Introduction
Periodontal disease is a heterogeneous infection of
the tissues supporting the teeth, primarily characterized by
gingival inflammation (1), and may
lead to tooth loss and destruction of the tissues surrounding and
supporting the teeth (2,3). It is established that certain
microorganisms serve important roles in the development of
periodontal disease, and the disease has been associated with a
number of infectious diseases, as well as chronic diseases
including cancer (4). There are two
types of periodontitis disease, namely aggressive (localized and
generalized) and chronic, both of which involve loss of supporting
structures of the teeth (5).
Aggressive periodontitis is characterized by fast rates of
progression, tissue attachment-loss and tissue destruction
(6). Due to its increasing prevalence
worldwide, aggressive periodontitis is regarded as a major health
problem. The pathogenesis and development of periodontitis disease
are associated with an interaction of immunological, genetic and
environmental variables (7). In
particular, genetic background has been significantly associated
with susceptibility to periodontitis disease (3,8). There
have been several studies on genetic factors implicated in the
pathogenesis and severity of the diseases (3,9,10), and a number of key genes have been
demonstrated to be involved in disease susceptibility and severity,
or in determining the clinical course (11). The process of apoptosis serves a
critical role in the development of periodontitis disease;
therefore, genes related to this process are common candidates in
genetic analyses regarding possible associations with disease
course (12).
Apoptosis is among the main physiological types of
cell death that is involved in different physiological and
pathological functions including maintenance of tissue homeostasis,
regulating cell numbers and eliminating unwanted or potentially
dangerous cells during organism development (13). Therefore, alterations in apoptotic
pathways and abnormal regulation of apoptosis contributes to the
pathogenesis of various human diseases including cancer and
inflammatory diseases (14). In
previous reports, there has been focus on the
FAS/FASL axis as a key and major pathway involved in
apoptosis induction, in the prevention of autoimmune diseases and
in the maintenance of immune tolerance (12,15,16). The
FAS and FASL genes are located on chromosome 10q24.1
and 1q23, respectively. FAS also known as tumor necrosis factor
(TNF) receptor superfamily member 6, cluster of differentiation
(CD)95 or apoptosis antigen 1 is a type I cell membrane receptor
protein that is expressed in a variety of tissues and serves a
critical role in apoptotic signaling in many cell types (17). FASL (also known as TNF superfamily
member 6 or CD95 ligand), as a natural ligand of FAS, is a type II
transmembrane protein belonging to the TNF superfamily that can
trigger cell death signal cascades by cross-linking with FAS
(18). Several previous studies have
reported that decreased expression of FAS and/or elevated
expression of FASL may be associated with numerous types of
human cancer and immunological diseases (13,19–21).
Substitution of A to G at position −670 in the
promoter region of FAS and C to T substitution at position
−844 in the promoter region of FASL are among the major
single-nucleotide polymorphisms to have been identified in the
genes (22). The FAS
polymorphism −670A/G is situated within the signal transducers and
activators of transcription 1 transcription factor binding sites;
thus, presence of the −670G allele diminishes promoter activity and
decreases FAS gene expression (21,23). The
FASL 844C/T polymorphism is located in a putative binding
motif of CAAT/enhanced-binding protein β element (24).
Due to the association of apoptosis with aggressive
periodontitis and FAS/FASL, the aim of the present study was to
evaluate whether the FAS-670A/G and FASL-844C/T
polymorphisms were associated with disease course in a Kurdish
population with generalized aggressive periodontitis (GAP) in
Western Iran.
Materials and methods
Materials
The DNA extraction kit, agarose, Taq polymerase and
dNTPs used for polymerase chain reaction (PCR) experiments were
purchased from Zagros Bioidea Co. (Incubator Center of Razi
University, Kermanshah, Iran). The MvaI and BsrDI
restriction enzymes were supplied by New England BioLabs, Inc.
(Ipswich, MA, USA). All other chemicals and reagents of analytical
grade were supplied by Merck KGaA (Darmstadt, Germany). Forward and
reverse primers were synthesized by SinaClon Bioscience Co.
(Tehran, Iran).
Sample collection
A total of 25 patients with GAP were enrolled,
comprising of 87% females and 13% males with a mean age of 21.0±3.5
years (3). All subjects were enrolled
from the School of Dentistry, Qazvin University of Medical Sciences
(Qazvin, Iran) from November, 2011 to May, 2012. As a control
group, 110 men with the same ethnic background as patients, who
were free from signs of periodontitis (mean age, 33.91±7.43 years),
were also enrolled. All patients and control individuals were free
of general and genetics diseases. Periodontal disease was diagnosed
according to clinical and radiographic parameters that included
bleeding on probing, probing pocket depth and clinical attachment
loss. Aggressive periodontitis was defined by inter-proximal
attachment loss affecting at least three teeth that were either
first molars or incisors (3,6,25). For the
patient and control groups, the exclusion criteria were a history
of HIV and/or hepatitis infection, diabetes, usage of
anti-inflammatory and/or immunosuppressive drugs, pregnancy or
lactation, and smoking. The current research procedures were
approved by the ethics committee of Kermanshah University of
Medical Sciences (Kermanshah, Iran). All subjects enrolled in the
study were informed of the aim and procedures of the research and
provided written informed consent in accordance with the Helsinki
II declaration.
DNA extraction
From each subject, 5-ml blood samples were collected
in micro-tubes containing EDTA. DNA was extracted from whole blood
using the DNA isolation kit (Zagros Bioidea Co.) (26). The purified genomic DNA was analyzed
and verified by electrophoresis on 1% agarose gel. The DNA
concentration and purity were assessed with a NanoDrop
Spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE,
USA) at wavelengths of 260 and 280 nm (27,28).
Genotyping
FAS and FASL gene polymorphisms were
assessed with a PCR-based restriction fragment length polymorphism
(PCR-RFLP) method (19).
The PCR was conducted to amplify the FAS and
FASL polymorphisms with the following primers: For
FAS-670A/G, forward, 5′-GGCTGTCCATGTTGTGGCTGC-3′ and
reverse, 5′-CTACCTAAGAGCTATCTACCGTTC-3′ (22); and for FASL-844C/T, forward,
5′-CAGCTACTCAGGAGGCCAAG-3′ and reverse, 5′-GCTCTGAGGGGAGAGACCAT-3′
(20). The PCR thermal cycling
conditions were as follows: For FAS, one cycle of
denaturation at 94°C for 5 min followed by 30 cycles at 94°C for 30
sec, 61°C for 30 sec and 72°C for 30 sec, with a final extension at
72°C for 6 min; and for FASL, one cycle of denaturation at
94°C for 5 min followed by 30 cycles at 94°C for 30 sec, 67°C for
30 sec and 72°C for 30 sec, with a final extension at 72°C for 6
min. PCR was performed in a total volume of 25 µl containing 0.5 µM
of each primer, 2 µl DNA (100–300 ng) as the template, 0.2 mM
dNTPs, 1.5 mM MgCl2, 1 U Taq DNA polymerase and 2.5 µl
10X PCR buffer (200 mM Tris HCl, pH 8.4, 500 mM KCl).
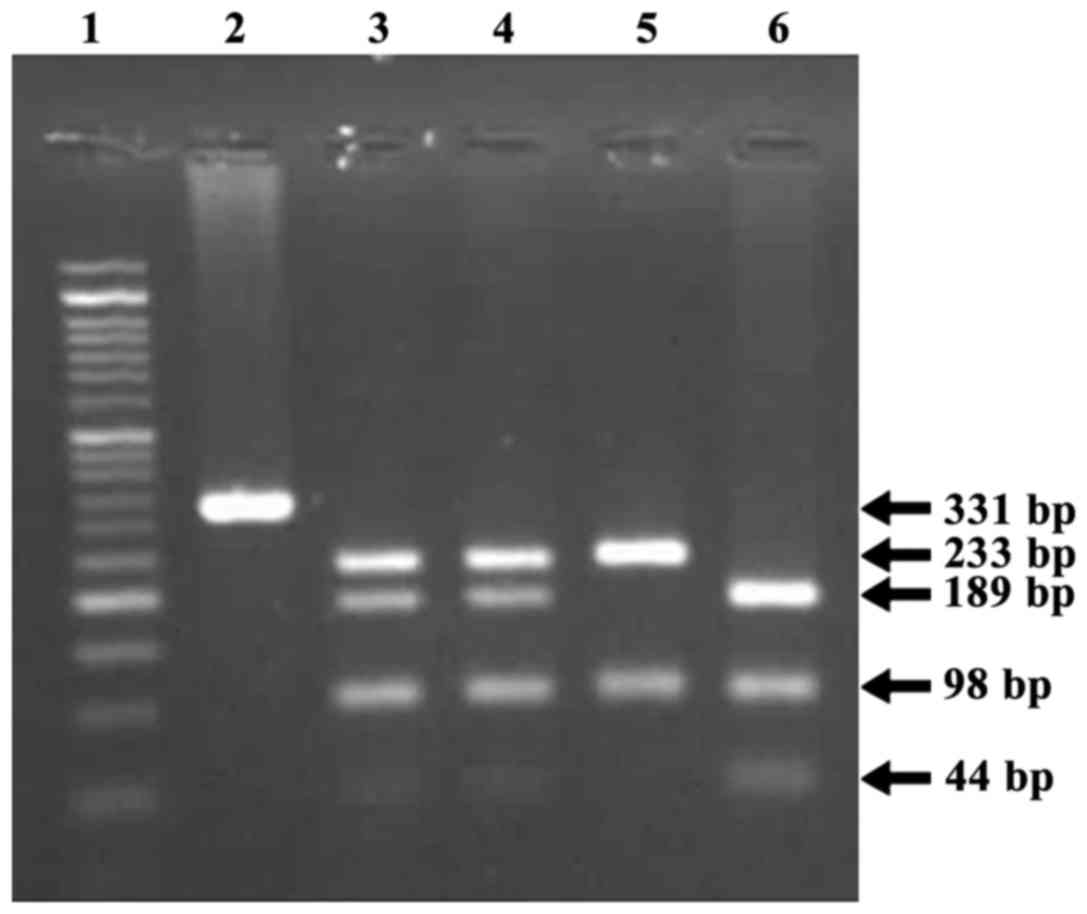
For FAS-670A/G, the obtained PCR product (331
bp) was digested with MvaI at 37°C for 15 h. The resulting
fragment sizes were as follows: 189, 98 and 44 bp for the −670AA
genotype, and 233 and 98 bp for the −670GG genotype. For
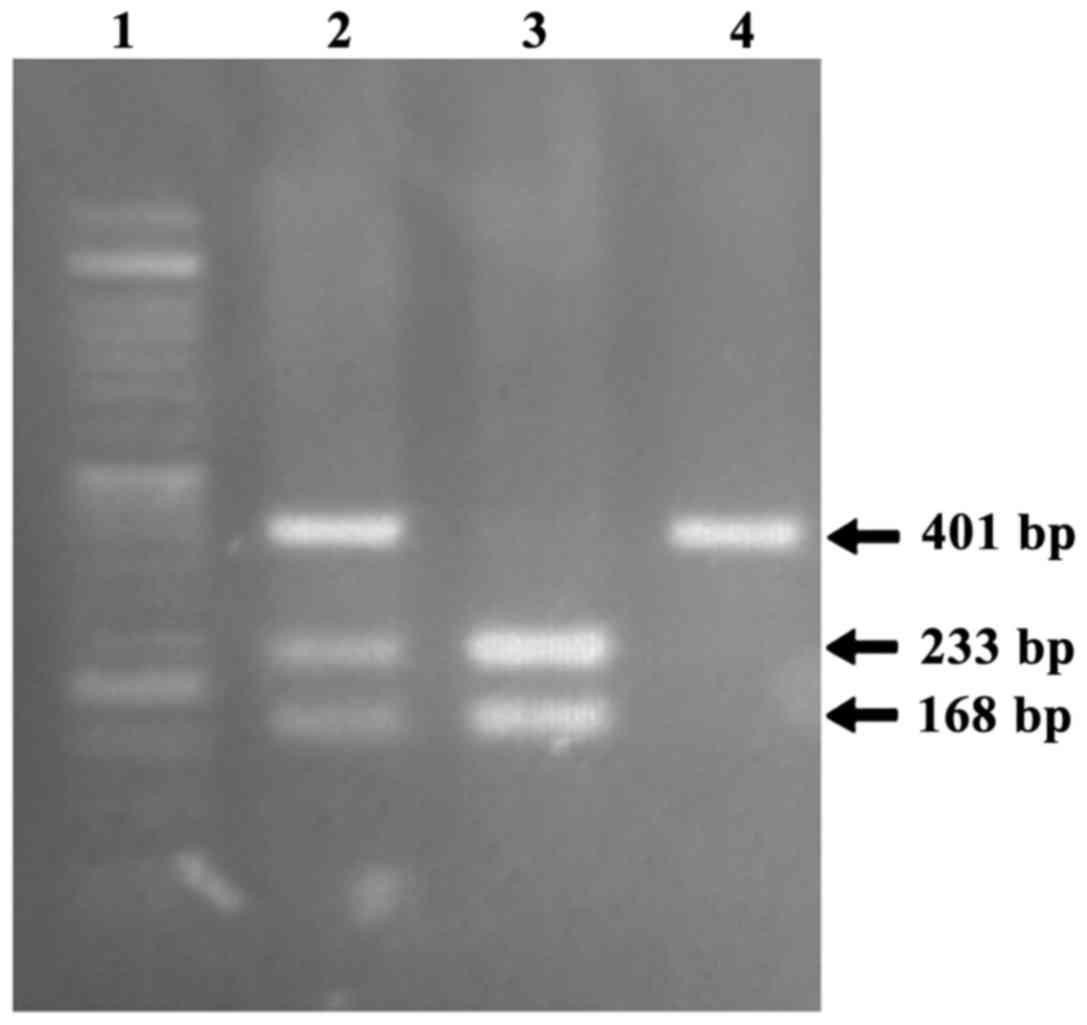
FASL-844C/T, the obtained PCR product (401 bp) was digested
with BsrDI at 37°C for 15 h. In the presence of the −844CC
genotype, the PCR product was digested into two fragments of 233
and 168 bp, while in the presence of the −844T allele a fragment of
401 bp was observed (19). Digested
PCR products were identified on a 2% agarose gel stained with Gel
Red DNA stain under ultraviolet light.
Statistical analysis
The Pearson's χ2 test was used to compare
differences in frequency distributions of alleles and genotypes of
the FAS and FASL polymorphisms. All statistical tests
were two-sided, and P<0.05 was assumed to indicate statistical
significance. SPSS statistical software version 21.0 (IBM Corp.,
Armonk, NY, USA) was used for all statistical analyses.
Results
FAS-670A/G and FASL-844C/T
products
Fragments of 331 and 401 bp in size corresponding to
the FAS and FASL genes, respectively, were amplified
by PCR. Figs. 1 and 2 depict the fragment separation pattern of
the FAS-670A/G and FASL-844C/T genotypes,
respectively, following agarose gel electrophoresis.
FAS-670A/G genotype frequencies
The allele frequencies of the FAS and
FASL genotypes were in Hardy-Weinberg equilibrium. The
frequencies of the FAS-670A/G genotypes and alleles among
the GAP and control subjects are listed in Table I. A higher frequency of the combined
genotype (AG+GG) was observed in the GAP patients (96.0%) compared
with the control subjects (90.9%), though the difference was not
significant [χ2=0.705, degrees of freedom (df)=1,
P=0.401]. Similarly, the prevalence of the G allele was
non-significantly higher in the GAP group (62.0%) compared with
that in the controls (60.0%; χ2=0.012, df=1,
P=0.913).
 | Table I.Frequency of FAS-670A/G
genotypes and alleles in GAP patients and controls. |
Table I.
Frequency of FAS-670A/G
genotypes and alleles in GAP patients and controls.
| Genotype/alleles, n
(%) | GAP group
(n=25) | Control group
(n=110) |
|---|
| −670A/G
genotype |
|
|
| AA | (4.0) 1 | 10 (9.1) |
| AG | 17 (68.0) | 68
(61.8) |
| GG | 7 (28.0) | 32
(29.1) |
|
(χ2=0.501, df=2,
P=0.489) |
|
|
|
AG+GG | 24 (96.0) | 100 (90.9) |
|
(χ2=0.705, df=1,
P=0.401) |
|
|
| −670A/G
alleles |
|
|
| A | 19 (38.0) | 88
(40.0) |
| G | 31 (62.0) | 132 (60.0) |
|
(χ2=0.012, df=1,
P=0.913) |
|
|
FASL-844C/T genotype frequencies
For FASL-844C/T genotype, as indicated in
Table II, the frequency of the TT
genotype was non-significantly higher in the GAP group (24.0%) when
compared with the control subjects (10.0%; χ2=3.897,
df=2, P=0.142). Additionally, the frequency of the combined
genotype (CT+TT) was higher in the GAP group (96.0%) compared with
the controls (91.8%); however, its association was not
statistically significant (χ2=0.519, df=1, P=0.471). The
frequency of the T allele was 60.0% in the GAP group and 50.9% in
the controls, though was deemed to not differ significantly between
the groups (χ2=3.627, df=1, P=0.057).
 | Table II.Frequency of FASL-844C/T genotypes
and alleles in GAP patients and controls. |
Table II.
Frequency of FASL-844C/T genotypes
and alleles in GAP patients and controls.
| Genotype/alleles, n
(%) | GAP group
(n=25) | Control group
(n=110) |
|---|
| −844C/T
genotype |
|
|
| CC | 1 (4.0) | 9
(8.2) |
| CT | 18 (72.0) | 90
(81.8) |
| TT | 6
(24.0) | 11
(10.0) |
|
(χ2=3.897, df=2,
P=0.142) |
|
|
|
CT+TT | 24 (96.0) | 101 (91.8) |
|
(χ2=0.519, df=1,
P=0.471) |
|
|
| −844C/T
alleles |
|
|
| C | 20 (40.0) | 108 (49.1) |
| T | 30 (60.0) | 112 (50.9) |
|
(χ2=3.627, df=1,
P=0.057) |
|
|
Discussion
The present study assessed the potential association
between two known polymorphisms in cell death pathway genes
(FAS-670A/G and FASL-844C/T) and the presentation of
GAP in Western Iranian patients. The absence of a significant
association between the FAS-670A/G polymorphism with GAP
risk was observed. Additionally, the study presented here confirmed
no significant associations between −844C/T alleles of FASL
polymorphism with GAP. Therefore, the study indicated that −844C/T
variants of FASL and −670A/G variants of FAS were not
associated with GAP in the Iranian population studied.
Periodontal disease as an inflammatory illness is a
primary cause of tooth loss. GAP, as a sub-type of periodontitis
disease, affects the supporting tissue of the teeth with
inflammatory infection (3).
Aggressive periodontitis as local or general-type generally
presents in individuals between the ages of 20 and 35 years old, as
damage to the first incisive and molar alveolar bone (9). Previous studies have demonstrated that
microbial agents, certain environmental risk factors including
living conditions and dietary habits, and genetic susceptibility
are necessary for the pathogenesis and development of periodontal
disease (10,29). Additionally, previous findings also
indicated that genetic factors were significantly associated with
progression of periodontal disease in various ethnic populations
(29).
The present study indicated that there were no
significant associations between the FAS-670A/G and
FASL-844C/T polymorphisms and the presentation of GAP in
patients when compared with control individuals.
Apoptosis, as a form of programmed cell death, is a
fundamental biological process that occurs in multi-cellular
organisms to maintain tissue turnover (17). Interactions of FAS/FASL may serve a
crucial role in preventing autoimmune diseases and maintaining
immune tolerance (17,30). FAS and FASL, as type-II transmembrane
proteins that belong to the TNF family, have been identified in
inflammatory infiltrates in patients with periodontitis disease
(19). In general, FASL has been
implicated in the induction of T-cell apoptosis (11). Alterations in the regulation of
FAS/FASL may contribute to the pathogenesis of various human
diseases including cancer and inflammatory diseases such as
periodontitis (31).
A number of studies have been conducted to analyze
the potential relationship between the main polymorphisms of
candidate genes with susceptibility to periodontitis diseases. For
instance, Kazemi et al (10)
reported an association between manganese superoxide dismutase
(Val-9Ala) genotype with risk of GAP. Ayazi et al (3) identified a positive association between
interleukin-1β gene polymorphism and risk of periodontitis disease.
Darvishi et al (9) determined
that there was no significant association between the
TNF-α-1031(T/C) genotypes and GAP in an Iranian population.
Furthermore, Menezes and Colombo (32) in Brazil and Heidari et al
(33) in Iran reported that there was
no significant association between chronic periodontitis with TNF-α
(−308G/A) gene polymorphism. Certain studies have reported an
association of the major polymorphisms FAS-670A/G and
FASL-844C/T with different human diseases including
prostate, bladder, gastric and cervical cancers, esophageal
squamous cell carcinoma, systemic lupus erythematosus and
infertility disorders (11,19,20). Wu
et al (21) observed that
FAS-670A/G gene polymorphism was associated with the
severity of villous atrophy in coeliac disease. Sun et al
(20) reported that polymorphisms in
the FAS and FASL genes were associated with an
increased risk of developing esophageal squamous-cell carcinoma in
Chinese individuals. Wu et al (24) identified the major polymorphism
−844C/T in the promoter region of FASL in African American
patients with systemic lupus erythematosus. Additionally, they
observed that −844C genotype may lead to increased risk of
autoimmunity with enhanced expression levels of FASL, which
led them to conclude that differences in the regulation of
FASL expression may contribute to the development of the
autoimmune phenotype (24).
To the best of our knowledge, there are no previous
studies on the association of FAS and FASL
polymorphisms with risk of periodontitis disease. However,
Wohlfahrt et al (11) reported
no significant association between selected candidate gene
polymorphisms and severe chronic periodontitis in white North
American individuals. Additionally, Brozovic et al (34) reported that Porphyromonas
gingivalis, a Gram-negative anaerobe associated with severe
periodontal disease, enhanced FASL expression through
up-regulation of nuclear factor-κB-mediated gene transcription and
induced apoptotic cell death in human gingival epithelial
cells.
In conclusion, the current study, to the best of our
knowledge, was the first to analyze the putative association of
FAS-670A/G and FASL-844C/T polymorphisms with risk of
GAP in an Iranian population; however, the results indicated that
there were no significant associations of these polymorphisms with
the presentation of GAP in patients when compared with normal
individuals. Nevertheless, the present results may be used as a
basis for studies on other major polymorphisms of candidate genes
related to periodontitis disease. The results should now be
confirmed by studies on larger groups in the future.
References
|
1
|
Tsuchida S, Satoh M, Takiwaki M and Nomura
F: Ubiquitination in Periodontal Disease: A Review. Int J Mol Sci.
18:14762017. View Article : Google Scholar
|
|
2
|
Prakash P and Victor D: Interleukin-1b
gene polymorphism and its association with chronic periodontitis in
South Indian population. Int J Genet Mol Biol. 2:179–183. 2010.
|
|
3
|
Ayazi G, Pirayesh M and Yari K: Analysis
of interleukin-1β gene polymorphism and its association with
generalized aggressive periodontitis disease. DNA Cell Biol.
32:409–413. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang S, Wang F, Shi X, Dai J, Peng Y, Guo
X, Wang X, Shen H and Hu Z: Association between manganese
superoxide dismutase (MnSOD) Val-9Ala polymorphism and cancer risk
- A meta-analysis. Eur J Cancer. 45:2874–2881. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Armitage GC: Development of a
classification system for periodontal diseases and conditions. Ann
Periodontol. 4:1–6. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guzeldemir E, Gunhan M, Ozcelik O and
Tastan H: Interleukin-1 and tumor necrosis factor-alpha gene
polymorphisms in Turkish patients with localized aggressive
periodontitis. J Oral Sci. 50:151–159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brett PM, Zygogianni P, Griffiths GS,
Tomaz M, Parkar M, D'Aiuto F and Tonetti M: Functional gene
polymorphisms in aggressive and chronic periodontitis. J Dent Res.
84:1149–1153. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Droździk A, Kurzawski M, Safronow K and
Banach J: Polymorphism in interleukin-1beta gene and the risk of
periodontitis in a Polish population. Adv Med Sci. 51 Suppl
1:13–17. 2006.PubMed/NCBI
|
|
9
|
Darvishi E, Aziziaram Z, Yari K, Bagheri
Dehbaghi M, Kahrizi D, Karim H, Vaziri S, Zargooshi J, Ghadiri K,
Muhammadi S, et al: Lack of association between the
TNF-α-1031genotypes and generalized aggressive periodontitis
disease. Cell Mol Biol (Noisy-le-grand). 62:63–66. 2016.PubMed/NCBI
|
|
10
|
Kazemi E, Moradi MT, Yari K, Mousavi SA
and Kahrizi D: Association between Manganese Superoxide Dismutase
(MnSOD Val-9Ala) genotypes with the risk of generalized aggressive
periodontitis disease. Cell Mol Biol (Noisy-le-grand). 61:49–52.
2015.PubMed/NCBI
|
|
11
|
Wohlfahrt JC, Wu T, Hodges JS, Hinrichs JE
and Michalowicz BS: No association between selected candidate gene
polymorphisms and severe chronic periodontitis. J Periodontol.
77:426–436. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Galbraith GMP, Hendley TM, Sanders JJ,
Palesch Y and Pandey JP: Polymorphic cytokine genotypes as markers
of disease severity in adult periodontitis. J Clin Periodontol.
26:705–709. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hashemi M, Fazaeli A, Ghavami S,
Eskandari-Nasab E, Arbabi F, Mashhadi MA, Taheri M, Chaabane W,
Jain MV and Łos MJ: Functional polymorphisms of FAS and FASL gene
and risk of breast cancer - pilot study of 134 cases. PLoS One.
8:e530752013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Darby IA, Bisucci T, Hewitson TD and
MacLellan DG: Apoptosis is increased in a model of
diabetes-impaired wound healing in genetically diabetic mice. Int J
Biochem Cell Biol. 29:191–200. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ji G, Gu A, Hu F, Wang S, Liang J, Xia Y,
Lu C, Song L, Fu G and Wang X: Polymorphisms in cell death pathway
genes are associated with altered sperm apoptosis and poor semen
quality. Hum Reprod. 24:2439–2446. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nagata S: Fas ligand-induced apoptosis.
Annu Rev Genet. 33:29–55. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Elmore S: Apoptosis: A review of
programmed cell death. Toxicol Pathol. 35:495–516. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Locksley RM, Killeen N and Lenardo MJ: The
TNF and TNF receptor superfamilies: Integrating mammalian biology.
Cell. 104:487–501. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Asgari R, Mansouri K, Bakhtiari M,
Bidmeshkipour A, Yari K, Shaveisi-Zadeh F and Vaisi-Raygani A:
Association of FAS-670A/G and FASL-844C/T polymorphisms with
idiopathic azoospermia in Western Iran. Eur J Obstet Gynecol Reprod
Biol. 218:55–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun T, Miao X, Zhang X, Tan W, Xiong P and
Lin D: Polymorphisms of death pathway genes FAS and FASL in
esophageal squamous-cell carcinoma. J Natl Cancer Inst.
96:1030–1036. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu J, Alizadeh BZ, Veen TV, Meijer JW,
Mulder CJ and Pena AS: Association of FAS (TNFRSF6)-670 gene
polymorphism with villous atrophy in coeliac disease. World J
Gastroenterol. 10:717–720. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang QR, Morris D and Manolios N:
Identification and characterization of polymorphisms in the
promoter region of the human Apo-1/Fas (CD95) gene. Mol Immunol.
34:577–582. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li M, Sun D, Li C, Zhang Z, Gao L, Li K,
Li H and Gao T: Functional polymorphisms of the FAS gene associated
with risk of vitiligo in Chinese populations: A case-control
analysis. J Invest Dermatol. 128:2820–2824. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu J, Metz C, Xu X, Abe R, Gibson AW,
Edberg JC, Cooke J, Xie F, Cooper GS and Kimberly RP: A novel
polymorphic CAAT/enhancer-binding protein β element in the FasL
gene promoter alters Fas ligand expression: A candidate background
gene in African American systemic lupus erythematosus patients. J
Immunol. 170:132–138. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khocht A, Heaney K, Janal M and Turner B:
Association of interleukin-1 polymorphisms with periodontitis in
Down syndrome. J Oral Sci. 53:193–202. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moradi MT, Yari K and Khodarahmi R: A
novel, efficient, fast and inexpensive DNA extraction protocol from
whole blood applicable for studying drug-DNA interaction. J Rep
Pharm Sci. 3:80–84. 2014.
|
|
27
|
Yari K, Afzali S, Mozafari H, Mansouri K
and Mostafaie A: Molecular cloning, expression and purification of
recombinant soluble mouse endostatin as an anti-angiogenic protein
in Escherichia coli. Mol Biol Rep. 40:1027–1033. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yari K and Rahimi Z, Moradi MT and Rahimi
Z: The MMP-2 −735 C allele is a risk factor for susceptibility to
breast cancer. Asian Pac J Cancer Prev. 15:6199–6203. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Michalowicz BS, Diehl SR, Gunsolley JC,
Sparks BS, Brooks CN, Koertge TE, Califano JV, Burmeister JA and
Schenkein HA: Evidence of a substantial genetic basis for risk of
adult periodontitis. J Periodontol. 71:1699–1707. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Voss M, Lettau M, Paulsen M and Janssen O:
Posttranslational regulation of Fas ligand function. Cell Commun
Signal. 6:112008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lima L, Morais A, Lobo F, Calais-da-Silva
FM, Calais-da-Silva FE and Medeiros R: Association between FAS
polymorphism and prostate cancer development. Prostate Cancer
Prostatic Dis. 11:94–98. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Menezes NG and Colombo AP: Lack of
association between the TNF-α −308 (G/A) genetic polymorphism and
periodontal disease in Brazilians. Braz Oral Res. 22:322–327. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Heidari Z, Mahmoudzadeh-Sagheb H, Hashemi
M and Rigi-Ladiz MA: Stereological analysis of interdental gingiva
in chronic periodontitis patients with tumor necrosis factor-alpha
(−308 G/A) gene polymorphisms. Gen Cell Tis. 1:e183152014.
|
|
34
|
Brozovic S, Sahoo R, Barve S, Shiba H,
Uriarte S, Blumberg RS and Kinane DF: Porphyromonas
gingivalis enhances FasL expression via up-regulation of
NFkappaB-mediated gene transcription and induces apoptotic cell
death in human gingival epithelial cells. Microbiology.
152:797–806. 2006. View Article : Google Scholar : PubMed/NCBI
|