Introduction
Cancer is the main cause of mortality in the world
(8.2 million mortalities in 2012). Gastric cancer (GC) is estimated
to be the fifth most common cancer worldwide, with 952,000 new
cases annually. In 2012, GC was the third cause of mortality due to
cancer in men and women; 70% of cases were reported in developing
countries, with one-half of these in East Asia. In Central and
South America, high mortality rates have been reported (1). In Mexico, according to the statistics of
the National Institute of Statistics and Geography, the principal
causes of mortalities due to neoplasms are those of the digestive
tract organs (32.52 for every 100,000 inhabitants 20 years of age)
(2).
GC comprises any malignant neoplasm that originates
from the region between the gastroesophageal junction and the
pylorus. A total of 95% of tumors developed in the stomach are of
epithelial origin; thus, these are denominated adenocarcinomas of
the stomach. The multifocal and polyclonal origin of the tumors
renders a morphologically based histological classification
complex. However, diverse classification systems have been
established (3), among which the study
by Lauren (4) described two types of
GC based on histological type and growth pattern: Intestinal and
diffuse. Intestinal- (or differentiated)-type GC (IGC) is
characterized by expansive and localized growth; generally, this
type of tumor type localizes in regions where an intestinal
metaplasia has developed previously that, in a number of cases, is
preceded by a precancerous cascade and a prior infection with
Helicobacter pylori (5).
Diffuse GC (DGC) possesses an infiltrating pattern, is an
undifferentiated adenocarcinoma and presents disperse cells with an
individual, or group, invasive capacity (3).
Gastric cancer and heredity
Approximately 1–3% of GC cases are associated with
heredity. Different syndromes have been determined of
hereditary-type GC as follows: i) Lynch syndrome; ii) familial
adenomatous polyposis; iii) Li-Fraumeni syndrome; iv) Peutz-Jeghers
syndrome; v) juvenile polyposis syndrome; and vi) hereditary-DGC
(HDGC) (6). The latter is one of the
better characterized types, with 80% penetrance (7). HDGC is an autosomal-type, dominant
syndrome in which 40% of cases are carriers of diverse mutations of
the CDH1 gene, which encodes for the cadherin protein
(8). The International Gastric Cancer
Linkage Consortium (IGCLC), based on diverse studies, has
established a direct association between the mutations of the
CDH1 gene and DGC heredity; the IGCLC has additionally
established various guidelines for its diagnosis, which comprise
the following: i) Familial history, which is the presence of >2
cases of GC in first- or second-degree consanguinity (one confirmed
case in a person aged >50 years of age), >3 cases of GC in
first- or second degree consanguinity, the latter is
age-independent; diagnosis of cancer <40 years of age, and a
personal or familial history of DGC and/or of lobular breast cancer
with an age of <50 years; ii) histopathological study; iii)
detection of the mutation by genetic analysis; and iv)
pathogenicity analysis (in vitro and in silico)
(7). However, van der Post et
al (9) postulated a risk
population of individuals with familiar antecedents of two or more
cases of bilateral lobular breast cancer, one of these in a
relative with confirmed DGC presenting cleft palate and the finding
in situ of signet ring cells, whether or not these are
propagating.
Diffuse gastric cancer
Today, DGC comprises one of the most aggressive
cancers, without defined molecular markers that permit its accurate
and timely diagnosis and/or prognosis. However, CDH1 gene
mutations are associated with the development of HDGC, one-half of
cases are negative for these. Thus, investigations are continuing
to identify genes that are associated with HDGC. The presence of
mutations has been observed in diverse genes, listed as follows:
Mutations reported in RHOA (10,11) are
associated with the initial stages of cancer and its progression to
metastasis; CTNNA1 mutated in HDGC acts as a
tumor-suppressor gene (12) and
changes in MAP3K6 generate an alteration in inflammation
pathways and apoptosis. In addition, it has been found that these
are predisposed to develop GC (13)
and that changes in the INSR gene (14) exert an effect on the insulin signaling
pathway, giving rise to changes in the pattern of glycosylation of
the E-cadherin protein, destabilization of cellular membranes and
the appearance of a mesenchymal phenotype. Contrary to previously
described, the roles of the mutations in the genes FBXO24,
DOT1L (14) and TGF-β
(15) have not yet been studied
(Table I). Some of the mutations
identified pertain to genes encoding regulatory elements or
proteins that form complexes with protein E-cadherin; as a
consequence of these, the loss of cellular adhesion can present due
to deregulation of the complex. However, the majority of the genes
postulated require studies of genetic penetrance and pathogeny in
order for the genes to be established as markers.
 | Table I.Mutations identified in patients who
were negative for mutations in the CDH1 gene. |
Table I.
Mutations identified in patients who
were negative for mutations in the CDH1 gene.
| Gene | Protein encoded | Mutation | Refs. |
|---|
| RHOA | GTPase | c.50G>A | (10,11) |
|
|
| c.14G>A |
|
| CTNNA1 | A-cadherin | c.76delGA | (12) |
|
|
| c.598G>T |
|
|
|
| c.620T>G |
|
| MAP3K6 | Serine/threonine | c.2837C>T | (13) |
|
| protein kinase | c.2872C>A |
|
|
|
| c.2544delC |
|
| DOT1L | Histone | c.3437C>T | (14) |
|
|
methyltransferase |
|
|
| FBXO24 | Protein 24 with F
box | c.242G>C | (14) |
| INSR | Insulin receptor | c.3937G>A | (14) |
| TGF-β | TGF-β | c.29C>G | (15) |
MicroRNA in HDGC
MicroRNA are non-encoded, single-chain RNA molecules
(17–25 nucleotides). These molecules regulate the majority of
cellular functions at the post-transcriptional level. The
regulation carried out by these molecules is complex in that they
have >1 target mRNA. However, analysis of these interactions by
means of Systems Biology, has allowed the understanding of complex
and heterogeneous diseases, such as cancer (16).
MicroRNA are transcribed from genes and introns,
individually or in groups. These molecules are ubiquitous; however,
their expression can be specific in different tissues either
temporarily or permanently, depending on the stage of the cell
(17). Due to the participation of
microRNA in the processes of cellular proliferation, cell cycle
control, apoptosis, differentiation and metabolism, these have been
indicated in the development of cancerous processes, by observing
specific patterns of expression in different neoplasms (18–20),
including GC (21–23), in which the microRNA expression profile
is different in samples of non-cancerous versus cancerous tissues.
A difference has been identified in the expression patterns of DGC
and IGC (23). However, the role of
microRNA in HDGC has not yet been established. There are studies
that describe the participation of microRNA in the regulation of
genes CDH1, RHOA, CTNNA1, INSR, and
TGF-β in different neoplasms (8,10–15).
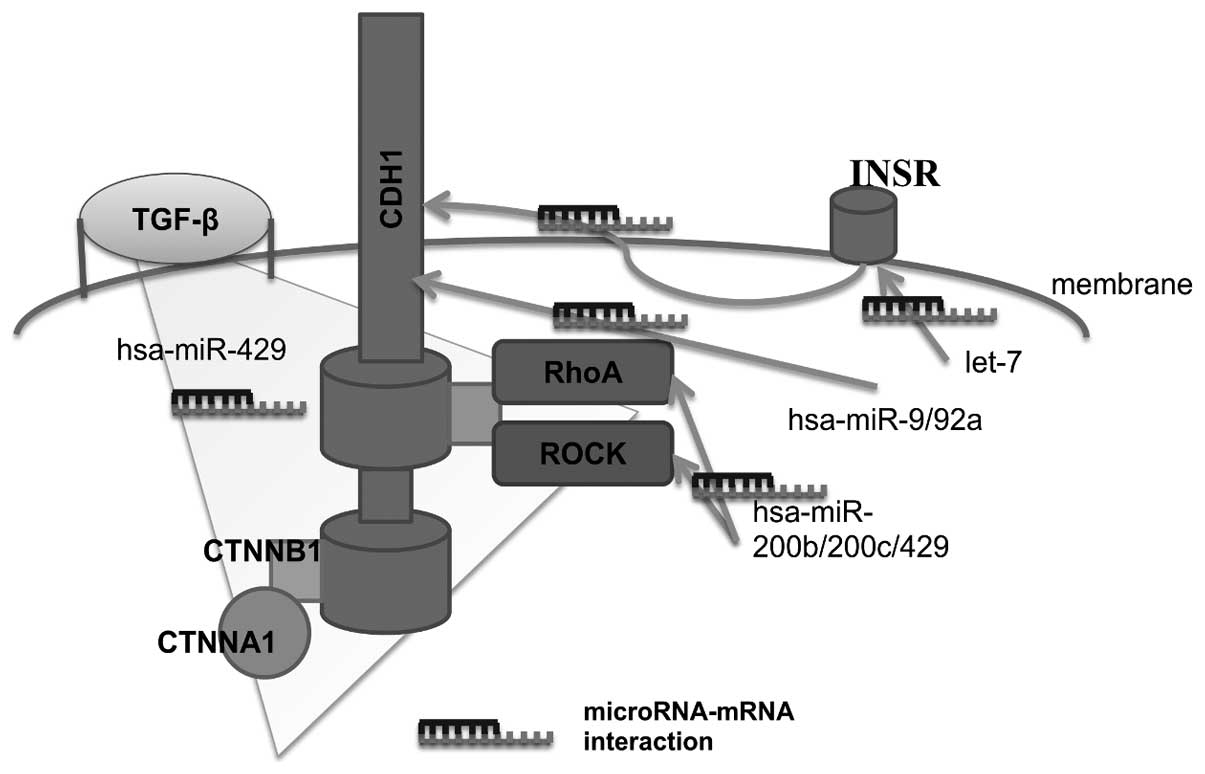
HDGC and genes
Beginning with the gene CDH1, in certain
studies of microRNA functionality, it has been reported that
hsa-miR-9 and hsa-miR-92a are associated with the development of
metastasis in esophageal squamous cell carcinoma, their binding
with the 3′-untranslated region of the CDH1 gene, impeding
the translation of the latter and induction in the
epithelial-mesenchymal transition (EMT) (24,25). The
cadherin complex is formed of signaling-charged E-cadherin,
catenins and proteins, and the whole complex participates in the
EMT. One of the genes comprising the cadherin complex is
CTNNA1, which encodes for catenin-α. Sun et al
(26) observed that there is a
difference in the expression profile of the CDH1,
CTNNA1, CTNNB1, CD44 and MMP2 genes
when EMT is induced, mediated by TGF-β, which is regulated
by hsa-miR-429, a member of the miR-200 family that has been
previously associated with the EMT, on regulating zinc finger E-box
binding homeobox 1/2 transcriptional repressors. Cadherin-complex
signaling-mediated GTPases have regulatory elements, including RhoA
and ROCK. In hepatocellular carcinoma, it was demonstrated that
these two proteins are targets of the hsa-miR-200b/200c/429
subfamily; all underexpressed in the samples of analyzed tumors. By
means of in vivo assays, it was demonstrated that
deregulation of the expression profile of this miR-200b subfamily
is involved in the EMT and in the development of the metastasis of
this carcinoma (27). The
glycosylation of E-cadherin mediated by insulin receptors (INSR) is
associated with the increase in the capacity of tumor cell
invasion, as well as the induction of a mesenchymal phenotype in
cancerous cells (28), and it has been
observed that the INSR gene is regulated by one of the
microRNA suppressor families of tumors involved in the development
of numerous neoplasms, such as let-7 (29). The regulation of the MAP3K6,
FBXO24 and DOT1L genes mediated by microRNA remains
to be elucidated; however, cooperation of the protein DOT1L with
c-Myc and an acetyltransferase has been described for activation of
the EMT in the initiation and progression of breast cancer to
metastasis (30).
Conclusion
In conclusion, the majority of mutations identified
in patients with HDGC, such as hsa-miR-9/92 microRNA, the miR-200
family and the let-7 family, which regulate these genes, are
associated with the induction of the EMT (Fig. 1). Thus, investigating the events that
trigger this process in patients with HDGC is of significant
importance for the establishment of genes and/or of microRNA that
can be employed for the diagnosis and prognosis of this neoplasm in
patients who are negative for mutations in the CDH1
gene.
Acknowledgements
The authors are grateful for the support of the
present study from the Coordinación de Investigación en
Salud-Instituto Mexicano del Seguro Social and Secretaría de
Investigación y Posgrado-Instituto Politécnico Nacional-México.
MCS-A acknowledges the scholarship and financial support provided
by Consejo Nacional de Ciencia y Tecnología (CONACyT no. 576518).
The present study constitutes partial fulfillment of the Graduate
Program of Master Degree in Sciences, Biomedicine and Molecular
Biotechnology, Escuela Nacional de Ciencias Biológicas, Instituto
Politécnico Nacional, Mexico City, Mexico.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
INEGI: Estadísticas a propósito del Día
Mundial contra el Cáncer. http://www.inegi.org.mx/saladeprensa/aproposito/2016/cancer2016_0.pdfAccessed.
March 06–2016
|
|
3
|
Hu B, El Hajj N, Sittler S, Lammert N,
Barnes R and Meloni-Ehrig A: Gastric cancer: Classification,
histology and application of molecular pathology. J Gastrointest
Oncol. 3:251–261. 2012.PubMed/NCBI
|
|
4
|
Lauren P: The two histological main types
of gastric carcinoma: Diffuse and so-called intestinal-type
carcinoma. An attempt at a histo-clinical classification. Acta
Pathol Microbiol Scand. 64:31–49. 1965.PubMed/NCBI
|
|
5
|
Piazuelo MB, Epplein M and Correa P:
Gastric cancer: An infectious disease. Infect Dis Clin North Am.
24:853–869, vii. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tan RYC and Ngeow J: Hereditary diffuse
gastric cancer: What the clinician should know. World J
Gastrointest Oncol. 7:153–160. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fitzgerald RC, Hardwick R, Huntsman D,
Carneiro F, Guilford P, Blair V, Chung DC, Norton J, Ragunath K,
Van Krieken JH, et al: International Gastric Cancer Linkage
Consortium: Hereditary diffuse gastric cancer: updated consensus
guidelines for clinical management and directions for future
research. J Med Genet. 47:436–444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Corso G, Marrelli D, Pascale V, Vindigni C
and Roviello F: Frequency of CDH1 germline mutations in gastric
carcinoma coming from high- and low-risk areas: Metanalysis and
systematic review of the literature. BMC Cancer. 12:82012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van der Post RS, Vogelaar IP, Carneiro F,
Guilford P, Huntsman D, Hoogerbrugge N, Caldas C, Schreiber KE,
Hardwick RH, Ausems MG, et al: Hereditary diffuse gastric cancer:
Updated clinical guidelines with an emphasis on germline CDH1
mutation carriers. J Med Genet. 52:361–374. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kakiuchi M, Nishizawa T, Ueda H, Gotoh K,
Tanaka A, Hayashi A, Yamamoto S, Tatsuno K, Katoh H, Watanabe Y, et
al: Recurrent gain-of-function mutations of RHOA in diffuse-type
gastric carcinoma. Nat Genet. 46:583–587. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi
ST, Siu HC, Deng S, Chu KM, Law S, et al: Whole-genome sequencing
and comprehensive molecular profiling identify new driver mutations
in gastric cancer. Nat Genet. 46:573–582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Majewski IJ, Kluijt I, Cats A, Scerri TS,
de Jong D, Kluin RJ, Hansford S, Hogervorst FB, Bosma AJ, Hofland
I, et al: An α-E-catenin (CTNNA1) mutation in hereditary diffuse
gastric cancer. J Pathol. 229:621–629. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gaston D, Hansford S, Oliveira C,
Nightingale M, Pinheiro H, Macgillivray C, Kaurah P, Rideout AL,
Steele P, Soares G, et al: Germline mutations in MAP3K6 are
associated with familial gastric cancer. PLoS Genet.
10:e10046692014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Donner I, Kiviluoto T, Ristimäki A,
Aaltonen LA and Vahteristo P: Exome sequencing reveals three novel
candidate predisposition genes for diffuse gastric cancer. Fam
Cancer. 14:241–246. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Yue ZC, Zhang YY, Bai J, Meng XN,
Geng JS and Fu SB: Elevated serum level and gene polymorphisms of
TGF-beta1 in gastric cancer. J Clin Lab Anal. 22:164–171. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Prasasya RD, Tian D and Kreeger PK:
Analysis of cancer signaling networks by systems biology to develop
therapies. Semin Cancer Biol. 21:200–206. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Munker R and Calin GA: MicroRNA profiling
in cancer. Clin Sci (Lond). 121:141–158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Volinia S, Calin GA, Liu C-G, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang C, Chen X, Alattar M, Wei J and Liu
H: MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis
of gastric cancer. Cancer Gene Ther. 22:291–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shrestha S, Hsu SD, Huang WY, Huang HY,
Chen W, Weng SL and Huang HD: A systematic review of microRNA
expression profiling studies in human gastric cancer. Cancer Med.
3:878–888. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ueda T, Volinia S, Okumura H, Shimizu M,
Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: A microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Song Y, Li J, Zhu Y, Dai Y, Zeng T, Liu L,
Li J, Wang H, Qin Y, Zeng M, et al: MicroRNA-9 promotes tumor
metastasis via repressing E-cadherin in esophageal squamous cell
carcinoma. Oncotarget. 5:11669–11680. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen Y, Kingham K, Ford JM, Rosing J, Van
Dam J, Jeffrey RB, Longacre TA, Chun N, Kurian A and Norton JA: A
prospective study of total gastrectomy for CDH1-positive hereditary
diffuse gastric cancer. Ann Surg Oncol. 18:2594–2598. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun Y, Shen S, Liu X, Tang H, Wang Z, Yu
Z, Li X and Wu M: miR-429 inhibits cells growth and invasion and
regulates EMT-related marker genes by targeting Onecut2 in
colorectal carcinoma. Mol Cell Biochem. 390:19–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wong CM, Wei L, Au SL, Fan DN, Zhou Y,
Tsang FH, Law CT, Lee JM, He X, Shi J, et al: MiR-200b/200c/429
subfamily negatively regulates Rho/ROCK signaling pathway to
suppress hepatocellular carcinoma metastasis. Oncotarget.
6:13658–13670. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
de-Freitas-Junior JC, Carvalho S, Dias AM,
Oliveira P, Cabral J, Seruca R, Oliveira C, Morgado-Díaz JA, Reis
CA and Pinho SS: Insulin/IGF-I signaling pathways enhances tumor
cell invasion through bisecting GlcNAc N-glycans modulation. an
interplay with E-cadherin. PLoS One. 8:e815792013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu H, Shyh-Chang N, Segrè AV, Shinoda G,
Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG,
et al: DIAGRAM Consortium; MAGIC Investigators: The Lin28/let-7
axis regulates glucose metabolism. Cell. 147:81–94. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cho MH, Park JH, Choi HJ, Park MK, Won HY,
Park YJ, Lee CH, Oh SH, Song YS, Kim HS, et al: DOT1L cooperates
with the c-Myc-p300 complex to epigenetically derepress CDH1
transcription factors in breast cancer progression. Nat Commun.
6:78212015. View Article : Google Scholar : PubMed/NCBI
|