Introduction
Over the last 30 years, the development of
monoclonal antibodies (mAbs) for treatment of various types of
disease, such as cancer or chronic inflammatory disease, has
increased markedly (1). Consequently,
using mAbs as specific binders to specific targets for potential
use in the clinic is continuously being improved and evaluated. The
identification and characterization of signaling pathways
responsible for the pathogenesis often guides target selection.
Furthermore, the identification of various cellular receptors, such
as the epidermal growth factor receptor (EGFR), or ligands, such as
the vascular EGF (VEGF), improved the understanding of how proteins
induce cancer growth via different biological signaling pathways.
These findings in turn enabled the construction of proteins capable
of interacting and inhibiting their function, such as the
development of cetuximab, an mAb against EGFR, and bevacizumab, an
mAb against VEGF (2–6).
The development of novel mAbs and conjugation
approaches, for example the conjugation of mAbs with toxins,
radionuclides or nanoparticles, aims to strengthen their
therapeutic potential against cancer cells (7,8). In such a
situation, it is important to understand the behavior of the
binding between the antibody and its target to potentially enhance
the binding behavior. Investigation of the affinity, and the on-
and off-rate provides information regarding the speed at which
molecules bind to a receptor, how likely they are to bind, and how
stably and strongly they bind. This information explains the
differences in cellular responses to facilitate and improve the
development of mAb design (9,10).
Various techniques, such as small-angle X-ray
scattering, electron microscopy, Förster resonance energy transfer
and nuclear magnetic resonance, allow the determination of
molecular structure and are fundamental for providing information
about the protein-protein interactions. However, kinetic and
thermodynamic studies of ligand-receptor interactions are required
for understanding the underlying mechanism of receptor activation.
For this purpose, techniques, such as surface plasmon resonance
enables the measurement of protein-protein interactions in
real-time, where the target is evenly exposed in a pure form to the
ligand (11). However, a receptor or a
protein in a cell has different conformations or is surrounded by
different molecules that interfere with the interaction of the
ligand that, overall, represents the cellular environment. The
measurement of protein interactions on living cells in real-time
allows the continuous detection of the quantity of cell-bound
ligands, resulting in a binding curve. Binding curves make it
possible to estimate the affinity, and the on- and off-rates of the
ligand-receptor interaction in a living system (12,13).
Spiegelberg et al (14),
demonstrated that real-time in vitro assays predict the
behavior of radio-immunotargeting compounds in in vivo
studies. This may lead to a more appropriate selection of potential
compounds for in vivo studies, which in turn may reduce the
development costs of a drug or imaging compounds.
Another aspect to consider in manual end-point and
real-time measurements is that the majority of the ligand-binding
studies are performed only at room temperature, which does not
allow estimation of thermodynamic coefficients, such as enthalpy
and entropy, that are dependent on temperature, and that provide
additional important mechanistic information about the biological
interaction between the ligand and the receptor. In living
material, certain additional challenges occur when binding is
evaluated close to 37°C, as the cell metabolizes and performs
processes, including internalization and degradation of the
antibody (15,16). However, when the measurements in living
materials are conducted at temperatures close to 4°C, viability and
adherence may be an issue.
Unfortunately, the effects of pH and temperature on
binding characteristics are commonly ignored in cell-based assays.
Variations in temperature (18–35°C) or pH (5.5–8.5) has been shown
to affect the measurement of the equilibrium dissociation constant
(KD) by a factor of 2 or 10, respectively (10,17–20), excluding effects due to metabolism.
In the present study, the effect of temperature
changes on the binding ability of the mAbs, cetuximab and
pertuzumab to their specific receptors in living cancer cells, EGFR
and human epidermal growth factor receptor 2 (HER2), respectively
were evaluated. Binding was measured in real-time using a
LigandTracer® instrument where the affinity and kinetics
of the antibody-receptor interactions were derived at three
temperatures. The measurement in real-time at different
temperatures introduces a unique understanding of the importance of
temperature in standard assays, thermodynamics and the true
behavior of ligands in living systems.
Materials and methods
Cell lines
The human SKOV3 ovarian cancer cell line (obtained
from the European Collection of Cell Cultures; Public Health
England, Salisbury, UK) was cultured in RPMI-1640 (cat. no. F1215;
Merck Sharp & Dohme Ltd., Hoddesdon, UK), and the human A431
squamous carcinoma cell line was cultured in Ham's F10 cell culture
medium (cat. no. F0715; Merck Sharp & Dohme Ltd.). The two
types of cell culture media were supplemented with 10% fetal bovine
serum (cat. no. F6765; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany), 2 mM L-glutamine (cat. no. K0283; Merck Sharp & Dohme
Ltd.), 100 IU penicillin and 100 µg/ml streptomycin (cat. no.
A2213; Merck Sharp & Dohme Ltd.). LigandTracer studies were
performed as described by Björkelund et al (21). In brief, cells were seeded on a local
area of a 10 cm petri dish (Nunclon™; cat. no. 150350; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and placed in an
incubator at 37°C, and used within 3 days after seeding.
Antibody labeling
Pertuzumab (purified from Omnitarg™; Genentech,
Inc., South San Francisco, CA, USA) was labeled with Texas
Red® label (Invitrogen; Thermo Fisher Scientific, Inc.),
the targets of which are primary amines, i.e., lysines. For this
purpose, the clinical mAb was labeled as described by Bondza et
al (22). Additionally, cetuximab
(purified from Erbitux®; Merck KGaA) was labeled with
fluorescein isothiocyanate (FITC; cat. no. F3651; Sigma-Aldrich;
Merck KGaA) as described by Stenberg et al (13). The labeled proteins were purified
through a NAP-5 column (GE Healthcare Life Sciences, Little
Chalfont, UK) for the removal of unbound fluorophore.
Analyzing antibody-receptor
interactions in real-time at different temperatures
The cetuximab-EGFR and pertuzumab-human epidermal
growth factor receptor 2 (HER2) interactions in living cells were
measured in real time using LigandTracer Green (Ridgeview
Instruments AB, Vänge, Sweden) at 8, 15, 21 and 37°C. The
LigandTracer technology measures the signal in a specific area of a
Petri dish, the interaction between a target (in this case HER2 and
EGFR expressed on SKOV3 and A431 cells, respectively) and a
specific ligand, which is either radiolabeled or labeled with
fluorescence (in this case cetuximab and pertuzumab were labeled
with the fluorescent dye, FITC and Texas Red®,
respectively). This technology has been used to measure real time
interactions on living cells in various previous studies (23–25). All
measurements in the present study were performed using two
detectors: The yellow (590 nm) to red (632 nm) detector (for the
Texas Red label) and the blue (488 nm) to green (535 nm) detector
(for the FITC label).
For the temperature measurements, the LigandTracer
instrument was placed either in an incubator (37°C) for cell
culture, at room temperature (~21ºC), in a Styrofoam box equipped
with a Peltier cooling element tuned to maintaining the temperature
at ~15°C for 24 h (thermobox), and in a cold room maintained at
~8°C. The instrument was placed in each condition for at least 2 h
before starting the assay to allow the temperature equilibrate. The
assay temperature was continuously monitored in the instrument
during measurements.
The interaction between Texas Red-pertuzumab and
HER2 was characterized in LigandTracer Green, where Texas
Red-pertuzumab was added to a Petri dish with seeded SKOV3 cells
containing RPMI cell culture medium (3 ml). The cells were
incubated with a first concentration of Texas Red-pertuzumab (4 nM)
for 1.5 h, followed by the second addition of Texas Red-pertuzumab
(12 nM) for a further 3 h incubation. Subsequent to the last
addition, RPMI medium was replaced with fresh medium and
measurements were performed to evaluate the dissociation process.
This protocol was performed at all incubation temperatures (8, 15,
21 and 37°C).
An alternative experimental setup was also tested,
where the association and dissociation were detected separately. In
the case of the association phase, an initial measurement was
obtained where the cells were incubated with 4 nM Texas
Red-pertuzumab for the first 3 h of the association phase, followed
by 12 nM Texas Red-pertuzumab for the second 3 h association phase.
The dissociation process was evaluated in a separate measurement by
the saturation of the cell receptor system where first, the
detection of rapid HER2 saturation with Texas Red-pertuzumab (100
nM) was performed during 30 min of measurement, followed by the
replacement of the incubation medium with fresh medium to measure
the dissociation process.
The interaction between FITC-cetuximab and EGFR was
characterized by adding a specific volume of FITC-cetuximab to the
final concentration of 9 nM in A431 cells in a Petri dish, and
containing the incubation medium (3 ml). This approach was used for
all investigated temperature points. For this purpose, the cells
were incubated with FITC-cetuximab (3 nM) for 3 h initially.
Subsequently, the addition of the next portion of FITC-cetuximab
(to a total of 9 nM) was added followed by 3 h incubation at the
different temperatures. When the association phase was complete,
the incubation medium containing the labeled mAb was replaced with
fresh medium (3 ml), allowing detection of the dissociation process
in the same measurement.
Statistical analysis
The shapes of the real-time binding curves produced
by LigandTracer Green were compared using the evaluation software,
TraceDrawer 1.7.1 (Ridgeview Instruments AB). The curves were
normalized to compensate for any differences in signal magnitude
caused by, for example, variations in cell number and labeling
efficiency. Normalization enabled a clear visual presentation of
differences in binding kinetics, observed as variations in
curvature. The signal at the end of the first incubation phase was
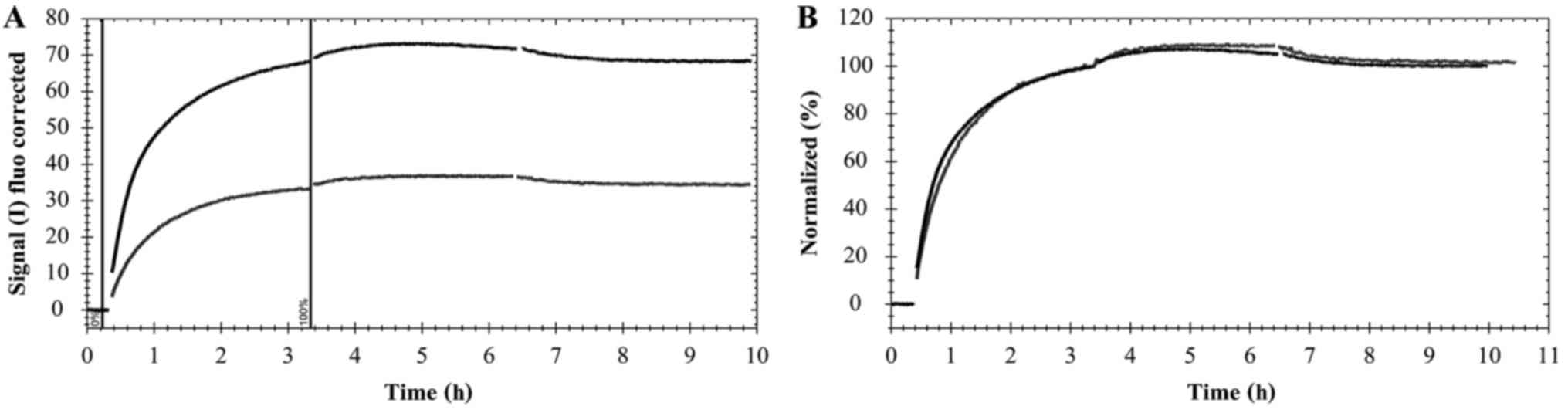
set to 100% and the curves were scaled according to Fig. 1.
Data fitting was conducted essentially, as
previously described by Bondza et al (22) for the pertuzumab-HER2 interaction and
by Barta et al (26) for
cetuximab-EGFR, to obtain information regarding the equilibrium
dissociation constant, KD (corresponding to the
affinity), the association rate constant, ka and the
dissociation rate constant, kd.
Results
Analysis of pertuzumab-HER2
interaction in SKOV3 cells
Attempts to evaluate the interactions (for SKOV3 and
A431) at ~8°C failed approximately every second assay due to cells
detaching from the cell dish shortly after initiating measurement.
Characterization of the interactions between Texas Red-pertuzumab
and HER2 at different temperatures were performed by two-step
incubation with two different mAb concentrations (4 and 12 nM;
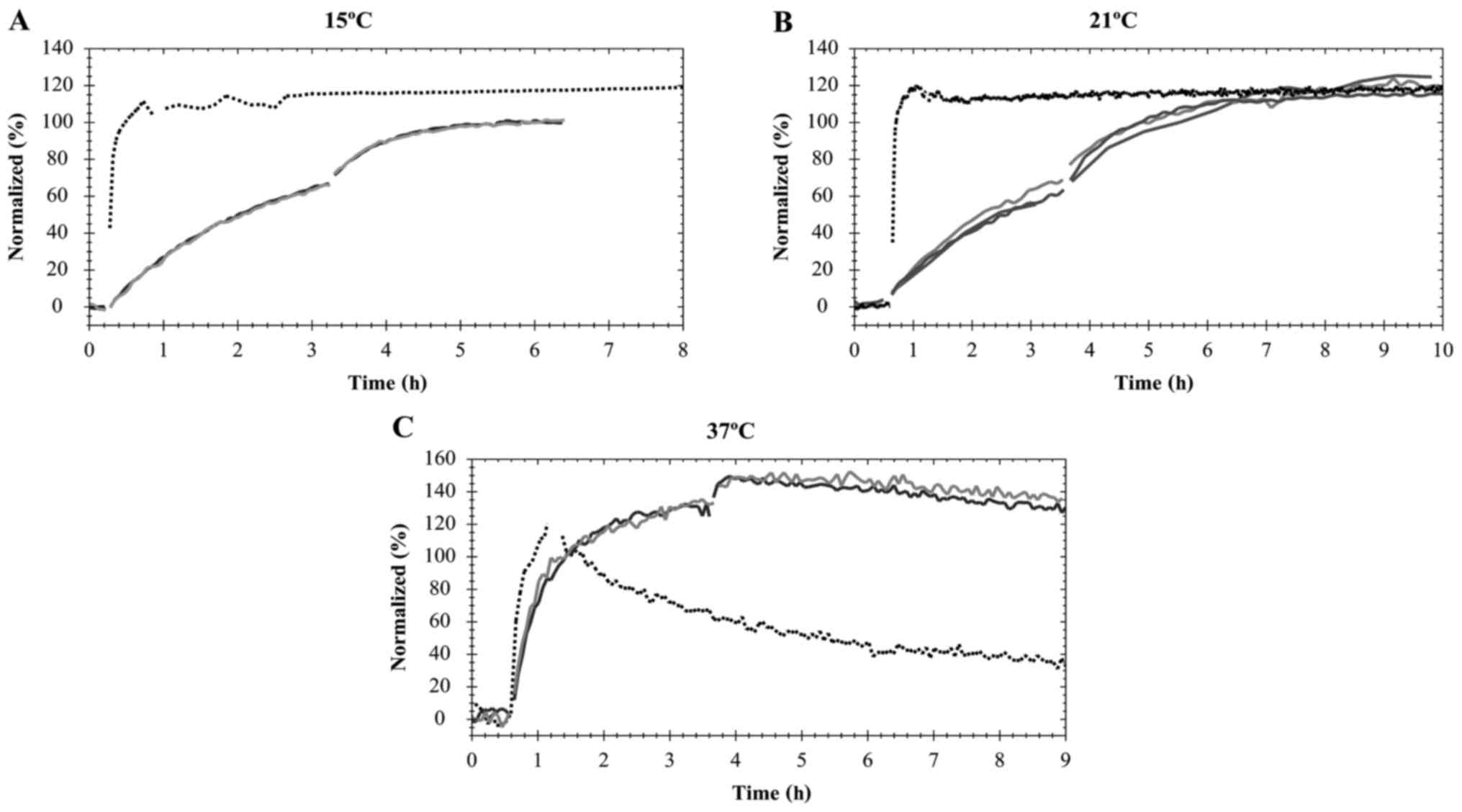
Fig. 2).
The interaction of Texas Red-pertuzumab with HER2
reached equilibrium at different times for different temperatures
(Fig. 2). At 37°C, when the Texas
Red-pertuzumab was added to a final concentration of 4 nM, the
interaction required ~3 h to approach equilibrium (Fig. 2C). However, the interaction was
significantly slower at 15 and 21°C, requiring ~5 h for the
concentration of 12 nM to approach equilibrium (Fig. 2A and B). Following 6 h of incubation
with Texas Red-pertuzumab at 37°C, the signal began to drop
marginally, indicating that Texas Red-pertuzumab began to be
internalized and degraded in the cells (Fig. 2C; time >6 h). After 7–8 h, the
incubation solution was replaced with fresh cell culture medium,
allowing detection of the dissociation process.
Table I summarizes the
kinetic constants and affinities presented with standard error (SE)
obtained at 15°C (n=2), 37°C (n=2) or 21°C (n=3) for the Texas
Red-pertuzumab interaction with HER2 in the SKOV3 cells. As
kd was difficult to determine, KD is not
displayed (15°C). The temperature range demonstrates the minimum
and maximum temperatures registered during the assays (excluding
short term temperature disturbances in conjunction with accessing
the instrument to change concentrations).
 | Table I.Summary of the kinetic constants
(ka and kd) and affinities (KD)
presented with SE obtained at 15ºC (n=2), 37ºC (n=2) or 21ºC (n=3)
for the interaction Texas Red-pertuzumab with human epidermal
growth factor receptor 2 in SKOV3 cells. |
Table I.
Summary of the kinetic constants
(ka and kd) and affinities (KD)
presented with SE obtained at 15ºC (n=2), 37ºC (n=2) or 21ºC (n=3)
for the interaction Texas Red-pertuzumab with human epidermal
growth factor receptor 2 in SKOV3 cells.
| Temperature,
°Ca | ka
(M−1s−1) ± SE | kd
(s−1) ± SE | KD (M) ±
SE |
|---|
| 15 (14.8
−16.2)b |
2.7×104±0.07×104 | – | – |
| 21 (20.5–21.6) |
2.42×104±0.21×104 | ~10−6 |
~10−10 |
| 37 (36.5–37.7) |
1.35×105±0.11×105 |
4.63×105±0.92×10−5 |
3.43×10−10±0.97×10−10 |
As observed in Fig. 2,
it was not possible to investigate association and dissociation
processes in one run. Initial attempts to evaluate the association
and dissociation with just one experiment failed to estimate the
kd value, irrespective of temperature (data not shown).
Therefore, a set of separate experiments was conducted to
investigate the dissociation processes alone. The two curves were
evaluated simultaneously via global fitting to estimate the
association and dissociation rate constants at the different
temperatures, as represented in Table
I. Going from 21 to 37°C resulted in a five-fold increase of
the association rate constant of the pertuzumab-HER2 interaction.
By contrast, no increase in ka was observed when
increasing from 15 to 21°C. In the current study, the kd
for the pertuzumab-HER2 interaction at 15°C was smaller than the
detection limit for LigandTracer (set at ~1×10−6
s−1 for the assay conditions in the present study).
Additionally, at 21°C, the kd could not be calculated
with precision, but it was possible to estimate that it was
<3×10−6 s−1 and ~10−6. Hence,
the affinity value could not be accurately derived, but is
estimated to be 10−10 M.
Analysis of cetuximab-EGFR interaction
in A431 cells
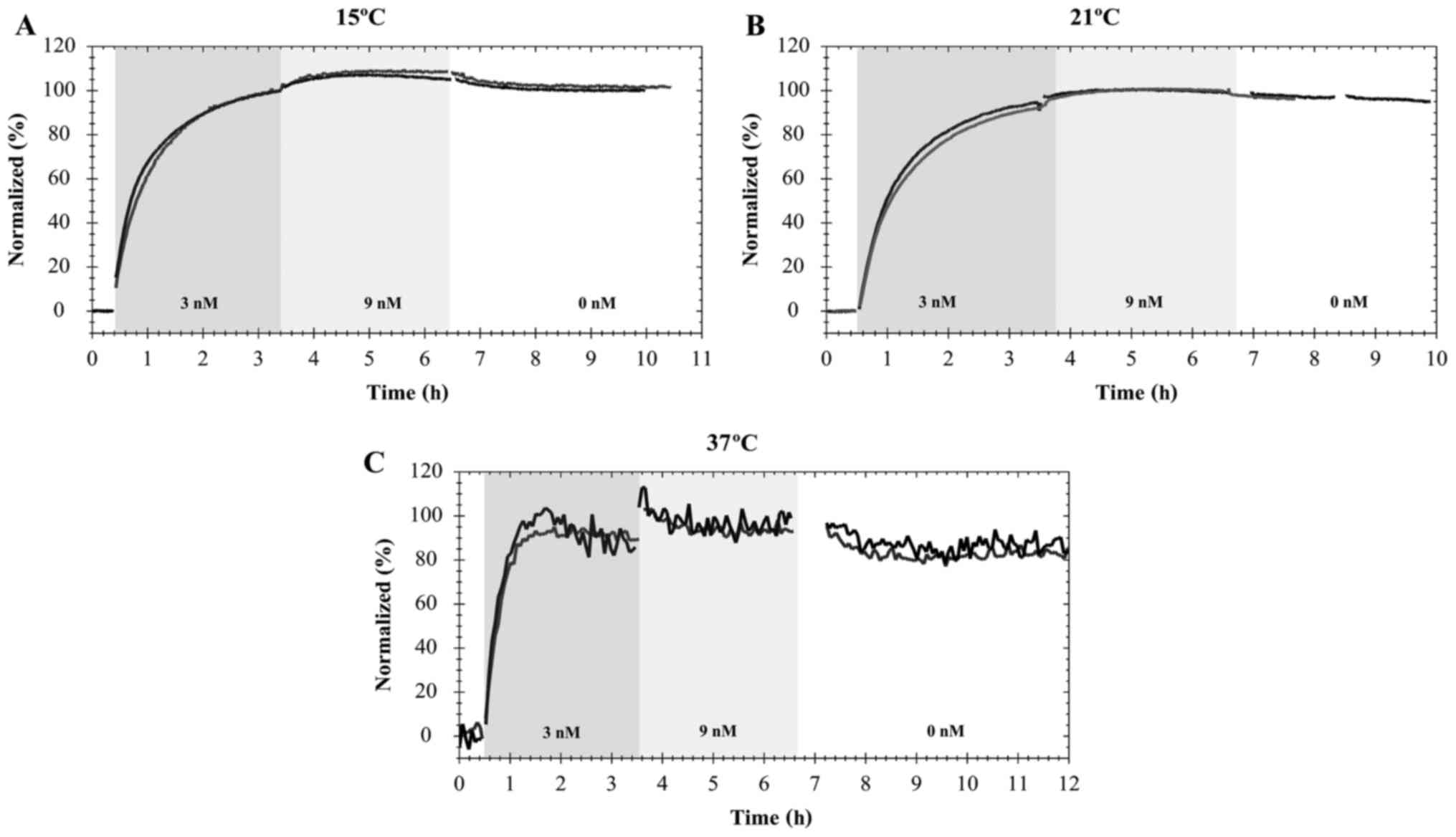
Furthermore, it was possible to observe that the
interaction of FITC-cetuximab with EGFR in A431 cells was similar
to that of pertuzumab-HER2. Namely, temperature influences the time
that the interaction requires to approach equilibrium (Fig. 3). When A431 cells were exposed to the
first concentration of FITC-cetuximab (3 nM), equilibrium was
approached within 1 h at 37°C (Fig.
3C). This was not observed at 21 or 15°C, when the interaction
required ~5 h of incubation at 9 nM to approach equilibrium
(Fig. 3A and B). After 7–8 h, the
incubation solution was replaced with fresh cell culture medium,
allowing detection of the dissociation process. The estimations of
the association and dissociation rate constants as the affinity at
different temperatures are presented in the Table II.
 | Table II.Summary of the kinetic constants
(ka and kd) and affinities (KD),
presented with SE for the interaction fluorescein
isothiocyanate-cetuximab with epidermal growth factor receptor in
A431 cells at different temperatures (n=2). |
Table II.
Summary of the kinetic constants
(ka and kd) and affinities (KD),
presented with SE for the interaction fluorescein
isothiocyanate-cetuximab with epidermal growth factor receptor in
A431 cells at different temperatures (n=2).
| Temperature,
°Ca | ka
(M−1s−1) ± SE | kd
(s−1) ± SE | KD (M) ±
SE |
|---|
| 15 (14.8–16.4) |
1.37×105±0.01×105 |
7.01×10−6±0.08×10−6 |
5.12×10−11±1.16×10−11 |
| 21 (20.2–21.3) |
1.16×105±0.03×105 |
9.79×10−6±0.05×10−6 |
8.44×10−11±0.27×10−11 |
| 37 (36.9–37.8) |
3.33×105±0.09×105 |
7.9×10−6±2.70×10−6 |
2.37×10−11±0.87×10−11 |
Discussion
In the present study, the variation in binding
behavior of clinical mAbs interacting with targets expressed on
living cells under the influence of temperature was investigated.
In traditional cell-based assays, the affinity measurements of
protein interactions are typically performed at room temperature or
at 4–8°C, with little or no reflection of the potential impact of
the temperature. It is well known that higher temperatures supply
more energy, potentially increasing the reaction rate, which often
leads to an accelerated interaction between ligands and targets.
Therefore, temperature variation should always be accounted for to
better predict the interaction behavior. The present study aimed to
estimate an approximate level of influence of temperature variation
on ligand-receptor interaction in living cells.
It is important to understand the possible influence
of temperature on protein-protein interactions in living cells, to
approach a real-life scenario, where the living cell system
provides crucial information regarding the behavior of the
therapeutic antibody. However, numerous challenges were identified
when working with adherent living cells at different temperatures,
such as viability, metabolism and cell adherence. At temperatures
of ~8°C, the cells began to lose their attachment properties and,
eventually, their viability (data not shown). At 37°C, the cells
were active and viable, with normal metabolism, potentially leading
to the internalization and, subsequently, degradation of the
antibody. On the technical side, the temperature and temperature
variations influence the signal detected. The LigandTracer
instrument was placed in different temperature equilibrated
conditions and considerable time was required to stabilize the
signal (to reduce drift) and reach the target temperature (for
example ~5 h at 37°C). Therefore, it is important to monitor the
temperature of the device when performing the experiment to
guarantee improved data. To avoid possible error, the instrument
was placed in temperature equilibrated conditions overnight prior
the measurements inside of the incubator at 37°C. A temperature
change of 1–2°C in the instrument may destabilize the signal for a
short period, making it essential to rapidly execute the manual
steps of the experiment (such as the addition of ligand during
which the incubator or thermobox are opened). Additionally, but not
equally critical, condensation is formed on the optical detector
surface leading to noise increase, which alters the aspect of the
curve and increases the number of spikes in the final signal.
However, an increase of noise to some extent does not necessarily
disturb the measurement. Thus, taking into account the biological
limitations, it is possible to measure ligand-receptor interactions
at different temperatures in real-time, which provides a foundation
for further investigation of thermodynamics on the interaction of
mAbs in living cells.
In the current study, it was possible to observe in
living cells that temperature influenced the time of incubation
required to approach equilibrium. When cells were incubated at
37°C, the interaction approached equilibrium faster than at either
15 or 21°C. In contrast to manual assays, which are blind with the
exception of the single measurement per well conducted following a
wash, measuring in real-time reduces blind-points and enables more
precise elucidation of the duration of incubation required to
approach equilibrium, which may have been missed using end-point
assays. Furthermore, the increase of the association rate with
temperature increase, from 15 to 37°C was observed during the
antibody interactions that were investigated. The differences in
the ka at 37°C were three- to five-fold higher than at
either 21 or 15°C. However, these differences were not as apparent
from 15 to 21°C. The differences between 37 and 21°C are explained
by the temperature increase and the fact that when cells are
incubated with optimal temperature for normal metabolic activity,
they are able to promote the interaction between the antibody and
the receptors. Conversely, it was not possible to precisely
evaluate the off-rate kd for pertuzumab, as the
dissociation process was so slow that obtaining consistent data was
not possible using a standard real-time experiment. This
demonstrates that the regular assay design was not good enough for
characterizing the pertuzumab-HER2 interaction.
A modified assay design was proposed and implemented
for pertuzumab. The standard experiment was replaced with two
separate experiments. Initially, the association process was
evaluated by incubating cells with two different concentrations
that were selected to obtain a clear estimation on curvature.
Subsequently, in a second experiment, the HER2 system was saturated
with an abundance of Texas Red-pertuzumab aiming to achieve greater
sensitivity of the signal and a longer measurement duration of the
dissociation process. It was possible to evaluate the two sets of
experiments in parallel using TraceDrawer software, where certain
variables were estimated independently (such as Bmax, associated
with cell number and, thus, differs between measurements) and
certain variables were estimated globally (such as the interaction
parameters ka, kd and KD). The
modified assay design enabled the estimation of the affinity and
kinetics in situations where the regular assay design was
insufficient for evaluating the interaction with precision.
However, accurate calculation of the dissociation
rate of Texas Red-pertuzumab for 15°C and 21°C was not performed,
as pertuzumab in conjugation with Texas Red has a particularly slow
dissociation process. Fluorescent labels alter the normal function
of the target protein and Texas Red has previously been
demonstrated to slow down the dissociation process in a study where
Texas Red was compared against other conjugates (22). In the same study, the values obtained
for cetuximab, at room temperature with other conjugates, were
similar to the values obtained in the present study with
FITC-cetuximab at 21°C.
The smaller temperature variations observed in the
current study, on on-rate (ka) and off-rate
(kd), result in larger equilibrium coefficient
(ka/kd) variations, and may be an interesting
starting point for a more detailed calculation of the thermodynamic
coefficients to provide a more complete mechanistic understanding
of the biologics involved for clinical antibodies in humans. A
deeper thermodynamic interpretation of the interactions requires
numerous additional temperatures under steady state thermostable
conditions and is beyond the scope of this particular study. The
aim of the present study was to investigate the impact of
temperature for the purpose of properly designing the assay.
However, a study that performs a deeper thermodynamic
interpretation of the interactions in living systems is planned for
the future, as it may reveal interesting mechanistic details, as
well as provide a more complete census and understanding of the
biology in the interactions. In an earlier such investigation, the
thermodynamic functions (ΔG°, ΔH° and ΔS°) were derived at each
site for the interaction between the optical isomers (R and S) of
the β-blocker, propranolol and the protein, cellobiohydrolase Cel7A
(27) revealing there are at least two
defined type of interactions; one interaction where R and S
propranolol behaved in a similar manner and one accounting for a
chiral (optically active) selective interaction (28,29).
Similarly, detailed findings may be possible in a living cell
interacting with biologics, although the experiment would be
particularly complex with metabolism involved.
In conclusion, the present study observed that it
was possible to measure the affinity and kinetics in living cells
in real-time at different temperatures, allowing better
understanding of the real behavior of protein-protein interactions
in a living system and, to a certain extent, introduced the
possibility of thermodynamic analysis of these interactions.
Therefore, as a potential rule-of-thumb, when measuring interaction
data acquired at different temperatures, the kinetics may increase
(or decrease) by less than 10-fold when comparing the physiological
temperature of 37°C with room temperature (20–25°C). The incubation
time required to reach equilibrium also varies by, approximately,
up to a factor 10, which has a direct impact on the design and
analysis of traditional end-point assays.
Acknowledgements
The authors would like to thank to Marika Nestor and
Hanna Björkelund for their assistance with the manuscript. The
study received funding from the European Union's Framework
Programme for Research and Innovation Horizon 2020 (grant no.
2014-2020) under the Marie Skłodowska Curie Grant (agreement no.
675555; Accelerated Early staGe drug dIScovery). Further support
was provided by Rector's Mobility Fund of Charles University 2016
and by Charles University (grant no. PRVOUK P40), and the Swedish
Knowledge Foundation in a KK HÖG 15 project (grant no.
20150233).
References
|
1
|
Liu JKH: The history of monoclonal
antibody development-Progress, remaining challenges and future
innovations. Ann Med Surg (Lond). 3:113–116. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Martinelli E, De Palma R, Orditura M, De
Vita F and Ciardiello F: Anti-epidermal growth factor receptor
monoclonal antibodies in cancer therapy. Clin Exp Immunol. 158:1–9.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Olsson AK, Dimberg A, Kreuger J and
Claesson-Welsh L: VEGF receptor signalling - in control of vascular
function. Nat Rev Mol Cell Biol. 7:359–371. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Niu G and Chen X: Vascular endothelial
growth factor as an anti-angiogenic target for cancer therapy. Curr
Drug Targets. 11:1000–1017. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bennouna J, Sastre J, Arnold D, Österlund
P, Greil R, Van Cutsem E, von Moos R, Viéitez JM, Bouché O, Borg C,
et al: ML18147 Study investigators: Continuation of bevacizumab
after first progression in metastatic colorectal cancer (ML18147):
A randomised phase 3 trial. Lancet Oncol. 14:29–37. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li S, Schmitz KR, Jeffrey PD, Wiltzius
JJW, Kussie P and Ferguson KM: Structural basis for inhibition of
the epidermal growth factor receptor by cetuximab. Cancer Cell.
7:301–311. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van Dongen GA, Visser GWM, Lub-de Hooge
MN, de Vries EG and Perk LR: Immuno-PET: A navigator in monoclonal
antibody development and applications. Oncologist. 12:1379–1389.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Arruebo M, Valladares M and
González-Fernández Á: Antibody-conjugated nanoparticles for
biomedical applications. J Nanomater. 2009:2009. View Article : Google Scholar
|
|
9
|
Andersson K: Bringing time into molecular
and cellular biology. J Anal Oncol. 2:65–68. 2013.
|
|
10
|
Kastritis PL and Bonvin AMJJ: On the
binding affinity of macromolecular interactions: Daring to ask why
proteins interact. J R Soc Interface. 10:201208352013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Renaud JP, Chung CW, Danielson UH, Egner
U, Hennig M, Hubbard RE and Nar H: Biophysics in drug discovery:
Impact, challenges and opportunities. Nat Rev Drug Discov.
15:679–698. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Björke H and Andersson K: Measuring the
affinity of a radioligand with its receptor using a rotating cell
dish with in situ reference area. Appl Radiat Isot. 64:32–37. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stenberg J, Spiegelberg D, Karlsson H and
Nestor M: Choice of labeling and cell line influences interactions
between the Fab fragment AbD15179 and its target antigen CD44v6.
Nucl Med Biol. 41:140–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Spiegelberg D, Stenberg J, Haylock AK and
Nestor M: A real-time in vitro assay as a potential predictor of in
vivo tumor imaging properties. Nucl Med Biol. 43:12–18. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nath N, Godat B, Zimprich C, Dwight SJ,
Corona C, McDougall M and Urh M: Homogeneous plate based antibody
internalization assay using pH sensor fluorescent dye. J Immunol
Methods. 431:11–21. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Perera RM, Zoncu R, Johns TG, Pypaert M,
Lee FT, Mellman I, Old LJ, Toomre DK and Scott AM: Internalization,
intracellular trafficking, and biodistribution of monoclonal
antibody 806: A novel anti-epidermal growth factor receptor
antibody. Neoplasia. 9:1099–1110. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Winquist J, Geschwindner S, Xue Y,
Gustavsson L, Musil D, Deinum J and Danielson UH: Identification of
structural-kinetic and structural-thermodynamic relationships for
thrombin inhibitors. Biochemistry. 52:613–626. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shuman CF, Hämäläinen MD and Danielson UH:
Kinetic and thermodynamic characterization of HIV-1 protease
inhibitors. J Mol Recognit. 17:106–119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Geitmann M and Danielson UH: Additional
level of information about complex interaction between
non-nucleoside inhibitor and HIV-1 reverse transcriptase using
biosensor-based thermodynamic analysis. Bioorg Med Chem.
15:7344–7354. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Reverberi R and Reverberi L: Factors
affecting the antigen-antibody reaction. Blood Transfus. 5:227–240.
2007.PubMed/NCBI
|
|
21
|
Björkelund H, Gedda L and Andersson K:
Comparing the epidermal growth factor interaction with four
different cell lines: Intriguing effects imply strong dependency of
cellular context. PLoS One. 6:e165362011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bondza S, Stenberg J, Nestor M, Andersson
K and Björkelund H: Conjugation effects on antibody-drug
conjugates: Evaluation of interaction kinetics in real time on
living cells. Mol Pharm. 11:4154–4163. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gedda L, Björkelund H and Andersson K:
Real-time immunohistochemistry analysis of embedded tissue. Appl
Radiat Isot. 68:2372–2376. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ekerljung L, Wållberg H, Sohrabian A,
Andersson K, Friedman M, Frejd FY, Ståhl S and Gedda L: Generation
and evaluation of bispecific affibody molecules for simultaneous
targeting of EGFR and HER2. Bioconjug Chem. 23:1802–1811. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pa̧zik R, Andersson R, Kȩpiński L, Nedelec
J-M, Kessler VG and Seisenbaeva GA: Surface functionalization of
the metal oxide nanoparticles with biologically active molecules
containing phosphonate moieties. Case study of BaTiO 3. J Phys Chem
C. 115:9850–9860. 2011. View Article : Google Scholar
|
|
26
|
Barta P, Malmberg J, Melicharova L,
Strandgård J, Orlova A, Tolmachev V, Laznicek M and Andersson K:
Protein interactions with HER-family receptors can have different
characteristics depending on the hosting cell line. Int J Oncol.
40:1677–1682. 2012.PubMed/NCBI
|
|
27
|
Fornstedt T: Characterization of
adsorption processes in analytical liquid-solid chromatography. J
Chromatogr A. 1217:792–812. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ståhlberg J, Henriksson H, Divne C,
Isaksson R, Pettersson G, Johansson G and Jones TA: Structural
basis for enantiomer binding and separation of a common β-blocker:
Crystal structure of cellobiohydrolase Cel7A with bound
(S)-propranolol at 1.9 A resolution. J Mol Biol. 305:79–93. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fornstedt T and Guiochon G: Nonlinear
effects in LC and Chiral LC. Anal Chem. 73:608A–617A. 2001.
View Article : Google Scholar : PubMed/NCBI
|