Introduction
Degenerative lumbar scoliosis (DLS) is defined as a
spinal deformity with a Cobb angle of >10°, which develops after
skeletal maturity without a previous history of scoliosis. It
generally presents in the lumbar or thoracolumbar part of the spine
and may be associated with severe back pain (1,2). The
prevalence of DLS increases with age and its incidence in patients
>50 years of age was reported to be ∼6% (3). The etiology of DLS has not been
clearly established. However, the most commonly proposed causes of
DLS were ost eoporosis and degenerative diseases of the spinal
column (3). Several factors,
including life-style, intrinsic mediators and hormonal or genetic
factors, are likely to affect the development or the acceleration
of DLS (4,5).
Rab3-interacting molecules (RIMs) are presynaptic
active zone proteins that perform an essential function in
neurotransmitter release (6,7). RIMs
were identified as putative effectors for Rab3, which is a synaptic
vesicle protein that regulates neurotransmitter release (6,7). RIMs
have been shown to interact with multiple synaptic proteins, such
as UNC-13 homolog B (C. elegans) (UNC13B, Munc13) (8), ELKS/Rab6-interacting/CAST family
member 1 (ERC1, ELKS) (9,10), RIM-binding proteins (RIM-BPs)
(11), α-liprins and synaptotagmin
1 (12–14). RIMs are encoded by four regulating
synaptic membrane exocytosis genes (RIMS1–4), of which RIMS1, RIMS3
and RIMS4 express a single isoform (RIM1α, 3γ and 4γ,
respectively), whereas RIMS2 (RAB3IP3, OBOE) expresses three
isoforms (RIM2α, RIM2β and RIM2γ) via independent internal
promoters (15).
Two single-nucleotide polymorphisms (SNPs)
[rs2028945 (Gln1200Gln) and rs10461 (Ala1327Ala)] in the coding
region of RIMS2 in DLS were investigated in this study.
Subjects and methods
Subjects and clinical phenotypes
A total of 51 DLS patients and 145 control subjects
were enrolled in this study. DLS patients were recruited from the
Spine Center of Kyung Hee University East-West Neo Medical Center
(Seoul, Korea) and the National Medical Center (Seoul, Korea) and
were selected among ∼10,000 patients per year during a 3-year
period. Each patient was diagnosed by a specialized spinal surgeon
and fulfilled all physical examination and radiographic criteria.
The DLS group comprised 44 females and 7 males and the mean age was
68.67±8.00 years [mean ± standard deviation (SD)]. The Cobb angle
in the DLS group was 19.36±7.38° and the lateral listhesis was
4.84±4.24 mm. A left convex curve was observed more frequently
compared to a right convex curve (31 and 20 patients, respectively)
(Table I). DLS patients were
divided into two subgroups according to clinical characteristics,
including Cobb angle (≤30° vs. >30°), lateral listhesis (≤5 vs.
>5 mm) and curve direction (left vs. right) (16,17).
The control group included 145 patients (124 females and 21 males)
who were recruited from a general health check-up program after it
was confirmed that they had no clinical evidence of scoliosis or
any other severe disorders. The gender and age of the control group
were matched with those of the DLS group. This study was conducted
according to the Declaration of Helsinki guidelines. Written
informed consent was obtained from each individual. The research
protocol was approved by the Ethics Review Committee of the Medical
Research Institute, Kyung Hee University Medical Center, Seoul,
Korea.
 | Table I.Demographic characteristics of the
study subjects. |
Table I.
Demographic characteristics of the
study subjects.
| Characteristic | Control group | DLS group |
|---|
| Number of subjects
(n) | 145 | 51 |
| Male/female (n) | 124/21 | 44/7 |
| Age (mean ± SD,
years) | 65.57±8.65 | 68.67±8.00 |
| Cobb angle
(degrees) | | 19.36±7.38 |
| Lateral listhesis
(mm) | | 4.84±4.24 |
| Curve direction
(left/right) | | 31/20 |
SNP selection and genotyping
We investigated SNPs in the coding region of RIMS2.
The related information was obtained from the SNP database
(www.ncbi.nlm.nih.gov/SNP, dbSNP BUILD131). The
SNPs with unknown heterozygosity, minor allele frequencies <10%
and no data regarding Asian populations (rs17854256, rs76323676 and
rs61753732) were excluded. Consequently, rs2028945 (Gln1200Gln) and
rs10461 (Ala1327Ala) were selected for this investigation. Genomic
DNA was extracted from the blood sample of each subject using
Qiagen DNA Extraction kit (Qiagen, Tokyo, Japan) following the
manufacturer’s instructions and was amplified using the primers for
each SNP of RIMS2 (Table II). PCR
products were sequenced by an ABI PRISM 3730×l DNA Analyzer
(Applied Biosystems Inc., Foster City, CA, USA). Sequence data were
analyzed using SeqMan II software (DNASTAR, Inc., Madison, WI,
USA).
 | Table II.Primer sequences used for each SNP in
RIMS2. |
Table II.
Primer sequences used for each SNP in
RIMS2.
| SNP | Sequence (5′-3′) | Product size
(bp) |
|---|
| rs2028945 | | |
| Sense |
TCCTCACTGAACACTCATTCAGG | 543 |
| Antisense |
CTCAGCCCAGTGGAATCTTTAAC | |
| rs10461 | | |
| Sense |
AGATCATCGTCTGGGGAGATTA | 343 |
| Antisense |
CCCTAGAAACAGGCTCAGAAGA | |
Statistical analysis
The Hardy-Weinberg equilibrium (HWE) was assessed
using SNPStats (http://bioinfo.iconcologia.net/index.php) (18). The allele frequencies of each SNP
were compared by the Pearson’s Chi-square test. The effect of SNP
genotypes was analyzed using the codominant, dominant and recessive
models. Logistic regression analysis was used to calculate the odds
ratio (ORs), 95% confidence intervals (CIs) and P-values with
controlling age and gender as covariables. The linkage
disequilibrium (LD) was assessed using Haploview software version
4.1 (Broad Institute, Cambridge, MA, USA). LD block was constructed
using the Gabriel method (19,20).
The association of SNPs and haplotypes was analyzed using SNPStats,
HapAnalyzer version 1.0 (http://hap.ngri.go.kr/), and HelixTree (Golden Helix
Inc., Bozeman, MT, USA) software.
Statistical analysis was performed using the package
of SPSS Statistics software version 17.0 (SPSS Inc., Chicago, IL,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
The clinical characteristics of control subjects and
DLS patients are summarized in Table
I. The mean age of the control and the DLS group was 65.57±8.65
(mean ± SD) and 68.67±8.00 years, respectively. In the DLS group,
the average Cobb angle was 19.36±7.38° and the length of the
lateral listhesis was 4.84±4.24 mm. Patients with a left convex
curve outnumbered those with a right convex curve (31 and 20,
respectively) (Table I).
The genotype distributions of rs2028945 and rs10461
were in HWE equilibrium (P>0.05). In an analysis of the allele
frequencies on two SNPs, the C allele of rs10461 was more
frequently encountered in the DLS group compared to the control
group (OR=1.857; 95% CI, 1.174–2.937 and P=0.008) (Table III). In an analysis of the genotype
distributions, the rs10461 was associated with DLS (OR=1.93; 95%
CI, 1.20–3.10 and P=0.006 in the codominant model; OR=2.62; 95% CI,
1.12–6.13 and P=0.018 in the dominant model, and OR=2.25; 95% CI,
1.10–4.61 and P=0.029 in the recessive model). The rs2028945 was
not significantly different between the control and the DLS groups
(Table IV). One LD block was
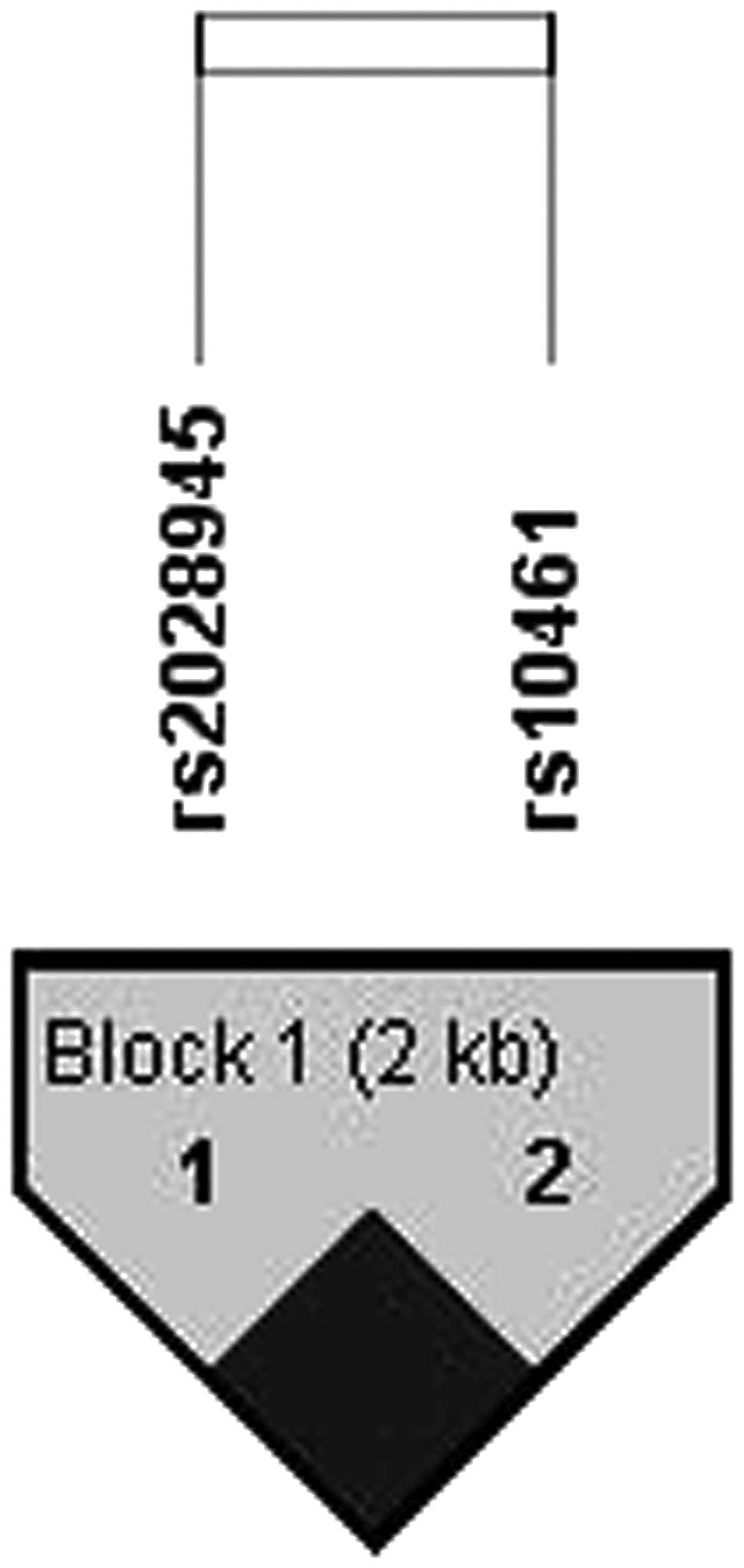
constructed between rs2028945 and rs10461 with the Gabriel method
(Fig. 1). Of the haplotypes in the
block, the CC and CT haplotypes were associated with the risk of
DLS (CC haplotype: P=0.009 in the codominant model, P=0.038 in the
dominant model and P=0.030 in the recessive model; CT haplotype:
P=0.041 in the codominant model and P=0.021 in the dominant model)
(Table IV).
 | Table III.Genotype and allele frequencies of
RIMS2 polymorphisms in the control and DLS groups. |
Table III.
Genotype and allele frequencies of
RIMS2 polymorphisms in the control and DLS groups.
| SNP | Type | Control
| DLS
| Model | OR (95% CI) | P-value |
|---|
| No. | (%) | No. | (%) |
|---|
rs2028945
Gln1200Gln | Genotype | | | | | | | |
| C/C | 49 | 33.8 | 20 | 39.2 | Codominant | 0.72
(0.45–1.16) | 0.18 |
| C/T | 69 | 47.6 | 26 | 51.0 | Dominant | 0.79
(0.40–1.53) | 0.48 |
| T/T | 27 | 18.6 | 5 | 9.8 | Recessive | 0.44
(0.16–1.23) | 0.09 |
| Allele | | | | | | | |
| C | 167 | 57.6 | 66 | 64.7 | | 1 | |
| T | 123 | 42.4 | 36 | 35.3 | | 0.74
(0.46–1.18) | 0.21 |
rs10461
Ala1327Ala | Genotype | | | | | | | |
| T/T | 45 | 31.0 | 8 | 15.7 | Codominant | 1.93
(1.20–3.10) | 0.006 |
| T/C | 71 | 49.0 | 25 | 49.0 | Dominant | 2.62
(1.12–6.13) | 0.018 |
| C/C | 29 | 20.0 | 18 | 35.3 | Recessive | 2.25
(1.10–4.61) | 0.029 |
| Allele | | | | | | | |
| T | 161 | 55.5 | 41 | 40.2 | | 1 | |
| C | 129 | 44.5 | 61 | 59.8 | | 1.857
(1.174–2.937) | 0.008 |
 | Table IV.Haplotype distributions of RIMS2
polymorphisms (rs2028945 and rs10461) in the control and DLS
groups. |
Table IV.
Haplotype distributions of RIMS2
polymorphisms (rs2028945 and rs10461) in the control and DLS
groups.
| Haplotype | Control
| DLS
| Model | OR (95% CI) | P-value |
|---|
| No. | (%) | Freq | No. | (%) | Freq |
|---|
| HAP1 (CC) | | | | | | | | | |
| HH | 29 | 20.0 | | 29 | 20.0 | | Codominant | 1.86
(1.16–2.96) | 0.009 |
| H - | 71 | 49.0 | 0.44 | 71 | 49.0 | 0.60 | Dominant | 2.42
(1.05–5.56) | 0.038 |
| - - | 45 | 31.0 | | 45 | 31.0 | | Recessive | 2.18
(1.08–4.41) | 0.030 |
| HAP2 (TT) | | | | | | | | | |
| HH | 27 | 18.6 | | 27 | 18.6 | | Codominant | 0.74
(0.46–1.18) | 0.208 |
| H - | 69 | 47.6 | 0.42 | 69 | 47.6 | 0.35 | Dominant | 0.79
(0.41–1.53) | 0.486 |
| - - | 49 | 33.8 | | 49 | 33.8 | | Recessive | 0.48
(0.17–1.31) | 0.150 |
| HAP3 (CT) | | | | | | | | | |
| HH | 4 | 2.8 | | 4 | 2.8 | | Codominant | 0.38
(0.15–0.96) | 0.041 |
| H - | 30 | 20.7 | 0.13 | 30 | 20.7 | 0.05 | Dominant | 0.28
(0.09–0.83) | 0.021 |
| - - | 111 | 76.6 | | 111 | 76.6 | | Recessive | 0.70
(0.08–6.46) | 0.757 |
The results obtained in this study were compared
with different ethnic populations by searching the human SNP
database (www.ncbi.nlm.nih.gov/SNP; dbSNP BUILD
131), which includes the genotype frequencies of each SNP. Genotype
distributions in our control subjects were similar to those in
Asian populations, particularly the Chinese population (Table V).
 | Table V.Genotype distributions of RIMS2
polymorphisms among ethnic populations. |
Table V.
Genotype distributions of RIMS2
polymorphisms among ethnic populations.
| SNP | Populations
|
|---|
| Korean | European | Chinese | Japanese | Sub-Saharan
African |
|---|
| rs2028945 | | | | | |
| C/C | 0.338 | 0.833 | 0.267 | 0.182 | 0.850 |
| C/T | 0.476 | 0.167 | 0.489 | 0.341 | 0.133 |
| T/T | 0.186 | - | 0.244 | 0.477 | 0.017 |
| rs10461 | | | | | |
| C/C | 0.200 | 0.414 | 0.222 | 0.159 | 0.153 |
| C/T | 0.490 | 0.483 | 0.356 | 0.341 | 0.610 |
| T/T | 0.103 | 0.103 | 0.422 | 0.500 | 0.237 |
Discussion
DLS develops de novo in adulthood and
presents more frequently in the elderly (3). With increasing life expectancy, DLS
may complicate degenerative spondylolisthesis, lateral listhesis or
spinal stenosis, in which the neural elements are compressed by
bone and soft tissue, leading to nerve root ischemia (2). Therefore, patients with accelerated
DLS may develop severe back pain and neurological deficits
(3). Despite a variety of studies
on DLS, the etiology remains unknown. Therefore, the potential of
RIMS2 as a candidate gene was evaluated. Our results demonstrated
that rs10461 (Ala1327Ala) was significantly associated with DLS. It
was hypothesized that the C allele of rs10461 may be a risk factor
in the development of DLS (Table
III). In addition, two haplotypes (CC and CT) exhibited
significant differences between the control and DLS groups. The
results indicated that RIMS2 may be associated with DLS. However,
RIMS2 polymorphisms were not associated with the clinical features
of DLS (Cobb angle, lateral listhesis and curve direction) (data
not shown).
RIMS comprise four members, RIMS1–4. RIMS1 and RIMS2
are 512.7 and 747.9 kb in size, respectively, whereas RIMS3 and
RIMS4 are 15.4 and 54.5 kb, respectively (8,15).
RIMS1 expresses RIM1α; the gene may encode a single isoform. By
contrast, RIMS2 encodes three isoforms (RIM2α, RIM2β and RIM2γ);
besides the exon for the α isoform, two additional separate exons
in RIMS2 may encode the N-termini of RIM2β and RIM2γ through β- and
γ-specific promoters, respectively. RIMS3 and RIMS4 express a
single isoform, RIM3γ and RIM4γ, respectively (8,15). The
α-RIMs (RIM1α and RIM2α) contain the full complement of domains
which are the N-terminal Zn2+-finger domain, the central
PDZ and C2A domains and the C-terminal C2B
domain (15). They are able to bind
to Rab3 and Munc13 proteins (13,21,22)
and closely interact with α-liprins (13), which in turn regulate the active
zone structure (23,24). Therefore, α-RIMs are considered to
be essential for regulating neurotransmitter releases (8,13,25,26),
some of which are involved in the neurobiology of schizophrenia
(27,28).
The exocytosis of neurotransmitter-filled synaptic
vesicles is under tight regulation in presynaptic nerve terminals.
RIMs may mediate the regulation of exocytosis via interacting with
multiple synaptic proteins, such as Rab3 (6,7),
Munc13 (8), ELKS (9,10),
RIM-BPs (11), α-liprins and
synaptotagmin 1 (12–14). In addition, RIMs are indirectly
connected with the active zone proteins Piccolo and Bassoon via
ELKS (29). In a previous study,
the experimental loss of α-RIMs resulted in the severe impairment
of motor control, profound defects of synaptic transmission at
neuromuscular junctions and increment of irregularly distributed
motor synapses in skeletal muscle fibers (8). These results suggested that RIMs may
affect the function of the neuromuscular system. However, no
published studies assessing the potential role of RIMs in bone
degeneration or the genetic association of RIMS polymorphisms in
DLS are available. We demonstrated that RIMS2, known as a
regulatory gene for synaptic membrane exocytosis in the central
nervous system, may be a candidate gene associated with the risk of
DLS in the Korean population. However, additional studies including
a larger number of subjects and different populations are required
to confirm these findings. In conclusion, the rs10461 in RIMS2 was
associated with DLS in the Korean population. This finding suggests
that RIMS2 may affect the development of DLS.
Abbreviations:
|
RIMS2
|
regulating synaptic membrane
exocytosis 2
|
|
SNP
|
single-nucleotide polymorphism
|
|
HWE
|
Hardy-Weinberg equilibrium
|
|
LD
|
linkage disequilibrium
|
|
DLS
|
degenerative lumbar scoliosis
|
References
|
1.
|
Aebi M: The adult scoliosis. Eur Spine J.
14:925–948. 2005. View Article : Google Scholar
|
|
2.
|
Ploumis A, Transfledt EE and Denis F:
Degenerative lumbar scoliosis associated with spinal stenosis.
Spine J. 7:428–436. 2007. View Article : Google Scholar
|
|
3.
|
Oskouian RJ Jr and Shaffrey CI:
Degenerative lumbar scoliosis. Neurosurg Clin N Am. 17:299–315.
2006. View Article : Google Scholar
|
|
4.
|
Ferrari S, Rizzoli R and Bonjour JP:
Heritable and nutritional influences on bone mineral mass. Aging
(Milano). 10:205–213. 1998.PubMed/NCBI
|
|
5.
|
Kawaguchi Y, Kanamori M, Ishihara H,
Ohmori K, Matsui H and Kimura T: The association of lumbar disc
disease with vitamin-D receptor gene polymorphism. J Bone Joint
Surg Am. 84-A:2022–2028. 2002.PubMed/NCBI
|
|
6.
|
Dresbach T, Qualmann B, Kessels MM, Garner
CC and Gundelfinger ED: The presynaptic cytomatrix of brain
synapses. Cell Mol Life Sci. 58:94–116. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Lonart G: RIM1: an edge for presynaptic
plasticity. Trends Neurosci. 25:329–332. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Schoch S, Mittelstaedt T, Kaeser PS, et
al: Redundant functions of RIM1α and RIM2α in
Ca2+-triggered neurotransmitter release. EMBO J.
25:5852–5863. 2006.
|
|
9.
|
Ohtsuka T, Takao-Rikitsu E, Inoue E, et
al: Cast: a novel protein of the cytomatrix at the active zone of
synapses that forms a ternary complex with RIM1 and munc13-1. J
Cell Biol. 158:577–590. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Wang Y, Liu X, Biederer T and Südhof TC: A
family of RIM-binding proteins regulated by alternative splicing:
implications for the genesis of synaptic active zones. Proc Natl
Acad Sci USA. 99:14464–14469. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Wang Y, Sugita S and Südhof TC: The
RIM/NIM family of neuronal C2domain proteins.
Interactions with Rab3 and a new class of Src homology 3 domain
proteins. J Biol Chem. 275:20033–20044. 2000.PubMed/NCBI
|
|
12.
|
Coppola T, Magnin-Luthi S, Perret-Menoud
V, et al: Direct interaction of the Rab3 effector RIM with
Ca2+channels, SNAP-25, and synaptotagmin. J Biol Chem.
276:32756–32762. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Schoch S, Castillo PE, Jo T, et al:
RIM1alpha forms a protein scaffold for regulating neurotransmitter
release at the active zone. Nature. 415:321–326. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Schoch S and Gundelfinger ED: Molecular
organization of the presynaptic active zone. Cell Tissue Res.
326:379–391. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Wang Y and Südhof TC: Genomic definition
of RIM proteins: evolutionary amplification of a family of synaptic
regulatory proteins. Genomics. 81:126–137. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Pritchett JW and Bortel DT: Degenerative
symptomatic lumbar scoliosis. Spine (Phila Pa 1976). 18:700–703.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Chin KR, Furey C and Bohlman HH: Risk of
progression in de novo low-magnitude degenerative lumbar curves:
natural history and literature review. Am J Orthop (Belle Mead NJ).
38:404–409. 2009.PubMed/NCBI
|
|
18.
|
Solé X, Guinó E, Valls J, Iniesta R and
Moreno V: SNPStats: a web tool for the analysis of association
studies. Bioinformatics. 22:1928–1929. 2006.
|
|
19.
|
Gabriel SB, Schaffner SF, Nguyen H, et al:
The structure of haplotype blocks in the human genome. Science.
296:2225–2229. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Betz A, Thakur P, Junge HJ, et al:
Functional interaction of the active zone proteins Munc13-1 and
RIM1 in synaptic vesicle priming. Neuron. 30:183–196. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Wang X, Hu B, Zimmermann B and Kilimann
MW: Rim1 and rabphilin-3 bind Rab3-GTP by composite determinants
partially related through N-terminal alpha-helix motifs. J Biol
Chem. 276:32480–32488. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Zhen M and Jin Y: The liprin protein SYD-2
regulates the differentiation of presynaptic termini in C.
elegans. Nature. 401:371–375. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Kaufmann N, DeProto J, Ranjan R, et al:
Drosophila liprin-alpha and the receptor phosphatase Dlar
control synapse morphogenesis. Neuron. 34:27–38. 2002. View Article : Google Scholar
|
|
25.
|
Koushika SP, Richmond JE, Hadwiger G, et
al: A post-docking role for active zone protein Rim. Nat Neurosci.
4:997–1005. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Castillo PE, Schoch S, Schmitz F, et al:
RIM1alpha is required for presynaptic long-term potentiation.
Nature. 415:327–330. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Weidenhofer J, Bowden NA, Scott RJ and
Tooney PA: Altered gene expression in the amygdala in
schizophrenia: up-regulation of genes located in the cytomatrix
active zone. Mol Cell Neurosci. 31:243–250. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Weidenhofer J, Scott RJ and Tooney PA:
Investigation of the expression of genes affecting cytomatrix
active zone function in the amygdala in schizophrenia: effects of
antipsychotic drugs. J Psychiatr Res. 43:282–290. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Takao-Rikitsu E, Mochida S, Inoue E, et
al: Physical and functional interaction of the active zone
proteins, CAST, RIM1, and Bassoon, in neurotransmitter release. J
Cell Biol. 164:301–311. 2004. View Article : Google Scholar : PubMed/NCBI
|