Introduction
Echinococcosis, also termed cystic echinococcosis,
is a type of zoonosis caused by the larval stages of
Echinococcus granulosus (E. granulosus). This disease is
endemic and widespread throughout the world (1–4). In China,
it is mainly endemic in large pastoral areas of northwestern
regions, such as Xinjiang, Gansu, Qinghai, and Inner Mongolia, as
well as in southwestern areas, such as Sichuan and Tibet (5). The life cycle of this cestode involves
dogs, wolves and other canids as definitive hosts, and intermediate
hosts are usually sheep, cattle, goats and pigs. However, humans
may become accidental intermediate hosts when food and water
sources are contaminated by the eggs of E. granulosus.
Echinococcosis causes significant harm to human health and hampers
the development of local animal husbandry. At present, many actions
have been taken to cure this disease; however, few are sufficiently
effective to cure it completely. Surgery in combination with
chemotherapy has been identified as most efficacious and has become
the first choice of therapy. However, this therapeutic strategy
inevitably entails surgical risk and requires considerable labour,
material and financial resources (6).
An earlier diagnosis correlates to more effective surgery and
chemotherapy. Therefore, early diagnosis is of great importance to
the treatment of echinococcosis. However, early diagnosis continues
to be an obstacle for doctors, as it is difficult to confirm the
larvae using imaging diagnosis (7–10).
Immunological methods are more promising, however, to the best of
our knowledge, no studies regarding the early diagnosis of the
E. granulosus infection have been performed. Therefore,
there is an urgent requirement to develop effective preventative
and treatment strategies.
A study by Lightowlers et al (11) revealed that the Eg95 recombinant
antigen induced a 95–100% protective immune effect against
oncospheres in sheep. Thus, immune prevention may be an effective
measure that could be taken to prevent cystic echinococcosis.
Strohmaier et al (12)
identified the immunogenic amino acid sites of the foot and mouth
disease virus and thereby designed novel epitope vaccines. With the
rapid development of molecular biology and immunology, epitope
vaccines have become the focus of research on molecular vaccines
(13). To prevent infection by
Echinococcus multilocularis (E. multilocularis), Kouguchi
et al (14) developed the
Emy162 recombinant antigen, which induced a 74.3% protective rate
in rats against E. multilocularis. In addition, Katoh et
al (15) developed a vaccine based
on the Em95 protein, and the protective efficiency of the vaccine
was 78.5–82.9%. These results suggest that the prevention of
echinococcosis using a molecular vaccine is feasible.
Antigen 5 (Ag5) is one of the dominant antigens of
E. granulosus cyst fluids. It is a dimeric protein composed
of 22- and 38-kDa subunits linked by a disulphide bridge, with the
two subunits bearing an N-glycan modification (16,17). The
38-kDa subunit is closely associated with serine proteases of the
trypsin family (16). It has been
confirmed that Ag5 is expressed in all stages of the life cycle of
E. granulosus (18). Ag5 is
strongly expressed in the protoscolex tegument, the embryonic
membrane of eggs, and the surface of oncospheres; it is also
expressed weakly in the adult tegument. As a result of this, the
present study hypothesized that Ag5 may have diagnostic value and
serve as a candidate antigen for a vaccine against cystic
echinococcosis. To the best of our knowledge, recent studies
regarding Ag5 have primarily focused on the diagnostic evaluation
of cystic echinococcosis (16–19). Yarzabal et al (20) demonstrated the occurrence of
cross-reactions in ELISA detection between Ag5 and other parasites,
such as E. multilocularis and certain other helminthes. This
result revealed the low specificity of this protein, thus Ag5 could
not be widely applied in clinical diagnosis. However, whether Ag5
may be a candidate vaccine for cystic echinococcosis remains
unclear.
In the current study, computer technology and online
molecular biology software were used to predict various
characteristics of Ag5, particularly the B and T cell epitopes, to
analyze its immunogenicity and lay the theoretical foundation for
further investigation of epitope vaccines against cystic
echinococcosis.
Materials and methods
Amino acid sequence of Ag5
The nucleotide sequence of Ag5 was selected from
GenBank (NIH, Bethesda, MD, USA; GenBank no. JF970202). The amino
acid sequence of Ag5 was predicted by DNAman software
(LynnonBiosoft, San Ramon, CA, USA).
Prediction of the Ag5 protein
secondary structure
The secondary structure of Ag5 protein was predicted
using the Self-Optimized Prediction method With Alignment (SOPMA)
Server (https://npsa-prabi.ibcp.fr/cgibin/npsa_automat.pl?page=npsa_sopma.html)
(21). The sequence of Ag5 protein was
input, then four conformational states were predicted. The
transmembrane structure of Ag5 protein was predicted by TMHMM
Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) (22). The sequence of Ag5 protein was input,
and three regions, including inside, transmembrane and outside
regions, were analyzed.
Prediction of the Ag5 protein tertiary
structure
The tertiary structure of Ag5 protein was predicted
using the online prediction server 3DLigandSite (http://www.sbg.bio.ic.ac.uk) (23). To improve the accuracy of the
prediction, the tertiary structure was also predicted using an
additional online software application, Center for Biological
Sequence Analysis (CBS) Prediction Servers (http://www.cbs.dtu.dk/services) (24). The 3DLigandSite software used homology
modelling to predict the tertiary structure, as did the CBS
Prediction Servers. When the sequence of Ag5 was input, a high
homology model was output.
Prediction of Ag5 protein
epitopes
Prediction of the Ag5 protein B cell
epitopes
The B cell epitopes of the Ag5 protein were
predicted using the online prediction software Immune Epitope
Database (IEDB; http://tools.immuneepitope.org/bcell/) (25). The sequence of Ag5 protein was input,
and then the thresholds were all set to 1.0. The other parameters
were not changed.
Prediction of the Ag5 protein T cell
epitopes
Major histocompatibility complex (MHC)-I human
leukocyte antigen (HLA)-A*0201-restricted T cell epitopes were
predicted by the online prediction software BioInformatics and
Molecular Analysis Section (BIMAS; http://www-bimas.cit.nih.gov/molbio/hla_bind/)
(26) together with SYFPEITHI
(http://www.syfpeithi.de/bin/MHCServer.dll/EpitopePrediction.htm)
(27). The amino acid sequence of Ag5
protein was input, the HLA molecule was set as HLA-A*0201, and the
nonamers were set to 9. The remaining parameters were not
altered.
Prediction of various physicochemical properties
of the Ag5 protein
Various physicochemical properties of the Ag5
protein were predicted by the online prediction software ProtParam
(http://web.expasy.org/protparam/)
(28). The sequence of Ag5 protein was
input and five properties, including molecular weight, theoretical
isoelectric point (pI), the instability index, the aliphatic index
and the grand average of hydropathicity (GRAVY) were analyzed.
Results
Amino acid sequence encoded by the Ag5
gene
The open reading frame of Ag5 is 1,455 bp in length
and encodes 484 amino acids, as predicted by DNAman software
(Fig. 1).
Secondary structure of the Ag5
protein
To analyze the immunogenicity of the Ag5 protein,
its secondary structure was predicted using SOPMA Server, and the
results revealed that α-helixes, β-turns, random coils and extended
strands accounted for 23.35, 10.95, 41.32, and 24.38% of the
secondary structure, respectively. The predicted secondary
structure results for the Ag5 protein are displayed in Fig. 2. The high proportion of random coils
and extended strands in the structure of Ag5 protein suggest that
the protein is likely to form antigenic epitopes.
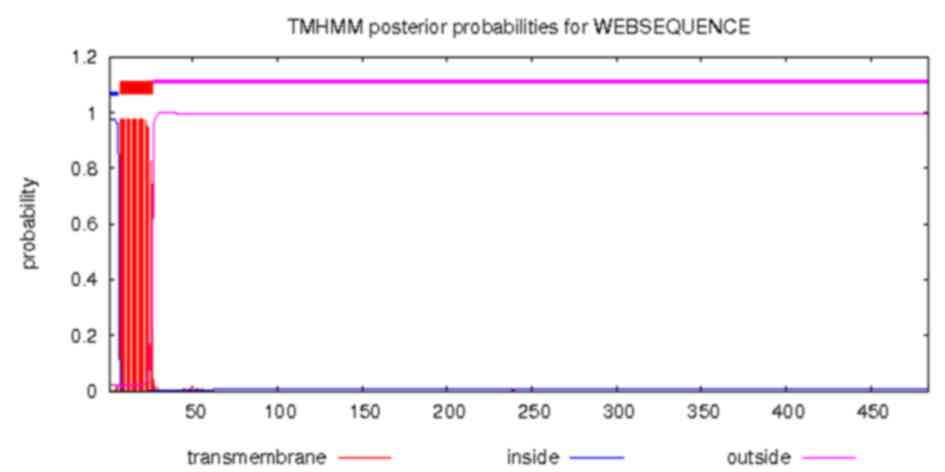
The transmembrane structure of Ag5 protein was
predicted using the online CBS prediction software TMHMM Server
version 2.0. The inside, transmembrane and outside regions of Ag5
were located at positions 1–6, 7–26 and 27–484, respectively. The
results are displayed in Fig. 3.
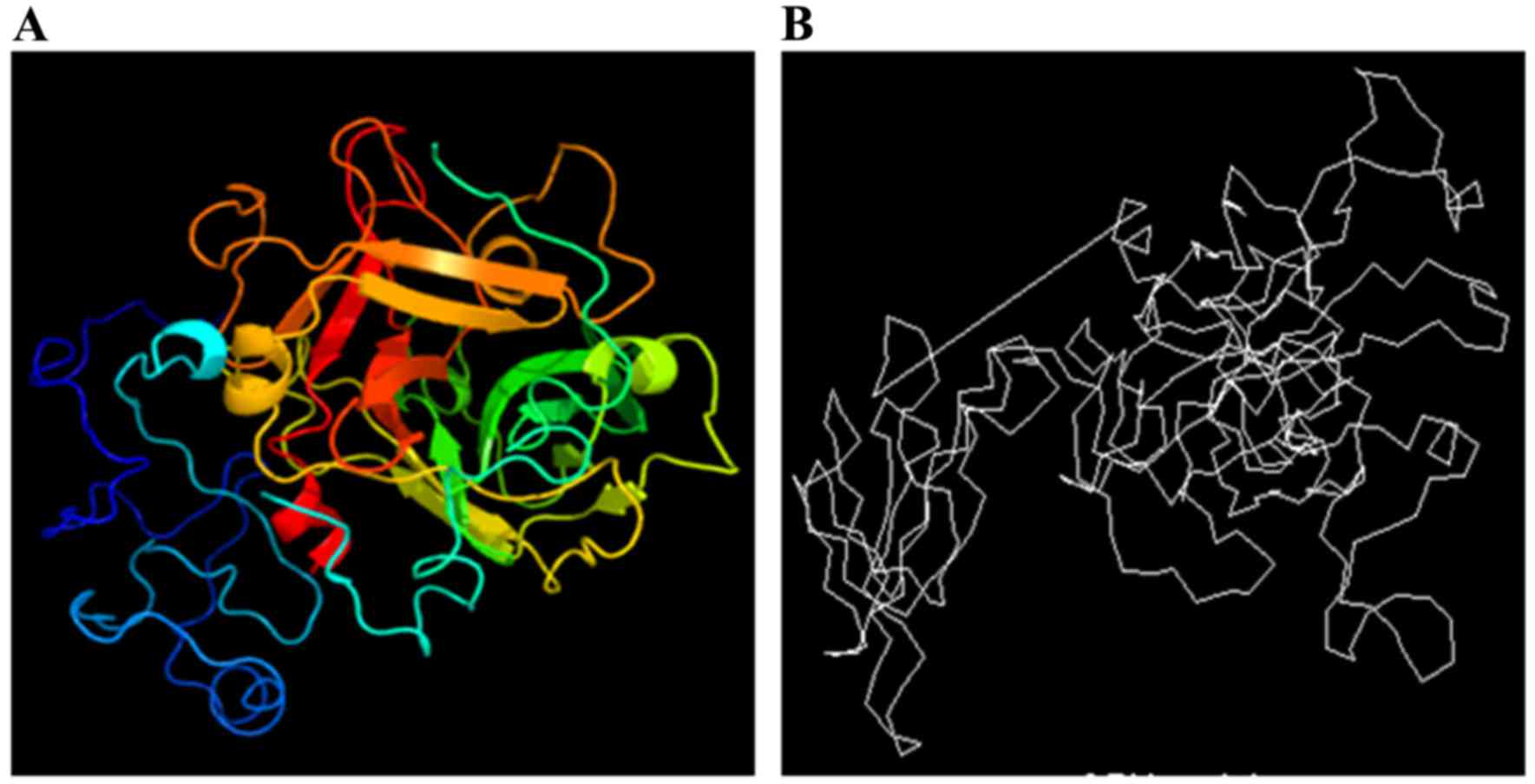
Tertiary structure of the Ag5
protein
The tertiary structure of the Ag5 protein was
predicted using the online prediction server 3DLigandSite and
compared with the structure from the CBS Prediction Servers. The
results are displayed in Fig. 4.
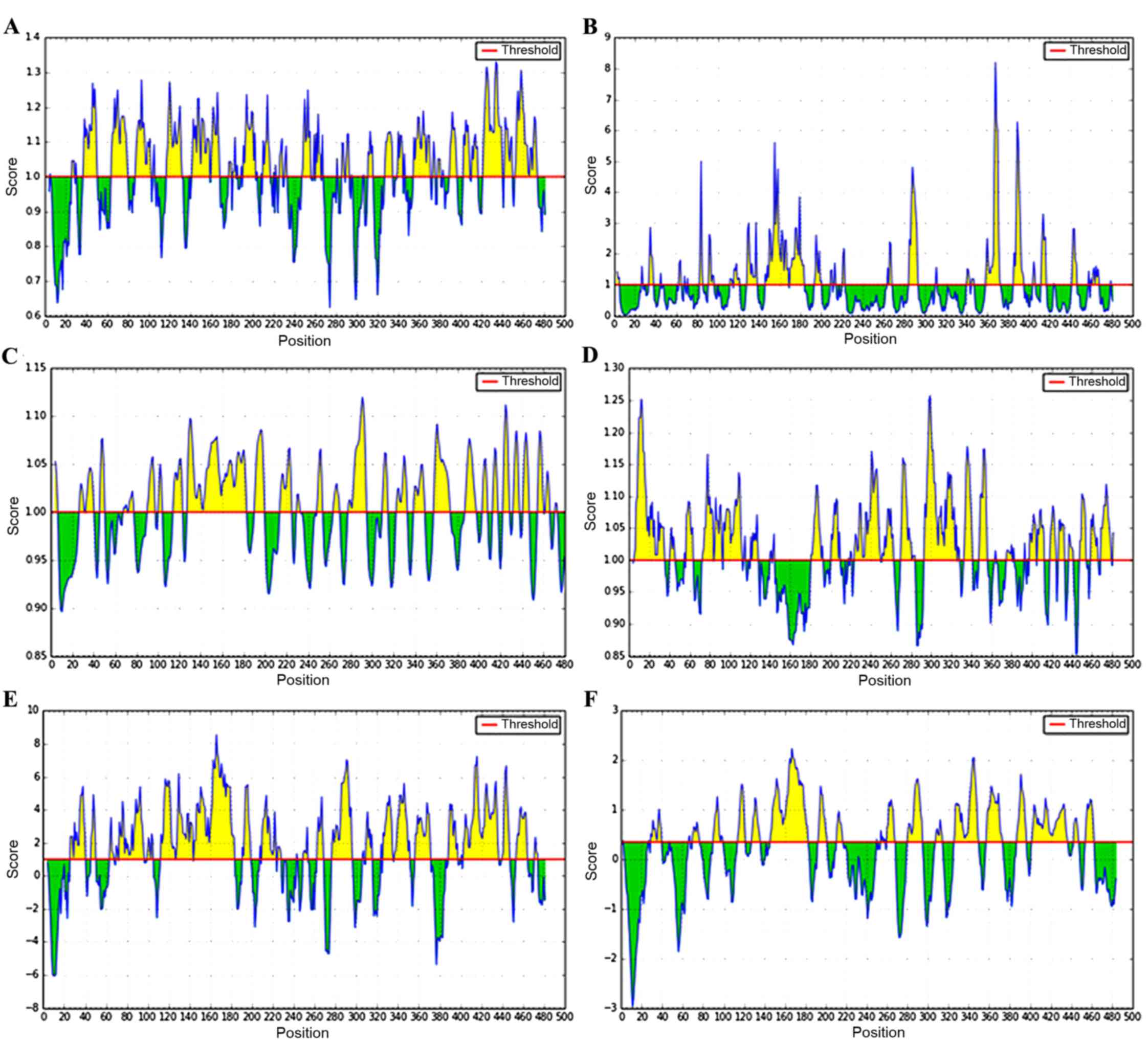
Epitopes of the Ag5 protein
B cell epitopes of the Ag5
protein
B cell epitopes of Ag5 protein were predicted using
the online prediction software IEDB (Fig.
5). High-scoring regions of β-turn were located at positions
26–30, 36–50, 64–80, 87–102, 117–131, 142–168, 177–202, 210–216,
248–267, 327–346, 355–396, 403–416, and 422–473, and the accessible
surface areas were located at positions 91–96, 116–121, 128–138,
146–183, 195–200, 284–293, 340–347, 359–373, 385–395, 412–417, and
460–468. The flexible regions were 27–40, 46–50, 68–77, 90–104,
114–123, 127–182, 190–199, 214–225, 249–253, 264–269, 278–295,
310–314, 323–333, 339–349, 358–373, 387–397, 403–408, 413–417,
423–428, 423–428, 432–447, and 455–473, and the possible antigenic
regions proved to be 5–35, 57–63, 73–83, 85–92, 94–112, 121–128,
181–192, 201–209, 224–249, 252–262, 270–284, 293–325, 333–341,
348–355, 375–380, 398–404, 406–412, 418–424, 427–432, 448–454,
459–465, and 469–479. Areas that had high hydrophilicity were
located at positions 26–40, 45–50, 64–67, 70–96, 100–105, 113–183,
191–198, 210–228, 249–253, 263–268, 277–296, 310–315, 326–351,
357–371, 389–397, 403–436, 440–447, 454–464, and 471–474, and the
linear epitopes were demonstrated to be 29–39, 70–76, 90–99,
114–121, 128–136, 146–182, 193–201, 211–216, 260–267, 280–294,
308–314, 326–349, 357–372, 387–396, 402–436, 443–447, and
454–462.
A combination of the results predicted by the
different parameters indicated that the B cell epitopes were
located at positions 27–39, 70–80, 117–130, 146–168, 250–262,
284–293, 339–349, 359–371, 403–412, and 454–462.
T cell epitopes of the Ag5
protein
To increase the accuracy of the prediction, T cell
epitopes were predicted using the online prediction software BIMAS
together with SYFPEITHI. There were 20 high-scoring regions, and
the results are presented in Table I.
These two software programs use different predicting systems. The
BIMAS software resulted in high scores ranging from 3.932 to
1,737.776; however, the high scores predicted by the SYFPEITHI
software ranged between 19 and 27. These high-scoring regions had
strong potential of forming epitopes regardless of the
difference.
 | Table I.Major histocompatibility
complex-Inonamer T cell epitope using BIMAS and SYFPEITHI. |
Table I.
Major histocompatibility
complex-Inonamer T cell epitope using BIMAS and SYFPEITHI.
|
| BIMAS software | SYFPEITHI
software |
|---|
|
|
|
|
|---|
| No. | Initiation site | Amino acid
sequence | Score site | Initiation
sequence | Amino acid | Score |
|---|
| 1 | 52 | LMFNRSLFV | 1737.776 | 395 | SLNEIRVSI | 27 |
| 2 | 467 | WLYNSVGSV | 306.118 | 467 | WLYNSVGSV | 27 |
| 3 | 318 | ALLRLETPV | 257.342 | 318 | ALLRLETPV | 25 |
| 4 | 476 | IQWINRYAV | 99.501 | 18 | AALGLELTL | 24 |
| 5 | 231 | TLITPRWVL | 65.689 | 273 | LLVRAGDTV | 24 |
| 6 | 273 | LLVRAGDTV | 57.937 | 231 | TLITPRWVL | 23 |
| 7 | 9 | IVFVCLFAT | 54.713 | 16 | ATAALGLEL | 22 |
| 8 | 395 | SLNEIRVSI | 42.774 | 52 | LMFNRSLFV | 22 |
| 9 | 237 | WVLTAAHCL | 31.814 | 57 | SLFVYRENI | 22 |
| 10 | 299 | LVVIHPDWV | 27.882 | 182 | RLPYSLKIL | 22 |
| 11 | 182 | RLPYSLKIL | 20.145 | 213 | NASEGLTRL | 22 |
| 12 | 57 | SLFVYRENI | 18.915 | 50 | ATLMFNRSL | 21 |
| 13 | 8 | WIVFVCLFA | 17.282 | 226 | IICGGTLIT | 21 |
| 14 | 272 | FLLVRAGDT | 16.488 | 6 | PLWIVFVCL | 20 |
| 15 | 442 | NEEDGRWYV | 14.638 | 24 | LTLDPDELV | 20 |
| 16 | 24 | LTLDPDELV | 12.207 | 225 | NIICGGTLI | 20 |
| 17 | 471 | SVGSVIQWI | 11.548 | 247 | PIFGSSNAL | 20 |
| 18 | 11 | FVCLFATAA | 7.599 | 327 | NIESDGAGV | 20 |
| 19 | 6 | PLWIVFVCL | 7.411 | 330 | SDGAGVACV | 20 |
| 20 | 230 | GTLITPRWV | 3.932 | 189 | ILGGKSAKS | 19 |
Comparing the results predicted by the two online
programs, seven high-scoring T cell epitopes were found to be
located at positions 52–60, 57–65, 182–190, 231–239, 273–281,
318–326 and 467–475.
Physicochemical properties of the Ag5
protein
The molecular weight of Ag5 is 54,874.8 Da, and its
theoretical pI is 6.36. The instability index is computed to be
55.78, classifying the protein as unstable. The aliphatic index and
GRAVY were found to be 69.32 and −0.525, respectively.
Discussion
In China, cystic echinococcosis is a public health
problem requiring a prompt solution. Furthermore, the infection
region is gradually expanding (29).
Currently, surgery in combination with chemotherapy is the first
choice as a treatment strategy for cystic echinococcosis; however,
it is not completely effective and the disease recurs. Immune
prevention may be an effective measure to prevent the epidemic of
cystic echinococcosis. Thus, developing a novel and effective
vaccine is considered to be important.
It has been confirmed that Ag5 is expressed in all
stages of the life cycle of E. granulosus (18), such as the tegument of the protoscolex,
the embryonic membrane of eggs, the surface of oncospheres and
adults. Thus, Ag5 presents as a promising vaccine for cystic
echinococcosis.
Obtaining information regarding antigen epitopes
will facilitate the development of epitope vaccines. Previously,
epitope prediction was performed using a single parameter and its
accuracy was limited. Now, as a result of the development of
bioinformatics, the prediction of epitopes is accurate and simple.
Making predictions using a multi-parameter and multi-method
analysis improves the accuracy of epitope prediction greatly. In a
study by Li et al (30),
HLA-A*0201, HLA-A*1101 and HLA-A*2401 cytotoxic T lymphocyte
restricted epitopes of platelet membrane glycoprotein IIb/IIIa (GP
IIb/IIIa) antibody of human and mice were predicted using
SYFPEITHI, RANKPEP, BIMAS, SVMHC, PREDEP, MHCPRED and PROPRED
software, and the T cell epitopes of GP IIb/IIIa antibody were
predicted. In a different study conducted by Shen et al
(31), the secondary structure and
surface characteristics of the follicle stimulating hormone
receptor (FSHR) extracellular domain were analyzed using DNAStar
Protean software, and B cell epitope prediction was conducted using
alternative online software. The possible antigenic epitopes of the
FSHR extracellular domain were predicted, the peptides of the
epitopes were synthesized and the immunogenicity of these peptides
was determined.
In the current study, the transmembrane structure of
Ag5 was predicted and inside, transmembrane and outside regions
were identified. The transmembrane region was stable and it could
not alter or form epitopes easily. The outside region, which
constituted the majority of the structure, was located at position
27–484. The flexible areas usually appeared here and this region
could potentially form epitopes.
The secondary structure of the Ag5 protein was
predicted, as it was closely associated with antigenic features.
The α-helixes and extended strands are regular structures in the
secondary structure of the Ag5 protein. They are not deformed
readily, as hydrogen bonds maintain the stability of their
structure. However, ligand binding is difficult as the α-helixes
and extended strands are located inside the protein. By contrast,
random coils and β-turns are located at the surface of Ag5, and
they are suitable for ligand binding. Therefore, it is possible for
these to form epitopes. As predicted by the SOPMA Server software,
the proportion of α-helixes was 23.35% and that of the extended
strands was 24.38%. Thus, it was inferred from the results that the
Ag5 protein has good stability. Furthermore, random coils and
β-turns, which represent potential epitope regions, account for
41.32 and 10.95% of the protein, respectively.
The tertiary structure is based on the secondary
structure of the protein, and it is a three-dimensional globular
structure composed of further coiling and folding of secondary
structure elements, such as α-helixes, β-turns, random coils and
extended strands. The tertiary structure clarified that the random
coils and β-turns are located at the surface of Ag5, and are
suitable for ligand binding. These regions form epitopes easily.
Therefore, the tertiary structure prediction was a necessary
supplement to the prediction of Ag5 antigenic epitopes.
To improve the accuracy of the B cell epitope
prediction, a multi-parameter analysis was utilized. The β-turn
parameter prediction indicated that the β-turn is one of the normal
types in the secondary structure of the protein. Hydrogen bonds
alter the direction of the peptide chains, similarly to a U-model
structure. Epitopes are able to easily form in these regions.
Accessibility reflects the possibility of being found on the
surface; the higher the accessibility, the more likely epitopes are
to form. The flexibility parameter prediction demonstrates the
ability of the protein to bend and fold. With a greater
flexibility, the polypeptide skeleton of a protein has a strong
capacity to bend and fold, thus promoting the formation of a
secondary structure. The antigenic propensity demonstrated the
immunogenic regions of the Ag5 protein. Those regions with high
antigenic propensity are closely associated with the epitopes of
the protein. The hydrophilicity parameter prediction explains the
position of hydrophilic residues in the amino acid sequence of the
protein. The hydrophilic residues are located on the surface of the
protein and are suitable for ligand binding. The dominant epitopes
are more likely to be located in regions with a high
hydrophilicity. In combination with these parameters, the B cell
epitopes of the Ag5 protein were predicted using IEDB software.
Potential epitopes were revealed and were located at positions
27–39, 70–80, 117–130, 146–168, 250–262, 284–293, 339–349, 359–371,
403–412 and 454–462.
In the Chinese population, HLA-A*0201 is the most
common HLA-I molecule (32). The
accuracy of the MHC-I epitope prediction in the prediction of the T
cell epitopes is declared to be ≤90%. To increase the accuracy of
the prediction, the T cell epitopes were predicted using BIMAS and
SYFPEITHI software. Taken together, the T cell epitopes of the Ag5
protein were found to be located at positions 52–60, 57–65,
182–190, 231–239, 273–281, 318–326, and 467–475.
The aim of the present study was to obtain the
bioinformatic characteristics of the Ag5 protein and analyze its
immunogenicity. The results of the secondary structure prediction
demonstrated that there are potential epitopes in the protein. The
B cell epitopes were found to be located at positions 27–39, 70–80,
117–130, 146–168, 250–262, 284–293, 339–349, 359–371, 403–412, and
454–462; whereas the T cell epitopes were found to be located at
positions 52–60, 57–65, 182–190, 231–239, 273–281, 318–326, and
467–475. These regions possess great potential for forming
epitopes.
In conclusion, the current study predicts
significant biological data for E. granulosus Ag5 to provide
a theoretical basis for investigating its antigenicity and provides
a theoretical foundation for epitope vaccine development for cystic
echinococcosis.
Acknowledgements
Project support was provided, in part, by the
training Programs of Innovation and Entrepreneurship for College
Students in Jiangsu Province (grant no. 201510313017Z), the
National Natural Science Foundation of China (grant no. 81501762),
the Talents Scientific Research Foundation of Xuzhou Medical
University (grant no. D2015004), the Natural Science Foundation of
the Jiangsu Higher Education Institutions (grant no. 15KJB310025)
and the Jiangsu Planned Projects for Postdoctoral Research Funds
(grant no. 1501061A).
References
|
1
|
Wang K, Zhang X, Jin Z, Ma H, Teng Z and
Wang L: Modeling and analysis of the transmission of Echinococcosis
with application to Xinjiang Uygur Autonomous Region of China. J
Theor Biol. 333:78–90. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torgerson PR: The emergence of
echinococcosis in central Asia. Parasitology. 140:1667–1673. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moldovan R, Neghina AM, Calma CL, Marincu
I and Neghina R: Human cystic echinococcosis in two south-western
and central-western Romanian counties: A 7-year epidemiological and
clinical overview. Acta Trop. 121:26–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siracusano A, Delunardo F, Teggi A and
Ortona E: Host-parasite relationship in cystic echinococcosis: An
evolving story. Clin Dev Immunol. 2012:6393622012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu CY, Ma XM, Ding JB and Shen HM:
Different host status of echinococcus infection in China. Chin J
Zoonoses. 25:586–588. 2009.
|
|
6
|
Nasrieh MA, Abdel-Hafez SK, Kamhawi SA,
Craig PS and Schantz PM: Cystic echinococcosis in Jordan:
Socioeconomic evaluation and risk factors. Parasitol Res.
90:456–466. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gonlugur U, Ozcelik S, Gonlugur TE and
Celiksoz A: The role of Casoni's skin test and indirect
haemagglutination test in the diagnosis of hydatid disease.
Parasitol Res. 97:395–398. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kalantari E, Bandehpour M, Pazoki R,
Taghipoor-Lailabadi N, Khazan H, Mosaffa N, Nazaripouya MR and
Kazemi B: Application of recombinant Echinococcus granulosus
antigen B to ELISA kits for diagnosing hydatidosis. Parasitol Res.
106:847–851. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tawfeek GM, Elwakil HS, El-Hoseiny L,
Thabet HS, Sarhan RM, Awad NS and Anwar WA: Comparative analysis of
the diagnostic performance of crude sheep hydatid cyst fluid,
purified antigen B and its subunit (12 Kda), assessed by ELISA, in
the diagnosis of human cystic echinococcosis. Parasitol Res.
108:371–376. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wen H, Aji T and Shao YM: Diagnosis and
management against the complications of human cystic
echinococcosis. Front Med China. 4:394–398. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lightowlers MW, Lawrence SB, Gauci CG,
Young J, Ralston MJ, Maas D and Heath DD: Vaccination against
hydatidosis using a defined recombinant antigen. Parasite Immunol.
18:457–462. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Strohmaier K, Franze R and Adam KH:
Location and characterization of the antigenic portion of the FMDV
immunizing protein. J Gen Virol. 59:295–306. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kouguchi H, Matsumoto J, Katoh Y, Oku Y,
Suzuki T and Yagi K: The vaccination potential of EMY162 antigen
against Echinococcus multilocularis infection. Biochem Biophys Res
Commun. 363:915–920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kouguchi H, Matsumoto J, Yamano K, Katoh
Y, Oku Y, Suzuki T and Yagi K: Echinococcus multilocularis:
Purification and characterization of glycoprotein antigens with
serodiagnostic potential for canine infection. Exp Parasitol.
128:50–56. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Katoh Y, Kouguchi H, Matsumoto J, Goto A,
Suzuki T, Oku Y and Yagi K: Characterization of emY162 encoding an
immunogenic protein cloned from an adult worm-specific cDNA library
of Echinococcus multilocularis. Biochim Biophys Acta. 1780:1–6.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lorenzo C, Salinas G, Brugnini A,
Wernstedt C, Hellman U and González-Sapienza G: Echinococcus
granulosus antigen 5 is closely related to proteases of the trypsin
family. Biochem J. 369:191–198. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lightowlers MW, Liu DY, Haralambous A and
Rickard MD: Subunit composition and specificity of the major cyst
fluid antigens of Echinococcus granulosus. Mol Biochem Parasitol.
37:171–182. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Xu H, Chen J, Gan W, Wu W, Wu W and
Hu X: Gene cloning, expression, and localization of antigen 5 in
the life cycle of Echinococcus granulosus. Parasitol Res.
110:2315–2323. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Paul M and Stefaniak J: Detection of
specific Echinococcus granulosus antigen 5 in liver cyst bioptate
from human patients. Acta Trop. 64:65–77. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yarzabal LA, Bout DT, Naquira FR and
Capron AR: Further observations on the specificity of antigen 5 of
Echinococcus granulosus. J Parasitol. 63:495–499. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Geourjon C and Deléage G: SOPMA:
Significant improvements in protein secondary structure prediction
by consensus prediction from multiple alignments. Comput Appl
Biosci. 11:681–684. 1995.PubMed/NCBI
|
|
22
|
Melén K, Krogh A and von Heijne G:
Reliability measures for membrane protein topology prediction
algorithms. J Mol Biol. 327:735–744. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wass MN, Kelley LA and Sternberg MJ:
3DLigandSite: Predicting ligand-binding sites using similar
structures. Nucleic Acids Res. 38:W469–W473. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nielsen M, Lundegaard C, Lund O and
Petersen TN: CPHmodels-3.0-remote homology modeling using
structure-guided sequence profiles. Nucleic Acids Res.
38:W576–W581. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vita R, Zarebski L, Greenbaum JA, Emami H,
Hoof I, Salimi N, Damle R, Sette A and Peters B: The immune epitope
database 2.0. Nucleic Acids Res. 38:(Database). D854–D862. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Parker KC, Bednarek MA and Coligan JE:
Scheme for ranking potential HLA-A2 binding peptides based on
independent binding of individual peptide side-chains. J Immunol.
152:163–175. 1994.PubMed/NCBI
|
|
27
|
Rammensee H, Bachmann J, Emmerich NP,
Bachor OA and Stevanović S: SYFPEITHI: Database for MHC ligands and
peptide motifs. Immunogenetics. 50:213–219. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gasteiger E, Gattiker A, Hoogland C,
Ivanyi I, Appel RD and Bairoch A: ExPASy: The proteomics server for
in-depth protein knowledge and analysisNucleic. Acids Res.
31:3784–3788. 2003. View Article : Google Scholar
|
|
29
|
Li YJ, Wang J, Zhao H, Jia HY, Li B, Ma
XM, Wen H and Ding JB: Bioinformatics prediction on Eg95 antigen
epitopes of Echinococcus granulosus. Chin J Zoonoses. 27:892–895.
2011.
|
|
30
|
Li Z, Zhang M, Hu H, Liu S and Lu Z: On
predicting the T cell and B cell epitopes of platelet membrane
glycoprotein II b/III a antibody from human and mice]. Sheng Wu Yi
Xue Gong Cheng Xue Za Zhi. 27:1146–1151. 2010.(In Chinese).
PubMed/NCBI
|
|
31
|
Shen ZG, Yan P, He W, Chen Z, He H, Zhang
J, Yang X, Wu Y, Liang Z and Li J: Prediction of the secondary
structure and the B cell epitope of the extracellular domin of
FSHR. J Chongqing Med Univ. 35:1317–1320. 2010.
|
|
32
|
Yan C, Wang R, Li J, Deng Y, Wu D, Zhang
H, Zhang H, Wang L, Zhang C, Sun H, et al: HLA-A gene polymorphism
defined by high-resolution sequence-based typing in 161 Northern
Chinese Han people. Genomics Proteomics Bioinformatics. 1:304–309.
2003. View Article : Google Scholar : PubMed/NCBI
|