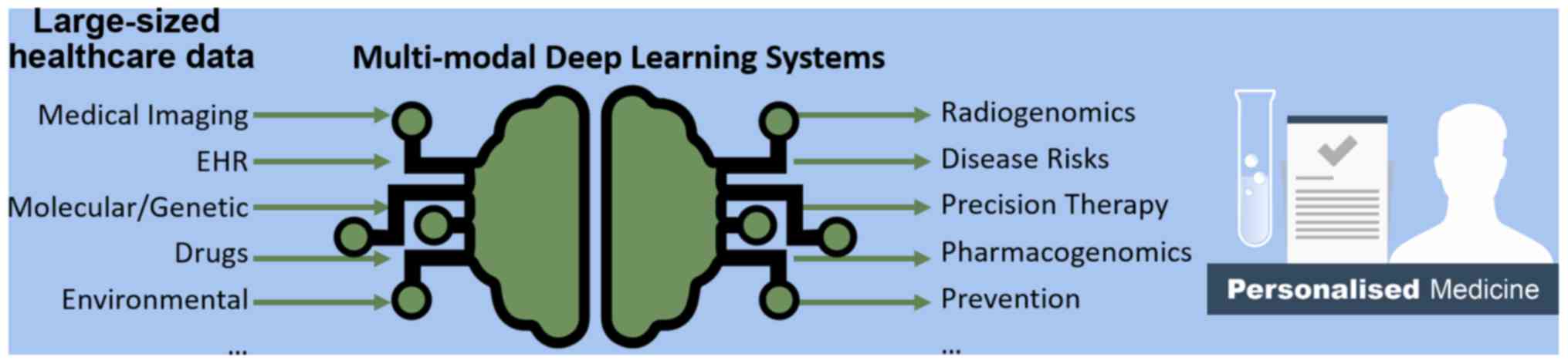
1. Introduction
Deep learning (DL) is a specialized machine learning
approach based on multiple-layered structures (algorithms) of
artificial neurons, which are able to process information and learn
by adjusting the weights at each synapse, enabling the performance
of an intelligent task with high precision. Furthermore, these deep
networks are able to ‘learn’ from large amounts of data and,
similar to the human brain, can generalize concepts and apply these
to new data with high accuracy. This ‘intelligence’ property has
been expanded towards deep multimodal learning by integrating
different sources of information (images, text, speech etc.) for
further improving high-level multimodal decision support and
reasoning (1). As a result, current
DL systems are able to integrate and model heterogeneous data from
an individual patient, across modalities and time, allowing better
predictions and recommending treatment options tailored to each
patient's individual characteristics and needs (2). This concept is illustrated in Fig. 1, highlighting the potential of DL to
transform large-sized healthcare data into useful tools for
advancing personalized medicine. DL algorithms are well-suited for
large amounts of healthcare data and perform better than
traditional statistical models at the expense of sacrificing
interpretability for predictive power.
2. Current developments
Accurate diagnosis is a milestone for personalized
medicine. Ehteshami Bejnordi et al reported that 7 DL
algorithms trained to detect metastases in hematoxylin- and
eosin-stained tissue sections of lymph nodes of women with breast
cancer, outperformed a panel of 11 pathologists with an area under
the curve (AUC) of 0.994 (best algorithm) vs. 0.884 (best
pathologist) (3). Madani et al
presented a DL echocardiography system for the automated diagnosis
of cardiac disease to address the issue of echocardiographic
assessment inaccuracy (up to 30%) of echo reports, achieving an 80%
accuracy in echocardiographic view classification and a 92.3%
accuracy for left ventricular hypertrophy classification (4). Recently, Ding et al proposed a DL
system demonstrating an improved early prediction of the final
diagnosis of Alzheimer's disease (AD) (82% specificity at 100%
sensitivity, on average of 75.8 months prior to the final
diagnosis), utilizing 18F-fludeoxyglucose positron
emission tomography (PET) of the brain, and therefore enhancing
opportunities for early therapeutic interventions (5). Finally, another recent study presented a
DL algorithm (CheXNeXt) that matched the expert human radiologist
performance in diagnosing 11 lung pathologies (6).
DL has begun to revolutionize decision support
systems in oncology due to the plethora of the available data and
the heterogeneity of various malignancies, which require algorithms
able to decipher hidden phenotype and genotype patterns, as well as
their associations. Recently, Causey et al reported a DL
system (NoduleX) which achieved an impressive accuracy (AUC of
~0.99) in nodule malignancy classification (>1,000 nodules)
based on CT scans, matching the performance of experienced
radiologists (7). Previously, Li
et al reported a DL brain tumor radiogenomic system able to
predict with high accuracy the IDH1 mutation status solely from
multi-modal magnetic resonance imaging (MRI) data, achieving an
impressive 95% AUC (8). Using a
similar approach, Akkus et al demonstrated a DL system
capable of predicting the codeletion of 1p/19q chromosome arms with
high accuracy (87.7%) in 159 patients with low-grade glioma
(9). In both cases, DL seems to
enable the discovery of hidden imaging phenotype properties, which
are associated with molecular/genetic information of high
prognostic value.
The in silico prediction of therapy
optimization is of utmost importance for personalized medicine.
Bibault et al presented a DL radiomics system capable of
predicting complete response following neo-adjuvant chemoradiation
for locally advanced rectal cancer with the ultimate goal of
identifying patients who would benefit more from conservative
treatment rather than radical resections (10). From a bioinformatics standpoint,
Yousefi et al developed the SurvivalNet framework based on
DL and Bayesian optimization for predicting clinical outcomes from
large amounts of data, which was generated by diverse genomic
platforms (11). In a recent study on
768 patients, DL outperformed the widely used Cox proportional
hazard regression model as well as other widely used models for
survival analysis indicating that it can enhance the role of
survival prediction in individualized clinical decision making
(12). Reliable health predictors
based on patient's heterogeneous electronic health records (EHRs)
data integration is another important application field of DL. In
this direction, Miotto et al developed the ‘Deep Patient’
prediction system trained on 700,000 EHRs and demonstrated high
disease prediction performance for severe diabetes, schizophrenia
and various malignancies (13).
Drug discovery for the personalization of therapy is
another area that can benefit from the flexible architecture of DL
systems towards de novo molecular design, reaction and
bioactivity predictions (14). In
this direction, IBM and Pfizer have joined forces for advancing
immuno-oncology research with Watson for Drug Discovery (www.pfizer.com/news/press-release/press-releasedetail/ibm_and_pfizer_to_accelerate_immuno_oncology_research_with_watson_for_drug_discovery).
Moreover, predicting environmental factors that influence health
status is an integral part of preventive, personalized medicine.
Ong et al proposed a time-series forecasting DL system for
predicting the concentration of fine particulate matter PM2.5
(interfering with both human health and climate), in Japan, using
exclusively real and publicly available sensor data, outperforming
currently used methods (15).
3. Conclusions and future prospects
DL architectures have already demonstrated
significant technological advances across the personalized medicine
informatics application spectrum, due to their versatility towards
integrating multi-modal clinical data, discovering hidden
properties and successfully generalizing results in unseen
datasets. The number of DL applications for personalized medicine
will continue to grow due to the high demand for such technologies
and their anticipated socioeconomic impact. Although this process
is facilitated by the widely available DL open computational
frameworks and the abundance of large amounts of healthcare data,
there is still skepticism regarding the clinical adoption of DL
technologies emanating from the lack of causality and their
‘black-box’ nature. The inability to understand why an algorithm
achieves generalization and performs so well, may be a critical
factor inhibiting the clinical translation of DL technologies. To
address this issue, several researchers are focusing on explaining
the inner organization of DL networks. Shwartz-Ziv and Tishby, and
Tishby and Zaslavsky have proposed an influential theory based on
the ‘information bottleneck’ concept, according to which the
network compresses input data as if by squeezing the information
through a bottleneck, retaining only the features most relevant to
the general concepts of the learning task (16,17). This
compression process is pronounced at the higher layers of the
network for promoting better predictions via preserving information
relevant to the output labels at the expense of input data
information, which is gradually ‘forgotten’. The confirmation of
such theoretical explanatory models in conjunction with large-sized
data validation across clinical sites and patient data registries,
is a sine qua non condition for accelerating the clinical
translation of DL personalized medicine decision support
systems.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the current
study.
Authors' contributions
GZP and KM conceived and designed the study. GZP and
KM researched the literature, and wrote the manuscript. AHK, MΤ, AΤ
and DAS, substantially contributed to the conception and design of
the study, and to the selection of the data included in the study,
revised it critically for important intellectual content, and
finally approved the version to be published. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
References
|
1
|
Noda K, Arie H, Suga Y and Ogata T:
Multimodal integration learning of robot behavior using deep neural
networks. Robot Auton Syst. 62:721–736. 2014. View Article : Google Scholar
|
|
2
|
Chen JH and Asch SM: Machine Learning and
Prediction in Medicine - Beyond the Peak of Inflated Expectations.
N Engl J Med. 376:2507–2509. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ehteshami Bejnordi B, Veta M, Johannes van
Diest P, van Ginneken B, Karssemeijer N, Litjens G, van der Laak
JA, Hermsen M, Manson QF, Balkenhol M, et al: the CAMELYON16
Consortium: Diagnostic assessment of deep learning algorithms for
detection of lymph node metastases in women with breast cancer.
JAMA. 318:2199–2210. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Madani A, Ong JR, Tibrewal A and Mofrad
MRK: Deep echocardiography: Data-efficient supervised and
semi-supervised deep learning towards automated diagnosis of
cardiac disease. NPJ Digit Med. 1(59)2018. View Article : Google Scholar
|
|
5
|
Ding Y, Sohn JH, Kawczynski MG, Trivedi H,
Harnish R, Jenkins NW, Lituiev D, Copeland TP, Aboian MS, Mari
Aparici C, et al: A Deep learning model to predict a diagnosis of
alzheimer disease by using 18F-FDG PET of the brain.
Radiology. 290:456–464. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Rajpurkar P, Irvin J, Ball RL, Zhu K, Yang
B, Mehta H, Duan T, Ding D, Bagul A, Langlotz CP, et al: Deep
learning for chest radiograph diagnosis: A retrospective comparison
of the CheXNeXt algorithm to practicing radiologists. PLoS Med.
15(e1002686)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Causey JL, Zhang J, Ma S, Jiang B, Qualls
JA, Politte DG, Prior F, Zhang S and Huang X: Highly accurate model
for prediction of lung nodule malignancy with CT scans. Sci Rep.
8(9286)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Li Z, Wang Y, Yu J, Guo Y and Cao W: Deep
Learning based Radiomics (DLR) and its usage in noninvasive IDH1
prediction for low grade glioma. Sci Rep. 7(5467)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Akkus Z, Ali I, Sedlář J, Agrawal JP,
Parney IF, Giannini C and Erickson BJ: Predicting deletion of
chromosomal arms 1p/19q in low-Grade gliomas from MR images using
machine intelligence. J Digit Imaging. 30:469–476. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bibault JE, Giraud P, Housset M, Durdux C,
Taieb J, Berger A, Coriat R, Chaussade S, Dousset B, Nordlinger B,
et al: Deep Learning and Radiomics predict complete response after
neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci
Rep. 8(12611)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yousefi S, Amrollahi F, Amgad M, Dong C,
Lewis JE, Song C, Gutman DA, Halani SH, Velazquez Vega JE, Brat DJ,
et al: Predicting clinical outcomes from large scale cancer genomic
profiles with deep survival models. Sci Rep.
7(11707)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Matsuo K, Purushotham S, Jiang B,
Mandelbaum RS, Takiuchi T, Liu Y and Roman LD: Survival outcome
prediction in cervical cancer: Cox models vs deep-learning model.
Am J Obstet Gynecol. Dec 21, 2018 (Epub ahead of print).
2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Miotto R, Li L, Kidd BA and Dudley JT:
Deep Patient: An Unsupervised representation to predict the future
of patients from the electronic health records. Sci Rep.
6(26094)2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen H, Engkvist O, Wang Y, Olivecrona M
and Blaschke T: The rise of deep learning in drug discovery. Drug
Discov Today. 23:1241–1250. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ong BT, Sugiura K and Zettsu K:
Dynamically pre-trained deep recurrent neural networks using
environmental monitoring data for predicting PM2.5. Neural Comput
Appl. 27:1553–1566. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Shwartz-Ziv R and Tishby N: Opening the
Black Box of Deep Neural Networks via Information. Intel
Collaborative Research institute for Computational Intelligence
(ICRI-CI). arXiv:1703.00810. 2017.
|
|
17
|
Tishby N and Zaslavsky N: Deep learning
and the information bottleneck principle. In 2015 IEEE Information
Theory Workshop (ITW). pp1-5. 2015. View Article : Google Scholar
|