genes, functional dysbiosis of the intestinal
microbiome, and the determination of interactions and co-evolution
between microbiota and host (134,135). Although the complete bacterial
repertoire of the human microbiome is unidentified, the study by
Almeida et al (136)
provided information of a vast diversity of uncultured gut
bacteria, and supports the expansion of the gut microbiome
repertoire of the human gut microbiota. Metagenomics also enables
the profiling of whole gene repertoire and plays a vital role in
the pan-genomic analyses and also enables the identification of
whole genome content from a sample and determination of closely
related taxa (137).
Metatranscriptomics involves the sequencing of the
complete transcriptome of the microbial community and determines
the genes that are expressed by the microbiome and helps determine
the functional profile of the community under different conditions
(138). In brief, metagenomics
helps to identify the composition of a microbial community under
different conditions, while metatranscriptomics helps identify the
genes that are collectively expressed under different conditions
(139). A metatranscriptome
experiment involves the isolation of total RNA from a sample and
its sequencing followed by annotation, assembly and bioinformatics
analysis (140), as illustrated
in Fig. 2B.
Other novel omics approaches, such as
metatranscriptomics (141),
metaproteomics (142) and
metabolomics (143) also
complement the study of the human gut microbiome and support the
identification of the microbiota community. Metaproteomics provides
the knowledge of the entire protein complement of the microbiota
communities, provides insight into the genes expressed and helps in
the identification of the key metabolic activities (144). Metabolomics provides details of
the metabolite composition of the microbiota community, and
provides an understanding of the functional dynamics of the
microbiome community and the host interactions (145). A brief comparison of the
different omics techniques, including advantages and disadvantages
to understand the choices and limitations of each approach is
provided in Table III.
 | Table IIIA brief comparison of the different
omics techniques. |
Table III
A brief comparison of the different
omics techniques.
| Omics
techniques | What is
analyzed | Advantages | Limitations |
|---|
| Metagenomics | DNA (microbial
genomes) | Identifies
microbial composition and potential functional genes. | Does not indicate
active gene expression; unable to assess actual function. |
|
Metatranscriptomics | RNA (microbial
transcripts) | Captures gene
expression and functional activity under specific conditions. | RNA is unstable;
requires careful sample handling; highly dynamic and
contextual. |
| Metaproteomics | Proteins (microbial
proteome) | Provides direct
evidence of expressed proteins and metabolic function. | Technically
challenging; limited by protein extraction and database
completeness. |
| Metabolomics | Metabolites (small
molecule end-products) | Reflects functional
output of microbiome and host-microbe interactions. | Difficult to link
metabolites to specific microbes; complex data interpretation. |
8. Machine learning for the development of
microbiome therapeutics
Machine learning is an artificial intelligence (AI)
technique that focuses on developing the intelligence to imitate
humans for analyzing a large volume of data. The technique is
widely applied to answer various biological and biomedical queries
including the microbiome field. A simple boolean search using the
query ‘gut microbiome’ and ‘machine learning’ retrieved 513 PubMed
articles (on May 31, 2025). The first article was published in 2013
and the study focused on predicting the genes in metagenomic
fraction by using support vector machines (SVM), one of the
standard machine learning approaches for classification (146). The authors of that study
presented a novel gene prediction method called MetaGUN and
included three different stages: i) classifying the fragments into
different phylogenetic groups with k-mer based sequence binding
approach; ii) identifying protein coding sequences from each
phylogenetic group using SVM that uses entropy density profiles,
translation initiation site, and open reading frame length as input
patterns; and iii) adjusting the translation initiation site
(146).
Human health is greatly affected by the gut
microbiota, which affects immune responses, metabolism and disease
prevention. Numerous diseases, including cancer, are associated
with microbial imbalance, or dysbiosis, a key observation that can
be utilized for the early detection of disease. AI methods are well
suited for large-scale biomedical big data mining, including
microbiome data mining. Foundation models and transfer learning (ML
and DL models) have been shown to have immense success with
microbiome-based classification and prediction (147). Random Forest (RF), an ensemble
learning technique, has been used for feature selection and
classification in microbiome datasets to identify key microbial
taxa associated with diseases such as type 2 diabetes and
inflammatory bowel disease (148). SVMs and other machine learning
techniques have markeldy enhanced the predictive power of cancer
diagnosis and prognosis, especially in research involving gut
microbiota. When trying to understand the complex interaction
between the gut microbiota and cancer prevention, the SHapley
Additive exPlanations (SHAP) algorithm is a crucial part of the
eXplainable Artificial Intelligence (XAI) architecture (146,149). Machine learning models such as
Ridge regression, Elastic Net, LASSO, Random Forest, Ridge
regression and LASSO were constructed using the SIAMCAT v_2.0 and
v_2.10 toolbox and have been used to accurately classify patients
with Parkinson's diasease, with an average AUC of 71.9%. However,
it was found that the trained models were study-specific and do not
generalize well to datasets from other neurodegenerative diseases
research (150). The increasing
application of these varied algorithms highlights their potential
for improving integrative multi-omics analysis, precision medicine
and microbiome-based diagnostics.
Recent research has focused on disease prediction
(151-153),
the association between the gut microbiome and disease (e.g.
obesity, constipation, type 2 diabetes, cardiovascular diseases)
(154-157),
the epigenetic regulation of brain disorders (158), and the role of the gut microbiome
in promoting precision medicine for cancer (159). The interaction between the gut
microbiome and the intestinal cells is well known. Recently, the
interaction between the gut microbiome and nervous system,
particularly the enteric nervous system and central nervous system
has gained increasing attention among researchers and neurologists
(158). Studies have shown the
involvement of the gun microbiome in anxiety (160), autism (161), depression (162), and a number of neurodegenerative
diseases (163-167).
The influence of the gut microbiome on the host metabolism by
activating epigenetic regulators plays a crucial role in
understanding the pathogenesis of numerous neurological disorders,
including brain disorders (143).
The integration of multi-omics data, including genomics,
metabolomics, connectomics and gut microbiome multi-omics can
provide a path for moving forward towards the prevention of disease
and for the development of personalized therapeutics, as opposed to
merely the prediction of diseases. Both supervised and unsupervised
machine learning approaches can be used for such studies (158). Thus, machine learning approaches
find a wide range of applications in gut microbiome research.
9. Conclusion and future perspectives
Respiratory diseases are markedly affected both by
genetic and external factors. Where the genetic factors are
difficult to control, the immunological response to the
environmental factors can be mediated. In this regard, the human
microbiota serves as a source towards this approach. The health of
the gut microbiota is sustained with fibrous food intake,
probiotics, etc. This cascades the immunological response in
chronic respiratory diseases. The omics approach provides details
of the metabolite composition and provides an understanding of the
functional dynamics of the microbiome community and host
interactions. The association between diet and the gut microbiome
is analyzed using machine learning approaches, and much of the
hidden information in literature can be explored using text mining
approaches. This survey highlights the diagnostic aspects of
personalized medicine based on the gut-lung axis microbiome of an
individual by integrating various areas of research, such as
molecular biology, immunology, omics, machine learning and text
mining.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
All the authors (SM, ORI, AP, BB and KR) were
involved in the conceptualization of the study. All the authors
(SM, ORI, AP, BB and KR) were also involved in the formal analysis
(collecting the relevant data and articles from the literature), in
the writing and preparation of the original draft of the
manuscript, and in the writing, review and editing of the
manuscript. SM, ORI and AP were involved in visualization
(preparation of the figures). KR supervised the study. All authors
have read and agreed to the published version of the manuscript.
Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Miedema I, Feskens EJ, Heederik D and
Kromhout D: Dietary determinants of long-term incidence of chronic
nonspecific lung diseases. The zutphen study. Am J Epidemiol.
138:37–45. 1993.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wilson DO, Donahoe M, Rogers RM and
Pennock BE: Metabolic rate and weight loss in chronic obstructive
lung disease. JPEN J Parenter Enteral Nutr. 14:7–11.
1990.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Han SN and Meydani SN: Vitamin E and
infectious diseases in the aged. Proc Nutr Soc. 58:697–705.
1999.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Tabak C, Feskens EJ, Heederik D, Kromhout
D, Menotti A and Blackburn HW: Fruit and fish consumption: A
possible explanation for population differences in COPD mortality
(The Seven Countries Study). Eur J Clin Nutr. 52:819–825.
1998.PubMed/NCBI View Article : Google Scholar
|
|
5
|
DeMeo MT, Van de Graaff W, Gottlieb K,
Sobotka P and Mobarhan S: Nutrition in acute pulmonary disease.
Nutr Rev. 50:320–328. 1992.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ellis JA: The immunology of the bovine
respiratory disease complex. Vet Clin North Am Food Anim Pract.
17:535–550, vi-vii. 2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Furrie E: Probiotics and allergy. Proc
Nutr Soc. 64:465–469. 2005.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Behrouzi A, Nafari AH and Siadat SD: The
significance of microbiome in personalized medicine. Clin Transl
Med. 8(16)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hou K, Wu ZX, Chen XY, Wang JQ, Zhang D,
Xiao C, Zhu D, Koya JB, Wei L, Li J and Chen ZS: Microbiota in
Health and Diseases. Signal Transduct Target Ther.
7(135)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Liu XF, Shao JH, Liao YT, Wang LN, Jia Y,
Dong PJ, Liu ZZ, He DD, Li C and Zhang X: Regulation of short-chain
fatty acids in the immune system. Front Immunol.
14(1186892)2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Brittain HK, Scott R and Thomas E: The
rise of the genome and personalised medicine. Clin Med (Lond).
17:545–551. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lin BM, Grinde KE, Brody JA, Breeze CE,
Raffield LM, Mychaleckyj JC, Thornton TA, Perry JA, Baier LJ, de
las Fuentes L, et al: Whole genome sequence analyses of eGFR in
23,732 people representing multiple ancestries in the NHLBI
trans-omics for precision medicine (TOPMed) consortium.
EBioMedicine. 63(103157)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kowalski MH, Qian H, Hou Z, Rosen JD,
Tapia AL, Shan Y, Jain D, Argos M, Arnett DK, Avery C, et al: Use
of >100,000 NHLBI trans-omics for precision medicine (TOPMed)
consortium whole genome sequences improves imputation quality and
detection of rare variant associations in admixed African and
hispanic/latino populations. PLoS Genet.
15(e1008500)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Eichler EE: Genetic variation, comparative
genomics, and the diagnosis of disease. N Engl J Med. 381:64–74.
2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Khan I, Bai Y, Zha L, Ullah N, Ullah H,
Shah SRH, Sun H and Zhang C: Mechanism of the gut microbiota
colonization resistance and enteric pathogen infection. Front Cell
Infect Microbiol. 11(716299)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Arapidi GP, Urban AS, Osetrova MS, Shender
VO, Butenko IO, Bukato ON, Kuznetsov AA, Saveleva TM, Nos GA,
Ivanova OM, et al: Non-human peptides revealed in blood reflect the
composition of small intestine microbiota. BMC Biol.
22(178)2024.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Davis OK and Rosenwaks Z: Personalized
medicine or ‘one size fits all’? Fertil Steril. 101:922–923.
2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
NIH HMP Working Group. Peterson J, Garges
S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE,
Wetterstrand KA, et al: The NIH human microbiome project. Genome
Res. 19:2317–2323. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sender R, Fuchs S and Milo R: Revised
estimates for the number of human and bacteria cells in the body.
PLoS Biol. 14(e1002533)2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Martinez FJ, Erb-Downward JR and Huffnagle
GB: Significance of the microbiome in chronic obstructive pulmonary
disease. Ann Am Thorac Soc. 10 (Suppl 1):S170–S179. 2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kamel M, Aleya S, Alsubih M and Aleya L:
Microbiome dynamics: A paradigm shift in combatting infectious
diseases. J Pers Med. 14(217)2024.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Druszczynska M, Sadowska B, Kulesza J,
Gąsienica-Gliwa N, Kulesza E and Fol M: The intriguing connection
between the gut and lung microbiomes. Pathogens.
13(1005)2024.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Belaid A, Roméo B, Rignol G, Benzaquen J,
Audoin T, Vouret-Craviari V, Brest P, Varraso R, von Bergen M, Hugo
Marquette C, et al: Impact of the lung microbiota on development
and progression of lung cancer. Cancers (Basel).
16(3342)2024.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Borges RM, Colby SM, Das S, Edison AS,
Fiehn O, Kind T, Lee J, Merrill AT, Merz KM Jr, Metz TO, et al:
Quantum chemistry calculations for metabolomics. Chem.Rev.
121:5633–5670. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Natalini JG, Singh S and Segal LN: The
dynamic lung microbiome in health and disease. Nat Rev Microbiol.
21:222–235. 2023.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kennedy MS and Chang EB: The microbiome:
Composition and locations. Prog Mol Biol Transl Sci. 176:1–42.
2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Budden KF, Shukla SD, Rehman SF, Bowerman
KL, Keely S, Hugenholtz P, Armstrong-James DPH, Adcock IM,
Chotirmall SH, Chung KF and Hansbro PM: Functional effects of the
microbiota in chronic respiratory disease. Lancet Respir Med.
7:907–920. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ritchie AI and Wedzicha JA: Definition,
causes, pathogenesis, and consequences of chronic obstructive
pulmonary disease exacerbations. Clin Chest Med. 41:421–438.
2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang L, Cai Y, Garssen J, Henricks PAJ,
Folkerts G and Braber S: The bidirectional gut-lung axis in chronic
obstructive pulmonary disease. Am J Respir Crit Care Med.
207:1145–1160. 2023.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Rastogi S, Mohanty S, Sharma S and
Tripathi P: Possible role of gut microbes and host's immune
response in gut-lung homeostasis. Front Immunol.
13(954339)2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Liu Y, Teo SM, Méric G, Tang HHF, Zhu Q,
Sanders JG, Vázquez-Baeza Y, Verspoor K, Vartiainen VA, Jousilahti
P, et al: The gut microbiome is a significant risk factor for
future chronic lung disease. J Allergy Clin. Immunol. 151:943–952.
2023.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Koh A, De Vadder F, Kovatcheva-Datchary P
and Bäckhed F: From dietary fiber to host physiology: Short-chain
fatty acids as key bacterial metabolites. Cell. 165:1332–1345.
2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wu W, Sun M, Chen F, Cao AT, Liu H, Zhao
Y, Huang X, Xiao Y, Yao S, Zhao Q, et al: Microbiota metabolite
short-chain fatty acid acetate promotes intestinal IgA response to
microbiota which is mediated by GPR43. Mucosal Immunol. 10:946–956.
2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Deguen S, Amuzu M, Simoncic V and
Kihal-Talantikite W: Exposome and social vulnerability: An overview
of the literature review. Int J Environ Res Public Health.
19(3534)2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wiertsema SP, van Bergenhenegouwen J,
Garssen J and Knippels LMJ: The interplay between the gut
microbiome and the immune system in the context of infectious
diseases throughout life and the role of nutrition in optimizing
treatment strategies. Nutrients. 13(886)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yang L, Yuan TJ, Wan Y, Li WW, Liu C,
Jiang S and Duan JA: Quorum sensing: A new perspective to reveal
the interaction between gut microbiota and host. Future Microbiol.
17:293–309. 2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Xiao Y, Zou H, Li J, Song T, Lv W, Wang W,
Wang Z and Tao S: Impact of quorum sensing signaling molecules in
gram-negative bacteria on host cells: Current understanding and
future perspectives. Gut Microbes. 14(2039048)2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kho ZY and Lal SK: The human gut
microbiome-A potential controller of wellness and disease. Front
Microbiol. 9(1835)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Dubourg G, Lagier JC, Armougom F, Robert
C, Audoly G, Papazian L and Raoult D: High-level colonisation of
the human gut by verrucomicrobia following broad-spectrum
antibiotic treatment. Int J Antimicrob Agents. 41:149–155.
2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Markus V, Paul AA, Teralı K, Özer N, Marks
RS, Golberg K and Kushmaro A: Conversations in the Gut: The role of
quorum sensing in normobiosis. Int J Mol Sci.
24(3722)2023.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Tomova A, Bukovsky I, Rembert E, Yonas W,
Alwarith J, Barnard ND and Kahleova H: The Effects of vegetarian
and vegan diets on gut microbiota. Front Nutr. 6(47)2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Nagata N, Takeuchi T, Masuoka H, Aoki R,
Ishikane M, Iwamoto N, Sugiyama M, Suda W, Nakanishi Y,
Terada-Hirashima J, et al: Human gut microbiota and its metabolites
impact immune responses in COVID-19 and its complications.
Gastroenterology. 164:272–288. 2023.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Fusco W, Lorenzo MB, Cintoni M, Porcari S,
Rinninella E, Kaitsas F, Lener E, Mele MC, Gasbarrini A, Collado
MC, et al: Short-chain fatty-acid-producing bacteria: Key
components of the human gut microbiota. Nutrients.
15(2211)2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Maslowski KM and Mackay CR: Diet, gut
microbiota and immune responses. Nat Immunol. 12:5–9.
2011.PubMed/NCBI View Article : Google Scholar
|
|
45
|
den Besten G, van Eunen K, Groen AK,
Venema K, Reijngoud DJ and Bakker BM: The role of short-chain fatty
acids in the interplay between diet, gut microbiota, and host
energy metabolism. J Lipid Res. 54:2325–2340. 2013.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Miller TL and Wolin MJ: Pathways of
acetate, propionate, and butyrate formation by the human fecal
microbial flora. Appl Environ Microbiol. 62:1589–1592.
1996.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Martino C, Zaramela LS, Gao B, Embree M,
Tarasova J, Parker SJ, Wang Y, Chu H, Chen P, Lee KC, et al:
Acetate reprograms gut microbiota during alcohol consumption. Nat
Commun. 13(4630)2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Priyadarshini M, Kotlo KU, Dudeja PK and
Layden BT: Role of short chain fatty acid receptors in intestinal
physiology and pathophysiology. Compr Physiol. 8:1091–1115.
2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Nogal A, Louca P, Zhang X, Wells PM,
Steves CJ, Spector TD, Falchi M, Valdes AM and Menni C: Circulating
levels of the short-chain fatty acid acetate mediate the effect of
the gut microbiome on visceral fat. Front Microbiol.
12(711359)2021.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Louis P and Flint HJ: Formation of
propionate and butyrate by the human colonic microbiota. Environ
Microbiol. 19:29–41. 2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Canani RB, Costanzo MD, Leone L, Pedata M,
Meli R and Calignano A: Potential beneficial effects of butyrate in
intestinal and extraintestinal diseases. World J Gastroenterol.
17:1519–1528. 2011.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Peredo-Lovillo A, Romero-Luna HE and
Jiménez-Fernández M: Health promoting microbial metabolites
produced by gut microbiota after prebiotics metabolism. Food Res
Int. 136(109473)2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Slavin J: Fiber and prebiotics: Mechanisms
and health benefits. Nutrients. 5:1417–1435. 2013.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Ramirez-Olea H, Reyes-Ballesteros B and
Chavez-Santoscoy RA: Potential application of the probiotic as an
adjuvant in the treatment of diseases in humans and animals: A
systematic review. Front Microbiol. 13(993451)2022.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Markowiak P and Śliżewska K: Effects of
probiotics, prebiotics, and synbiotics on human health. Nutrients.
9(1021)2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Luo Y, De Souza C, Ramachandran M, Wang S,
Yi H, Ma Z, Zhang L and Lin K: Precise oral delivery systems for
probiotics: A review. J Control Release. 352:371–384.
2022.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Cui L, Morris A, Huang L, Beck JM, Twigg
HL III, von Mutius E and Ghedin E: The microbiome and the lung. Ann
Am Thorac Soc. 11 (Suppl 4):S227–S232. 2014.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Vijay A and Valdes AM: Role of the gut
microbiome in chronic diseases: A narrative review. Eur J Clin
Nutr. 76:489–501. 2022.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Chen S, Kuhn M, Prettner K, Yu F, Yang T,
Bärnighausen T, Bloom DE and Wang C: The global economic burden of
chronic obstructive pulmonary disease for 204 countries and
territories in 2020-50: A health-augmented macroeconomic modelling
study. Lancet Glob Health. 11:e1183–e1193. 2023.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Xiong T, Bai X, Wei X, Wang L, Li F, Shi H
and Shi Y: Exercise rehabilitation and chronic respiratory
diseases: effects, mechanisms, and therapeutic benefits. Int J
Chron Obstruct Pulmon Dis. 18:1251–1266. 2023.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Mattiuzzi C and Lippi G: Worldwide asthma
epidemiology: Insights from the global health data exchange
database. Int Forum Allergy Rhinol. 10:75–80. 2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Halpin DMG, Faner R, Sibila O, Badia JR
and Agusti A: Do chronic respiratory diseases or their treatment
affect the risk of SARS-CoV-2 Infection? Lancet Respir Med.
8:436–438. 2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Weinberg F, Dickson RP, Nagrath D and
Ramnath N: The lung microbiome: A central mediator of host
inflammation and metabolism in lung cancer patients? Cancers
(Basel). 13(13)2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Millen BE, Abrams S, Adams-Campbell L,
Anderson CA, Brenna JT, Campbell WW, Clinton S, Hu F, Nelson M,
Neuhouser ML, et al: The 2015 dietary guidelines advisory committee
scientific report: Development and major conclusions. Adv Nutr.
7:438–444. 2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Cheng CW, Biton M, Haber AL, Gunduz N, Eng
G, Gaynor LT, Tripathi S, Calibasi-Kocal G, Rickelt S, Butty VL, et
al: Ketone body signaling mediates intestinal stem cell homeostasis
and adaptation to diet. Cell. 178:1115–1131.e15. 2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Tan-Shalaby J: Ketogenic diets and cancer:
Emerging evidence. Fed Pract. 34 (Suppl 1):37S–42S. 2017.PubMed/NCBI
|
|
67
|
Vieira RS, Castoldi A, Basso PJ, Hiyane
MI, Câmara NOS and Almeida RR: Butyrate attenuates lung
inflammation by negatively modulating Th9 cells. Front Immunol.
10(67)2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Zhao H and Jin X: Causal associations
between dietary antioxidant vitamin intake and lung cancer: A
mendelian randomization study. Front Nutr. 9(965911)2022.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Hagihara M, Kato H, Yamashita M, Shibata
Y, Umemura T, Mori T, Hirai J, Asai N, Mori N and Mikamo H: Lung
cancer progression alters lung and gut microbiomes and lipid
metabolism. Heliyon. 10(e23509)2023.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Gui Q, Li H, Wang A, Zhao X, Tan Z, Chen
L, Xu K and Xiao C: The association between gut butyrate-producing
bacteria and non-small-cell lung cancer. J Clin Lab Anal.
34(e23318)2020.PubMed/NCBI View Article : Google Scholar
|
|
71
|
O'Shaughnessy M, Sheils O and Baird AM:
The lung microbiome in COPD and lung cancer: Exploring the
potential of metal-based drugs. Int J Mol Sci.
24(12296)2023.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Goto T: Airway microbiota as a modulator
of lung cancer. Int J Mol Sci. 21(3044)2020.PubMed/NCBI View Article : Google Scholar
|
|
73
|
An J, Kim H and Yang KM: An aqueous
extract of a species induces apoptosis and inhibits invasiveness of
non-small cell lung cancer cells. J Microbiol. Biotechnol.
30:885–893. 2020.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Takada K, Shimokawa M, Takamori S,
Shimamatsu S, Hirai F, Tagawa T, Okamoto T, Hamatake M,
Tsuchiya-Kawano Y, Otsubo K, et al: Clinical impact of probiotics
on the efficacy of anti-PD-1 monotherapy in patients with nonsmall
cell lung cancer: A multicenter retrospective survival analysis
study with inverse probability of treatment weighting. Int J
Cancer. 149:473–482. 2021.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Tomita Y, Ikeda T, Sakata S, Saruwatari K,
Sato R, Iyama S, Jodai T, Akaike K, Ishizuka S, Saeki S and
Sakagami T: Association of probiotic therapy with survival and
response to immune checkpoint blockade in patients with lung
cancer. Cancer Immunol Res. 8:1236–1242. 2020.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Xiao X, Cao Y and Chen H: Profiling and
characterization of microRNAs responding to sodium butyrate
treatment in A549 cells. J Cell Biochem. 119:3563–3573.
2018.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Corrêa RO, Castro PR, Moser R, Ferreira
CM, Quesniaux VFJ, Vinolo MAR and Ryffel B: Butyrate: Connecting
the gut-lung axis to the management of pulmonary disorders. Front
Nutr. 9(1011732)2022.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Gokulan K, Joshi M, Khare S and Bartter T:
Lung microbiome, gut-lung axis and chronic obstructive pulmonary
disease. Curr Opin Pulm Med. 28:134–138. 2022.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Song Q, Chen P and Liu XM: The role of
cigarette smoke-induced pulmonary vascular endothelial cell
apoptosis in COPD. Respir Res. 22(39)2021.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Ding K, Chen J, Zhan W, Zhang S, Chen Y,
Long S and Lei M: Microbiome links cigarette smoke-induced chronic
obstructive pulmonary disease and dietary fiber via the gut-lung
axis: A narrative review. COPD. 19:10–17. 2021.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Cabrera-Rubio R, Garcia-Núñez M, Setó L,
Antó JM, Moya A, Monsó E and Mira A: Microbiome diversity in the
bronchial tracts of patients with chronic obstructive pulmonary
disease. J Clin Microbiol. 50:3562–3568. 2012.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Dickson RP, Erb-Downward JR, Freeman CM,
McCloskey L, Beck JM, Huffnagle GB and Curtis JL: Spatial variation
in the healthy human lung microbiome and the adapted Island model
of lung biogeography. Ann Am Thorac Soc. 12:821–830.
2015.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Karakasidis E, Kotsiou OS and
Gourgoulianis KI: Lung and Gut Microbiome in COPD. J Pers Med.
13(804)2023.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Wedzicha JA and Seemungal TA: COPD
Exacerbations: Defining their cause and prevention. Lancet.
370:786–796. 2007.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Wang Z, Bafadhel M, Haldar K, Spivak A,
Mayhew D, Miller BE, Tal-Singer R, Johnston SL, Ramsheh MY, Barer
MR, et al: Lung microbiome dynamics in COPD exacerbations. Eur
Respir J. 47:1082–1092. 2016.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Wedzicha JA and Donaldson GC:
Exacerbations of chronic obstructive pulmonary disease. Respir
Care. 48:1204–1213; discussion 1213-1215. 2003.PubMed/NCBI
|
|
87
|
Xue Q, Xie Y, He Y, Yu Y, Fang G, Yu W, Wu
J, Li J, Zhao L, Deng X, et al: Lung microbiome and cytokine
profiles in different disease states of COPD: A cohort study. Sci
Rep. 13(5715)2023.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Varraso R, Chiuve SE, Fung TT, Barr RG, Hu
FB, Willett WC and Camargo CA: Alternate healthy eating index 2010
and risk of chronic obstructive pulmonary disease among US women
and men: Prospective study. BMJ. 350(h286)2015.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Yang L, Li N, Yi X and Wang Z: The
translational potential of the lung microbiome as a biomarker and a
therapeutic target for chronic obstructive pulmonary disease.
Interdiscip. Med. 1(e20230023)2023.
|
|
90
|
Loverdos K, Bellos G, Kokolatou L,
Vasileiadis I, Giamarellos E, Pecchiari M, Koulouris N, Koutsoukou
A and Rovina N: Lung microbiome in asthma: Current perspectives. J
Clin Med Res. 8(1967)2019.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Valverde-Molina J and García-Marcos L:
Microbiome and asthma: Microbial dysbiosis and the origins,
phenotypes, persistence, and severity of asthma. Nutrients.
15(486)2023.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Aziz N and Benjamin B: Activation of
natural killer cells by probiotics. For Immunopathol Dis Therap.
7:41–55. 2016.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Mazziotta C, Tognon M, Martini F,
Torreggiani E and Rotondo JC: Probiotics mechanism of action on
immune cells and beneficial effects on human health. Cells.
12(184)2023.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Salva S, Villena J and Alvarez S:
Immunomodulatory activity of lactobacillus rhamnosus strains
isolated from goat milk: impact on intestinal and respiratory
infections. Int J Food Microbiol. 141:82–89. 2010.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Kalliomäki M, Salminen S, Poussa T and
Isolauri E: Probiotics during the first 7 years of life: A
cumulative risk reduction of eczema in a randomized,
placebo-controlled trial. J Allergy Clin Immunol. 119:1019–1021.
2007.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Hougee S, Vriesema AJM, Wijering SC,
Knippels LM, Folkerts G, Nijkamp FP, Knol J and Garssen J: Oral
treatment with probiotics reduces allergic symptoms in
ovalbumin-sensitized mice: A bacterial strain comparative study.
Int Arch Allergy Immunol. 151:107–117. 2010.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Zuany-Amorim C, Sawicka E, Manlius C, Le
Moine A, Brunet LR, Kemeny DM, Bowen G, Rook G and Walker C:
Suppression of airway eosinophilia by killed mycobacterium
vaccae-induced allergen-specific regulatory T-cells. Nat Med.
8:625–629. 2002.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Mortaz E, Adcock IM, Folkerts G, Barnes
PJ, Paul Vos A and Garssen J: Probiotics in the management of lung
diseases. Mediators Inflamm. 2013(751068)2013.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Karim A, Muhammad T, Shahid Iqbal M and
Qaisar R: A multistrain probiotic improves handgrip strength and
functional capacity in patients with COPD: A randomized controlled
trial. Arch Gerontol Geriatr. 102(104721)2022.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Vasconcelos JA, Mota AS, Olímpio F, Rosa
PC, Damaceno-Rodrigues N, de Paula Vieira R, Taddei CR and Aimbire
F: Lactobacillus rhamnosus modulates lung inflammation and
mitigates gut dysbiosis in a murine model of asthma-COPD overlap
syndrome. Probiotics Antimicrob Proteins. 17:588–605.
2025.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Bikov A, Dragonieri S, Csoma B, Mazzuca C,
Finamore P, Rocchi G, Putignani L, Guarino M and Scarlata S: The
role of gut bacteriome in asthma, chronic obstructive pulmonary
disease and obstructive sleep apnoea. Microorganisms.
10(2457)2022.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Lai HC, Lin TL, Chen TW, Kuo YL, Chang CJ,
Wu TR, Shu CC, Tsai YH, Swift S and Lu CC: Gut microbiota modulates
COPD pathogenesis: Role of anti-inflammatory lipopolysaccharide.
Gut. 71:309–321. 2022.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Maniar K, Singh V, Moideen A,
Bhattacharyya R, Chakrabarti A and Banerjee D: Inhalational
supplementation of metformin butyrate: A strategy for prevention
and cure of various pulmonary disorders. Biomed Pharmacother.
107:495–506. 2018.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Venzon M, Bernard-Raichon L, Klein J,
Axelrad JE, Hussey GA, Sullivan AP, Casanovas-Massana A, Noval MG,
Valero-Jimenez AM, Gago J, et al: Gut microbiome dysbiosis during
COVID-19 is associated with increased risk for bacteremia and
microbial translocation. Res Sq [Preprint] rs.3.rs-726620,
2021.
|
|
105
|
Zhang W, Zhao Y, Zhang F, Wang Q, Li T,
Liu Z, Wang J, Qin Y, Zhang X, Yan X, et al: The use of
anti-inflammatory drugs in the treatment of people with severe
coronavirus disease 2019 (COVID-19): The perspectives of clinical
immunologists from China. Clin. Immunol. 214(108393)2020.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Karst SM: The influence of commensal
bacteria on infection with enteric viruses. Nat Rev Microbiol.
14:197–204. 2016.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Viana SD, Nunes S and Reis F: ACE2
imbalance as a key player for the poor outcomes in COVID-19
patients with age-related comorbidities-role of gut microbiota
dysbiosis. Ageing Res Rev. 62(101123)2020.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Zhang H, Penninger JM, Li Y, Zhong N and
Slutsky AS: Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2
receptor: Molecular mechanisms and potential therapeutic target.
Intensive Care Med. 46:586–590. 2020.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Domínguez-Díaz C, García-Orozco A,
Riera-Leal A, Padilla-Arellano JR and Fafutis-Morris M: Microbiota
and its role on viral evasion: Is it with us or against us? Front
Cell Infect Microbiol. 9(256)2019.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Lee H and Ko G: Antiviral effect of
vitamin a on norovirus infection via modulation of the gut
microbiome. Sci Rep. 6(25835)2016.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Steed AL, Christophi GP, Kaiko GE, Sun L,
Goodwin VM, Jain U, Esaulova E, Artyomov MN, Morales DJ, Holtzman
MJ, et al: The microbial metabolite desaminotyrosine protects from
influenza through type I interferon. Science. 357:498–502.
2017.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Wang Z, MacLeod DT and Di Nardo A:
Commensal bacteria lipoteichoic acid increases skin mast cell
antimicrobial activity against vaccinia viruses. J Immunol.
189:1551–1558. 2012.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Gonzalez-Perez G and Lamousé-Smith ES:
Gastrointestinal microbiome dysbiosis in infant mice alters
peripheral CD8 T cell receptor signaling. Front Immunol.
8(265)2017.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Belkacem N, Serafini N, Wheeler R, Derrien
M, Boucinha L, Couesnon A, Cerf-Bensussan N, Gomperts Boneca I, Di
Santo JP, Taha MK and Bourdet-Sicard R: Lactobacillus paracasei
feeding improves immune control of influenza infection in mice.
PLoS One. 12(e0184976)2017.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Aan FJ, Glibetic N, Montoya-Uribe V and
Matter ML: COVID-19 and the microbiome: The gut-lung connection.
Comprehensive Gut Microbiota. 442–458. 2022.
|
|
116
|
Malik JA, Ahmed S, Yaseen Z, Alanazi M,
Alharby TN, Alshammari HA and Anwar S: Association of SARS-CoV-2
and polypharmacy with gut-lung axis: From pathogenesis to
treatment. ACS Omega. 7:33651–33665. 2022.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Youssef M, Ahmed HY, Zongo A, Korin A,
Zhan F, Hady E, Umair M, Shahid Riaz Rajoka M, Xiong Y and Li B:
Probiotic supplements: Their strategies in the therapeutic and
prophylactic of human life-threatening diseases. Int J Mol Sci.
22(11290)2021.PubMed/NCBI View Article : Google Scholar
|
|
118
|
So D, Whelan K, Rossi M, Morrison M,
Holtmann G, Kelly JT, Shanahan ER, Staudacher HM and Campbell KL:
Dietary fiber intervention on gut microbiota composition in healthy
adults: A systematic review and meta-analysis. Am J Clin Nutr.
107:965–983. 2018.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Sadremomtaz A, Al-Dahmani ZM, Ruiz-Moreno
AJ, Monti A, Wang C, Azad T, Bell JC, Doti N, Velasco-Velázquez MA,
de Jong D, et al: Synthetic peptides that antagonize the
angiotensin-converting enzyme-2 (ACE-2) interaction with SARS-CoV-2
receptor binding spike protein. J Med Chem. 65:2836–2847.
2022.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Ceccarelli G, Marazzato M, Celani L,
Lombardi F, Piccirilli A, Mancone M, Trinchieri V, Pugliese F,
Mastroianni CM and d'Ettorre G: Oxygen sparing effect of
bacteriotherapy in COVID-19. Nutrients. 13(2898)2021.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Lim WS: Pneumonia-Overview. In:
Encyclopedia of Respiratory Medicine. Elsevier, Oxford, PH,
pp185-197, 2022.
|
|
122
|
Ding W, Cheng Y, Liu X, Zhu Z, Wu L, Gao
J, Lei W, Li Y, Zhou X, Wu J, et al: Harnessing the human gut
microbiota: An emerging frontier in combatting multidrug-resistant
bacteria. Front Immunol. 16(1563450)2025.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Patangia DV, Anthony Ryan C, Dempsey E,
Paul Ross R and Stanton C: Impact of antibiotics on the human
microbiome and consequences for host health. Microbiologyopen.
11(e1260)2022.PubMed/NCBI View Article : Google Scholar
|
|
124
|
Dongre DS, Saha UB and Saroj SD: Exploring
the role of gut microbiota in antibiotic resistance and prevention.
Ann Med. 57(2478317)2025.PubMed/NCBI View Article : Google Scholar
|
|
125
|
de Steenhuijsen Piters WA, Huijskens EG,
Wyllie AL, Biesbroek G, van den Bergh MR, Veenhoven RH, Wang X,
Trzciński K, Bonten MJ, Rossen JW, et al: Dysbiosis of upper
respiratory tract microbiota in elderly pneumonia patients. ISME J.
10:97–108. 2016.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Jenior ML, Leslie JL, Young VB and Schloss
PD: Alters the structure and metabolism of distinct cecal
microbiomes during initial infection to promote sustained
colonization. mSphere. 3:e00261–18. 2018.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Wang X, Qi Y and Zheng H: Dietary
polyphenol, gut microbiota, and health benefits. Antioxidants
(Basel). 11(1212)2022.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Turnbaugh PJ, Hamady M, Yatsunenko T,
Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA,
Affourtit JP, et al: A core gut microbiome in obese and lean twins.
Nature. 457:480–484. 2009.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Schloissnig S, Arumugam M, Sunagawa S,
Mitreva M, Tap J, Zhu A, Waller A, Mende DR, Kultima JR, Martin J,
et al: Genomic variation landscape of the human gut microbiome.
Nature. 493:45–50. 2012.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Armour CR, Nayfach S, Pollard KS and
Sharpton TJ: A metagenomic meta-analysis reveals functional
signatures of health and disease in the human gut microbiome.
mSystems. 4:e00332–18. 2019.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Ye SH, Siddle KJ, Park DJ and Sabeti PC:
Benchmarking metagenomics tools for taxonomic classification. Cell.
178:779–794. 2019.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Oulas A, Pavloudi C, Polymenakou P,
Pavlopoulos GA, Papanikolaou N, Kotoulas G, Arvanitidis C and
Iliopoulos I: Metagenomics: Tools and insights for analyzing
next-generation sequencing data derived from biodiversity studies.
Bioinform Biol Insights. 9:75–88. 2015.PubMed/NCBI View Article : Google Scholar
|
|
133
|
de Vries JJC, Brown JR, Couto N, Beer M,
Le Mercier P, Sidorov I, Papa A, Fischer N, Oude Munnink BB,
Rodriquez C, et al: Recommendations for the introduction of
metagenomic next-generation sequencing in clinical virology, part
II: Bioinformatic analysis and reporting. J Clin Virol.
138(104812)2021.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Wang WL, Xu SY, Ren ZG, Tao L, Jiang JW
and Zheng SS: Application of metagenomics in the human gut
microbiome. World J Gastroenterol. 21:803–814. 2015.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Ventura M, Turroni F, Canchaya C, Vaughan
EE, O'Toole PW and van Sinderen D: Microbial diversity in the human
intestine and novel insights from metagenomics. Front Biosci
(Landmark Ed). 14:3214–3221. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
136
|
Almeida A, Mitchell AL, Boland M, Forster
SC, Gloor GB, Tarkowska A, Lawley TD and Finn RD: A new genomic
blueprint of the human gut microbiota. Nature. 568:499–504.
2019.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Gmiter D, Nawrot S, Pacak I, Zegadło K and
Kaca W: Towards a better understanding of the bacterial pan-genome.
Acta universitatis lodziensis. Folia Biologica Et Oecologica.
17:84–96. 2021.
|
|
138
|
Shakya M, Lo CC and Chain PSG: Advances
and challenges in metatranscriptomic analysis. Front Genet.
10(904)2019.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Heintz-Buschart A and Wilmes P: Human gut
microbiome: Function matters. Trends Microbiol. 26:563–574.
2018.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Wang Y, Hu Y, Liu F, Cao J, Lv N, Zhu B,
Zhang G and Gao GF: Integrated metagenomic and metatranscriptomic
profiling reveals differentially expressed resistomes in human,
chicken, and pig gut microbiomes. Environ Int.
138(105649)2020.PubMed/NCBI View Article : Google Scholar
|
|
141
|
Aguiar-Pulido V, Huang W, Suarez-Ulloa V,
Cickovski T, Mathee K and Narasimhan G: Metagenomics,
metatranscriptomics, and metabolomics approaches for microbiome
analysis. Evol Bioinform Online. 12 (Suppl 1):S5–S16.
2016.PubMed/NCBI View Article : Google Scholar
|
|
142
|
Lai LA, Tong Z, Chen R and Pan S:
Metaproteomics study of the gut microbiome. Methods Mol Biol.
1871:123–132. 2019.PubMed/NCBI View Article : Google Scholar
|
|
143
|
Larsen PE and Dai Y: Metabolome of human
gut microbiome is predictive of host dysbiosis. Gigascience.
4(42)2015.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Verberkmoes NC, Russell AL, Shah M, Godzik
A, Rosenquist M, Halfvarson J, Lefsrud MG, Apajalahti J, Tysk C,
Hettich RL and Jansson JK: Shotgun metaproteomics of the human
distal gut microbiota. ISME J. 3:179–189. 2009.PubMed/NCBI View Article : Google Scholar
|
|
145
|
Ursell LK, Haiser HJ, Van Treuren W, Garg
N, Reddivari L, Vanamala J, Dorrestein PC, Turnbaugh PJ and Knight
R: The Intestinal Metabolome: An Intersection between Microbiota
and Host. Gastroenterology. 146:1470–1476. 2014.PubMed/NCBI View Article : Google Scholar
|
|
146
|
Liu Y, Guo J, Hu G and Zhu H: Gene
prediction in metagenomic fragments based on the SVM Algorithm. BMC
Bioinformatics. 14 (Suppl 5)(S12)2013.PubMed/NCBI View Article : Google Scholar
|
|
147
|
Han J, Zhang H and Ning K: Techniques for
learning and transferring knowledge for microbiome-based
classification and prediction: Review and assessment. Brief
Bioinform. 26(bbaf015)2024.PubMed/NCBI View Article : Google Scholar
|
|
148
|
Bakir-Gungor B, Temiz M, Jabeer A, Wu D
and Yousef M: microBiomeGSM: The identification of taxonomic
biomarkers from metagenomic data using grouping, scoring and
modeling (G-S-M) approach. Front Microbiol.
14(1264941)2023.PubMed/NCBI View Article : Google Scholar
|
|
149
|
Hemmati MA, Monemi M, Asli S, Mohammadi S,
Foroozanmehr B, Haghmorad D, Oksenych V and Eslami M: Using new
technologies to analyze gut microbiota and predict cancer risk.
Cells. 13(1987)2024.PubMed/NCBI View Article : Google Scholar
|
|
150
|
Romano S, Wirbel J, Ansorge R, Schudoma C,
Ducarmon QR, Narbad A and Zeller G: Machine learning-based
meta-analysis reveals gut microbiome alterations associated with
Parkinson's disease. Nat Commun. 16(4227)2025.PubMed/NCBI View Article : Google Scholar
|
|
151
|
Vilne B, Ķibilds J, Siksna I, Lazda I,
Valciņa O and Krūmiņa A: Could artificial intelligence/machine
learning and inclusion of diet-gut microbiome interactions improve
disease risk prediction? Case study: Coronary artery disease. Front
Microbiol. 13(627892)2022.PubMed/NCBI View Article : Google Scholar
|
|
152
|
Curry KD, Nute MG and Treangen TJ: It
takes guts to learn: Machine learning techniques for disease
detection from the gut microbiome. Emerg Top Life Sci. 5:815–827.
2021.PubMed/NCBI View Article : Google Scholar
|
|
153
|
Manandhar I, Alimadadi A, Aryal S, Munroe
PB, Joe B and Cheng X: Gut microbiome-based supervised machine
learning for clinical diagnosis of inflammatory bowel diseases. Am
J Physiol Gastrointest Liver Physiol. 320:G328–G337.
2021.PubMed/NCBI View Article : Google Scholar
|
|
154
|
Liu W, Fang X, Zhou Y, Dou L and Dou T:
Machine learning-based investigation of the relationship between
gut microbiome and obesity status. Microbes Infect.
24(104892)2022.PubMed/NCBI View Article : Google Scholar
|
|
155
|
Chen Y, Wu T, Lu W, Yuan W, Pan M, Lee YK,
Zhao J, Zhang H, Chen W, Zhu J and Wang H: Predicting the role of
the human gut microbiome in constipation using machine-learning
methods: A meta-analysis. Microorganisms. 9(2149)2021.PubMed/NCBI View Article : Google Scholar
|
|
156
|
Gou W, Ling CW, He Y, Jiang Z, Fu Y, Xu F,
Miao Z, Sun TY, Lin JS, Zhu HL, et al: Interpretable machine
learning framework reveals robust gut microbiome features
associated with type 2 diabetes. Diabetes Care. 44:358–366.
2021.PubMed/NCBI View Article : Google Scholar
|
|
157
|
Aryal S, Alimadadi A, Manandhar I, Joe B
and Cheng X: Machine learning strategy for gut microbiome-based
diagnostic screening of cardiovascular disease. Hypertension.
76:1555–1562. 2020.PubMed/NCBI View Article : Google Scholar
|
|
158
|
Kaur H, Singh Y, Singh S and Singh RB: Gut
microbiome-mediated epigenetic regulation of brain disorder and
application of machine learning for multi-omics data analysis1.
Genome. 64:355–371. 2021.PubMed/NCBI View Article : Google Scholar
|
|
159
|
Cammarota G, Ianiro G, Ahern A, Carbone C,
Temko A, Claesson MJ, Gasbarrini A and Tortora G: Gut microbiome,
big data and machine learning to promote precision medicine for
cancer. Nat Rev Gastroenterol Hepatol. 17:635–648. 2020.PubMed/NCBI View Article : Google Scholar
|
|
160
|
Lee HJ, Hong JK, Kim JK, Kim DH, Jang SW,
Han SW and Yoon IY: Effects of probiotic NVP-1704 on mental health
and sleep in healthy Adults: An 8-week randomized, double-blind,
placebo-controlled trial. Nutrients. 13(2660)2021.PubMed/NCBI View Article : Google Scholar
|
|
161
|
Kang DW, Adams JB, Vargason T, Santiago M,
Hahn J and Krajmalnik-Brown R: Distinct fecal and plasma
metabolites in children with autism spectrum disorders and their
modulation after microbiota transfer therapy. mSphere. 5:e00314–20.
2020.PubMed/NCBI View Article : Google Scholar
|
|
162
|
Tarutani S, Omori M, Ido Y, Yano M,
Komatsu T and Okamura T: Effects of 4G-Beta-D-galactosylsucrose in
patients with depression: A randomized, double-blinded,
placebo-controlled, parallel-group comparative study. J Psychiatr.
Res. 148:110–120. 2022.PubMed/NCBI View Article : Google Scholar
|
|
163
|
Philip Mani A, Balasubramanian B, Mali LA,
Joseph KS, Meyyazhagan A, Pappuswamy M and Joseph BV: The role of
the gut microbiota in neurodegenerative diseases. Microbiol Res.
15:489–507. 2024.
|
|
164
|
Zhu X, Li B, Lou P, Dai T, Chen Y, Zhuge
A, Yuan Y and Li L: The relationship between the gut microbiome and
neurodegenerative diseases. Neurosci Bull. 37:1510–1522.
2021.PubMed/NCBI View Article : Google Scholar
|
|
165
|
Zhang H, Chen Y, Wang Z, Xie G, Liu M,
Yuan B, Chai H, Wang W and Chen P: Implications of gut microbiota
in neurodegenerative diseases. Front Immunol.
13(785644)2022.PubMed/NCBI View Article : Google Scholar
|
|
166
|
Ma YY, Li X, Yu JT and Wang YJ:
Therapeutics for neurodegenerative diseases by targeting the gut
microbiome: from bench to bedside. Transl Neurodegener.
13(12)2024.PubMed/NCBI View Article : Google Scholar
|
|
167
|
Jain A, Madkan S and Patil P: The role of
gut microbiota in neurodegenerative diseases: Current insights and
therapeutic implications. Cureus. 15(e47861)2023.PubMed/NCBI View Article : Google Scholar
|