Introduction
Promyelocytic leukemia protein (PML; also known as
TRIM19) belongs to the family of tripartite motif (TRIM) proteins
(1). The PML gene, which was
found in the majority of acute promyelocytic leukemia, was first
noted at the breakpoint of the t(15;17) chromosomal translocation
(2). PML is mainly expressed in the
nucleus, where it forms dynamic structures known as PML nuclear
bodies (PML-NBs). PML-NBs recruit many other proteins. A plethora
of proteins have been shown to be recruited by PML within PML-NBs,
either permanently, such as the death domain-associated protein
(Daxx), SP100, and SUMO, or transiently, such as the p53 or CAMP
response element-binding protein. PML-NBs are involved in many cell
processes, including cell cycle progression, DNA damage response,
transcriptional regulation, and apoptosis (3,4). PML is
reported to retain functionally critical oncogenic properties and
to play a key role in leukemogenesis (5). It mediates several complex downstream
signaling pathways. The determinant function of PML in
tumorigenesis and cancer progression has kindled research interest
in determining its involvement in many types of cancer.
Human PML can regulate alternative splicing through
a variety of transcripts. These isoforms have the same identical
N-terminal region containing the ring, B-box, and coiled-coil motif
but differ in their C-termini (6). In
PML-NBs, PML has proved to be the organizing center, taking
responsibility for recruiting various proteins via mechanisms
involving SUMO modifications and interactions (7). The best-known posttranslational
modification of PML is sumoylation, whereby PML directly binds SUMO
and the SUMO-conjugating enzyme (8).
Sumoylation of PML is necessary for the formation of PML-NBs
because a PML mutant that cannot be modified by SUMO fails to
recruit classical PML-NB components such as SP100, a protein
involved in transcriptional regulation, and DAXX, a transcriptional
repressor (9). PML is an important
factor in the regulation of p53-dependent and p53-independent
apoptotic pathways, and DAXX pro-apoptotic or anti-apoptotic
activity in the PML-NBs may be cell-type specific (10). CCAAT/enhancer-binding protein β is
negatively regulated by the transcriptional co-repressor Daxx
(11). A variety of regulatory factors
is located in PML-NBs, and PML plays an important role in apoptosis
regulation, for example, findings show that cells from
PML-deficient mice reveal severe apoptotic defects (12). However, the role of PML with regard to
proliferation, apoptosis, and DNA damage in ovarian cancer cells
remains to be determined.
Therefore, we investigated the effect of PML on
growth, clone formation and DNA damage in ovarian cancer cells.
Materials and methods
Reagents and cell culture
The ovarian cell line OV2008 was purchased from the
American Type Culture Collection (CRL-1740; Rockville, MD, USA). At
37°C in a humidified 5% CO2 incubator, the OV2008cells
were cultured in DMEM medium (Invitrogen Life Technologies,
Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Hyclone,
Logan, UT, USA) and 1% penicillin/streptomycin solution (Invitrogen
Life Technologies). Cells seeded at the same density and cultured
at the same time in complete medium were used as the control.
Plasmids and RNA interference
For the construction and identification of the
pSRG-short hairpin RNA (shRNA) expression vector, sense and
antisense strands of shRNA were produced. The 100 µl reaction mix
contained nuclease-free sterile water, 20 µl annealing buffer
(Biyuntian, Shanghai, China), and 20 µl each of 50 µM shRNA sense
and antisense strands. The annealing conditions were: 95°C for 2
min, followed by a decreasing temperature gradient of 1°C per 90
sec until 25°C. Annealing products were stored at −20°C. The pSRG
empty vector (1 µg) was treated with the enzymes SalI and
BglII separately at 37°C for 3 h. The digestion products
were electrophoresed, and the shRNA was incubated with the
digestion products The products were transformed by DH5α competent
cells at 16°C overnight, coated on LB plates containing Amp
resistance, and cultured overnight at 37°C for 8–10 h to generate
monoclonal colonies. The bacterial liquid was then collected and
subjected to rapid plasmid miniprep DNA extraction. The plasmids
were digested with XhoI and EcoRI, and the digested
products were sent to Beijing Dingguo Biotechnology (Beijing,
China) for sequencing. Subsequently, the cells were transfected
with these plasmids.
PML-knockdown ovarian cancer cell
lines
Cells were transfected with 2 µg of pSRG-shCon or
pSRG-shPml plasmids using 200 µl solution A (Opti-MEM) in each
60-mm dish. This was followed by incubation at room temperature for
5 min. Solution B (6 µl Lipofectamine® 2000 and 200 µl
Opti-MEM) was added and incubated at room temperature for 5 min.
Solutions A and B were then mixed and incubated at room temperature
for 30 min. The mixture was added to the 6-well plates 6 h after
transfection, and the cell culture medium was replaced with normal
medium. The cells were cultured with puromycin (1 µg/ml) for 7–10
days and were selected by EGFP fluorescence.
RT-qPCR
Total RNA was extracted from PML-knockdown ovarian
cancer cells using TRIzol reagent (Invitrogen Life Technologies).
cDNA was produced as per the manufacturer's protocols (Bio-Rad,
Hercules, CA, USA), and the total RNA was reverse transcribed. The
primers used were: β-actin: 5′-GCTCTTTTCCAGCCTTCCTT-3′ and
5′-GTACTTGCGCTCAGGAGGAG-3′; PML: 5′-GCTGACCCCCAAGCAGAAGA-3′ and
5′-CTCAGAAAGCTGAGGAAGTGCTG-3′. RT-qPCR conditions used were: 50°C
for 5 min; denaturation at 95°C for 5 min; and 30 cycles of 95°C
for 30 sec, 60°C for 32 sec, and 72°C for 40 sec.
Cell growth assay
OV2008 cells (pSRG-shCon and pSRG-shPml) were grown
in 96-well plates for 24 h, and the number of cells in each well
was 3×103. The proliferative activity was tested by MTT
assay, followed by 4 h incubation of 20 µl MTT solution (5 mg/ml in
PBS). Absorbance was measured at 490 nm. The experiment was
repeated three times.
Clone formation experiment
After 24-h incubation, cells with a stable
expression of pSRG-shCon and pSRG-shPml were collected by
trypsinization to form monoplast suspension. The cells were
counted, plated in 6-cm culture dishes at 1×103 cells,
and cultured for 10–15 days at 37°C and 5% CO2. The
cells were stained with Coomassie brilliant blue, followed by three
rinses in tap water and drying at room temperature. The number of
clones (cell number >50 cells/colony) was counted, each with 4–5
replicates. The experiment was repeated three times.
Immunofluorescence
Cultured cells were seeded in 24-well plates with a
glass bottom and continued to culture for 24 h. The cells were
washed with PBS and then fixed with 4% paraformaldehyde. The
immobilized cells were then washed with PBS and permeabilized with
0.5% Triton X-100 (PBST). PBST containing 5% bovine serum albumin
was used as block buffer. After 1 h at room temperature, primary
antibodies (PML, Santa Cruz Biotechnology, Santa Cruz, CA, USA;
p-H2AX, Cell Signaling Technologies, Danvers, MA, USA; cleaved
caspase-3, Cell Signaling Technology) and Alexa Fluor 594- or
488-conjugated secondary antibodies (Molecular Probes; Jackson
Immunoresearch Laboratories, Inc., West Grove, PA, USA) were used.
The nuclei was observed by DAPI staining (DAPI; Vector
Laboratories, Inc., Burlingame, CA, USA). Images were observed
through a fluorescence microscope (Nikon Eclipse 80i; Nikon
Corporation, Tokyo, Japan).
Western blot analysis
The proteins separated by SDS-PAGE were transferred
to PVDF membranes (Millipore Corp., Bedford, MA, USA) and the
membranes were incubated with blocking liquid (5% non-fat dry milk)
for 1 h, and then probed with primary antibodies for 1 h at room
temperature. After washing with PBST, the membranes were incubated
with secondary antibodies labelled with horseradish peroxidase
(Cell Signaling Technologies) for 1 h at room temperature, and the
membranes washed with PBST. After hybridization reaction, ECL
Western Blot chemiluminescence reagent kit (Amersham Biosciences,
Uppsala, Sweden) was applied for developing. The primary antibodies
that were used included: PML (dilution, 1:1,000; mouse no.
sc-966;Santa Cruz Biotechnology), β-Actin (dilution, 1:1,000;
Rabbit.no. 4970S), p-H2AX (dilution, 1:1,000; Rabbit.no. 9718),
p-CHK1 (dilution, 1:1,000; Rabbit no. 2348), cleaved caspase-3
(dilution, 1:1,000; Rabbit. no. 9661) (Cell Signaling
Technologies).
Immunohistochemistry
Ovarian tumor tissues were provided by the Second
Hospital of Jiaxing and the Jiaxing Maternity and Child Health Care
Hospital (Jiaxing China). Paraffin sections were deparaffinized by
xylene (three times) and ethanol (three times), for 3 min each
time, and the sections were rehydrated. After the sections were
incubated in 0.3% H2O2, antigen retrieval was
carried out by 0.02 M sodium citrate at 95°C for 15 min. Ovarian
tumor tissues were incubated with blocking liquid and probed with
PML (dilution, 1:1,000; mouse. no. sc-966;Santa Cruz
Biotechnology), and then washed with PBST. The membranes were
incubated with secondary antibodies for 30 min at room temperature.
The ovarian tumor tissues were then washed with PBST and incubated
with ABC solution for 30 min at room temperature. The tissues were
again washed with PBST and developed using 3,3′-diaminobenzidine
(DAB). After being dehydrated, the tissues were mounted with
neutral resins.
Statistical analysis
The results were repeated three times. The analysis
of variance and t-test were applied in comparing the intergroup
difference of measurement data. GraphPad Prism statistical programs
(GraphPad Prism, San Diego, CA, USA) was used. P<0.05 was
considered to indicate a statistically significant difference.
Results
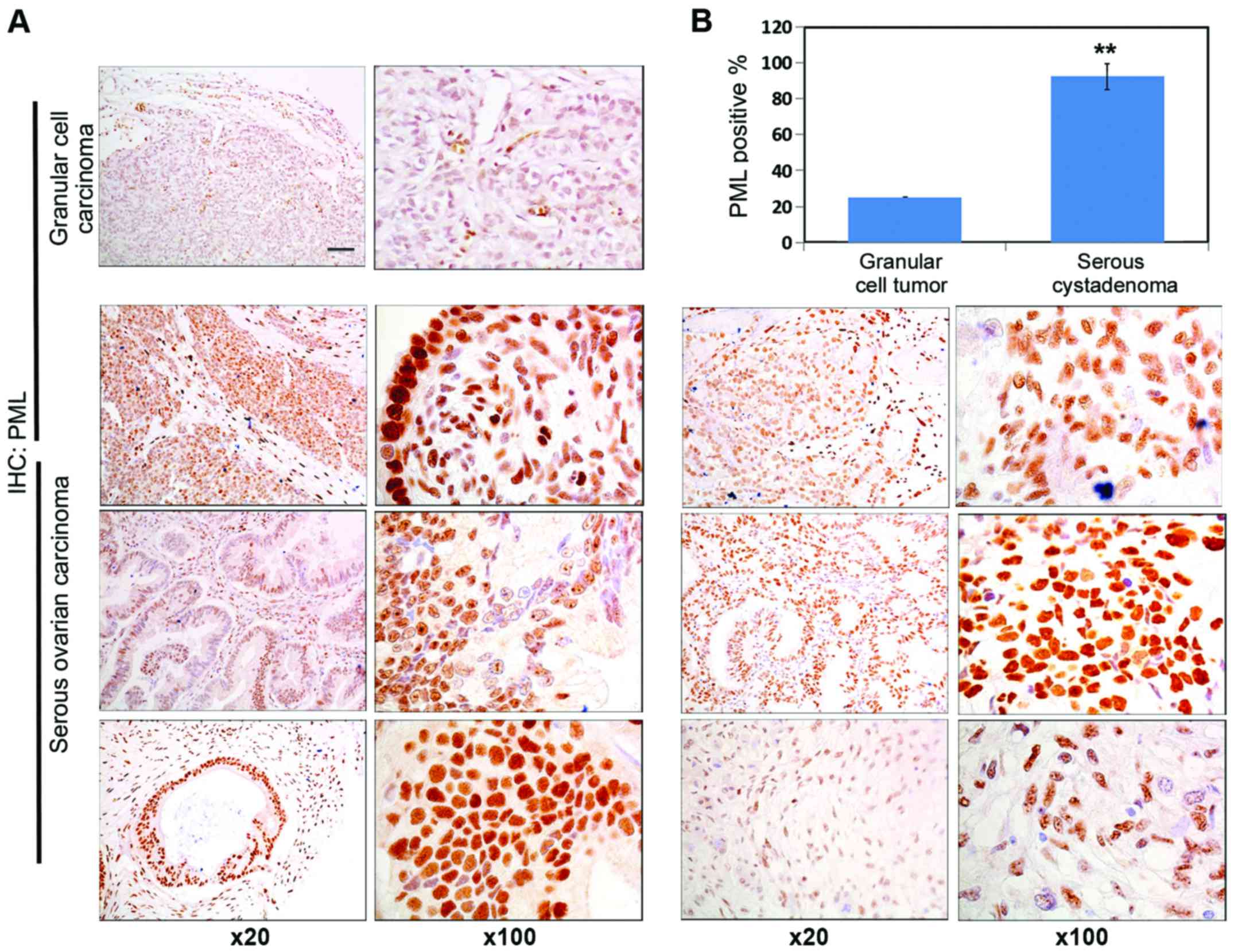
PML is highly expressed in human
ovarian cancer tissues
PML protein expression in human granular cell
carcinoma tissue and human ovarian carcinoma tissue was detected by
immunohistochemistry. As shown in Fig.
1A, significant PML staining intensity was detected in ovarian
cancer tissue. The PML expression level was significantly increased
in human ovarian carcinoma tissue, and the number of positive cells
in human ovarian carcinoma tissue was significantly higher than
that of granular cell carcinoma tissue (Fig. 1B).
Establishment and detection of stable
cell lines with PML silencing
We used EcoRI and XhoI to construct
and identify double-digested vectors. We successfully constructed
pSRG-shPml and the control vector pSRG-shCon (Fig. 2A). To detect the interference
efficiency of shRNA, we transfected the vectors pSRG-shCon and
pSRG-shPml into OV2008 cells. Total cell RNA was extracted after 24
h. . RT-qPCR was used to detect the RNA interference efficiency.
The shPml could markedly inhibit the expression of PML RNA in the
cells (Fig. 2B). We also validated
this result by immunofluorescence(Fig.
2C) and quantifcation of PML positive cells (Fig. 2D).
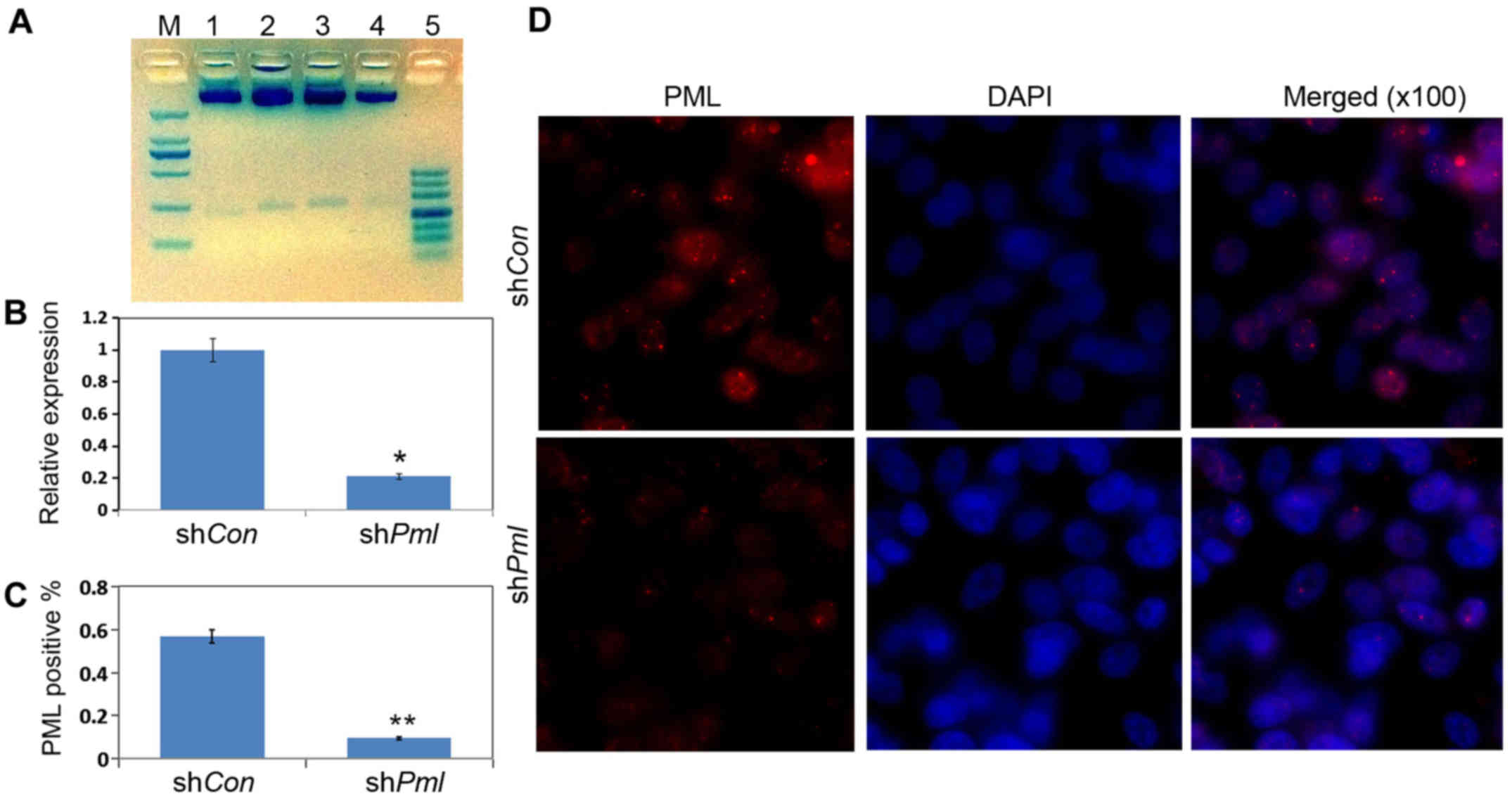 | Figure 2.RT-qPCR and immunofluorescence results
for the effects of promyelocytic leukemia (PML) silencing by
RNA interference. (A) Restriction enzyme digestion analyses of the
plasmids. Different plasmids were extracted and separated by 2.5%
agarose gel. M, 2,000 bp DNA marker; 1, pSRG/XhoI,
EcoRI; 2, pSRG-shCon/XhoI, EcoRI; 3–4,
pSRG-shPml/XhoI, EcoRI; 5, 500 bp DNA marker.
(B) OV2008 cells were transfected with pSRG-shCon and
pSRG-shPml; total mRNAs were isolated 24 h after
transfection. RT-qPCR for Pml mRNA expression levels in
cultured cells. (C) Quantification of PML-positive cells. (D)
Immunofluorescence analysis for PML protein expression levels. Red,
PML; blue, DAPI (magnification, ×100). |
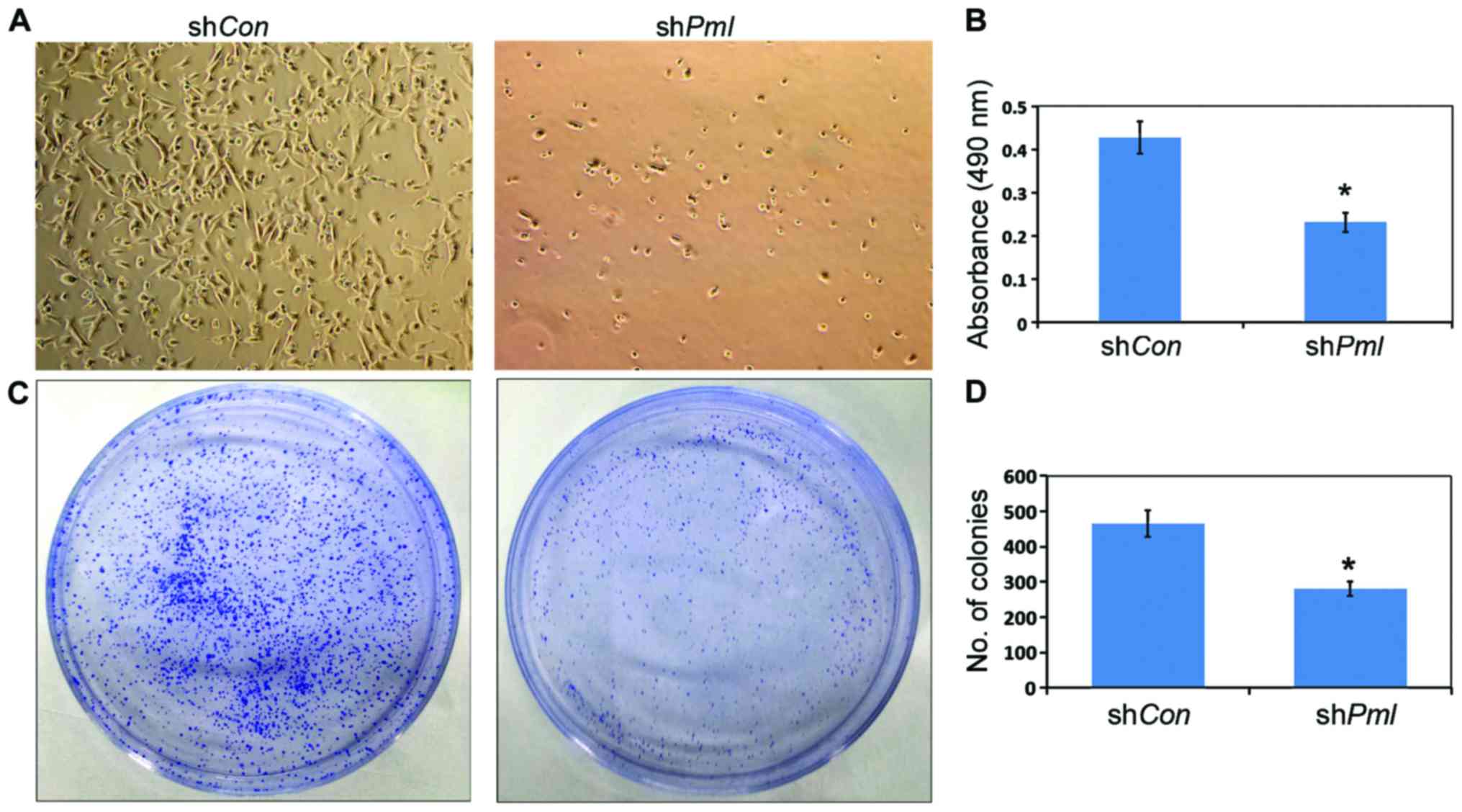
PML silencing induced inhibition of
the proliferation and colony formation of ovarian cancer cells
After PML silencing, the number of viable OV2008
cells decreased (Fig. 3A), and MTT
assay showed the cell proliferation ability decreased by 50%
(Fig. 3B). At the same time, the
ability of ovarian cancer cells to form monoclonal colonies
decreased (Fig. 3C and D).
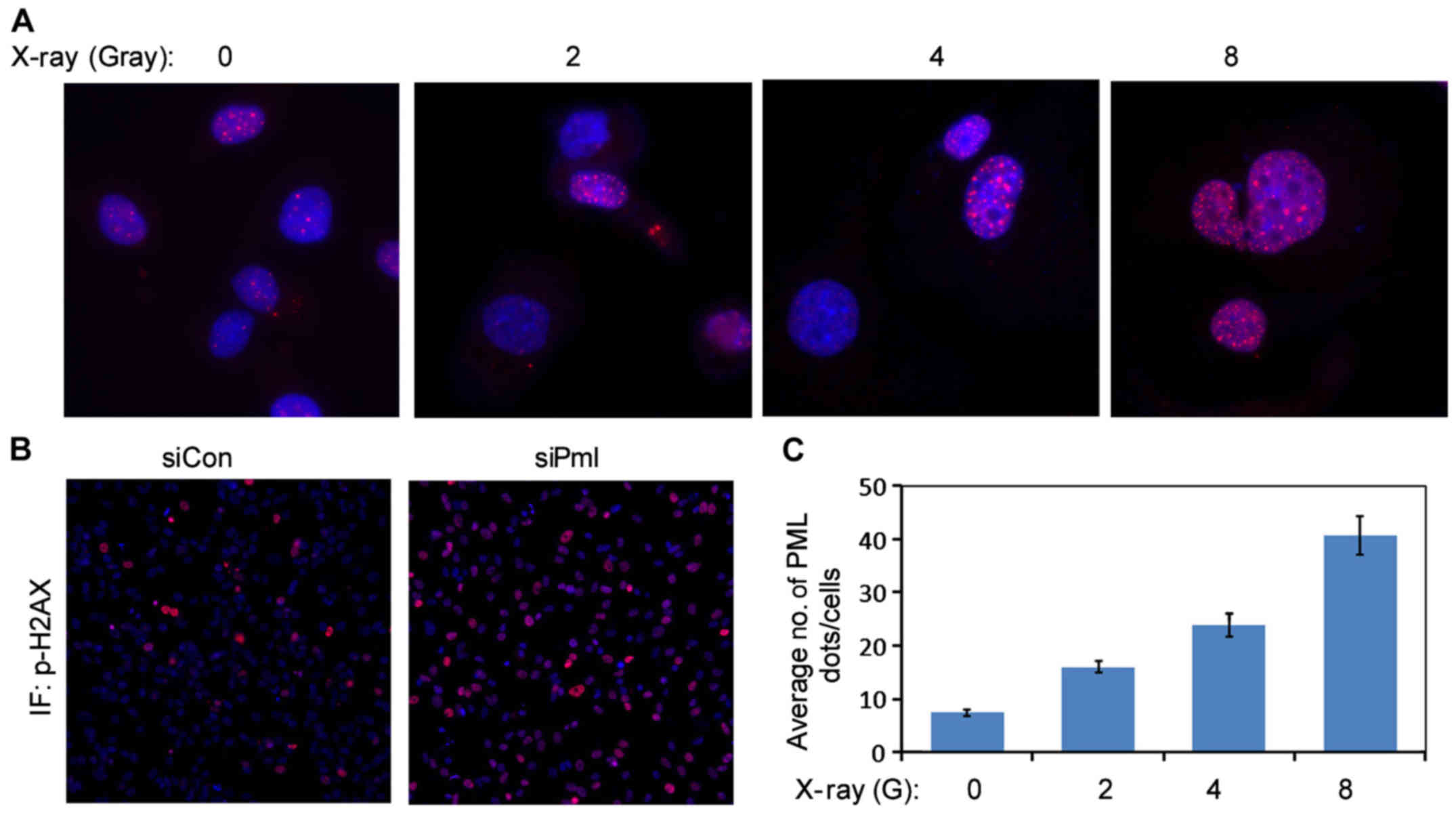
PML silencing promoted DNA damage in
ovarian cancer cells
X-rays can induce PML to form foci in the nuclei of
ovarian cancer cells (13). Using the
immunofluorescence assay, PML was found to accumulate in the
nucleus in response to X-ray irradiation. With the increase in the
irradiation dose, the number of foci increased gradually (Fig. 4A and C). We also found that the
expression of p-H2AX, a DNA damage protein, increased after
PML silencing (Figs. 4B and
5B). These results suggested that the
subcellular localization of PML protein can be affected by
radiotherapy, and decreased PML protein expression can induce DNA
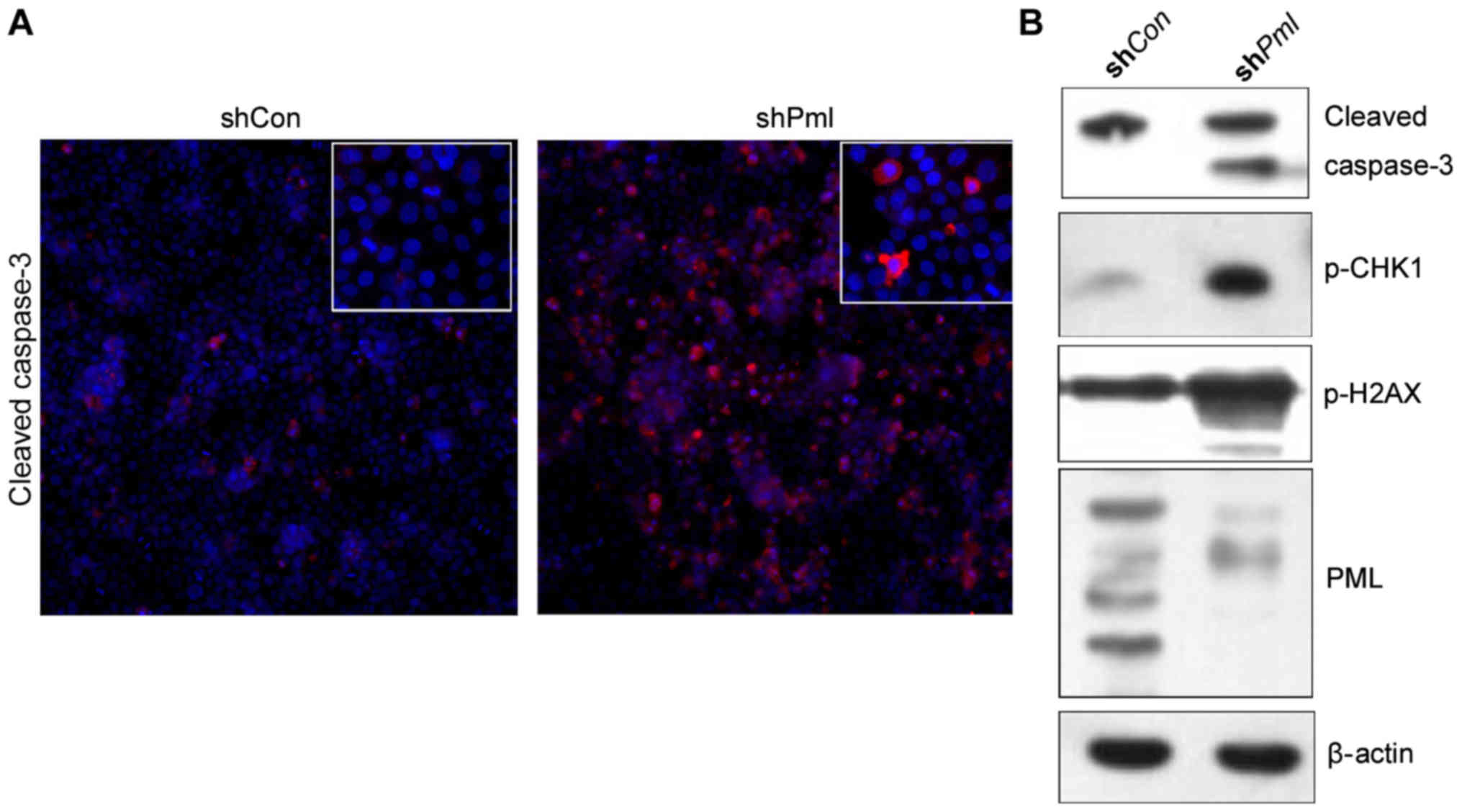
damage in ovarian cancer cells. PML silencing promoted
apoptosis of ovarian cancer cells. The expression of the apoptosis
protein cleaved caspase-3 increased over 24 h, peaking 48 h after
PML silencing (Fig. 5A and
B).
Discussion
PML is an important part of the PML-NB structure and
can recruit more than 30 different proteins, including Daxx, ATRX,
and small ubiquitin-like molecules such as SUMO to the PML-NB
region (14,15). PML acts as a tumor suppressor and is
involved in proliferation, apoptosis, senescence, DNA damage and
angiogenesis in tumors (16). Numerous
studies have demonstrated PML involvement in the regulation of
apoptosis (17). Studies show cells
from PML−/− mice showed a greater resistance to
apoptosis activated by either intrinsic or extrinsic apoptotic
pathways compared with wild type, such as thymocytes and embryonic
fibroblasts (18,19). By regulating acetylation of p53, PML
played a critical role in the cellular senescence of
V-H-Ras-induction. As T-box transcription factor, TBX2 is an E2F
target and regulates PML-induced senescence (20,21). KLHL20,
a BTB-family protein, targets the tumor suppressor PML and DAPK
(death-associated protein kinase) to its kelch-repeat domain for
ubiquitination and degradation. PML and DAPK stabilization has been
found to be mediated by KLHL39, a negative regulator of Cul3-KLHL20
ubiquitin ligase, in the process of colon cancer metastasis
(22).
Additionally, the p38-MNK1-PML network regulates
TNFα-induced apoptosis in breast cancer cells and TNFα-mediated
inhibition of migration and capillary tube formation in endothelial
cells (23). PML can also activate
fatty acid oxidation (FAO) and promote the self renewal of
hematopoietic stem cells (24). The
PML-FAO pathway can promote survival and proliferation in breast
cancer cells. A high PML expression level is related to the
prognosis of breast cancer. Cell metabolism mediated by PML is
related to asymmetric cell division and multipotency, and it can
promote the survival of cancer stem cells. PML deficiency leads to
an increased number of DNA lesions, which is accompanied by changes
in histone signature. 53BP1, the p53-binding protein, is associated
with DNA damage responses. 53BP1 protein has two mobile fractions
with distinct diffusion in wild-type cells; however, these
fractions are absent in PML-deficient cells. This phenomenon
indicated that PML plays key roles in the local motion of 53BP1 NBs
in response to irradiation (25).
Ovarian cancer is one of the highest mortality rates
that causes more deaths than any other cancer of the female
reproductive system (26). Multi-drug
resistance of ovarian cancer cells towards chemotherapeutic drugs
is the main reason for failure of chemotherapy (27). In order to improve ovarian cancer
chemotherapeutic effects to chemotherapy and survival of women,
further research to explore how to rise the chemotherapeutic
efficacy of ovarian cancer cells is needed. However, the role of
PML in the occurrence and drug resistance of ovarian cancer has not
been explored extensively.
To investigate the effect of the PML on ovarian
cancer cell proliferation, apoptosis and DNA damage, we assessed
the expression of PML protein in human ovarian cancer tissue by
immunohistochemical methods. The high PML levels in ovarian cancer
led us to surmise that PML is involved in the occurrence and drug
resistance of ovarian cancer. We constructed a eukaryotic
expression vector for PML silencing (pSRG-shPml) and transfected
ovarian cancer cells using the vector. Through the establishment of
stable cell lines with PML silencing, we found PML to be involved
in ovarian cancer cell proliferation and clone formation by cell
growth assays and clone formation experiments. We induced DNA
damage in ovarian cancer cells by irradiation and found that the
number of PML-NBs in the nuclei were elevated gradually with
increases in the radiation dose, suggesting that PML-NB nuclei are
highly sensitive to DNA damage.
However, the underlying mechanism of this phenomenon
requires further research. A decreased expression of PML can
promote apoptosis, which preliminarily showed that PML plays an
important role in the process of ovarian cancer cell apoptosis.
With the loss of PML expression, DNA damage in ovarian cancer cells
increased, and CHK1 and H2AX phosphorylation were significantly
elevated. This suggests that PMLNBs are also involved in the repair
of DNA damage in ovarian cancer cells. However, the exact mechanism
underlying the DNA damage occurring in ovarian cancer remains to be
ascertained. Our present results suggest the applicability of PML
as an early diagnostic marker for ovarian cancers. Targeting the
PML degradation pathway may therefore be a promising approach for
anticancer therapy.
Acknowledgements
The present study was supported by the Experimental
Animal Science and Technology program of Zhejiang Province Grants
(grant no. 2017C37114), Science and Technology Bureau Project of
Jiaxing (grant no. 2015AY23063), and a grant from the 12th
Five-year Plan for University Key Academic Subject (Pharmacology),
Zhejiang, China.
References
|
1
|
Dutrieux J, Maarifi G, Portilho DM, Arhel
NJ, Chelbi-Alix MK and Nisole S: PML/TRIM19-dependent inhibition of
retroviral reverse-transcription by Daxx. PLoS Pathog.
11:e10052802015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fagioli M, Alcalay M, Pandolfi PP,
Venturini L, Mencarelli A, Simeone A, Acampora D, Grignani F and
Pelicci PG: Alternative splicing of PML transcripts predicts
coexpression of several carboxy-terminally different protein
isoforms. Oncogene. 7:1083–1091. 1992.PubMed/NCBI
|
|
3
|
Sahin U, de Thé H and
Lallemand-Breitenbach V: PML nuclear bodies: assembly and oxidative
stress-sensitive sumoylation. Nucleus. 5:499–507. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bernardi R, Papa A and Pandolfi PP:
Regulation of apoptosis by PML and the PML-NBs. Oncogene.
27:6299–6312. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Collins SJ: Retinoic acid receptors,
hematopoiesis and leukemogenesis. Curr Opin Hematol. 15:346–351.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jensen K, Shiels C and Freemont PS: PML
protein isoforms and the RBCC/TRIM motif. Oncogene. 20:7223–7233.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shen TH, Lin HK, Scaglioni PP, Yung TM and
Pandolfi PP: The mechanisms of PML-nuclear body formation. Mol
Cell. 24:331–339. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Duprez E, Saurin AJ, Desterro JM,
Lallemand-Breitenbach V, Howe K, Boddy MN, Solomon E, de Thé H, Hay
RT and Freemont PS: SUMO-1 modification of the acute promyelocytic
leukaemia protein PML: Implications for nuclear localisation. J
Cell Sci. 112:381–393. 1999.PubMed/NCBI
|
|
9
|
Zhong S, Müller S, Ronchetti S, Freemont
PS, Dejean A and Pandolfi PP: Role of SUMO-1-modified PML in
nuclear body formation. Blood. 95:2748–2752. 2000.PubMed/NCBI
|
|
10
|
Bernardi R and Pandolfi PP: Structure,
dynamics and functions of promyelocytic leukaemia nuclear bodies.
Nat Rev Mol Cell Biol. 8:1006–1016. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wethkamp N and Klempnauer KH: Daxx is a
transcriptional repressor of CCAAT/enhancer-binding protein β. J
Biol Chem. 284:28783–28794. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hofmann TG and Will H: Body language: The
function of PML nuclear bodies in apoptosis regulation. Cell Death
Differ. 10:1290–1299. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pan WW, Zhou JJ, Liu XM, Xu Y, Guo LJ, Yu
C, Shi QH and Fan HY: Death domain-associated protein DAXX promotes
ovarian cancer development and chemoresistance. J Biol Chem.
288:13620–13630. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Salomoni P, Dvorkina M and Michod D: Role
of the promyelocytic leukaemia protein in cell death regulation.
Cell Death Dis. 3:e2472012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van Damme E, Laukens K, Dang TH and Van
Ostade X: A manually curated network of the PML nuclear body
interactome reveals an important role for PML-NBs in SUMOylation
dynamics. Int J Biol Sci. 6:51–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen RH, Lee YR and Yuan WC: The role of
PML ubiquitination in human malignancies. J Biomed Sci. 19:812012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen RH, Lee YR and Yuan WC: The role of
PML ubiquitination in human malignancies. J Biomed Sci. 19:812012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo A, Salomoni P, Luo J, Shih A, Zhong S,
Gu W and Pandolfi PP: The function of PML in p53-dependent
apoptosis. Nat Cell Biol. 2:730–736. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang ZG, Ruggero D, Ronchetti S, Zhong S,
Gaboli M, Rivi R and Pandolfi PP: PML is essential for multiple
apoptotic pathways. Nat Genet. 20:266–272. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pearson M, Carbone R, Sebastiani C, Cioce
M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S,
Pandolfi PP, et al: PML regulates p53 acetylation and premature
senescence induced by oncogenic Ras. Nature. 406:207–210. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martin N, Benhamed M, Nacerddine K,
Demarque MD, van Lohuizen M, Dejean A and Bischof O: Physical and
functional interaction between PML and TBX2 in the establishment of
cellular senescence. EMBO J. 31:95–109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen HY, Hu JY, Chen TH, Lin YC, Liu X,
Lin MY, Lang YD, Yen Y and Chen RH: KLHL39 suppresses colon cancer
metastasis by blocking KLHL20-mediated PML and DAPK ubiquitination.
Oncogene. 34:5141–5151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hsu KS, Guan BJ, Cheng X, Guan D, Lam M,
Hatzoglou M and Kao HY: Translational control of PML contributes to
TNFα-induced apoptosis of MCF7 breast cancer cells and decreased
angiogenesis in HUVECs. Cell Death Differ. 23:469–483. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ito K, Carracedo A, Weiss D, Arai F, Ala
U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH, et al: A
PML-PPAR-δ pathway for fatty acid oxidation regulates hematopoietic
stem cell maintenance. Nat Med. 18:1350–1358. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Legartová S, Sehnalová P, Malyšková B,
Küntziger T, Collas P, Cmarko D, Raška I, Sorokin DV, Kozubek S and
Bártová E: Localized movement and levels of 53BP1protein are
changed by γ-irradiation in PML deficient cells. J Cell Biochem.
117:2583–2596. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Van Colombo Ν, Gorp T, Parma G, Amant F,
Gatta G, Sessa C and Vergote I: Ovarian cancer. Crit Rev Oncol
Hematol. 60:159–179. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|