Introduction
Human papillomavirus (HPV) infection is a major
cause of cervical cancer and is also associated with the
development of oropharyngeal cancer and prognosis in affected
patients (1–3). To date, >170 HPV genotypes have been
reported, of which ~50 are known to infect the anogenital and oral
mucosa regions (4–6). As for oral cancer, a previous study
conducted by the present research group indicated that ~80% of
patients with HPV16-positive moderate epithelial dysplasia
progressed to oral squamous cell carcinoma (OSCC), indicating that
oral HPV16 infection may also be associated with the development of
oral cancer (7). However, few
epidemiologic studies have focused on oral HPV prevalence in
non-OSCC cases and the risk factors for oral HPV infection have not
been fully elucidated. Hence, we previously performed a
meta-analysis, which demonstrated that sexual behavior and smoking
are significant risk factors for oral HPV infection in cancer-free
individuals (8). In addition, other
factors, such as poor oral hygiene related to periodontitis and
gingivitis, have been indicated to be associated with oral HPV
infection (8).
In order to examine the correlation between HPV16
prevalence in the oral cavity and clinical factors, we also
investigated HPV16 prevalence using oral rinse samples obtained
from subjects without oral cancer (9). Notably, a significantly higher
prevalence was found in males than in females, indicating a
sex-specific susceptibility to HPV16 infection. As for the cervix,
increased HPV16 load may be predictive of HPV infection persistence
that eventually leads to malignancy (10). However, the relationship between oral
HPV16 viral load and oral hygiene status was not fully elucidated
in the previous study. In the present preliminary investigation,
HPV16 viral load in the oral cavity and its association with oral
bacteria count was examined. Furthermore, 16S ribosomal (r)RNA gene
sequencing for taxonomic classification of harbored bacteria was
performed in order to investigate the association of HPV16 viral
load with oral microbiome composition.
Patients and methods
Subjects
A total of 124 patients (48 males and 76 females;
mean age, 61.6 years; age range, 20–97 years) who visited the
Department of Oral and Maxillofacial Reconstructive Surgery of
Hiroshima University Hospital (Hiroshima, Japan) between November
2016 and August 2017 were enrolled in the present study. None of
the patients had evidence of oral cancer or pre-malignant lesions
(e.g., epithelial dysplasia or leukoplakia). The present study
design was approved by the Ethics Committee of Hiroshima University
and all participants signed an informed consent agreement.
Oral rinse sample processing and DNA
extraction
Briefly, the subjects were asked to rinse their
mouth with 10 ml saline for 30 sec and then expectorate into a
sterile 15-ml Falcon tube. Immediately after collection, all
samples were centrifuged at 3,000 × g for 10 min at 4°C, the
supernatant was decanted and the pellets were stored at −80°C until
further processing. Finally, DNA was extracted using a bead-beating
tube (cat. no. A29158; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), according to the manufacturer's protocol, and purified using
a PureLink™ Microbiome DNA Purification kit (Thermo Fisher
Scientific, Inc.), which enables purification of host and microbial
DNA from a wide variety of sample types.
Quantitation of human cell
numbers
The human endogenous retrovirus group 3 member 1
(ERV3-1) gene was employed to quantitate human cells using a
quantitative polymerase chain reaction (qPCR) assay. Using a
previously reported method (9),
serial 10-fold dilutions of the pUC57 vector inserted into the
ERV3-1 genome (Hokkaido System Science Co., Ltd., Sapporo, Japan)
were generated with copy numbers ranging from
100−109. Quantitation of DNA levels was
determined using a CFX connect real-time PCR detection system
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) and SYBR-Green PCR
Master mix (Toyobo Life Science, Osaka, Japan), with a reaction
mixture containing 1.0 µl DNA, 9.0 µl SYBR-Green mix and 10 µmol of
each pair of oligonucleotide primers. G3PDH was used as a reference
gene for PCR analysis. The primer sequences were as follows: ERV-3
sense, 5′-CATGGGAAGCAAGGGAACTAATG-3′ and antisense,
5′-CCCAGCGAGCAATACAGAATTT-3′; and G3PDH sense,
5′-ACCACAGTCCATGCCATCAC-3′ and antisense,
5′-TCCACCACCCTGTTGCTGTA-3′. Amplifications were performed with
initial melting at 95°C for 5 min, followed by 40 cycles of
denaturation at 95°C for 30 sec, annealing at 56°C for 30 sec and
extension at 72°C for 1 min. A standard curve indicating the cycle
threshold (CT) value vs. ERV3-1 copy number was obtained to
estimate the number of human cells in each sample.
HPV16 viral copy quantitation
qPCR assays were performed to determine HPV16 viral
load in the samples. Using a previously reported method (9), a 200-bp fragment within HPV16 E6 genome
was prepared, which was then cloned into a pUC57 vector (GenScript
Biotech Corporation, Piscataway, NJ, USA). Serial 10-fold dilutions
of the vector were made with copy numbers ranging from
103−109. Those 10-fold dilutions were used to
generate a standard curve, which indicated the CT value vs. copy
number of HPV16 E6. DNA samples extracted from 5,000–10,000 human
cells were prepared from each PCR mixture and the HPV16 sequence
was examined by PCR with type-specific primers. The primer
sequences for HPV16 E6 were as follows: Sense,
5′-AAGGGCGTAACCGAAATCGGT-3′ and antisense,
5′-GTTTGCAGCTCTGTGCATA-3′. A positive control containing HPV16 DNA
extracted from CaSki cells [American Type Culture Collection
(ATCC), Manassas, VA, USA] and a negative control containing DNA
purified from the HPV-negative cell line HTB31 (ATCC) were used for
the PCR reactions. Quantitation of DNA levels was determined using
a CFX connect real-time PCR detection system and SYBR-Green PCR
Master mix. Amplifications were performed with initial melting at
95°C for 5 min, followed by 40 cycles of denaturation at 95°C for
30 sec, annealing at 58°C for 30 sec and extension at 72°C for 1
min. G3PDH was used as a reference gene for PCR analysis.
Analysis of bacterial numbers
The number of oral bacteria was determined using a
Bacterial Counter (DU-AA01 NP-H; Panasonic Healthcare Co., Ltd.,
Tokyo, Japan), which is an oral bacteria detection device that
employs the dielectrophoretic impedance measurement method with a
microelectrode chip on which bacteria in liquids are captured by
dielectrophoresis (11). The
impedance change is converted to bacterial concentration/ml of each
sample (CFU/ml). Samples were obtained from the tongue surface
using a cotton swab (Panasonic Healthcare Co., Ltd.), according to
the manufacturer's protocol, before collecting the oral rinse
sample. Bacterial numbers were defined as follows: Level 1
(<105 CFU/ml), level 2 (≥105 and
<106 CFU/ml), level 3 (≥106 and
<106.5 CFU/ml), level 4 (≥106.5 and
<107.0 CFU/ml), level 5 (≥107.0 and
<107.5 CFU/ml), level 6 (≥107.5 and
<108.0 CFU/ml) and level 7 (≥108.0
CFU/ml).
16S rRNA gene sequencing for taxonomic
classification of bacteria
The V3-V4 region of the bacterial 16S rRNA gene was
amplified from DNA in the oral rinse samples using forward primer
341F (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′) and
reverse primer 806R
(5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACGVGGGTWTCTAAT-3′).
Amplicons were generated, cleaned, indexed and sequenced according
to the 16S Metagenomic Sequencing Library Preparation, part
15044223, Rev. B. Briefly, the initial PCR reaction contained 10.0
ng DNA, 2.5 µl 16S V3-V4 Primer mix (10X), 12.5 µl 2X Gflex PCR
Buffer and 0.5 µl Tks Gflex DNA Polymerase (Takara Biotechnology
Co., Ltd., Dalian, China) in a total volume of 25 µl.
Amplifications were performed with initial melting at 94°C for 1
min, followed by 28 cycles of denaturation at 98°C for 10 sec,
annealing at 50°C for 15 sec and extension at 68°C for 15 sec using
a Takara PCR Thermal Cycler Dice® Touch device (Takara
Biotechnology Co., Ltd.). The resultant PCR product was cleaned
using an Agencourt AMPure XP (Beckman Coulter, Inc., Brea, CA,
USA). Samples were multiplexed with a dual-index approach using a
Nextera XT Index kit (Illumina, Inc., San Diego, CA, USA). Briefly,
the second PCR reaction was composed of 2.0 µl PCR products, 2.5 µl
index primer 1
(5′-CAAGCAGAAGACGGCATACGAGATGTAGAGGAGTCTCGTGGGCTCGG-3′) and 2
(5′-ATGATACGGCGACCACCGAGATCTACACAGAGTAGATCGTCGGCAGCGTC-3′), 2.5 µl
2X Gflex PCR Buffer and 0.5 µl Tks Gflex DNA Polymerase in a total
volume of 25.0 µl. Amplifications were performed with initial
melting at 94°C for 1 min, followed by 8 cycles of denaturation at
98°C for 10 sec, annealing at 60°C for 15 sec and extension at 68°C
for 15 sec. The final library was subjected to paired-end
sequencing with a MiSeq Reagent kit v.3 on the Illumina MiSeq
platform (both from Illumina, Inc.). Sequencing data were denoised,
low quality sequences were removed and the sequences were clustered
into operational taxonomic units (OTUs) at 97% identity using the
CD-HIT-OTU pipeline (12).
Subsequently, taxonomy classification was analyzed using the
Quantitative Insights into Microbial Ecology pipeline, as
previously reported (13).
Statistical analysis
Data were presented as the mean ± standard deviation
of three independent experiments. Statistical analysis was
performed using SPSS version 24.0 (IBM Corp., Armonk, NY, USA).
Mann Whitney U or Kruskal-Wallis tests were used to evaluate
significant differences regarding HPV16 copy numbers and clinical
factors. Mann Whitney U tests with Bonferroni's correction were
employed following Kruskal-Wallis tests. P<0.05 was considered
to indicate a statistically significant difference.
Results
Quantitation of HPV16 E6 viral copy
number using qPCR
HPV16 viral copy number was determined in a total of
124 oral rinse samples using qPCR, then the number of HPV16 DNA
copies was evaluated as copy number/human cell using a standard
curve. The CT value was identified to be below the detection limit
in the standard curve in 30 of the 124 samples, which were assessed
as HPV16 DNA not determined and were not included in further
analysis. Thus, the association of HPV16 viral copy number with
clinical parameters were examined in 94 cases.
The average number of HPV16 E6 viral copies was
1.65±3.47 copies/cell (range, 0.07–25.3 copies/cell). The
association between the number of HPV16 viral copies and clinical
parameters were then examined; however, no statistically
significant association was identified (Table I). With regards to sex, there was no
significant difference in the number of HPV16 E6 viral copies
between males (1.52±3.11 copies) and females (1.75±3.72 copies).
Regarding age, subjects ≥80 years old demonstrated the highest mean
number of HPV16 compared with the other age groups. Notably,
individuals using a full denture were revealed to have a higher
mean number of HPV16 viral copies than those with a partial denture
or who did not use a denture. Although a difference was observed
between the groups for age and denture use, these differences were
not significant.
 | Table I.Association between oral HPV16 E6 copy
number and clinical parameters. |
Table I.
Association between oral HPV16 E6 copy
number and clinical parameters.
| Clinical
characteristics | n | HPV16 copy
number/cell | P-value |
|---|
| Sex |
|
|
|
| Male | 38 | 1.52±3.11 | 0.76 |
|
Female | 56 | 1.75±3.72 |
|
| Age, years |
|
|
|
|
20–29 | 9 | 2.27±2.12 | 0.39 |
|
30–39 | 8 | 0.53±0.37 |
|
|
40–49 | 4 | 2.64±2.67 |
|
|
50–59 | 12 | 0.60±0.52 |
|
|
60–69 | 22 | 1.15±1.29 |
|
|
70–79 | 27 | 1.20±1.93 |
|
|
80–89 | 12 | 4.64±8.43 |
|
| Number of remaining
teeth |
|
|
|
|
≥20 | 68 | 1.33±2.64 | 0.23 |
|
10–19 | 14 | 1.03±2.10 |
|
|
0–9 | 12 | 3.33±7.13 |
|
| Denture use |
|
|
|
|
Non-denture user | 72 | 1.43±2.57 | 0.43 |
| Partial
denture | 18 | 1.40±2.22 |
|
| Full
denture | 4 | 6.94±12.3 |
|
| Bacteria count |
|
|
|
| Level
2 | 6 | 0.62±0.47 | 0.14 |
| Level
3 | 24 | 0.83±1.07 |
|
| Level
4 | 41 | 1.92±4.15 |
|
| Level
5 | 20 | 1.53±1.97 |
|
| Level
6 | 3 | 7.48±10.1 |
|
Association between HPV16 E6 viral
copy number and oral bacteria
The mean number of oral bacteria was
8.25±9.58×106 CFU/ml (range,
2.1×105−5.95×107 CFU/ml). The number of
bacteria was classified as levels 1–7 and the numbers of viral
copies according to the different bacterial levels are summarized
in Table I. The number of HPV16 viral
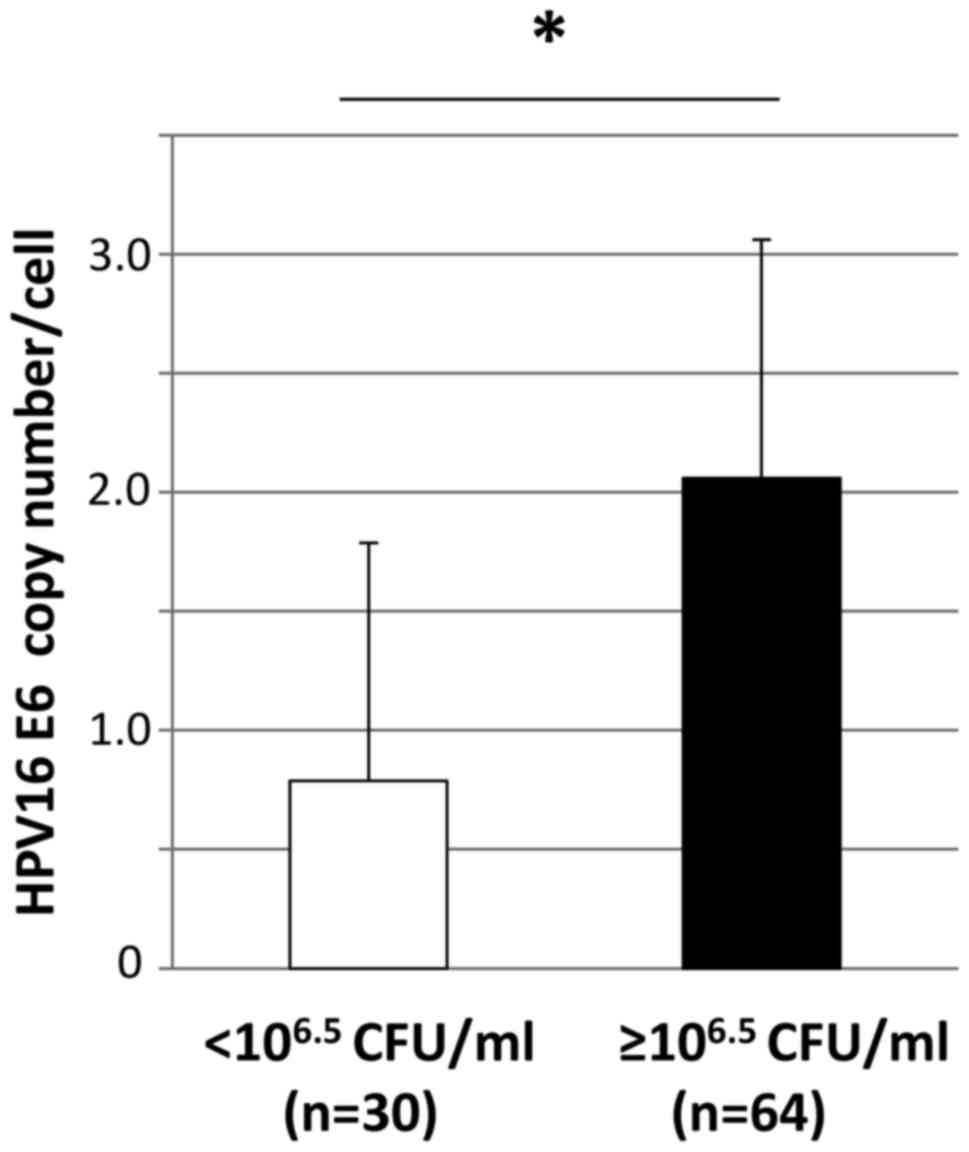
copies was significantly higher in individuals with a high level of
oral bacteria (≥106.5 CFU/ml; levels 4–6) than in those
with a low level (<106.5 CFU/ml; levels 2 and 3)
(0.79±0.98 vs. 2.06±4.11 copy numbers/cell; P=0.030; Fig. 1).
Furthermore, analysis of 16s rRNA in bacterial flora
was performed using the oral rinse samples to examine microbiome
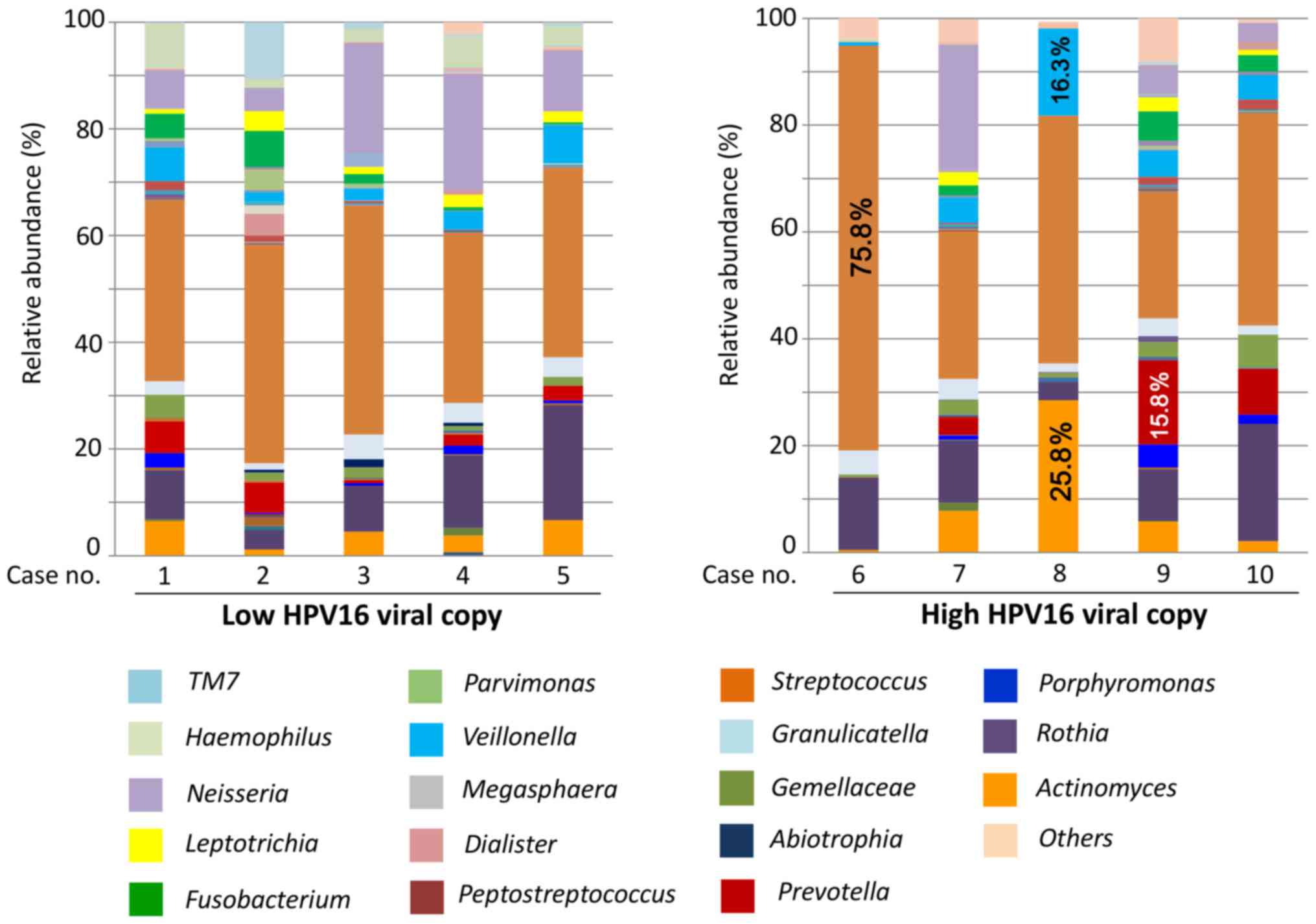
diversity in cases with a high number of HPV16 viral copies. The
present findings of 5 cases with high HPV16 viral copy numbers
(>5.0 HPV16 DNA copy/cell) were compared with those of 5 cases
with low HPV16 viral copy numbers (<0.5 HPV16 DNA copy/cell). At
the genus level, a total of 41 genera were identified in the
groups, of which Actinomyces, Streptococcus,
Prevotella, Veillonella, Gemella,
Fusobacterium, Neisseria, Rothia and
Haemophilus constituted >70% in both.
Streptococcus was most commonly detected in both groups.
Notably, except for Streptococcus, no other genus was found
in >25% of the cases with a low HPV16 viral copy number
(Fig. 2). In contrast,
Streptococcus comprised 75.8% in one case in the high HPV16
viral copy group, while Actinomyces (25.8%) and
Veillonella (16.3%) were frequently detected in another case
in the high HPV16 viral copy group (Fig.
2). Additionally, an increased percentage of gram-negative
anaerobic bacteria, such as Prevotella (15.8%), was
identified in a case in the group with a high number of HPV16 viral
copies (Fig. 2).
Discussion
We previously performed a meta-analysis of oral HPV
prevalence to clarify risk factors related to oral HPV infection in
healthy individuals. The results suggested that changes in the oral
environment due to sexual behavior (i.e., oral sex and
homosexuality in men) and smoking were significantly associated
with oral HPV infection, whereas alcohol consumption was not a
factor (8). As for smoking,
suppression of the oral immune system caused by chemical substances
in tobacco may be involved in oral HPV infection (14). Notably, female smokers have been
demonstrated to have a significantly higher prevalence of HPV
infection as compared to male smokers (15). However, a significant association
between smoking and HPV viral copy number was not identified in the
present study (data not shown). On the other hand, gingivitis and
periodontal disease are also potential risk factors for oral HPV
infection (16–18), indicating that poor oral hygiene may
induce the risk of HPV infection in gingival pockets. A comparison
of denture wearers and non-wearers demonstrated a higher HPV
infection rate in the former, indicating that mucosal injury or
poor dental hygiene associated with denture use is importantly
involved in HPV infection (8,9). In the present study, subjects who wore
full dentures exhibited a higher number of HPV copies than partial
denture and non-denture users; however, the difference was not
significant. Thus, full denture use may be a risk factor for oral
HPV infection and HPV DNA replication in elderly individuals.
The present study revealed that the levels of oral
bacteria in samples from the tongue dorsum were significantly
associated with increased HPV copy number. However, there was no
significant association between the levels of oral bacteria and
sex, age, remaining teeth or denture use (data not shown). In
addition, increased percentages of periodontal pathogens, including
Veillonella and Prevotella, were detected in
individuals with a high number of HPV copies as compared with those
with a low number. These findings indicate that increased HPV16
viral replication may be related to the development of an
unbalanced microbiome and increased pathogenic bacteria in the oral
cavity. It has been speculated that an inflamed periodontal pocket
serves as a reservoir for HPV and induces HPV infection (19). Whether periodontal disease is related
to oral HPV infection remains controversial (20–22),
though the presence of such inflammatory diseases may serve an
important role in the biology of HPV infection. On the other hand,
HPV may be involved in deterioration associated with periodontal
disease (i.e., increased alveolar bone loss) in accordance with
other pathogenic factors (18).
Similarly, the presence of human viruses, including the herpes
virus and Epstein-Barr virus, in periodontal sites may worsen
periodontal disease (23,24). It is thus considered that human viral
factors may serve a vital role in modulation of inflammatory
disease in the oral cavity, such as periodontitis, as well as
harbored bacteria.
The oral cavity harbors characteristic microbiomes
in different sites, including the gingiva, tongue surface, palate,
buccal mucosa and teeth (25,26). Thus, it is likely that the microbiome
in an oral rinse sample is composed of a variety of bacteria from
different oral regions. The tongue surface has been demonstrated to
exhibit a relatively stable bacterial microbiome with a composition
very similar to that of saliva (27,28). The
present study compared differences in bacterial composition between
oral rinse samples and different oral sites. Notably, the oral
rinse samples demonstrated a bacterial composition similar to that
of the tongue surface (data not shown), indicating that the
microbiome obtained by oral rinsing is predominantly associated
with that of the tongue surface. However, it remains uncertain
whether HPV virus load is affected by an unbalanced bacterial
composition and elevated levels of anaerobic bacteria. Additional
cluster analyses of microbial communities are essential, as such
findings should provide greater insight into the relationship
between specific oral bacteria and HPV.
We previously employed PCR assays to detect HPV16
DNA in oral rinse and gargle samples to examine differences between
the oral cavity and oropharynx regarding HPV16 prevalence (9). Notably, a higher prevalence of HPV16 in
gargle samples was observed than in the oral rinse samples in the
previous study, indicating that the oropharynx is more susceptible
to HPV16 infection. Furthermore, the number of HPV16 copies was
significantly higher in the gargle samples than in the oral rinse
samples from those subjects, indicating that the oropharynx is more
susceptible to HPV16 infection and HPV16 gene amplification is more
common here as compared with the oral cavity (9). HPV may be trapped in tonsillar crypts
and then invade basal cells in the reticulated epithelium,
resulting in HPV infection (8). In a
study on patients with head and neck squamous cell carcinoma, the
HPV16 genome copy number was much higher in samples from patients
with oropharyngeal cancer than in those with oral cancer,
supporting the notion that the oropharynx is a more favorable site
for HPV16 infection and replication as compared with other head and
neck regions (29). It is possible
that the variability of HPV16 infection rate among different sites
may be associated with the biological features of HPV-related
cancer.
In conclusion, the present results suggested that
HPV16 viral load may be associated with an increased bacterial
number in the oral cavity. Further investigations are required to
clarify the correlation between oral HPV load and oral hygiene
status. As oral HPV infection is considered to be associated with
poor oral hygiene, focus on oral health care and smoking cessation
may serve a vital role in its prevention. For prevention strategies
and treatment of HPV-related oral cancer, it is necessary to
clarify the biological features of oral HPV and its environmental
influence in the near future.
Acknowledgements
The present study was supported by a Grant-in-aid
for Scientific Research (C) (grant no. 15K11290) from the Ministry
of Education, Culture, Sports and Technology of Japan.
Glossary
Abbreviations
Abbreviations:
|
HPV
|
human papillomavirus
|
|
ERV3-1
|
human endogenous retrovirus group 3
member 1
|
References
|
1
|
Dürst M, Gissmann L, Ikenberg H and zur
Hausen H: A papillomavirus DNA from a cervical carcinoma and its
prevalence in cancer biopsy samples from different geographic
regions. Proc Natl Acad Sci USA. 80:pp. 3812–3815. 1983; View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ang KK, Harris J, Wheeler R, Weber R,
Rosenthal DI, Nguyen-Tân PF, Westra WH, Chung CH, Jordan RC, Lu C,
et al: Human papillomavirus and survival of patients with
oropharyngeal cancer. N Engl J Med. 363:24–35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smeets SJ, Braakhuis BJ, Abbas S, Snijders
PJ, Ylstra B, van de Wiel MA, Meijer GA, Leemans CR and Brakenhoff
RH: Genome-wide DNA copy number alterations in head and neck
squamous cell carcinomas with or without oncogene-expressing human
papillomavirus. Oncogene. 25:2558–2564. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ma Y, Madupu R, Karaoz U, Nossa CW, Yang
L, Yooseph S, Yachimski PS, Brodie EL, Nelson KE and Pei Z: Human
papillomavirus community in healthy persons, defined by
metagenomics analysis of human microbiome project shotgun
sequencing data sets. J Virol. 88:4786–4797. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
de Villiers EM: Cross-roads in the
classification of papillomaviruses. Virology. 445:2–10. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
IARC, . Human papillomaviruses. A review
of human carcinogens - part B: Biological agents. IARC Monogr Eval
Carcinog Risks Hum. 100B:1–475. 2011.
|
|
7
|
Sugiyama M, Bhawal UK, Dohmen T, Ono S,
Miyauchi M and Ishikawa T: Detection of human papillomavirus-16 and
HPV-18 DNA in normal, dysplastic, and malignant oral epithelium.
Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 95:594–600. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shigeishi H and Sugiyama M: Risk factors
for oral human papillomavirus infection in healthy individuals: A
systematic review and meta-analysis. J Clin Med Res. 8:721–729.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shigeishi H, Sugiyama M, Ohta K, Rahman MZ
and Takechi M: Higher prevalence and gene amplification of HPV16 in
oropharynx as compared to oral cavity. J Appl Oral Sci. 24:397–403.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Trevisan A, Schlecht NF, Ramanakumar AV,
Villa LL and Franco EL; Ludwig-McGill Study Group, : Human
papillomavirus type 16 viral load measurement as a predictor of
infection clearance. J Gen Virol. 94:1850–1857. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hamada R, Suehiro J, Nakano M, Kikutani T
and Konishi K: Development of rapid oral bacteria detection
apparatus based on dielectrophoretic impedance measurement method.
IET Nanobiotechnol. 5:25–31. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li W, Fu L, Niu B, Wu S and Wooley J:
Ultrafast clustering algorithms for metagenomic sequence analysis.
Brief Bioinform. 13:656–668. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Caporaso JG, Kuczynski J, Stombaugh J,
Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich
JK, Gordon JI, et al: QIIME allows analysis of high-throughput
community sequencing data. Nat Methods. 7:335–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kreimer AR, Pierce Campbell CM, Lin HY,
Fulp W, Papenfuss MR, Abrahamsen M, Hildesheim A, Villa LL,
Salmerón JJ, Lazcano-Ponce E and Giuliano AR: Incidence and
clearance of oral human papillomavirus infection in men: The HIM
cohort study. Lancet. 382:877–887. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gillison ML, Broutian T, Pickard RK, Tong
ZY, Xiao W, Kahle L, Graubard BI and Chaturvedi AK: Prevalence of
oral HPV infection in the United States, 2009–2010. JAMA.
307:693–703. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hormia M, Willberg J, Ruokonen H and
Syrjänen S: Marginal periodontium as a potential reservoir of human
papillomavirus in oral mucosa. J Periodontol. 76:358–363. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Engels EA, Biggar RJ, Hall HI, Cross H,
Crutchfield A, Finch JL, Grigg R, Hylton T, Pawlish KS, McNeel TS
and Goedert JJ: Cancer risk in people infected with human
immunodeficiency virus in the United States. Int J Cancer.
123:187–194. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tezal M, Sullivan Nasca M, Stoler DL,
Melendy T, Hyland A, Smaldino PJ, Rigual NR and Loree TR: Chronic
periodontitis-human papillomavirus synergy in base of tongue
cancers. Arch Otolaryngol Head Neck Surg. 135:391–396. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kines RC, Thompson CD, Lowy DR, Schiller
JT and Day PM: The initial steps leading to papillomavirus
infection occur on the basement membrane prior to cell surface
binding. Proc Natl Acad Sci USA. 106:pp. 20458–63. 2009; View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dayakar MM, Shipilova A and Gupta D:
Periodontal pocket as a potential reservoir of high risk human
papilloma virus: A pilot study. J Indian Soc Periodontol.
20:136–140. 2016.PubMed/NCBI
|
|
21
|
Fuster-Rossello L, Ribotta E, Cuffini C
and Fuster-Juan M: Human papilloma virus in oral mucosa and its
association with periodontal status of gynecologically infected
women. Acta Odontol Latinoam. 27:82–88. 2014.PubMed/NCBI
|
|
22
|
Wiener RC, Sambamoorthi U and Jurevic RJ:
Association of periodontitis and human papillomavirus in oral rinse
specimens: Results from the National Health and Nutrition Survey
2009–2012. J Am Dent Assoc. 146:382–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Idesawa M, Sugano N, Ikeda K, Oshikawa M,
Takane M, Seki K and Ito K: Detection of Epstein-Barr virus in
saliva by real-time PCR. Oral Microbiol Immunol. 19:230–232. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Saygun I, Kubar A, Sahin S, Sener K and
Slots J: Quantitative analysis of association between herpesviruses
and bacterial pathogens in periodontitis. J Periodontal Res.
43:352–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Segata N, Haake SK, Mannon P, Lemon KP,
Waldron L, Gevers D, Huttenhower C and Izard J: Composition of the
adult digestive tract bacterial microbiome based on seven mouth
surfaces, tonsils, throat and stool samples. Genome Biol.
13:R422012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bik EM, Long CD, Armitage GC, Loomer P,
Emerson J, Mongodin EF, Nelson KE, Gill SR, Fraser-Liggett CM and
Relman DA: Bacterial diversity in the oral cavity of 10 healthy
individuals. ISME J. 4:962–974. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou Y, Gao H, Mihindukulasuriya KA, La
Rosa PS, Wylie KM, Vishnivetskaya T, Podar M, Warner B, Tarr PI,
Nelson DE, et al: Biogeography of the ecosystems of the healthy
human body. Genome Biol. 14:R12013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Takeshita T, Yasui M, Tomioka M, Nakano Y,
Shimazaki Y and Yamashita Y: Enteral tube feeding alters the oral
indigenous microbiota in elderly adults. Appl Environ Microbiol.
77:6739–6745. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ndiaye C, Mena M, Alemany L, Arbyn M,
Castellsagué X, Laporte L, Bosch FX, de Sanjosé S and Trottier H:
HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck
cancers: A systematic review and meta-analysis. Lancet Oncol.
15:1319–1331. 2014. View Article : Google Scholar : PubMed/NCBI
|