Spandidos Publications style
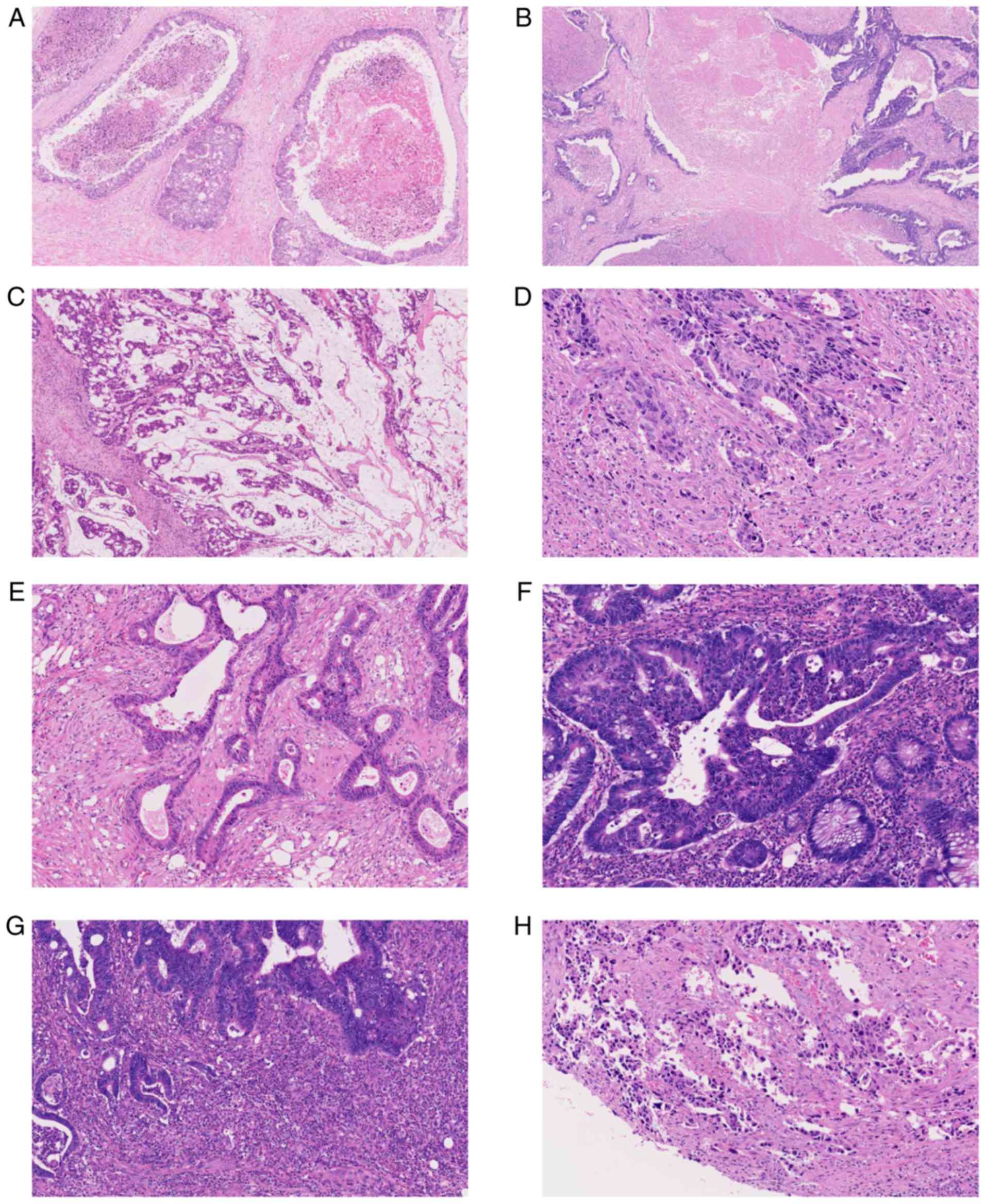
Fujii S, Yoshino T, Yamazaki K, Muro K, Yamaguchi K, Nishina T, Yuki S, Shinozaki E, Shitara K, Bando H, Bando H, et al: Histopathological factors affecting the extraction of high quality genomic DNA from tissue sections for next‑generation sequencing. Biomed Rep 11: 171-180, 2019.
APA
Fujii, S., Yoshino, T., Yamazaki, K., Muro, K., Yamaguchi, K., Nishina, T. ... Tsuchihara, K. (2019). Histopathological factors affecting the extraction of high quality genomic DNA from tissue sections for next‑generation sequencing. Biomedical Reports, 11, 171-180. https://doi.org/10.3892/br.2019.1235
MLA
Fujii, S., Yoshino, T., Yamazaki, K., Muro, K., Yamaguchi, K., Nishina, T., Yuki, S., Shinozaki, E., Shitara, K., Bando, H., Mimaki, S., Nakai, C., Matsushima, K., Suzuki, Y., Akagi, K., Yamanaka, T., Nomura, S., Esumi, H., Sugiyama, M., Nishida, N., Mizokami, M., Koh, Y., Abe , Y., Ohtsu, A., Tsuchihara, K."Histopathological factors affecting the extraction of high quality genomic DNA from tissue sections for next‑generation sequencing". Biomedical Reports 11.4 (2019): 171-180.
Chicago
Fujii, S., Yoshino, T., Yamazaki, K., Muro, K., Yamaguchi, K., Nishina, T., Yuki, S., Shinozaki, E., Shitara, K., Bando, H., Mimaki, S., Nakai, C., Matsushima, K., Suzuki, Y., Akagi, K., Yamanaka, T., Nomura, S., Esumi, H., Sugiyama, M., Nishida, N., Mizokami, M., Koh, Y., Abe , Y., Ohtsu, A., Tsuchihara, K."Histopathological factors affecting the extraction of high quality genomic DNA from tissue sections for next‑generation sequencing". Biomedical Reports 11, no. 4 (2019): 171-180. https://doi.org/10.3892/br.2019.1235