1. Introduction
MicroRNAs (miRNAs or miRs) have been identified as
an important class of endogenously expressed small RNAs with
critical regulatory functions that modulate protein synthesis rate
(1–3). The functional miRNA that targets
mature mRNA is single-stranded and typically 19–22-nt long after
being processed from double-stranded RNA. miRNA molecules regulate
gene expression mainly by binding to complementary sequences in the
3′-untranslated region of target mRNAs and the formation of
RNA-induced silencing complexes, suppressing mRNA translation or
degrading the miRNA-bound mRNA transcripts (4). The functional significance of miRNAs
was initially demonstrated in the developmental process of
Caenorhabditis elegans(5,6).
Additional studies confirmed a strong association between miRNA and
cancer (7–11). Cancer is characterized by
uncontrolled cell growth, high proliferation, anti-apoptosis,
acquisition of invasive properties to adjacent tissues and organs
and inappropriate cell survival (12–14).
Epigenetic change is crucial in tumorigenesis and miRNAs are key
components of epigenetic alterations (15). With the expansion of miRNA
expression profiling efforts, miRNA expression patterns have been
identified as unique signatures associated with diagnosis, staging,
progression, prognosis and response to treatment. In addition,
miRNAs are directly involved in cancer development by activating
oncogenic pathways or suppressing tumor suppressor gene pathways or
protein expression (16–19). Among these, miR-181b was identified
as one of the most important miRNAs contributing to tumor
initiation and progression.
2. miR-181b provides a critical link between
inflammation and cancer
miR-181 family members contain four highly conserved
mature miRNAs: miR-181a, -181b, -181c and -181d (20). These are independently derived from
six precursors located on three different chromosomes: miR-181a-1
and -181b-1 are located on chromosome 1; miR-181a-2 and -181b-2 are
located on chromosome 9; miR-181c and -181d are located on
chromosome 19; miR-181b-1 and -181b-2 are identical in their mature
sequences, but located on different chromosomes (21). Evidence indicates that miR-181s are
aberrantly expressed in tumor tissues, suggesting a potentially
important role in tumor development and/or progression (22,23).
miR-181b was previously shown to be highly expressed
in pancreatic (22,24), head and neck (25) and bladder cancer (26). By contrast, it was found to be
downregulated in gastric (27) and
prostate cancer (28). Those
results suggested that the function of miR-181b may be unique,
depending on the type of tumor and cellular context (29–31).
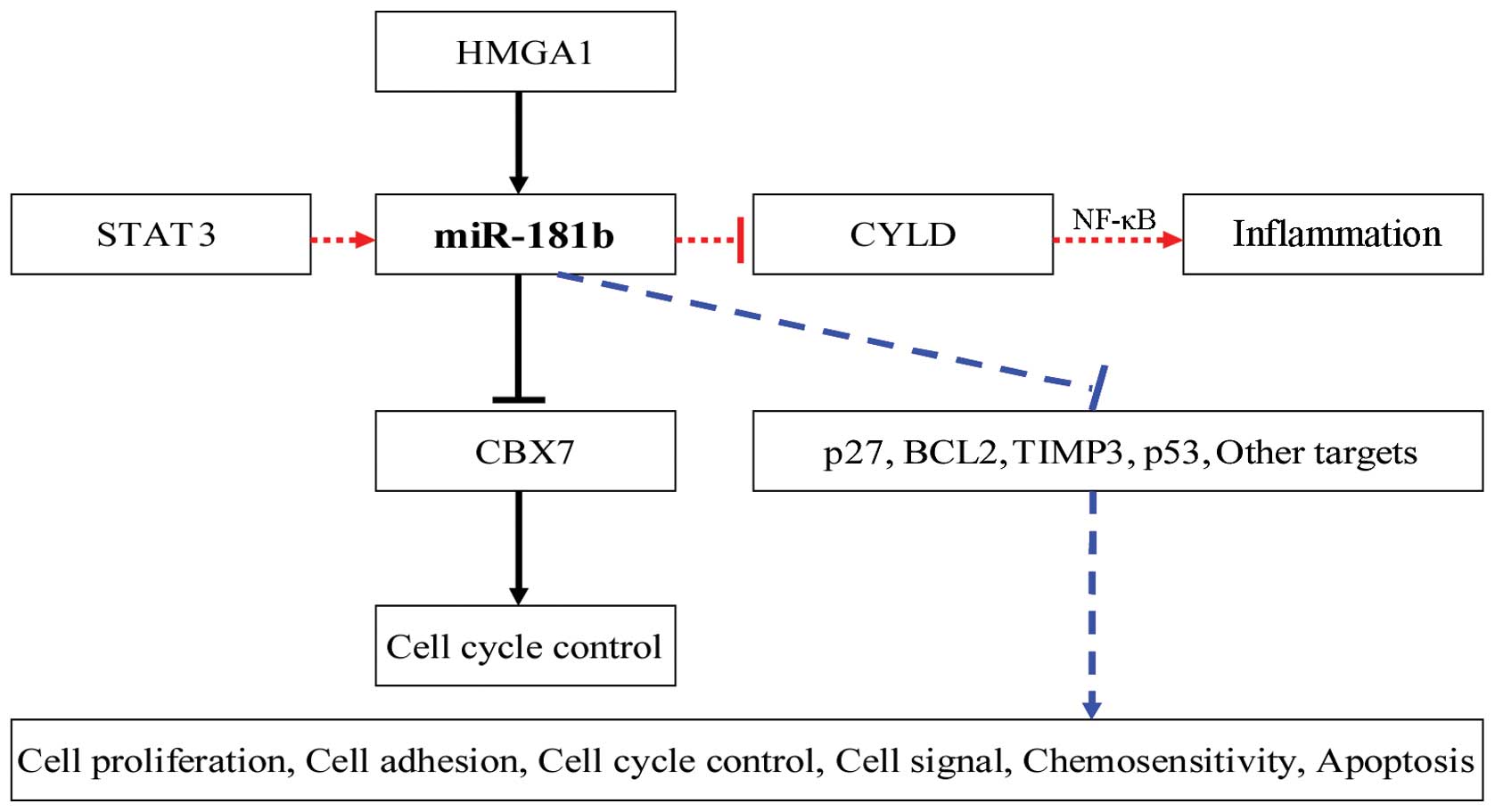
It was previously demonstrated that miR-181b is a critical link
between inflammation and malignant transformation (32). STAT3, a transcription factor
activated by IL-6 (33–35), directly activates miR-21 and -181b-1
(36). Of note, transient
expression of miR-181b induces the epigenetic switch and inhibits
cylindromatosis (CYLD) tumor suppressor, a target of miR-181b, from
negatively modulating NF-κB activity. Therefore, miR-181b and CYLD
are part of the positive feedback loop that underlies the
epigenetic switch between inflammation and cancer (36,37).
The expression status of miR-181b in different types of cancer and
underlying mechanisms are listed in Table I. Our current understanding of the
role of miR-181b in tumorigenesis is illustrated in Fig. 1.
 | Table IExpression of miR-181b in different
types of cancer and relevant mechanism. |
Table I
Expression of miR-181b in different
types of cancer and relevant mechanism.
| Type of cancer | Change of
miR-181b | Functions and
mechanisms | Refs. |
|---|
| Gastric | Downregulation | miR-181b plays a
role in the development of MDR in gastric cancer cell lines by
targeting BCL2
miR-181b modulates CREB1 expression in human gastric
adenocarcinoma | 23,40,41 |
| Lung | Downregulation | miR-181b targeting
BCL2 plays a role in the development of MDR in lung cancer cell
lines by modulation of apoptosis | 41 |
| Colon | Upregulation | p53 mutations or
deletions trigger oncogene activation | 16,38,39 |
| Pancreatic | Upregulation | Unclear | 22,24 |
| Head and neck | Upregulation | Unclear | 25 |
| Prostate | Downregulation | Unclear | 28 |
| HCC | Upregulation | TIMP3, a tumor
suppressor, is targeted by miR-181b
miR-181b enhances MMP2 and 9 activity and promotes growth,
clonogenic survival, migration and invasion of HCC cells | 21,36,42 |
| Breast | Upregulation | HMGA1, CBX7 and
miR-181b may play critical roles in cancer progression | 44,45 |
| Thyroid | Upregulation | miR-181b may
promote cell proliferation
c-Kit, a target of miR-181b, may also be involved | 17 |
| Oral carcinoma | Upregulation | miR-181b may play
an important role in malignant transformation | 32 |
| Osteosarcoma | Upregulation | Unclear | 46 |
| Retinoblastoma | Upregulation | miR-181b may
promote proliferation of retinoblastoma cells | 47 |
| Glioma | Downregulation | miR-181b acts as a
tumor suppressor that triggers growth inhibition, apoptosis and
invasion inhibition in glioma cells | 48–52 |
| AML | Upregulation | miR-181b targets
MLK2, contributing to cell proliferation, which plays a critical
role in the progression of AML | 55 |
| CLL | Downregulation | miR-181b inhibits
BCL2, MCL1 and X-linked inhibitor of apoptosis protein
miRNAs, including miR-181b, upregulate TCL1 associated with BMSCs,
protecting CLL cells from apoptosis | 8,56–58 |
3. Functional significance and clinical
relevance of miR-181b in cancer
miR-181b in colorectal cancer
One of the first clinical translational studies of
miR-181b was on colorectal cancer. Compared to normal colon tissue,
four highly expressed miRNAs, including miR-181b, were identified
in colon cancer specimens (38).
The expression level of miR-181b was significantly correlated with
chemoresponse to S-1 in colorectal cancer (39). S-1 is an oral fluorouracil
derivative, which comprises ftorafur, gimeracil and potassium
oxonate in a molar ratio of 1:0.4:1. A number of studies reported
that it is safe and effective to use S-1 in the treatment of
advanced colorectal cancer and late-stage gastric cancer patients
(40, 41). It was predicted that miR-181b is
able to modulate the expression of a number of genes at the
translational level, such as cytochrome c, ECIP-1, MAPPKKK1,
TEM6, E2F5, GATA6, PP2B and eIF5A. These genes may be active in
cell signaling, cell cycle control and chemosensitivity in the
majority of cells harbouring p53 mutations or deletions (38). In a recent study, miR-181b was shown
to be inversely correlated with colon cancer patient survival
(16).
miR-181b in gastric and lung cancer
The clinical significance of miR-181b was also
investigated in gastric cancer patients undergoing S-1 treatment.
Our recent studies demonstrated that miR-181b is overexpressed in
gastric cancer compared to normal tissues (42). The efficacy of S-1 in the treatment
of patients with advanced gastric cancer and patient survival were
inversely correlated with the expression of miR-181b (42). Thus, miR-181b exhibits great
potential as a prognostic biomarker in late-stage gastric
cancer.
Another study demonstrated that miR-181b was
downregulated in the multidrug-resistant (MDR) SGC7901/vincristine
human gastric cancer and A549/cisplatin lung cancer cell lines and
the downregulation of miR-181b was concurrent with the elevated
level of BCL2 protein in these two cell lines. Those findings
indicated that miR-181b may play a role in the development of MDR
through targeting BCL2 in gastric and lung cancer cell lines
(43).
miR-181b in hepatocellular carcinoma
(HCC)
miR-181b is transcriptionally activated by the
Wnt/β-catenin signaling pathway in HCC and the β-catenin binding
site is in the promoter region of the miR-181a-2 and -181b-2
transcripts. Thus, miR-181b is highly expressed as a result of the
activation of Wnt/β-catenin signaling (21). Another study reported that miRNAs
were deregulated during the early stages of hepatocarcinogenesis
induced by a choline- and amino acid-defined diet in C57BL/6 mice.
miR-181b was significantly upregulated in the livers of mice that
persisted at preneoplastic stage. TIMP3, a tumor suppressor and a
target of miR-181b, was significantly suppressed in the liver. By
contrast, miR-181b enhanced the activity of MMP2 and 9 and promoted
growth, clonogenic survival, migration and invasion of HCC cells.
It was also demonstrated that miR-181b was involved in the TGFβ
signaling pathway (43). The
overexpression of miR-181b enhanced the resistance of HCC cells to
doxorubicin. Increased expression of miR-181b was detecetd in
patients with non-alcoholic steatohepatitis and HCC (43). It was also recently demonstrated
that miR-181b is overexpressed in the serum of patients with liver
cirrhosis, targeting p27-regulated cell cycle (45). Those results suggested that miR-181b
may play an important role in hepatocarcinogenesis.
miR-181b in breast cancer
The efforts to elucidate the role of miRNAs in
breast cancer were initiated with expression profiling. In order to
investigate the association between clinicopathological
characteristics or patient survival and the expression profile of
miRNAs in primary breast cancer, the global expression profiling of
miRNAs in breast cancer was investigated by a microarray expression
profiling approach. The results revealed that nine miRNAs were
upregulated >2-fold in breast cancer compared to normal adjacent
tumor tissues, including miR-181b (46).
High-mobility group AT-hook 1 (HMGA1) modulates
miR-181b expression by direct transcriptional activation. HMGA1 is
frequently activated in several malignant tumors, including breast
carcinomas (29). miR-181b is
downregulated during cancer progression and targeted chromobox
homolog (CBX7) negatively regulates the cell cycle at the G1 phase.
In addition, HMGA1 negatively regulates CBX7 gene expression. Those
results suggested that there is a novel breast cancer progression
pathway, involving HMGA1, CBX7 and miR-181b (47).
miR-181b in papillary thyroid carcinoma
(PTC)
Expression analysis identified a subset of miRNAs
that is overexpressed in human PTC. miR-181b, -221 and -222 in
particular, were found to be highly expressed in the majority of
patients with PTC, suggesting that the increasing level of miR-181b
may represent a unique signature of human PTC, which may promote
cell proliferation. c-Kit, which is one of the critical targets of
miR-181b, may also be involved in this process (17).
miR-181b in oral carcinoma, osteosarcoma
and retinoblastoma
In oral carcinoma, miR-181b, -21 and -345 were found
to be consistently upregulated and were associated with the
increase in lesion severity during progression. Therefore, these
miRNAs may be valuable biomarkers for evaluating the risk of
malignant transformation (32).
miR-181b was found to be overexpressed in osteosarcoma and may
exhibit potential as a critical pre-treatment biomarker of
metastasis (48). Hypoxia is the
basic characteristic of retinoblastoma and facilitates poor
prognosis and resistance to traditional therapy. miR-181b was among
the highly expressed miRNAs under hypoxic conditions and inhibition
of miR-181b may suppress proliferation of retinoblastoma cells
(49). Thus, miR-181b may represent
a novel target for the future clinical treatment of
retinoblastoma.
miR-181b in glioma
Several miRNAs are involved in the modulation of
glioma development (50). miR-181b
was found to be downregulated in glioma tissue (50–52).
miR-181b acts as a tumor suppressor that causes growth inhibition,
apoptosis and invasion inhibition in glioma cells (52). Those findings suggested that
aberrantly downregulated miR-181b may be a critical factor for the
appearance in human malignant glioma.
Moreover, Kaplan-Meier survival analysis
demonstrated that low expression of miR-181b and -106a or
overexpression of miR-21 was significantly associated with poor
patient survival and Cox proportional hazard regression analysis
suggested that its prognostic impact is independent of other
clinicopathological factors (53).
A recent study demonstrated that the most significant change was
the downregulation of miR-181b in patients in response to
concomitant chemoradiotherapy with temozolomide (TMZ) (54). Olea europaea leaf extract
(OLE) was shown to exert an anti-proliferative effect on the T98G
cell line. miRNA target genes are involved in cell cycle and
apoptotic pathways. miR-181b, in particular, was markedly
overexpressed following treatment with TMZ and OLE (55). Thus, miR-181b may be a potential
biomarker for glioma and chemotherapy treatment outcome.
miR-181b in acute myeloid leukemia (AML)
and chronic lymphocytic leukemia (CLL)
miRNAs have been shown to modulate disease
progression in leukemia (56).
miR-181b was found to be highly expressed in AML and, by targeting
the mixed lineage kinase 2 (MLK2), caused cell proliferation, which
is crucial in the progression of AML and provides a useful
treatment biomarker (57). However,
miR-181b was downregulated in CLL, which exhibits potential as a
biomarker of CLL and is positively correlated with patient
survival. miR-181b was also shown to inhibit BCL2 and MCL1. In
cells containing wild-type p53, increasing the expression level of
miR-181b markedly accelerated apoptosis (8,58,59).
miRNAs upregulate TCL1 associated with bone marrow stromal cells
(BMSC), protecting CLL cells from apoptosis (60). Therefore, miR-181b may provide a
novel target in the treatment of CLL.
4. Conclusion
miRNAs have a broad range of regulatory functions in
the modulation of protein synthesis. Abnormal expression of miRNAs
has been shown to contribute to cancer development and/or
progression. Growing evidence suggests that miR-181b is one of the
main factors involved in tumorigenesis, affecting several tumor
types. As a result, it is crucial to further investigate the
functions and underlying molecular mechanisms of miR-181b in
malignant tumor transformation. By fully understanding the target
genes and regulatory networks of miR-181b using the systems biology
approach, we may be able to determine the potential of miR-181b as
biomarker and novel therapeutic target.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (nos. 81171653 and 30972703), the
Natural Science Foundation of Jiangsu province (nos. BK2011246 and
BK2011247), the Project of Six Batch of Major Talent Summit (no.
BRA2010037) and Society Development Plans, Department of Science
and Technology, Changzhou (nos. CJ20112020, CZ20110024 and
CS20102020).
References
|
1
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
|
|
2
|
Lagos-Quintana M, Rauhut R, Lendeckel W,
et al: Identification of novel genes coding for small expressed
RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee RC and Ambros V: An extensive class of
small RNAs in Caenorhabditis elegans. Science. 294:862–864.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Salaun B, Yamamoto T, Badran B, et al:
Differentiation associated regulation of microRNA expression in
vivo in human CD8+T cell subsets. J Transl Med.
9:442011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ooi AG, Sahoo D, Adorno M, et al:
MicroRNA-125b expands hematopoietic stem cells and enriches for the
lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci
USA. 107:21505–21510. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ghisi M, Corradin A, Basso K, et al:
Modulation of microRNA expression in human T-cell development:
targeting of NOTCH3 by miR-150. Blood. 117:7053–7062. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Visone R, Veronese A, Balatti V, et al:
MiR-181b: new perspective to evaluate disease progression in
chronic lymphocytic leukemia. Oncotarget. 3:195–202.
2012.PubMed/NCBI
|
|
8
|
Zhu DX, Miao KR, Fang C, et al: Aberrant
microRNA expression in Chinese patients with chronic lymphocytic
leukemia. Leuk Res. 35:730–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tavano F, di Mola FF, Piepoli A, et al:
Changes in miR-143 and miR-21 expression and clinicopathological
correlations in pancreatic cancers. Pancreas. 41:1280–1284. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
11
|
Michael MZ, O’ Connor SM, van Holst
Pellekaan NG, et al: Reduced accumulation of specific microRNAs in
colorectal neoplasia. Mol Cancer Res. 1:882–891. 2003.PubMed/NCBI
|
|
12
|
Lazebnik Y: What are the hallmarks of
cancer? Nat Rev Cancer. 10:232–233. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar
|
|
14
|
Lopez-Camarillo C, Marchat LA,
Arechaga-Ocampo E, et al: MetastamiRs: non-coding microRNAs driving
cancer invasion and metastasis. Int J Mol Sci. 13:1347–1379. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mendell JT and Olson EN: MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schetter AJ, Leung SY, Sohn JJ, et al:
MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pallante P, Visone R, Ferracin M, et al:
MicroRNA deregulation in human thyroid papillary carcinomas. Endocr
Relat Cancer. 13:497–508. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhai H and Ju J: Implications of microRNAs
in colorectal cancer development, diagnosis, prognosis, and
therapeutics. Front Genet. 2:000782011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Y, Gelfond J, McManus LM, et al:
Temporal microRNA expression during in vitro myogenic progenitor
cell proliferation and differentiation: regulation of proliferation
by miR-682. Physiol Genomics. 43:621–630. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu G, Min H, Yue S, et al: Pre-miRNA loop
nucleotides control the distinct activities of mir-181a-1 and
mir-181c in early T cell development. PLoS One. 3:e35922008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ji J, Yamashita T and Wang XW:
Wnt/beta-catenin signaling activates microRNA-181 expression in
hepatocellular carcinoma. Cell Biosci. 1:42011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ren Y, Gao J, Liu JQ, et al: Differential
signature of fecal microRNAs in patients with pancreatic cancer.
Mol Med Rep. 6:201–209. 2012.PubMed/NCBI
|
|
23
|
Chen L, Yang Q, Kong WQ, et al:
MicroRNA-181b targets cAMP responsive element binding protein 1 in
gastric adenocarcinomas. IUBMB Life. 64:628–635. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Panarelli NC, Chen YT, Zhou XK, et al:
MicroRNA expression aids the preoperative diagnosis of pancreatic
ductal adenocarcinoma. Pancreas. 41:685–690. 2012.PubMed/NCBI
|
|
25
|
Nurul-Syakima AM, Yoke-Kqueen C, Sabariah
AR, et al: Differential microRNA expression and identification of
putative miRNA targets and pathways in head and neck cancers. Int J
Mol Med. 28:327–336. 2011.PubMed/NCBI
|
|
26
|
Ratert N, Meyer HA, Jung M, et al:
Reference miRNAs for miRNAome analysis of urothelial carcinomas.
PLoS One. 7:e393092012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li X, Zhang Y, Zhang H, et al: miRNA-223
promotes gastric cancer invasion and metastasis by targeting tumor
suppressor EPB41L3. Mol Cancer Res. 9:824–833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schaefer A, Jung M, Mollenkopf HJ, et al:
Diagnostic and prognostic implications of microRNA profiling in
prostate carcinoma. Int J Cancer. 126:1166–1176. 2010.PubMed/NCBI
|
|
29
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gregory RI and Shiekhattar R: MicroRNA
biogenesis and cancer. Cancer Res. 65:3509–3512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Eis PS, Tam W, Sun L, et al: Accumulation
of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad
Sci USA. 102:3627–3632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cervigne NK, Reis PP, Machado J, et al:
Identification of a microRNA signature associated with progression
of leukoplakia to oral carcinoma. Hum Mol Genet. 18:4818–4829.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Frank DA: STAT3 as a central mediator of
neoplastic cellular transformation. Cancer Lett. 251:199–210. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu H, Kortylewski M and Pardoll D:
Crosstalk between cancer and immune cells: role of STAT3 in the
tumour microenvironment. Nat Rev Immunol. 7:41–51. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: a leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Iliopoulos D, Jaeger SA, Hirsch HA, et al:
STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are
part of the epigenetic switch linking inflammation to cancer. Mol
Cell. 39:493–506. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun X, Icli B, Wara AK, et al:
MicroRNA-181b regulates NF-kappaB-mediated vascular inflammation. J
Clin Invest. 122:1973–1990. 2012.PubMed/NCBI
|
|
38
|
Xi Y, Formentini A, Chien M, et al:
Prognostic values of microRNAs in colorectal cancer. Biomark
Insights. 2:113–121. 2006.PubMed/NCBI
|
|
39
|
Nakajima G, Hayashi K, Xi Y, et al:
Non-coding microRNAs hsa-let-7g and hsa-miR-181b are associated
with chemoresponse to S-1 in colon cancer. Cancer Genomics
Proteomics. 3:317–324. 2006.PubMed/NCBI
|
|
40
|
Xu R, Ma N, Wang F, Ma L, et al: Results
of a randomized and controlled clinical trial evaluating the
efficacy and safety of combination therapy with Endostar and S-1
combined with oxaliplatin in advanced gastric cancer. Onco Targets
Ther. 6:925–929. 2013.PubMed/NCBI
|
|
41
|
Watanabe K, Kawahara H, Enomoto H, et al:
Feasibility Study of Oxaliplatin with Oral S-1 or Capecitabine as
First-line Therapy for Patients with Metastases from Colorectal
Cancer. Anticancer Res. 33:4029–4032. 2013.PubMed/NCBI
|
|
42
|
Jiang J, Zheng X, Xu X, et al: Prognostic
significance of miR-181b and miR-21 in gastric cancer patients
treated with S-1/oxaliplatin or doxifluridine/oxaliplatin. PLoS
One. 6:e232712011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhu W, Shan X, Wang T, et al: miR-181b
modulates multidrug resistance by targeting BCL2 in human cancer
cell lines. Int J Cancer. 127:2520–2529. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang B, Hsu SH, Majumder S, et al:
TGFbeta-mediated upregulation of hepatic miR-181b promotes
hepatocarcinogenesis by targeting TIMP3. Oncogene. 29:1787–1797.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang B, Li W, Guo K, et al: miR-181b
promotes hepatic stellate cells proliferation by targeting p27 and
is elevated in the serum of cirrhosis patients. Biochem Biophys Res
Commun. 421:4–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yan LX, Huang XF, Shao Q, et al: MicroRNA
miR-21 overexpression in human breast cancer is associated with
advanced clinical stage, lymph node metastasis and patient poor
prognosis. RNA. 14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mansueto G, Forzati F, Ferraro A, et al:
Identification of a new pathway for tumor progression:
microRNA-181b up-regulation and CBX7 down-regulation by HMGA1
protein. Genes Cancer. 1:210–224. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jones KB, Salah Z, Del Mare S, et al:
miRNA signatures associate with pathogenesis and progression of
osteosarcoma. Cancer Res. 72:1865–1877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu X, Jia R, Zhou Y, et al:
Microarray-based analysis: Identification of hypoxia-regulated
microRNAs in retinoblastoma cells. Int J Oncol. 38:1385–1393.
2011.PubMed/NCBI
|
|
50
|
Shi L, Cheng Z, Zhang J, et al:
hsa-mir-181a and hsa-mir-181b function as tumor suppressors in
human glioma cells. Brain Res. 1236:185–193. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ciafre SA, Galardi S, Mangiola A, et al:
Extensive modulation of a set of microRNAs in primary glioblastoma.
Biochem Biophys Res Commun. 334:1351–1358. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Conti A, Aguennouz M, La Torre D, et al:
miR-21 and 221 upregulation and miR-181b downregulation in human
grade II–IV astrocytic tumors. J Neurooncol. 93:325–332.
2009.PubMed/NCBI
|
|
53
|
Zhi F, Chen X, Wang S, et al: The use of
hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators
of astrocytoma. Eur J Cancer. 46:1640–1649. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Slaby O, Lakomy R, Fadrus P, et al:
MicroRNA-181 family predicts response to concomitant
chemoradiotherapy with temozolomide in glioblastoma patients.
Neoplasma. 57:264–269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tunca B, Tezcan G, Cecener G, et al:
Olea europaea leaf extract alters microRNA expression in
human glioblastoma cells. J Cancer Res Clin Oncol. 138:1831–1844.
2012. View Article : Google Scholar
|
|
56
|
Calin GA, Dumitru CD, Shimizu M, et al:
Frequent deletions and down-regulation of micro- RNA genes miR15
and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad
Sci USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen H, Chen Q, Fang M, et al:
MicroRNA-181b targets MLK2 in HL-60 cells. Sci China Life Sci.
53:101–106. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhu DX, Zhu W, Fang C, et al: miR-181a/b
significantly enhances drug sensitivity in chronic lymphocytic
leukemia cells via targeting multiple anti-apoptosis genes.
Carcinogenesis. 33:1294–1301. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zanette DL, Rivadavia F, Molfetta GA, et
al: miRNA expression profiles in chronic lymphocytic and acute
lymphocytic leukemia. Braz J Med Biol Res. 40:1435–1440. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sivina M, Hartmann E, Vasyutina E, et al:
Stromal cells modulate TCL1 expression, interacting AP-1 components
and TCL1-targeting micro-RNAs in chronic lymphocytic leukemia.
Leukemia. 26:1812–1820. 2012. View Article : Google Scholar : PubMed/NCBI
|