Introduction
Family with sequence similarity 107 (FAM107) members
contain an N-terminal domain of unknown function (DUF1151) that is
conserved across species, including mammalian, Xenopus, fish
and Drosophila, with no homologous matches to other
functionally conserved domains (Fig.
1). This family includes several hypothetical eukaryotic
proteins with largely undetermined functions. Mammals have two
genes, FAM107A and FAM107B, which encode for proteins of 144 and
131 amino acids (aa), respectively. The C-terminal variable regions
of FAM107 members have a coiled-coil domain that has been
identified in several nuclear proteins, including transcription
factors, suggesting a role for FAM107 members in regulating gene
transcription. Protein-protein interaction analyses demonstrated
that FAM107A and FAM107B interact with transcriptional adaptor
(Tada)2α and 3α, respectively (1–3).
Tada2α and 3α are core proteins of the histone acetyltransferase
(HAT) complex, suggesting that FAM107 proteins modulate the
structure and function of HAT complexes, resulting in gene
transcriptional modifications and protein acetylation (4,5).
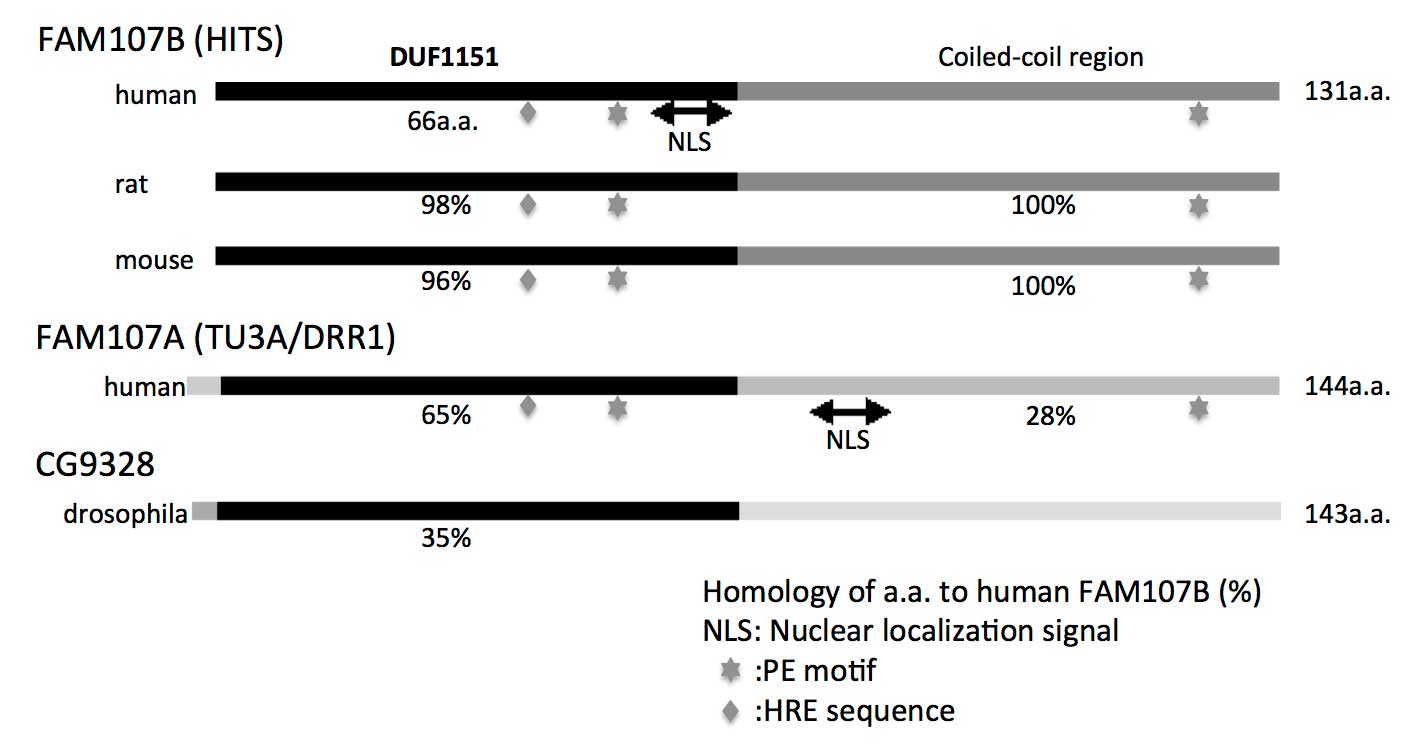 | Figure 1Comparison of the molecular structures
of FAM107 proteins among different species. For rat and mouse
FAM107B, human FAM107A and Drosophila CG9328, the homology
of each protein to the human FAM107B protein sequence is expressed
as a percentage (%). The NLS, PE and HRE motifs are indicated.
FAM107, family with sequence similarity 107; HITS, heat
shock-inducible tumor small protein; DUF1151, N-terminal domain of
unknown function; NLS, nuclear localization signal; TU3A, Tohoku
University cDNA clone A on chromosome 3; DRR1, downregulated in
renal cell carcinoma gene 1; PE, proline-glutamic acid; HRE,
histidine-arginine-glutamic acid sequence; a.a., amino acids. |
Although the major physiological functions of FAM107
proteins remain to be investigated, the present review aimed to
summarize the currently available biological information regarding
the role of FAM107 members in cancer and neural cells.
2. FAM107 in cancer
FAM107A has been designated Tohoku University cDNA
clone A on chromosome 3 (TU3A) and is also referred to as
downregulated in renal cell carcinoma gene 1 (DRR1). FAM107A is a
candidate tumor suppressor gene located on chromosome 3p21.1
(6,7). Several studies indicated that FAM107A
expression is downregulated in various types of cancer, such as
non-small-cell lung, renal cell and prostate cancers and
astrocytoma by epigenetic silencing, including promoter
hypermethylation (6–11). The forced expression of FAM107A was
shown to suppress tumor cell proliferation and induce apoptosis
(7,11–13).
Thus, FAM107A was considered as a tumor suppressor gene due to its
decreased expression in various types of cancer and since inducing
FAM107A expression suppresses cancer cell proliferation and induces
apoptosis. However, FAM107A was also found to be highly expressed
in the invasive component of gliomas and may drive tumor invasion
by modulating the cytoskeleton (14,15).
Thus, the physiological roles and functions of FAM107A in cancer
remain controversial.
Despite accumulating information regarding FAM107A,
the available biological data on FAM107B are currently limited. In
humans, the FAM107B protein is encoded by a gene on chromosome
10p13. This protein consists of 131 aa and its sequence is ~98%
identical with mouse and rat homologues (Fig. 1). FAM107A and FAM107B proteins
exhibit a 65% sequence similarity in their DUF1151 regions.
The most notable characteristic of FAM107B, unlike
FAM107A, is that the FAM107B gene has a promoter region with heat
shock transcription factor 1 (HSF1)-binding sites, and FAM107B
transcription is increased following heat-shock or hyperthermia
treatment. Thus, its protein was designated as heat shock-inducible
tumor small protein (HITS) (16).
Our preliminary investigation found that the level of HITS
expression in gastrointestinal cancer cells was significantly lower
compared to that in normal epithelial cells, although its
expression pattern and intensity varied among cancers of different
histological types. HITS expression was decreased during the
process underlying colorectal adenoma-to-carcinoma transition. In
addition, HITS expression was decreased in intestinal-type gastric
adenocarcinomas, but not in diffuse-type or mucinous
adenocarcinomas.
Multiple organ tissue microarray analyses revealed
that HITS expression was decreased in other tumor tissues, such as
breast, thyroid, testicular and uterine cervical in a histological
type-specific manner (17). HITS
expression intensity was found to be inversely correlated with
primary tumor size [T-value in tumor-node-metastasis (TNM) grading]
in breast and thyroid cancers, but not with lymph node metastasis
(N-value). For breast cancers, the statistical correlation analysis
for HITS expression and the clinicopathological parameters of human
epidermal growth factor 2 (HER2), estrogen receptor, progesterone
receptor (PR), Ki-67 and p53 revealed that HITS expression
intensity was positively correlated with the expression of HER2 and
Ki-67, but was inversely correlated with PR expression.
Accordingly, HITS expression was markedly lost in HER2-negative,
Ki-67-negative, PR-positive and desmoplastic reaction-negative type
of breast cancer, which is considered to be a non-aggressive or
indolent phenotype.
As regards uterine cervical diseases, HITS
expression was significantly lost in invasive squamous cell
carcinoma, but not in cervical intraepithelial neoplasia (CIN).
Infection by human papilloma virus (HPV) is known to induce the
development of cervical cancer, due to the strong causal
association between HPV infection, CIN and invasive carcinoma
(18). As certain CIN lesions may
progress to invasive cancer over a period of 10–20 years, our
findings suggest that HITS expression is lost during the
progression of CIN to invasive carcinoma.
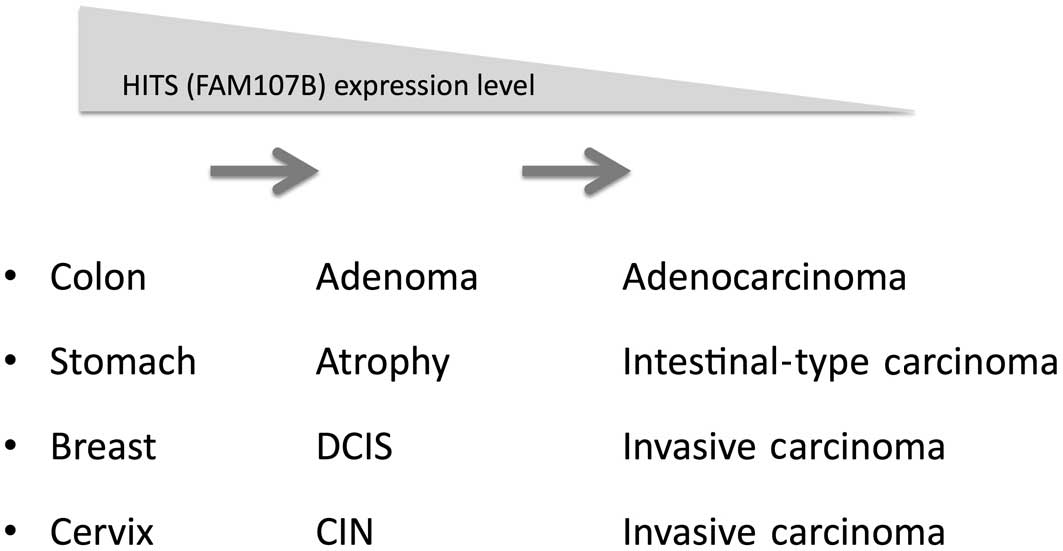
Considering that HITS expression was lost during the
course of tumor progression in terms of TNM grading T-values, we
hypothesize that HITS expression declines gradually during the
prolonged transition from preneoplastic or early neoplastic
lesions, such as ductal carcinoma in situ in the breast,
intestinal metaplasia in the stomach, tubular adenoma in the colon
and CIN in the uterine cervix, to invasive cancers (Fig. 2). By contrast, HITS expression is
preserved in aggressive types of cancers, such as scirrhous-type
gastric and breast cancers, which are characterized by distinct
genetic alterations and rapid growth or invasion.
Furthermore, it was reported that a point mutation
in the C-terminal region of HITS (chromosome 10: 14603968 C•G→T•A
transition) was frequently observed in genomic analyses of
basal-like breast cancer (19). The
protein sequence of the C-terminal coiled-coil region is unique to
HITS, which suggests its role in tumorigenicity through the
transcriptional regulation of oncogenes or tumor suppressor genes.
Therefore, it was hypothesized that, in an aggressive type of
breast cancer, such as basal-like or scirrhous-type, HITS
expression is relatively preserved, but its antioncogenic function
is lost by this genetic mutation.
In addition, forced expression of HITS was shown to
inhibit cancer cell proliferation in response to growth factors
in vitro and tumor xenograft growth in vivo (16,17).
HITS may be considered as a candidate tumor suppressor gene, since
loss of HITS expression was commonly observed in cancers of various
organs, resulting in tumor development and proliferation, similar
to FAM107A. We hypothesize that HITS expression affects the growth
of primary tumors during development, but does not affect invasion
or metastasis, such as scirrhous-type tumor spread or lymph node
metastasis. Consequently, HITS is a potential tumor suppressor
protein with the unique characteristic of its transcription being
induced by heat shock stimulation. This is a particularly distinct
characteristic of HITS, as other heat shock proteins and HSF1 are
considered to exhibit certain oncogenic activities (20,21).
3. FAM107 in neuron
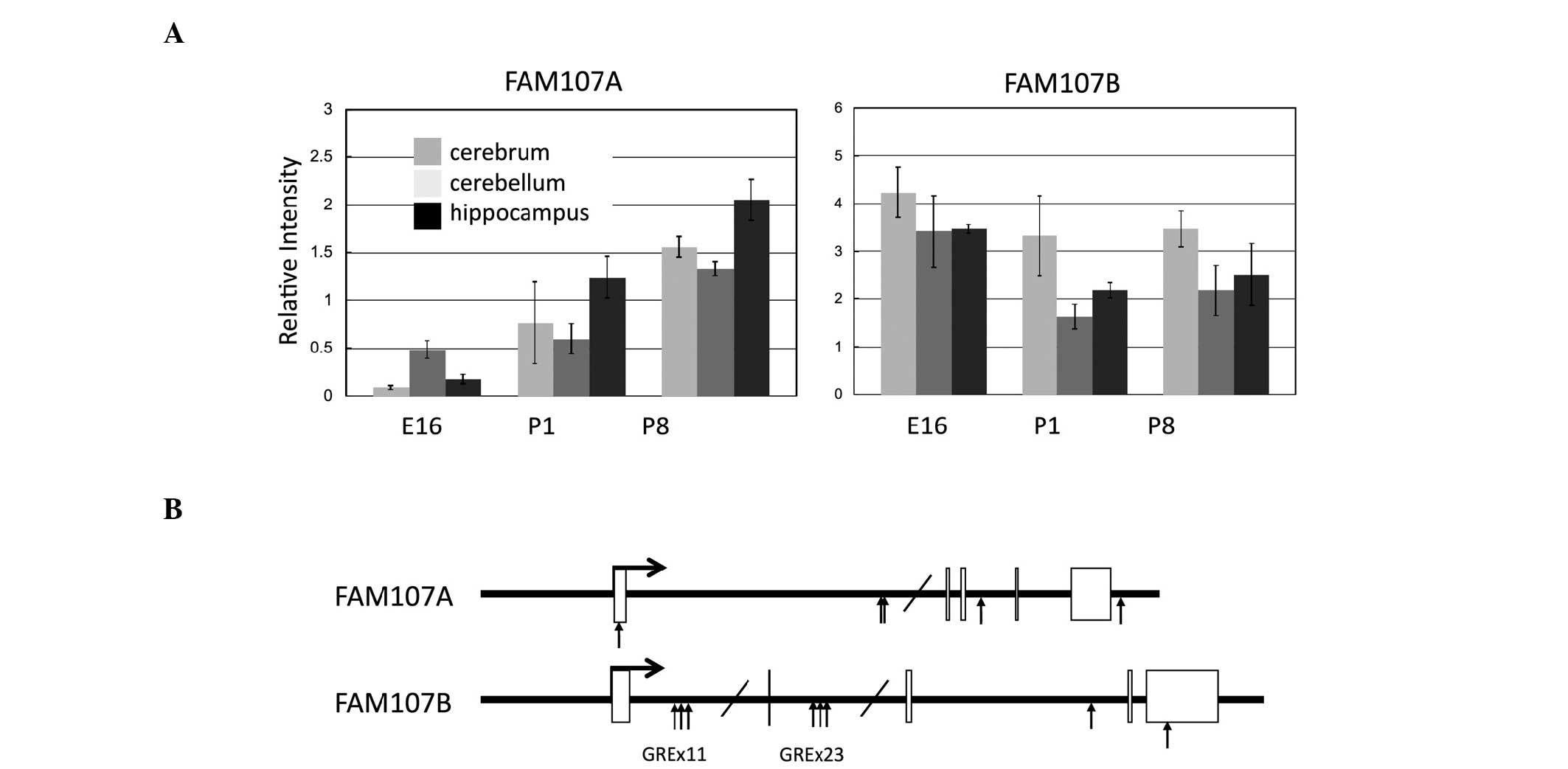
FAM107A and FAM107B are expressed in normal brain
tissues (Fig. 3A) and FAM107A
(DRR1) was shown to play critical roles in neural cell survival,
migration and spine formation (14,22,23).
Considering the molecular similarities between FAM107A and FAM107B,
FAM107B (HITS) may also play critical roles during brain
development.
FAM107A is widely expressed in various normal
tissues and its expression is particularly high in the brain
(7,13,14,22).
In primary cultures of rat fetal cerebral cortex, FAM107A was found
to be differentially expressed in neurons and enriched in axon
projections and cell nuclei (14,22). A
study using RNA interference knockdown indicated that FAM107A is
essential for neural cell survival (22).
Studies on glioma cells may help elucidate the
functions of FAM107A in neural cells. Le et al (14) indicated that FAM107A expression
co-localized with F-actin stress fiber focal adhesions (FAs) and
membrane ruffles in U251 glioma cells that were transfected with
FAM107A. The proline-glutamic acid (PE) motif (65–66/122–123 aa in
human FAM107A) is critical for the association of FAM107A with
F-actin and its intercellular localization (Fig. 1). The PE motif also binds to a light
chain subunit (LC2) of microtubule-associated protein 1 to modulate
microtubule assembly, which requires the
histidine-arginine-glutamic acid (HRE) sequence motif located in
the N-terminal region aa of FAM107A. In fact, these 2 motifs are
also conserved in human, mouse and rat FAM107B (Fig. 1). FAM107A overexpression was shown
to facilitate U251 cell invasion, whereas a PE or HRE motif
mutation suppressed the effect of overexpression. It is
hypothesized that FAM107A acts as an actin-microtubule cross-linker
to organize the cytoskeletons essential for FAs modulation and
glioma invasion. Similarly, FAM107A may regulate neural cell
migration in the developing brain. As regards modulation of the
cytoskeleton, Akt signaling may play a crucial role. FAM107A
overexpression was shown to increase Akt phosphorylation at the
Thr308 and Ser473 sites, which recruit Akt localization to FAs,
without affecting total Akt expression (15).
Schmidt et al (23) reported that FAM107A is a stress
response protein in the brain. The stress associated with maternal
separation (neonatal) or food deprivation (adult) significantly
increased FAM107A mRNA expression in the paraventricular nucleus
and CA3 regions of the hippocampus. Treating adult mice with
dexamethasone (DEX; a glucocorticoid agonist) also resulted in
increased FAM107A mRNA expression in the same brain regions.
Increased FAM107A expression induced by DEX was also reported in
cultured astrocytes (24). FAM107A
transcription may be directly regulated by glucocorticoid receptors
(5). In fact, a number of
glucocorticoid responsive elements are found in FAM107A genomic
regions, which is also the case for the FAM107B gene (Fig. 3B).
In the hippocampus, FAM107A expression was found to
be preferentially localized in the presynaptic regions of neuron
synapses and its overexpression reduced spine density. FAM107A
overexpression also caused a reduction in the magnitude of
long-term potentiation that improves cognitive behavior (23).
FAM107A may be associated with certain types of
human psychiatric disorders. FAM107A mRNA expression was increased
in postmortem RNA samples from the dorsolateral prefrontal cortex
(Brodman area 46) of patients suffering from schizophrenia and
bipolar disorder (25). A number of
studies suggested that human chromosome 3p14, the location of
FAM107A, is linked to psychiatric disorders, such as schizophrenia
(26–28), bipolar disorder (25,29–32)
and Asperger syndrome (33–35). Although there is currently no direct
evidence, FAM107A may be a candidate gene associated with these
psychiatric disorders.
4. Conclusion
The expression of FAM107A and FAM107B proteins is
prominent in neural cells, whereas their expression is
downregulated in cancer cells. The two proteins appear to affect
cytoskeleton rearrangements and are involved in cell migration and
expansion. The molecular mechanisms underlying the diverse
biological functions of FAM107 remain unclear. In particular, the
functions and the molecular interactions of the N-terminal
conserved domain (DUF1151) of FAM107 requires further investigation
as they may play crucial roles in the interactions with other
proteins for transducing cell signals and modulating gene
transcription, which may be common among FAM107 proteins. Further
investigations are required to provide evidence for the biological
importance of this well-conserved protein family in cancer and the
nervous system, which may lead to improved clinical diagnosis and
the development of therapeutic uses for FAM107 proteins.
Acknowledgements
This study was supported by Grants-in-Aid for
Scientific Research from the Japanese Ministry of Education,
Culture, Sports, Science and Technology.
References
|
1
|
Rual JF, Venkatesan K, Hao T, et al:
Towards a proteome-scale map of the human protein-protein
interaction network. Nature. 437:1173–1178. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ewing RM, Chu P, Elisma F, et al:
Large-scale mapping of human protein-protein interactions by mass
spectrometry. Mol Syst Biol. 3:892007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stelzl U, Worm U, Lalowski M, et al: A
human protein-protein interaction network: a resource for
annotating the proteome. Cell. 122:957–968. 2005.PubMed/NCBI
|
|
4
|
Wang YL, Faiola F, Xu M, Pan S and
Martinez E: Human ATAC is a GCN5/PCAF-containing acetylase complex
with a novel NC2-like histone fold module that interacts with the
TATA-binding protein. J Biol Chem. 283:33808–33815. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Frijters R, Fleuren W, Toonen EJ, et al:
Prednisolone-induced differential gene expression in mouse liver
carrying wild type or a dimerization-defective glucocorticoid
receptor. BMC Genomics. 11:3592010. View Article : Google Scholar
|
|
6
|
Yamato T, Orikasa K, Fukushige S, Orikasa
S and Horii A: Isolation and characterization of the novel gene,
TU3A, in a commonly deleted region on 3p14.3→p14.2 in renal cell
carcinoma. Cytogenet Cell Genet. 87:291–295. 1999.PubMed/NCBI
|
|
7
|
Wang L, Darling J, Zhang JS, Liu W, Qian
J, Bostwick D, Hartmann L, Jenkins R, Bardenhauer W, Schutte J,
Opalka B and Smith DI: Loss of expression of the DRR 1 gene at
chromosomal segment 3p21.1 in renal cell carcinoma. Genes
Chromosomes Cancer. 27:1–10. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van den Boom J, Wolter M, Blaschke B,
Knobbe CB and Reifenberger G: Identification of novel genes
associated with astrocytoma progression using suppression
subtractive hybridization and real-time reverse
transcription-polymerase chain reaction. Int J Cancer.
119:2330–2338. 2006.
|
|
9
|
Vanaja DK, Ballman KV, Morlan BW, et al:
PDLIM4 repression by hypermethylation as a potential biomarker for
prostate cancer. Clin Cancer Res. 12:1128–1136. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Awakura Y, Nakamura E, Ito N, Kamoto T and
Ogawa O: Methylation-associated silencing of TU3A in human cancers.
Int J Oncol. 33:893–899. 2008.PubMed/NCBI
|
|
11
|
Liu Q, Zhao XY, Bai RZ, et al: Induction
of tumor inhibition and apoptosis by a candidate tumor suppressor
gene DRR1 on 3p21.1. Oncol Rep. 22:1069–1075. 2009.PubMed/NCBI
|
|
12
|
Kholodnyuk ID, Kozireva S, Kost-Alimova M,
Kashuba V, Klein G and Imreh S: Down regulation of 3p genes, LTF,
SLC38A3 and DRR1, upon growth of human chromosome 3-mouse
fibrosarcoma hybrids in severe combined immunodeficiency mice. Int
J Cancer. 119:99–107. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao XY, Liang SF, Yao SH, et al:
Identification and preliminary function study of Xenopus
laevis DRR1 gene. Biochem Biophys Res Commun. 361:74–78. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Le PU, Angers-Loustau A, de Oliveira RM,
et al: DRR drives brain cancer invasion by regulating
cytoskeletal-focal adhesion dynamics. Oncogene. 29:4636–4647. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dudley A, Sater M, Le PU, et al: DRR
regulates AKT activation to drive brain cancer invasion. Oncogene.
Oct 21–2013.(Epub ahead of print). View Article : Google Scholar
|
|
16
|
Nakajima H, Ishigaki Y, Xia QS, et al:
Induction of HITS, a newly identified family with sequence
similarity 107 protein (FAM107B), in cancer cells by heat shock
stimulation. Int J Oncol. 37:583–593. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nakajima H, Koizumi K, Tanaka T, et al:
Loss of HITS (FAM107B) expression in cancers of multiple organs:
Tissue microarray analysis. Int J Oncol. 41:1347–1357.
2012.PubMed/NCBI
|
|
18
|
Bosch FX and de Sanjosé S: Chapter 1:
Human papillomavirus and cervical cancer - burden and assessment of
causality. J Natl Cancer Inst Monogr. 2003:3–13. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ding L, Ellis MJ, Li S, et al: Genome
remodelling in a basal-like breast cancer metastasis and xenograft.
Nature. 464:999–1005. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dai C, Whitesell L, Rogers AB and
Lindquist S: Heat shock factor 1 is a powerful multifaceted
modifier of carcinogenesis. Cell. 130:1005–1018. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Khalil AA, Kabapy NF, Deraz SF and Smith
C: Heat shock proteins in oncology: diagnostic biomarkers or
therapeutic targets? Biochim Biophys Acta. 1816:89–104.
2011.PubMed/NCBI
|
|
22
|
Asano Y, Kishida S, Mu P, Sakamoto K,
Murohara T and Kadomatsu K: DRR1 is expressed in the developing
nervous system and downregulated during neuroblastoma
carcinogenesis. Biochem Biophys Res Commun. 394:829–835. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schmidt MV, Schulke JP, Liebl C, et al:
Tumor suppressor down-regulated in renal cell carcinoma 1 (DRR1) is
a stress-induced actin bundling factor that modulates synaptic
efficacy and cognition. Proc Natl Acad Sci USA. 108:17213–17218.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Slezak M, Korostynski M, Gieryk A, et al:
Astrocytes are a neural target of morphine action via
glucocorticoid receptor-dependent signaling. Glia. 61:623–635.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shao L and Vawter MP: Shared gene
expression alterations in schizophrenia and bipolar disorder. Biol
Psychiatry. 64:89–97. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tastemir D, Demirhan O and Sertdemir Y:
Chromosomal fragile site expression in Turkish psychiatric
patients. Psychiatry Res. 144:197–203. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Paunio T, Arajarvi R, Terwilliger JD, et
al: Linkage analysis of schizophrenia controlling for population
substructure. Am J Med Genet B Neuropsychiatr Genet. 150B:827–835.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Greenwood TA, Swerdlow NR, Gur RE, et al:
Genome-wide linkage analyses of 12 endophenotypes for schizophrenia
from the Consortium on the Genetics of Schizophrenia. Am J
Psychiatry. 170:521–532. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cichon S, Schumacher J, Muller DJ, et al:
A genome screen for genes predisposing to bipolar affective
disorder detects a new susceptibility locus on 8q. Hum Mol Genet.
10:2933–2944. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Marcheco-Teruel B, Flint TJ, Wikman FP, et
al: A genome-wide linkage search for bipolar disorder
susceptibility loci in a large and complex pedigree from the
eastern part of Cuba. Am J Med Genet B Neuropsychiatr Genet.
141B:833–843. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Etain B, Mathieu F, Rietschel M, et al:
Genome-wide scan for genes involved in bipolar affective disorder
in 70 European families ascertained through a bipolar type I
early-onset proband: supportive evidence for linkage at 3p14. Mol
Psychiatry. 11:685–694. 2006. View Article : Google Scholar
|
|
32
|
Mathieu F, Dizier MH, Etain B, et al:
European collaborative study of early-onset bipolar disorder:
evidence for genetic heterogeneity on 2q14 according to age at
onset. Am J Med Genet B Neuropsychiatr Genet. 153B:1425–1433. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ylisaukko-oja T, Nieminen-von Wendt T,
Kempas E, et al: Genome-wide scan for loci of Asperger syndrome.
Mol Psychiatry. 9:161–168. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rehnstrom K, Ylisaukko-oja T, Nieminen-von
Wendt T, et al: Independent replication and initial fine mapping of
3p21–24 in Asperger syndrome. J Med Genet. 43:e62006.PubMed/NCBI
|
|
35
|
Salyakina D, Ma DQ, Jaworski JM, et al:
Variants in several genomic regions associated with asperger
disorder. Autism Autism Res. 3:303–310. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
So AY, Chaivorapol C, Bolton EC, Li H and
Yamamoto KR: Determinants of cell- and gene-specific
transcriptional regulation by the glucocorticoid receptor. PLoS
Genet. 3:e942007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kuo T, Lew MJ, Mayba O, Harris CA, Speed
TP and Wang JC: Genome-wide analysis of glucocorticoid
receptor-binding sites in myotubes identifies gene networks
modulating insulin signaling. Proc Natl Acad Sci USA.
109:11160–11165. 2012. View Article : Google Scholar : PubMed/NCBI
|