Introduction
Hepatitis B virus (HBV) infection is a major health
problem worldwide, especially in developing countries (1,2). The WHO
stated that in 2015, ~257 million people were living with an HBV
infection and 887,000 people died from this condition, mostly from
chronic liver disease (CLD) complications, such as liver cirrhosis
(LC) and hepatocellular carcinoma (HCC) (3). The progression of HBV infection can be
influenced by several factors, including viral, host and
environmental ones. Variations in host genes are associated with
the integration of mutated HBV genes that alter host genes and
cause CLD (4,5).
TNF-α is a multifunctional cytokine that regulates
inflammatory reactions and plays an important role in the
pathogenesis of liver disease, infectious diseases and inflammation
(5). The single-nucleotide
polymorphism (SNP)-238 G/A and -308 G/A are SNPs of TNF-α gene
promoters that have been a focus of several previous works, and
were reported to affect TNF-α production at the transcriptional
level (6,7). Another cytokine that contributes to the
progression of HBV infection is TGF-β1. The SNP-509 C/T in the
TGF-β1 promoter is reported to be associated with changes in the
plasma TGF-β1 concentration (8-10).
Genes that play a role in controlling the cell cycle, such as p53,
also play an important role in the pathogenesis of CLD (11,12). The
SNP Arg72Pro on the p53 gene disrupts the stability and function of
p53 by inhibiting the cell cycle and apoptosis (12-14).
Interactions between host and viral factors are
known to play an important role in the pathogenesis of HBV
infection (15,16). Mutated HBV X protein (HBx) increases
the progression and metastasis of cancer cells (17,18). When
the HBx mutant is formed, there is a transformation increase that
continuously affects the host genome (19). Specifically, the integration of HBx
into the host genome leads to the upregulation of certain important
genes, including NF-κB, TNF-α and TGF-β1, and the downregulation of
p53(20).
There is still an incomplete understanding of the
interactions of SNPs on the TNF-α, TGF-β1 and p53 genes in CLD
patients with chronic HBV infection. In the present study, to
obtain a deeper understanding of the pathogenesis of CLD, we
investigated the associations between these principal SNPs, HBV X
gene mutation and viral load in patients with chronic HBV
infection.
Materials and methods
Sampling
This work involved a cross-sectional study using 87
blood samples collected from patients with chronic HBV at the
inpatient/outpatient clinics of the Department of Internal
Medicine, Dr. Soetomo General Hospital (Surabaya, Indonesia)
between September 2016-May 2017 with a mean age of 47.0±12.5 years
(64 male and 23 female). Inclusion criteria were as follows: CLD
with chronic HBV infection; >16 years; willing to participate as
research subjects; conscious; not in an emergency state; never been
vaccinated for HBV; and HBV infection <10 years. Exclusion
criteria were as follows: Co-infection with hepatitis C virus or
HIV; and undergoing immunosuppressant therapy. This study received
approval from the Ethics Committee of Dr. Soetomo General Hospital
(approval no. 0949/KEPK/II/2019). Written informed consent was
provided by the participants, in accordance with the Declaration of
Helsinki. Patients were classified by CLD stage: (i) Chronic
hepatitis (CH); (ii) LC; and (iii) HCC. The classification was
determined by a hepatologist. CH was diagnosed based on positive
HBsAg for >6 months without any LC/HCC specific clinical
features, laboratory and radiology manifestation. LC was diagnosed
based on clinical features of portal hypertension, liver stiffness
by fibroscan and abnormal liver morphology by ultrasound. HCC was
diagnosed based on clinical manifestation, α-fetoprotein ≥200 ng/ml
and HCC-typical ultrasound findings. Alanine aminotransferase
(ALT), aspartate aminotransferase (AST), platelet and HBeAg were
obtained from medical records.
Genomic DNA extraction
Host DNA was extracted from peripheral blood
mononuclear cell isolated from blood samples and viral DNA was
extracted from 100 µl serum using QIAamp DNA Extraction kit (cat.
no. 51104; Qiagen, Inc.) in accordance with the manufacturer's
instructions.
HBV load measurement
Viral load was assessed in the extracted viral DNA
from all 87 blood samples using a Taqman quantitative (q)PCR method
for absolute quantification by the CFX96™ Real-Time PCR Detection
system (Bio-Rad Laboratories, Inc.) with the Master Mix Real-Time
PCR iTaq Universal Probes Supermix (cat. no. 1725130; Bio-Rad
Laboratories, Inc.) in accordance with the manufacturer's
instructions. The probes used was HBSP1
(FAM-5'-CAGAGTCTAGACTCGTGGTGGACTTC-3'-TAMRA). Primers included: SF1
forward, 5'-CACATCAGGATTCCTAGGACC-3' and SR1 reverse,
5'-GGTGAGTGATTGGAGGGTTG-3'. qPCR was performed with thermocycling
condition as follows: 50˚C for 2 min, 95˚C for 10 min, followed by
53 cycles of 95˚C for 20 sec and 60˚C for 1 min (21). Data reading was performed by Biorad
CFX Maestro software version number 4.1.2433.1219 (Bio-Rad
Laboratories, Inc.).
PCR restriction fragment length
polymorphism (RFLP) of TNF-α, TGF-β1 and p53 on host genome
PCR was carried out on host genome using PCR Master
Mix Solution (cat. no. 25027; iNtRON® Biotechnology,
Inc.). The primers, thermocycling conditions and restriction
enzymes used are listed in Table I.
All PCR products were assessed by 2% agarose gel electrophoresis
under UV light. All restriction enzyme incubation steps were
carried out overnight at 37˚C, except for BstUI for which
60˚C were applied.
 | Table IPrimers and restriction enzymes used
for host SNP and hepatitis B virus X gene detection. |
Table I
Primers and restriction enzymes used
for host SNP and hepatitis B virus X gene detection.
| SNP | Direction | Sequence
(5'-3') | Amplicon (bp) | Thermocycling
conditions | Restriction
enzyme |
|---|
| TNF-α-238 | Forward |
AGGCAATAGGTTTTGAGGG | 107 | 94˚C for 5 min; 40x
94˚C for 30 sec, | MspI |
| | | CCAT | | 60˚C for 30 sec and
72˚C for 40 sec; 72˚C for 7 min | |
| | Reverse |
TCCTCCCTGCTCCGATTCCG | | | |
| TNF-α-308 | Forward |
AGAAGACCCCCCTCGGAACC | 152 | 94˚C for 5 min; 40x
94˚C for 30 sec; | NcoI |
| | | | | 58.5˚C for 30 sec
and 72˚C for 40 sec; 72˚C for 7 min | |
| | Reverse |
ATCTGGAGGAAGCGGTAGTG | | | |
| TGF-β1-509 | Forward |
GGAGAGCAATTCTTACAGGTG | 120 | 94˚C for 3 min; 30x
94˚C for 30 sec, 60˚C | DdeI |
| | | | | for 30 sec and 72˚C
for 60 sec; 72˚C for 10 min | |
| | Reverse |
TAGGAGAAGGAGGGTCTGTC | | | |
| p53 Arg | Forward |
TCCCCCTTGCCGTCCCAA | 396 | 94˚C for 5 min; 30x
94˚C for 30 sec, | BstUI |
| 72 Pro | | | | 55˚C for 30 sec and
72˚C for 30 sec; 72˚C for 7 min | |
| | Reverse |
CGTGCAAGTCACAGACTT | | | |
| X gene | Forward |
CATGCGTGGAACCTTTGTG | 840 | 94˚C for 5 min; 35x
94˚C for 50 sec, | - |
| round 1 | | | | 50˚C for 50 sec and
72˚C for 60 sec; 72˚C for 7 min | |
| | Reverse |
CTTGCCTKAGTGCTGTATGG | | | |
| X gene | Forward | TCCTCTGCCGAT
CCATACTG | 684 | 94˚C for 5 min; 35x
94˚C for 30 sec, | - |
| round 2 | | | | 55˚C for 30 sec and
72˚C for 45 sec; 72˚C for 7 min | |
| | Reverse |
CAGAAGCTCCAAATTCTTTATA | | | |
Analysis of TNF-α, TGF-β1 and p53
SNPs
SNPs were identified via agarose gel (3.5% for TNF-α
SNP, 3% for TGF-β1 SNP and 2% for p53 SNP) electrophoresis analysis
based on the following criteria: (i) In the presence of an SNP at
position -238 on the TNF-α promoter, a 152-bp band (A allele) is
visible, while 133 and 19 bp fragments are identified for the
corresponding wild type (G allele). (ii) With an SNP at position
-308 on the TNF-α promoter, a 107-bp band (A allele) is visible,
while for the corresponding wild type (G allele) 87 and 20 bp
fragments are identified (22). (iii)
For an SNP at position -509 on the TGF-β1 gene, a 120-bp band (T
allele) is visible, while the corresponding wild type (C allele)
shows 74 and 46 bp fragments (23).
(iv) The Arg72Pro SNP on the p53 gene shows a 396-bp band (C
allele), while the corresponding wild type (G allele) displays 231
and 165 bp fragments (24).
Analysis of HBV X gene
Nested PCR on the HBV X gene was performed on the
previously extracted viral genome following the work of Lee et
al (25), as shown in Table I. It was performed using PCR Master
Mix Solution (cat. no. 25027; iNtRON® Biotechnology,
Inc.), following the procedure in the manufacturer's instructions.
PCR products were visualized on 2% agarose gels. DNA sequencing was
performed using an ABI Prism 310 Genetic Analyzer (PerkinElmer,
Inc.). The sequenced nucleotides were compared with a reference
strain nucleotide sequence that had previously been published in
GenBank [accession no., EF473977(26), AB219430(27) and D23678(28)] using Clone Manager 9 (Scientific &
Educational Software).
Statistical analysis
Statistical analyses were performed using SPSS 23
(SPSS, Inc.). Data are presented as the mean ± SD. Analysis was
repeated at least twice for each subject. χ2,
Mann-Whitney U, Kruskal Wallis with Dunn's post hoc test, or
one-way ANOVA with LSD post hoc tests were performed to assess
significance, depending on variable. Multinomial regression
analysis was performed to assess correlations. P<0.05 following
a two-tailed analysis was considered to indicate statistical
significance.
Results
Patient characteristics
For the 87 CLD patients included in this study, the
age range was 16-72 years, with mean ages of 45.0±14.0, 50.7±11.6
and 46.7±8.3 years for patients with CH, LC and HCC, respectively.
Male patients (73.6%) outnumbered females (26.4%), as shown in
Table II. In this study, CLD
patients were most often classed as CH (45/87; 51.7%), followed by
LC (27/87; 31.0%) and HCC (15/87; 17.3%). Each CLD stage was
dominated with male subjects (66.7, 74.7 and 93.0% for CH, LC and
HCC, respectively). AST levels in patients with HCC (211±204.1 U/l)
were significantly increased compared with those in patients with
LC (91.8±175.5 U/l) and CH (64.7±95.2 U/l).
 | Table IICharacteristics of patients with
chronic liver disease. |
Table II
Characteristics of patients with
chronic liver disease.
|
Characteristics | CH (n=45) | LC (n=27) | HCC (n=15) | Total (n=87) | P-value |
|---|
| Sex (male) | 30 (66.7) | 20 (74.7) | 14 (93.0) | 64 (73.6) | 0.128 |
| Age (years) | 45.0±14.0 | 50.7±11.6 | 46.7±8.3 | 47.0±12.5 | 0.167 |
| AST (U/l) | 64.7±95.2 | 91.8±175.5 | 211.0±204.1 | 98.3±153.5 | <0.001 |
| ALT (U/l) | 95.4±107.7 | 125.5±323.9 | 100.0±71.9 | 105.5±218.3 | 0.056 |
| Platelet
count/µl |
196,711.1±120,382.5 |
162,629.6±82,014.1 |
238,333.3±106,306.7 |
193,310.3±10,929.1 | 0.098 |
| HBeAg,
positive | 14 (31.1) | 9 (33.3) | 2 (13.3) | 25 (28.7) | 0.351 |
Genotype distributions of TNF-α,
TGF-β1 and p53 SNPs
The genotype distributions of TNF-α, TGF-β1 and p53
SNPs were determined by PCR-RFLP and are shown in Table III. No cases with the AA genotypes
(homozygous minor-mutant type) on the TNF-α promoter (-238/-308)
were identified. There was no difference in the distribution of
genotypes or SNP alleles of TNF-α, TGF-β1 and p53 in different CLD
groups (P>0.05).
 | Table IIIDistribution of host SNPs in patients
with CLD. |
Table III
Distribution of host SNPs in patients
with CLD.
| | CLD stage | |
|---|
| SNP genotype | CH | LC | HCC | Total |
|---|
| TNF-α-238 |
|
GG | 44 (98.0) | 24 (88.5) | 15(100) | 83 (95.4) |
|
GA | 1 (2.0) | 3 (11.5) | 0 | 4 (4.6) |
|
AA | 0 | 0 | 0 | 0 |
|
Total | 45(100) | 27(100) | 15(100) | 87(100) |
| TGF-β1-509 |
|
CC | 10 (22.2) | 5 (18.5) | 3 (20.0) | 18 (20.7) |
|
TC | 23 (51.1) | 14 (51.9) | 4 (26.7) | 41 (47.1) |
|
TT | 12 (26.7) | 8 (29.6) | 8 (53.3) | 28 (32.2) |
|
Total | 45(100) | 27(100) | 15(100) | 87(100) |
| Arg72Pro |
|
CC | 13 (28.9) | 6 (22.2) | 2 (13.3) | 21 (24.2) |
|
CG | 23 (51.1) | 16 (59.3) | 10 (66.7) | 49 (56.3) |
|
GG | 9 (20.0) | 5 (18.5) | 3 (20.0) | 17 (19.5) |
|
Total | 45(100) | 27(100) | 15(100) | 87(100) |
Analysis of HBV X gene mutations
Based on the multiple alignments, the main type of
HBV X gene mutation identified was amino acid substitutions, as
shown in Table IV. A total of 23%
(20/87) of samples had HBV X gene mutations with ten types of
previously reported substitution (29-32).
There was one sample that showed both the 32-nucleotide insertion
and the 20-nucleotide deletion. These types of mutation caused a
shift in the nucleotide reading frame, resulting in the formation
of a truncated HBx protein. X gene mutations were found in 17.8%
(8/45) of patients with CH, 25.9% (7/27) patients with LC and 33.3%
(5/15) patients with HCC. In addition to previously reported
mutations, we also found five variants of the X gene located in the
HBV functional region, including L37I, S43P, H86P/R, L98I and
T105A, which have not been reported in previous studies and may be
specific for Indonesian HBV.
 | Table IVHepatitis B virus X gene mutation
profiles and its association to chronic liver disease
progression. |
Table IV
Hepatitis B virus X gene mutation
profiles and its association to chronic liver disease
progression.
| | Samples with
mutation (%) | | |
|---|
| Mutation | CH (n=45) | LC (n=27) | HCC (n=15) | Total (n=87) | Functional region
affected | P-value |
|---|
| T36P | 0 | 3.7 | 0 | 1.2 | B-cell epitope | 0.483 |
| L37I | 0 | 3.7 | 0 | 1.2 | B-cell epitope | 0.483 |
| P38S | 2.2 | 0 | 0 | 1.2 | B-cell epitope | 1.000 |
| S43P | 6.7 | 0 | 13.3 | 5.8 | B-cell epitope | 0.147 |
| A44V/T | 2.2 | 3.7 | 6.7 | 3.4 | B-cell epitope | 0.748 |
| P46S | 2.2 | 3.7 | 0 | 2.3 | B-cell epitope | 1.000 |
| H86P/R | 2.2 | 7.4 | 0 | 3.4 | Core promoter,
EnhII | 0.576 |
| R87W/G | 6.7 | 7.4 | 6.7 | 6.9 | Core promoter,
EnhII | 1.000 |
| H94Y | 2.2 | 0 | 0 | 1.2 | Box α, C/EBP,
CCAAT/enhancing binding protein, core promoter, EnhII | 1.000 |
| L98I | 2.2 | 3.7 | 6.7 | 3.4 | Box α, C/EBP,
CCAAT/enhancing binding protein, core promoter, EnhII | 0.748 |
| T105A | 4.4 | 0 | 0 | 2.3 | Core promoter,
EnhII, BH3-like motif, T-cell epitope | 1.000 |
| L123S/V | 2.2 | 0 | 6.7 | 2.3 | BH3-like motif,
core promoter, NRE | 0.411 |
| I127L/T/N/S | 8.9 | 7.4 | 20 | 10.3 | BH3-like motif,
core promoter, NRE | 0.461 |
| Insertion (32
nt) | 2.2 | 0 | 0 | 1.2 | Box α, C/EBP,
CCAAT/enhancing binding protein, core promoter, EnhII, BH3-like
motif, T-cell epitope | 1.000 |
| Deletion (20
nt) | 2.2 | 0 | 0 | 1.2 | T-cell epitope,
BH3-like motif, core promoter | 1.000 |
| K130M | 4.4 | 25.9 | 26.7 | 14.9 | BH3-like motif,
core promoter | 0.006 |
| V131I | 4.4 | 25.9 | 26.7 | 14.9 | BH3-like motif,
core promoter | 0.006 |
The dominant mutations in the basal core promoter
(BCP) were K130M/V131I, as found in 12.6% (11/87) of CLD patients.
These two mutations were observed as double mutation in one sample.
There was an association between the K130M/V131I mutations and the
progression of CLD (P<0.05), but not for other X gene mutations
(P>0.05; Table IV). Multinomial
regression analysis confirmed that K130M/V131I mutations were
correlated with CLD progression (OR, 7.629; 95% CI, 1.578-36.884;
Table V). No association of HBV X
gene mutations with TNF-α (-238, -308), TGF-β1 or p53 gene SNPs
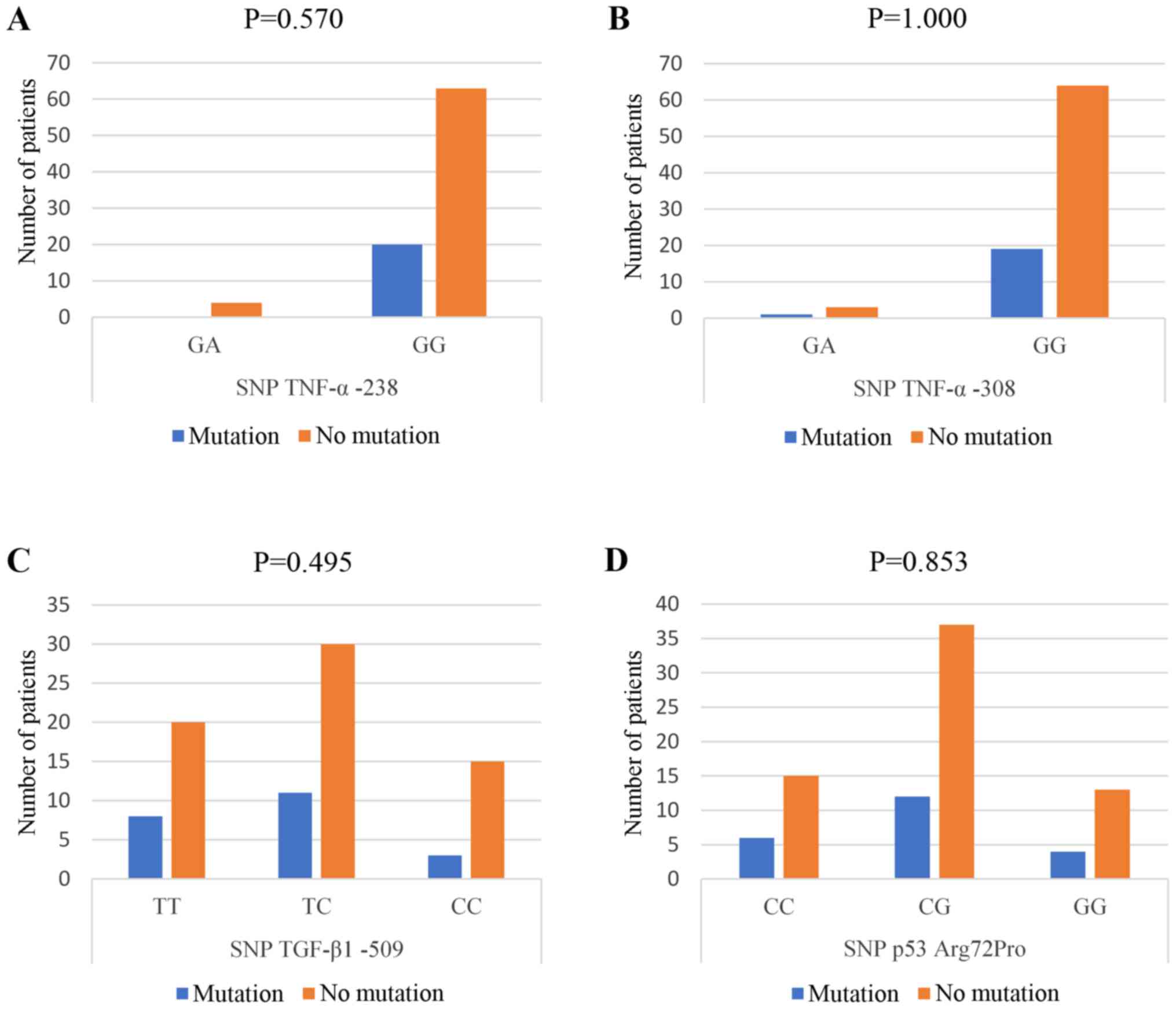
were found in CLD patients with HBV infection (Fig. 1).
 | Table VMultinomial regression analysis of
the K130M/V131I mutation in chronic liver disease progression. |
Table V
Multinomial regression analysis of
the K130M/V131I mutation in chronic liver disease progression.
| | Chi-square
value | P-value | OR | 95% CI |
|---|
| K130M/V131I | 8.71 | 0.011 | 7.629 | 1.578-36.884 |
HBV viral load measurement
The mean viral load was the highest in LC patients
(4.98±4.37 log copies/ml), followed by patients with HCC (3.49±2.63
log copies/ml) and patients with LC (2.95±3.59 log copies/ml).
Analysis showed no significant differences in viral load depending
on CLD stage (P>0.05; Table VI).
There were significant differences in viral load levels in
HBV-infected patients who had X gene mutations, as well as the
R87W/G, I127L/T/N/S and K130M/V131I mutation (P<0.05).
 | Table VIHepatitis B virus viral load in
patients with X gene mutations. |
Table VI
Hepatitis B virus viral load in
patients with X gene mutations.
| | Viral load (log
copies/ml) | |
|---|
| Mutation | Median | IQR | P-value |
|---|
| S43P | | | 0.137 |
|
Mutation | 5.74 | 1.90 | |
|
No
mutation | 2.50 | 6.95 | |
| A44 | | | 0.063 |
|
Mutation | 7.10 | 0.00 | |
|
No
mutation | 14.22 | 6.68 | |
| R87W/G | | | 0.029 |
|
Mutation | 6.78 | 1.36 | |
|
No
mutation | 2.14 | 6.78 | |
| I127L/T/N/S | | | 0.035 |
|
Mutation | 6.86 | 4.00 | |
|
No
mutation | 2.00 | 6.65 | |
| K130M/V131I | | | 0.004 |
|
Mutation | 6.86 | 2.53 | |
|
No
mutation | 1.04 | 5.90 | |
| X gene
mutation | | | <0.001 |
|
Mutation | 6.90 | 2.94 | |
|
No
mutation | 0.00 | 4.86 | |
Discussion
In the present study, 87 patients from Surabaya,
Indonesia, with HBV-associated CLD were enrolled. We found no
significant differences in the distribution of genotypes or alleles
of TNF-α-238 or -308 SNPs among the three CLD stages. Additionally,
we found no cases with the AA genotype for these SNPs. This result
was in line with reports from Banday et al (33) that showed that the frequency of A
alleles in Asia is decreased compared with other regions. A similar
study that was performed in China by Xu et al (6) also failed to identify patients with AA
genotypes. The SNPs in TNF-α at positions -238 and -308 are often
associated with various diseases, including severe inflammation,
infection and malignancy (34).
Research on SNPs of the TNF-α promoter in patients with HBV
infection has shown conflicting results regarding population- and
ethnic-specificity (35) and some but
not all studies showed a correlation between SNPs of TNF-α
promoters and HBV infection (36-38).
In this study, we found no differences in the
distribution of genotypes or alleles of TGF-β1 and p53 SNPs between
patients in the CLD groups. The frequency of SNPs in patients with
CLD was greater than occurrence of the wild type in this study.
Previous studies reported different results regarding the
TGF-β1-509 and Arg72Pro p53 SNPs among diverse populations. A
meta-analysis conducted by Guo et al (39) showed that in an Asian population, the
TGF-β1-509 SNP T allele was correlated with the incidence of HCC,
but this was not observed in Caucasian and African participants.
However, in a study conducted in China by Qi et al (40), the risk of HCC in patients with the TT
genotype was decreased compared with the CC genotype. Research
conducted in Turkey by Sümbül et al (41) showed a correlation between the
Arg72Pro SNP GG genotype and the incidence of HCC that was not
observed for the normal population without HBV infection. This was
not confirmed by research conducted in China by Cai et al
(42) that found no significant
differences between the Arg72Pro SNP in patients with HBV infection
and healthy controls. In addition to studies focusing on CLD,
studies on the association between the Arg72Pro p53 SNP and various
cancers types have also shown controversial results (43-45).
This is thought to be associated with the ethnic background of the
patients (41).
The present study found 12 types of X gene mutation,
among which the K130M/V131I mutations were significantly correlated
with CLD progression. These two mutations are located in the BCP,
which overlaps with the X gene. The presence of these double
mutations is thought to exacerbate the host's immune response,
increase viral replication and modify the coding region of the X
protein causing the progression of CLD (46). H94Y and I127L/T/N/S mutations have
been associated to K130M/V131I in previous studies (47,48). The
H94Y mutation causes changes in the α box, an element strongly
activating the EnhII/core promoter, and increase the binding
affinity of the α box and EnhII/core promoter (29,49). The
rate of the P38S mutation was reported to be significantly higher
in HCC than in asymptomatic carriers, but not significantly
different from the rates in CH and LC patients (49). The rate of the T36P mutation was
reported to be higher in HCC patients, presumably due to its
location at the B epitope, which affects the immune response
(50).
No associations of HBV X gene mutations with TNF-α
(-238 and -308), TGF-β1 and p53 SNPs were found in CLD patients. In
previous studies, several SNPs associated with HBV X gene mutations
were identified, particularly SNPs associated with HLA-DQ/DR
(rs9272105 and rs9277535) (51,52).
Research has also focused on the association between the SNP of the
STAT3 gene (rs1053004) and EnhII/BCP/PC mutations that overlap with
the X gene (53). However, to date,
no studies have linked SNPs to other genes with HBV mutations,
including TNF-α (-238 and -308), TGF-β1 and p53 gene SNPs, as it
was shown in this study.
In this study, the mean viral load was highest in LC
patients, followed by HCC and CH patients. Our results showed no
significant differences in viral load between the CLD stages.
Several studies have shown an association between HBV viral load
and liver damage, and high viral load is often associated with
severe liver damage (54,55). Although HBV viral load is often
associated with the risk of HCC, a study conducted by Harkisoen
et al (56) showed lower viral
load in LC patients. Similarly, a study conducted by Tseng et
al (57) showed no association
between viral load and HCC risk. This may be caused by the presence
of HBV DNA, which is integrated into hepatocytes more efficiently
compared with serum (58,59). In addition, there are other
influential factors for CLD progression to HCC, such as alcoholism,
cigarette smoking and HBV genotypes (56,57).
There were significant differences in viral load
levels between patients with X gene mutations, including R87W/G,
I127L/T/N/S and K130M/V131I mutations. X gene mutations affect HBV
viral load, as indicated by observations of a higher HBV
replication rate when featuring multiple mutations. This high rate
of HBV replication causes increased inflammation and viral invasion
(29,49). However, to date, conflicting findings
on the association between X gene mutations and HBV viral load have
been obtained (60). A study
conducted by Ghabeshi et al (61) showed no association between BCP
mutations and viral load. This may be due to BCP mutations reducing
viral secretion in the blood, but increasing viral load in the
liver, which directly causes liver damage. These differences in
results are influenced by differences in populations, such as HBV
genotypes, HBeAg status and clinical conditions (60).
In this study, the presence of K130M/V131I mutations
constituted an independent risk factor for CLD progression. X gene
mutations, including R87W/G, I127L/T/N/S and K130M/V131I mutations,
were correlated with HBV viral load. Additionally, we found no
association between SNPs of the TNF-α, TGF-β1 and p53 genes with X
gene mutation. While further studies are required to examine the
clinical value of these results, the presence of K130M/V131I
mutations may serve as a predictive marker for the progression of
CLD in Indonesia and may also be applicable for other countries
where chronic HBV infections are prevalent.
Acknowledgments
Not applicable.
Funding
The present study was supported by University of
Airlangga and the Indonesian Ministry of Research, Technology and
Higher Education (grant no. 122/SP2H/PTNBH/DRPM/2018) and in part
by a Grant-in-Aid from Dato' Sri Prof. Dr. Tahir for supporting
this research through the Tahir Professorship Program,
Indonesia.
Availability of data and materials
All data generated in the present study are included
in this article.
Authors' contributions
This study was conducted and designed by MIL, SS and
RH. CDKW, PBP, UK, UM and PBS performed sample collection. MA and
SENR performed the laboratory experiments. CDKW analyzed the data
and wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study received approval from the Ethics
Committee of Dr. Soetomo General Hospital (Surabaya, Indonesia)
(approval no. 0949/KEPK/II/2019). Written informed consent for
participation was obtained from each individual.
Patient consent for publication
Not applicable.
Competing interest
The authors declare that they have no competing
interests.
References
|
1
|
Xu HZ, Liu YP, Guleng B and Ren JL:
Hepatitis B virus-related hepatocellular carcinoma: Pathogenic
mechanisms and novel therapeutic interventions. Gastrointest
Tumors. 1:135–145. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zampino R, Boemio A, Sagnelli C, Alessio
L, Adinolfi LE, Sagnelli E and Coppola N: Hepatitis B virus burden
in developing countries. World J Gastroenterol. 21:11941–11953.
2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
World Health Organization: Hepatitis B.
2018.
|
|
4
|
Zeng Z: Human genes involved in hepatitis
B virus infection. World J Gastroenterol. 20:7696–7706.
2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mathew S, Abdel-Hafiz H, Raza A, Fatima K
and Qadri I: Host nucleotide polymorphism in hepatitis B virus
associated hepatocellular carcinoma. World J Hepatol. 8:485–498.
2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Xu XW, Lu MH and Tan DM: Association
between tumour necrosis factor gene polymorphisms and the clinical
types of patients with chronic hepatitis B virus infection. Clin
Microbiol Infect. 11:52–56. 2005.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Cheong JY, Cho SW, Hwang IL, Yoon SK, Lee
JH, Park CS, Lee JE, Hahm KB and Kim JH: Association between
chronic hepatitis B virus infection and interleukin-10, tumor
necrosis factor-α gene promoter polymorphisms. J Gastroenterol
Hepatol. 21:1163–1169. 2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hanafy SM and Abdo A: Impact of single
nucleotide polymorphism of TGF-β1 gene (SNP-Codon10) on
hepatocellular carcinoma risk in Egyptian patients following HCV
infection. Aust J Basic Appl Sci. 5:1814–1821. 2011.
|
|
9
|
Falleti E, Fabris C, Toniutto P, Fontanini
E, Cussigh A, Bitetto D, Fornasiere E, Avellini C, Minisini R and
Pirisi M: TGF-β1 genotypes in cirrhosis: Relationship with the
occurrence of liver cancer. Cytokine. 44:256–261. 2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Schon H and Weiskirchen R:
Immunomodulatory effects of transforming growth factor-β in the
liver. Hepatobiliary Surg Nutr. 3:386–406. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hussain SP, Schwank J, Staib F, Wang XW
and Harris CC: TP53 mutations and hepatocellular carcinoma:
Insights into the etiology and pathogenesis of liver cancer.
Oncogene. 26:2166–2176. 2007.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang Z, Gou W, Liu M, Sang W, Chu H and
Zhang W: Expression of P53 and HSP70 in chronic hepatitis, liver
cirrhosis, and early and advanced hepatocellular carcinoma tissues
and their diagnostic value in hepatocellular carcinoma: An
immunohistochemical study. Med Sci Monit. 21:3209–3215.
2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gouas DA, Villar S, Ortiz-Cuaran S, Legros
P, Ferro G, Kirk GD, Lesi OA, Mendy M, Bah E, Friesen MD, et al:
TP53 R249S mutation, genetic variations in HBX and risk of
hepatocellular carcinoma in The Gambia. Carcinogenesis.
33:1219–1224. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Hu S, Zhao L, Yang J and Hu M: The
association between polymorphism of P53 Codon72 Arg/Pro and
hepatocellular carcinoma susceptibility: Evidence from a
meta-analysis of 15 studies with 3,704 cases. Tumor Biol.
35:3647–3656. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Baumert TF, Thimme R and von Weizsäcker F:
Pathogenesis of hepatitis B virus infection. World J Gastroenterol.
13:82–90. 2007.PubMed/NCBI View Article : Google Scholar
|
|
16
|
You CR, Lee SW, Jang JW and Yoon SK:
Update on hepatitis B virus infection. World J Gastroenterol.
20:13293–13305. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Geng M, Xin X, Bi LQ, Zhou LT and Liu XH:
Molecular mechanism of hepatitis B virus X protein function in
hepatocarcinogenesis. World J Gastroenterol. 21:10732–10738.
2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang ZH, Wu CC, Chen XW, Li X, Li J and
Lu MJ: Genetic variation of hepatitis B virus and its significance
for pathogenesis. World J Gastroenterol. 22:126–144.
2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Guerrieri F, Belloni L, Pediconi N and
Levrero M: Molecular mechanisms of HBV-associated
hepatocarcinogenesis. Semin Liver Dis. 33:147–156. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ayub A, Ashfaq UA and Haque A: HBV induced
HCC: Major risk factors from genetic to molecular level. Biomed Res
Int. 2013(810461)2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Abe A, Kazuaki I, Take AT, Kato J,
Kajiyama N, Kawaguchi R, Tanaka S, Yoshiba M and Kohara M:
Quantification of hepatitis B virus genomic DNA by real-time
detection. J Clin Microbiol. 37:2899–2903. 1999.PubMed/NCBI
|
|
22
|
Jamil K, Jayaraman A, Ahmad J, Joshi S and
Yerra SK: TNF-alpha -308G/A and -238G/A polymorphisms and its
protein network associated with type 2 diabetes mellitus. Saudi J
Biol Sci. 24:1195–1203. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chou HT, Chen CH, Tsai CH and Tsai FJ:
Association between transforming growth factor-β1 gene C-509T and
T869C polymorphisms and rheumatic heart disease. Am Heart J.
148:181–186. 2004.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wu X, Zhao H, Amos CI, Shete S, Makan N,
Hong WK, Kadlubar FF and Spitz MR: p53 genotypes and haplotypes
associated with lung cancer susceptibility and ethnicity. J Natl
Cancer Inst. 94:681–690. 2002.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lee JH, Han KH, Lee JM, Park JH and Kim
HS: Impact of hepatitis B virus (HBV) X gene mutations on
hepatocellular carcinoma development in chronic HBV infection. Clin
Vaccine Immunol. 18:914–921. 2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Nurainy N, Muljono DH, Sudoyo H and
Marzuki S: Genetic study of hepatitis B virus in Indonesia reveals
a new subgenotype of genotype B in east Nusa Tenggara. Arch Virol.
153:1057–1065. 2008.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Nagasaki F, Niitsuma H, Cervantes JG,
Chiba M, Hong S, Ojima T, Ueno Y, Bondoc E, Kobayashi K, Ishii M
and Shimosegawa T: Analysis of the entire nucleotide sequence of
hepatitis B virus genotype B in the Philippines reveals a new
subgenotype of genotype B. J Gen Virol. 87:1175–1180.
2006.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Horikita M, Itoh S, Yamamoto K, Shibayama
T and Tsuda F: Differences in the entire nucleotide sequence
between hepatitis B virus genomes from carriers positive for
antibody to hepatitis B e antigen with and without active disease.
J Med Virol. 44:96–103. 1994.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kim H, Lee SA and Kim BJ: X region
mutations of hepatitis B virus related to clinical severity. World
J Gastroenterol. 22:5467–5478. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Fan W, Shi B, Wei H, Du G and Song S:
Comparison of hepatitis B X gene mutation between patients with
hepatocellular carcinoma and patients with chronic hepatitis B.
Virus Genes. 42:162–170. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lin X, Xu X, Huang QL, Liu YQ, Zheng DL,
Chen WN and Lin JY: Biological impacts of ‘hot-spot’ mutations of
hepatitis B virus X proteins are genotype B and C differentiated.
World J Gastroenterol. 11:4703–4708. 2005.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Fatimawali and Kepel B: Analisis
mutasi gen protein X virus HBV pada penderita hepatitis B akut di
manado. J LPPM Bid Sains Dan Teknol. 1:47–55. 2014.
|
|
33
|
Banday MZ, Balkhi HM, Hamid Z, Sameer AS,
Chowdri NA and Haq E: Tumor necrosis factor-α (TNF-α)-308G/A
promoter polymorphism in colorectal cancer in ethnic Kashmiri
population-A case control study in a detailed perspective. Meta
Gene. 9:128–136. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Li X, Liang C, Parkman V and Lv Z: The
association between TNF-α 238A/G and 308A/G polymorphisms and
juvenile idiopathic arthritis. Medicine (Baltimore).
97(e12883)2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Heidari Z, Moudi B, Mahmoudzadeh Sagheb H
and Moudi M: Association of TNF-α gene polymorphisms with
production of protein and susceptibility to chronic hepatitis B
infection in the South East Iranian population. Hepat Mon.
16(e41984)2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Jeng JE, Tsai JF, Chuang LY, Ho MS, Lin
ZY, Hsieh MY, Chen SC, Chuang WL, Wang LY, Yu ML, et al: Tumor
necrosis factor-α 308.2 polymorphism is associated with advanced
hepatic fibrosis and higher risk for hepatocellular carcinoma.
Neoplasia. 9:987–992. 2007.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Tavakolpour S and Sali S: Tumor necrosis
factor-α-308 G/A polymorphisms and risk of hepatocellular
carcinoma: A meta-analysis. Hepat Mon. 16(e33537)2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Xiao Q, Fu B, Chen P, Liu ZZ, Wang W and
Ye Q: Three polymorphisms of tumor necrosis factor-alpha and
hepatitis B virus related hepatocellular carcinoma: A
meta-analysis. Medicine (Baltimore). 95(e5609)2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Guo Y, Zang C, Li Y, Yuan L, Liu Q, Zhang
L and Li S: Association between TGF-β1 polymorphisms and
hepatocellular carcinoma risk: A meta-analysis. Genet Test Mol
Biomarkers. 17:814–820. 2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Qi P, Chen Y, Wang H, Fang M, Ji Q, Zhao
YP, Sun XJ, Liu Y and Gao CF: -509C>T polymorphism in the TGF-β1
gene promoter, impact on the hepatocellular carcinoma risk in
Chinese patients with chronic hepatitis B virus infection. Cancer
Immunol Immunother. 58:1433–1440. 2009.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Sümbül AT, Akkız H, Bayram S, Bekar A,
Akgöllü E and Sandıkçı M: p53 codon 72 polymorphism is associated
with susceptibility to hepatocellular carcinoma in the Turkish
population: A case-control study. Mol Biol Rep. 39:1639–1647.
2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Cai J, Cai Y, Ma Q, Chang F, Xu L, Zhang G
and Guo X: Association of p53 codon 72 polymorphism with
susceptibility to hepatocellular carcinoma in a Chinese population
from northeast Sichuan. Biomed Rep. 6:217–222. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Tian X, Dai S, Sun J, Jiang S and Jiang Y:
Association between TP53 Arg72Pro polymorphism and leukemia risk: A
meta-analysis of 14 case-control studies. Sci Rep.
6(24097)2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
He T, Wu J, Chen Y and Zhang J: TP 53
polymorphisms and melanoma: A meta-analysis. J Cancer Res Ther.
11:409–414. 2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lin YM, Shao J, Yin XH, Huang C, Jia XW,
Yuan YD, Wu CJ, Zhen EM, Yao ZX, Zeng XT and Liu RH: Meta-analysis
results on the association between TP53 codon 72 polymorphism with
the susceptibility to oral cancer. Front Physiol.
9(1014)2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chachá SGF, Gomes-Gouvêa MS, Malta FM,
Ferreira SDC, Villanova MG, Souza FF, Teixeira AC2 Passos ADDC,
Pinho JRR and Martinelli ALC: Basal core promoter and precore
mutations among hepatitis B virus circulating in Brazil and its
association with severe forms of hepatic diseases. Mem Inst Oswaldo
Cruz. 112:626–631. 2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Al-qahtani AA, Al-anazi MR, Nazir N, Ghai
R, Abdo AA, Sanai FM, Al-Hamoudi WK, Alswat KA, Al-Ashgar HI, Khan
MQ, et al: Hepatitis B virus (HBV) X gene mutations and their
association with liver disease progression in HBV-infected
patients. Oncotarget. 8:105115–105125. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Datta S, Ghosh A, Dasgupta D, Ghosh A,
Roychoudhury S, Roy G, Das S, Das K, Gupta S, Basu K, et al: Novel
point and combo-mutations in the genome of hepatitis B
virus-genotype D: Characterization and impact on liver disease
progression to hepatocellular carcinoma. PLoS One.
9(e110012)2014.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Kim HJ, Park JH, Jee Y, Lee SA, Kim H,
Song BC, Yang S, Lee M, Yoon JH, Kim YJ, et al: Hepatitis B virus X
mutations occurring naturally associated with clinical severity of
liver disease among Korean patients with chronic genotype C
infection. J Med Virol. 80:1337–1343. 2008.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Li W, Goto K, Matsubara Y, Ito S, Muroyama
R, Li Q and Kato N: The characteristic changes in hepatitis B virus
X region for hepatocellular carcinoma: A comprehensive analysis
based on global data. PLoS One. 10(e0125555)2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Wen J, Song C, Jiang D, Jin T, Dai J, Zhu
L, An J, Liu Y, Ma S, Qin N, et al: Hepatitis B virus genotype,
mutations, human leukocyte antigen polymorphisms and their
interactions in hepatocellular carcinoma: A multi-centre
case-control study. Nature. 5(16489)2015.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Zhang Q, Yin J, Zhang Y, Deng Y, Ji X, Du
Y, Pu R, Han Y, Zhao J, Han X, et al: HLA-DP polymorphisms affect
the outcomes of chronic hepatitis B virus infections, possibly
through interacting with viral mutations. J Virol. 87:12176–12186.
2013.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Xie J, Zhang Y, Zhang Q, Han Y, Yin J, Pu
R, Shen Q, Lu W, Du Y, Zhao J, et al: Interaction of signal
transducer and activator of transcription 3 polymorphisms with
hepatitis B virus mutations in hepatocellular carcinoma.
Hepatology. 57:2369–2377. 2013.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Ledesma MM, Galdame O, Bouzas B, Tadey L,
Livellara B, Giuliano S, Viaut M, Paz S, Fainboim H, Gadano A, et
al: Characterization of the basal core promoter and precore regions
in anti-HBe-positive inactive carriers of hepatitis B virus. Int J
Infect Dis. 15:e314–e320. 2011.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Chu CJ, Hussain M and Lok AS: Quantitative
serum HBV DNA levels during different stages of chronic hepatitis B
infection. Hepatology. 36:1408–1415. 2002.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Harkisoen S, Arends JE, van den Hoek JA,
Whelan J, van Erpecum KJ, Boland GJ and Hoepelman AI: Historic and
current hepatitis B viral DNA and quantitative HBsAg level are not
associated with cirrhosis in non-Asian women with chronic hepatitis
B. Int J Infect Dis. 29:133–138. 2014.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Tseng TC, Liu CJ, Yang HC, Su TH, Wang CC,
Chen CL, Kuo SF, Liu CH, Chen PJ, Chen DS and Kao JH: High levels
of hepatitis B surface antigen increase risk of hepatocellular
carcinoma in patients with low HBV load. Gastroenterology.
142:1140–1149.e3. 2012.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Tu T, Budzinska MA, Shackel NA and Urban
S: HBV DNA integration: Molecular mechanisms and clinical
implications. Viruses. 9(E75)2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Leung NW, Tam JS, Lau GT, Leung TW, Lau WY
and Li AK: Hepatitis B virus DNA in peripheral blood leukocytes a
comparison between hepatocellular carcinoma and Other hepatitis B
virus-related chronic liver diseases. Cancer. 73:1143–1148.
1994.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Barbini L, Tadey L, Fernandez S, Bouzas B
and Campos R: Molecular characterization of hepatitis B virus X
gene in chronic hepatitis B patients. Virol J.
9(131)2012.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Ghabeshi S, Sharifi Z, Hosseini SM and
Mahmoodian Shooshtari M: Correlation between viral load of HBV in
chronic hepatitis B patients and precore and basal core promoter
mutations. Hepat Mon. 13(e7415)2013.PubMed/NCBI View Article : Google Scholar
|