Introduction
Colorectal cancer is a disease localized to the
colon that may cause hematogenous liver metastasis, lymphogenous
lymph node metastasis and disseminated metastasis when it becomes
advanced. Among these, liver metastasis has the highest frequency
of occurrence and the strongest influence on cancer prognosis
(1). Currently the only effective
treatment for increasing the long-term survival rate of patients
with liver metastasis is hepatectomy (2,3).
However, the rate of recurrence in the remnant liver remains high,
possibly due to incomplete resection of the metastasis, which in
turn may be due to the existence of smaller cancer cell nests
(2,3). A standard approach to multi
detector-row computed tomography (MDCT) of the liver is to acquire
images with a slice thickness of 1 mm and perform multiphase
dynamic CT and multiplanar reformation. Magnetic resonance imaging
(MRI) of the liver consists of gadolinium ethoxybenzyl
diethylenetriamine pentaacetic acid-enhanced dynamic MRI combined
with diffusion-weighted MRI, requiring a slice thickness of 2–3 mm
(4). Advances in imaging techniques
have enabled the visualization of minute lesions that are
undetectable by conventional CT or ultrasonography. Incomplete
resection allows cancer cells to spread into the abdominal cavity
from cancer lesions penetrating the intestinal wall (5). Metastasis is not established by the
simple presence of cancer cells, as cancer cell engraftment in the
metastasis-target tissue is necessary. Therefore, the establishment
of metastasis is influenced by host factors including immune cells
and the blood flow environment, in addition to cancer cell factors,
such as high permeative and cytoadherent properties (6). Considering the influence of the host
side, the question of whether or not a single cell may cause
metastasis arises. Metastasis of single cancer cells that
metastasized to a lymph node have been detected by cytokeratin
staining (7), but no hematogenous
liver metastasis of single cells has been identified. It was
suggested that this may be due to rapid portal blood flow compared
with lymph flow, generating a difference in viability between
single cells floating in portal blood and lymph nodes (7). In addition, phagocytes, such as Kupffer
cells, are abundant in the liver and should inhibit cancer cell
engraftment Therefore, a specific number of cells that form a
cancer cell nest may be necessary to establish liver metastasis
(8); however, the number of cells
required to initiate metastasis is currently unknown. In addition,
difficulties lie in distinguishing single cells and/or ultra small
nests from the diverse cells present in the liver. Identifying a
single cell and/or an ultra small nest in the liver is expected to
be more difficult than detection in lymph nodes, which do not
possess epithelial-system cells (7).
The present study used a LacZ-labeled cancer cell line to visualize
intrahepatic metastasis of single cells or liver
micrometastasis.
Materials and methods
Cells
The human colon cancer cell line, KM12SM was
provided by Dr Nakajima at SBI Pharmaceuticals Co., Ltd., (Tokyo,
Japan) (9). Cells were cultured for
2 days at 37°C in Roswell Park Memorial Institute medium-1640
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented
with 10% fetal calf serum (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) and 1% penicillin and streptomycin (Thermo Fisher
Scientific, Inc.) within the total medium volume under a humidified
atmosphere containing 5% CO2.
Production of KM12SM-LacZ vectors
The production of a LacZ-manifested retrovirus was
quantified using TransIT-293 (Mirus Bio LLC, Madison, WI, USA),
according to the manufacturer's instructions. Briefly, the LacZ
expression vector was constructed using a pDON-5 DNA vector
(Clontech Laboratories, Inc., Mountain View, CA, USA) by inserting
a Kozak sequence (GCC CCA CC) and the lacZ coding region from a
pSV-β-Galactosidase control vector (all from Promega Corporation,
Madison, WI, USA). The lacZ coding region in the vector began with
the 7th amino acid of the wild-type lacZ gene (Full sequence for
pSV-beta-Galactosidase at https://www.addgene.org). G3T-hi cells (Takara
Biotechnology Co., Ltd., Kusatsu, Japan) were sprayed into 60 mm
collagen-coated plates (Cosmo Bio Co., Ltd., Tokyo, Japan) at a
density of 2–3×106 cells/dish and were cultured for 1
day at 37°C. Prior to cell transfection, G3T-hi cells were grown to
70–80% confluence in a petri dish. The following protocol was used
for cell transfection: A total of 250 µl Opti-MEMI Reduced-Serum
Medium (Thermo Fisher Scientific, Inc.) was added to a sterile
tube. Subsequently, 2 µl LacZ expression vector (1 µg/µl), 2 µl pGP
Vector (Takara Biotechnology Co., Ltd.), 1 µl pE-ampho Vector
(Takara Biotechnology Co., Ltd.) were added and a pipette was used
to gently mix all reagents completely. Then, 7.5 µl TransIT-293
reagent (Mirus Bio LLC) was warmed to room temperature and pipetted
into the mixture. The mixture (DNA-TransIT complex) was then
incubated at room temperature for 30 min. A total of 262.5 µl
DNA-TransIT complex, 62 µl 2 M calcium chloride and distilled water
were mixed in a 5 ml polystyrene round-bottom tube to 500 µl (DNA
mixture). The culture medium was prepared using 10% fetal bovine
serum (FBS; GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) and
Dulbecco's modified Eagle's medium high glucose (DMEM,
Sigma-Aldrich; Merck KGaA) with 25 mM chloroquine (1:1,000). The
supernatant was aspirated and discarded by an electric pipette from
the G3T-hi cells and 3 ml culture medium was added. The DNA mixture
was added slowly to 500 µl transfection buffer (Mirus Bio LLC). A
pipette was used to immediately mix this for 10–20 sec. Then, 1,000
µl DNA mixture was dropped evenly into a petri dish using an
electric pipette, then mixed within 1–2 min and cultured in 5% CO2.
A total of 24 h following transfection, the medium was replaced
with DMEM plus FBS. After a further 24 h, supernatant was filtered
through a 0.45-µm filter and the resulting solution was used as a
LacZ gene expression retrovirus vector solution. The retroviral
vector solution was aliquoted and stored at −80°C.
Construction of KM12SM-LacZ
KM12SM was seeded during the logarithmic growth
phase in a 6-well plate with 8×104 cells/well. Following
24 h, 1.2 ml LacZ expression vector and 1.2 µl polybrene solution
(8 mg/ml; Sigma-Aldrich; Merck KGaA) were added to 1 ml KM12SM in
each well after medium was removed. The medium was changed to a
medium containing 300 µg/ml of Geneticin (Thermo Fisher Scientific,
Inc.) 24 h after the virus infection. Cells resistant to geneticin
were harvested 14 d later and stored at −80°C in a Cellbanker 1
(1×106 cell/vial; Takara Biotechnology Co., Ltd.).
To confirm the expression of LacZ, X-gal staining
was completed. KM12SM-LacZ cells were cultured in the medium with
geneticin for 24 h at 37°C. To complete the X-gal staining, a
fixative agent was added to KM12SM-LacZ cells cultured on a
prepared slide, and incubated for 5 min at room temperature. The
composition of the fixative agent was 1% neutral formalin solution
(Muto Pure Chemicals Co., Ltd., Tokyo, Japan), 0.2% glutaraldehyde
(Wako Pure Chemical Industries, Ltd., Osaka, Japan), 0.02% Tergitol
solution (Sigma-Aldrich; Merck KGaA) and phosphate-buffered saline.
The staining solution was then added to the KM12SM-LacZ cells and
incubated for 3 h at 37°C. The stain solution was composed of 5 µm
hexacyanoferrate (II) potassium (Wako Pure Chemical Industries,
Ltd.), 5 µm hexacyanoferrate (III) potassium (Wako Pure Chemical
Industries, Ltd.), 0.02% Tergitol solution (Sigma-Aldrich; Merck
KGaA), 2 µm MgCl2 (Wako Pure Chemical Industries, Ltd.)
and 2 mg/ml X-gal (Promega Corporation).
Mouse liver metastasis model using
KM12SM-LacZ
A total of 12 athymic BALB/c male nude mice with a
body weight (BW) of 20–22 g (nu/nu; 6–8 weeks old) were obtained
from CLEA Japan, Inc. (Tokyo, Japan). Animals were kept at the
Animal Care and Use Facilities at Tokyo Medical University Hospital
(Tokyo, Japan) under specific pathogen-free conditions. Animals
were individually housed in cages at a temperature of 22–24°C and
relative humidity of 60–65% in a 12 h light/dark cycle. Mice feed
(CA-1; CLEA Japan, Inc.) was given ad libitum following
high-pressure steam sterilization. All experiments were approved by
the Animal Care and Ethics Committee of Tokyo Medical University.
For all operative procedures, animals were anesthetized with an
intraperitoneal injection of 1 µl saline supplemented with 0.75
mg/kg BW medetomidine (Asco Co., Ltd., Toyohashi, Japan), 0.75
mg/kg BW midazolam (Sandoz Inc., Princeton, NJ, USA) and 5.0 mg/kg
BW butorphanol (Asco Co., Ltd.).
KM12SM-lacZ was suspended using Hank's balanced salt
solution (Thermo Fisher Scientific, Inc.) to produce a
5×105 cells/50 µl per mouse cell suspension. The cell
suspension was placed in a 1 ml syringe kept on ice until use. The
mice underwent surgery to create a 1 cm incision above the spleen,
to move the spleen into the external abdominal cavity, following
anesthesia as described above. The cell suspension was slowly
injected under the splenic capsule using a 29-gauge needle. The
puncture hole was compressed using a cotton swab in order to avoid
leakage of the cell suspension and hemostasis. Following 1 week,
re-incision of each mouse was completed and the spleen was removed
under the aforementioned anesthesia. Mice were sacrificed by
intravenous administration (100–120 mg/kg) of pentobarbiturate
(Kyoritsu Seiyaku Co., Ltd., Inc., Tokyo, Japan) into a tail vein
1, 2 or 3 weeks following splenectomy and the liver was excised
(n=4 randomly selected mice per group). The excised liver was
initially examined by the naked eye to confirm the presence or
absence of macroscopic nodule. To avoid cancer cell contamination,
lobes with no observable nodule growth were set aside for DNA
sampling. The remaining lobes were then divided into two equal
groups. Lobes in one group were fixed for 2 d in 10% neutral
buffered formalin (Wako Pure Chemical Industries, Ltd.) at room
temperature and embedded in paraffin for histological diagnosis.
The remaining lobes were stored for 2 d at −80°C for X-gal
staining. Liver for pathological examination was cut in 2-mm
sections and stained using hematoxylin and eosin (H&E). Frozen
specimens, once cut into sections 10 µm thick were stained using
X-gal. To stain the nucleus and the cytoplasm, Nuclear Fast Red
stain (Vector Laboratories, Burlingame, CA, USA) was added.
Confirmation of liver metastasis
Grade-1 metastasis (DNA level), human DNA
(Chromosome 17-Specific Alfa Satellite DNA) (10,11) was
detected. Genomic DNA was isolated from liver tissue exhibiting no
observable tumor growth using a DNeasy® Blood &
Tissue kit (Qiagen GmbH, Hilden, Germany). The DNA from KM12SM
cells was used as a positive control and that from mouse liver
cells treated with distilled water was used as a negative control.
The target site in the isolated DNA was amplified by polymerase
chain reaction (PCR), which was performed using a total volume of
50 µl in the presence of 100 µg of isolated DNA, AmpliTaq
Gold® 360 Master mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and 0.25 µM of each primer. Primers for human
chromosome 17, detected in Grade-1 metastasis, were as follows:
Forward, 3′-GGGATAATTTCAGCTGACTAAACAG-5′ and reverse,
3′-TTCCGTTTAGTTAGGTGCAGTTATC-5′. The PCR conditions were as
follows: An initial denaturation step for 10 min at 95°C, then 35
cycles of denaturation for 1 min at 90°C, annealing/elongation for
1 min at 60°C and a final elongation step for 10 min at 72°C.
Samples were then stored at 4°C for 12 h prior to electrophoresis.
The target product (850 bp) was confirmed using a 1.5% agarose gel.
Grade-2 metastasis (metastasis of single cells) was confirmed using
X-gal staining (calculated as follows: X-gal-positive cell
area/liver area ×100). X-gal stained sections were imaged using a
Nano Zoomer-XR Digital slide scanner (Hamamatsu Photonics K.K.,
Shizuoka, Japan) and the image data was imported into the Tissue
Studio® version 4.0 (Definiens, Munich, Germany). A
total of 5 speculum images of the data from liver cross sections,
with the exception of the macroscopic liver metastasis were
randomly selected. X-gal positive cells were detected using the
image analyser. Grade-3 metastasis (histopathological
micrometastasis) was diagnosed using light microscopy on specimens
cut into 1-mm sections. Nodules were of various sizes (<1 mm to
several mm in diameter) and not exposed on the liver surface, and
could not be observed by the naked eye. Grade-4 metastasis (typical
metastasis) was detected macroscopically.
Results
Confirmation of KM12SM-LacZ
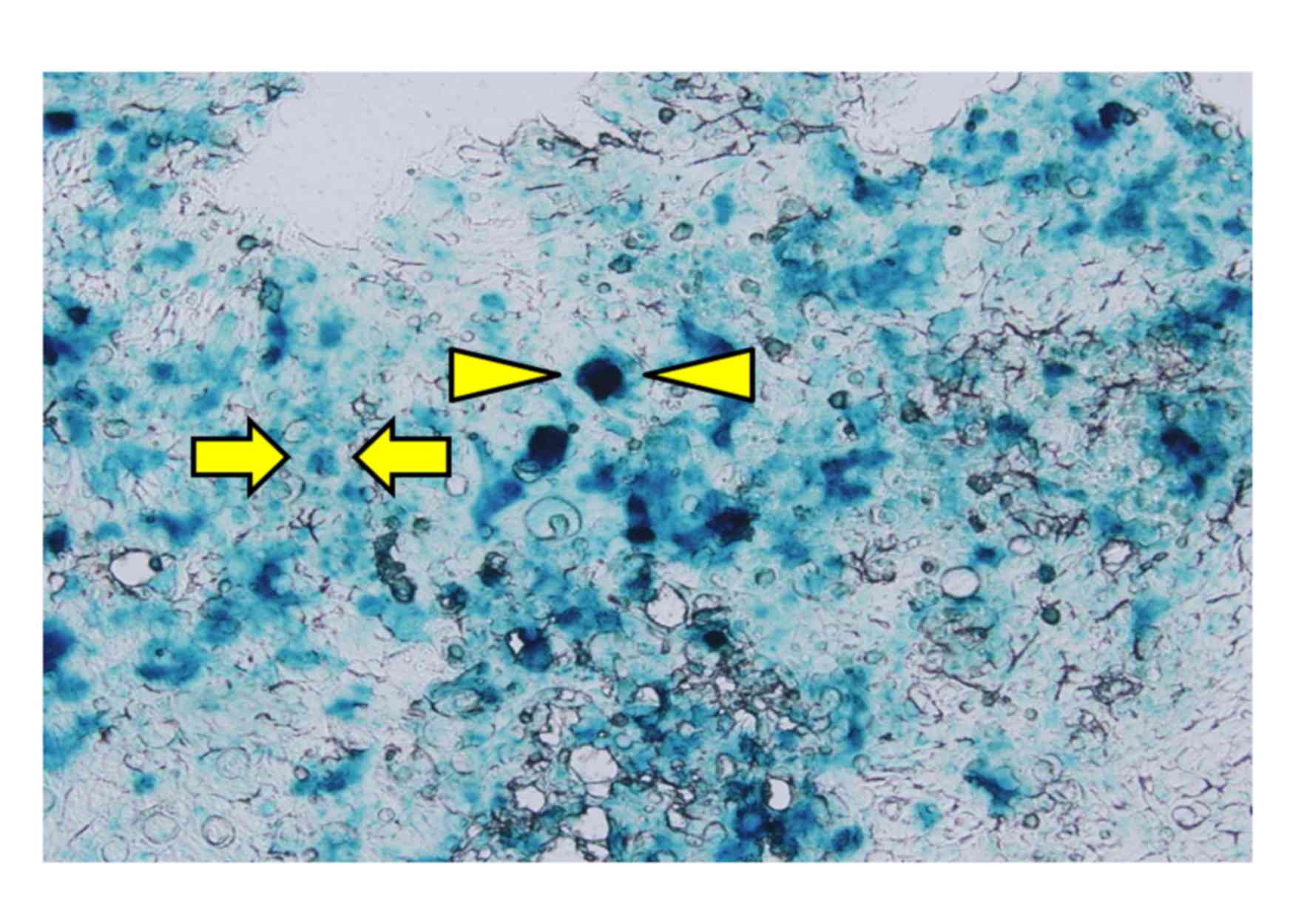
Histological analysis demonstrated that the
morphology of cells transfected with KM12SM-LacZ and KM12SM were
the same (data not shown). X-gal positive cells in the KM12SM-LacZ
group were identified in approximately half of cells (Fig. 1). No X-gal positive cells were
observed in the KM12SM group (data not shown).
Confirmation of liver metastasis
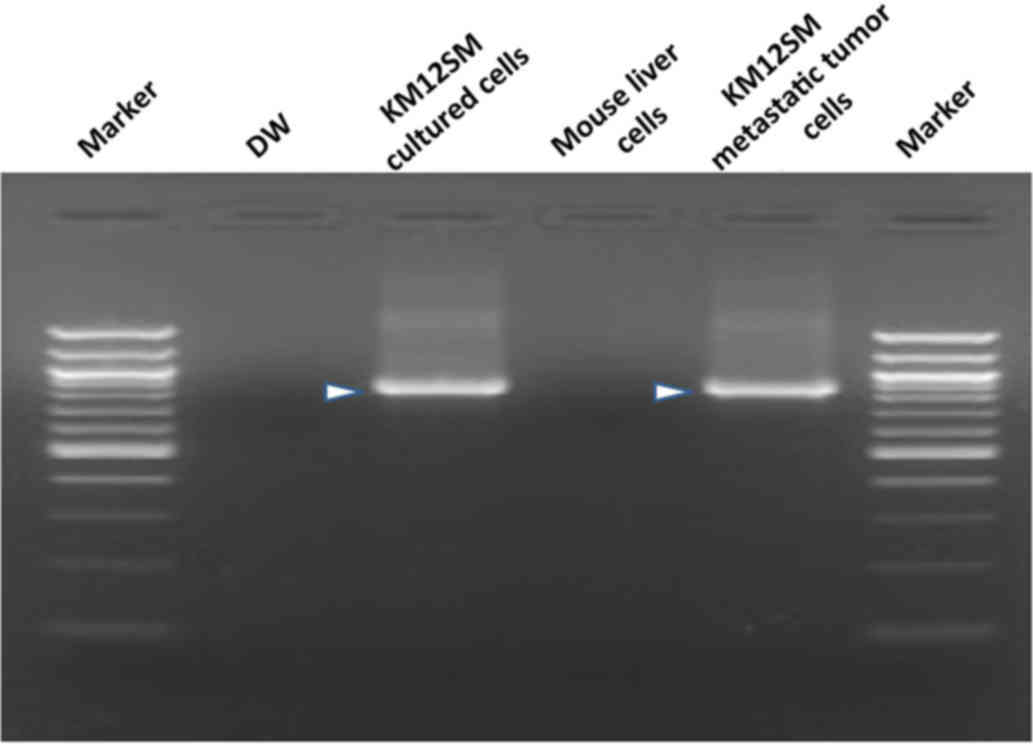
The presence of target bands (850 bp) observed in
the DNA extracted from cultured and metastatic KM12SM cells
(Fig. 2) indicated that metastasis
was present. Samples from non-tumorigenic mice liver lobes were
considered to be positive for grade 1 metastasis when bands of 850
bp were observed in PCR products. Detection rates at 1, 2 and 3
weeks post-splenectomy were 50, 100 and 100%, respectively.
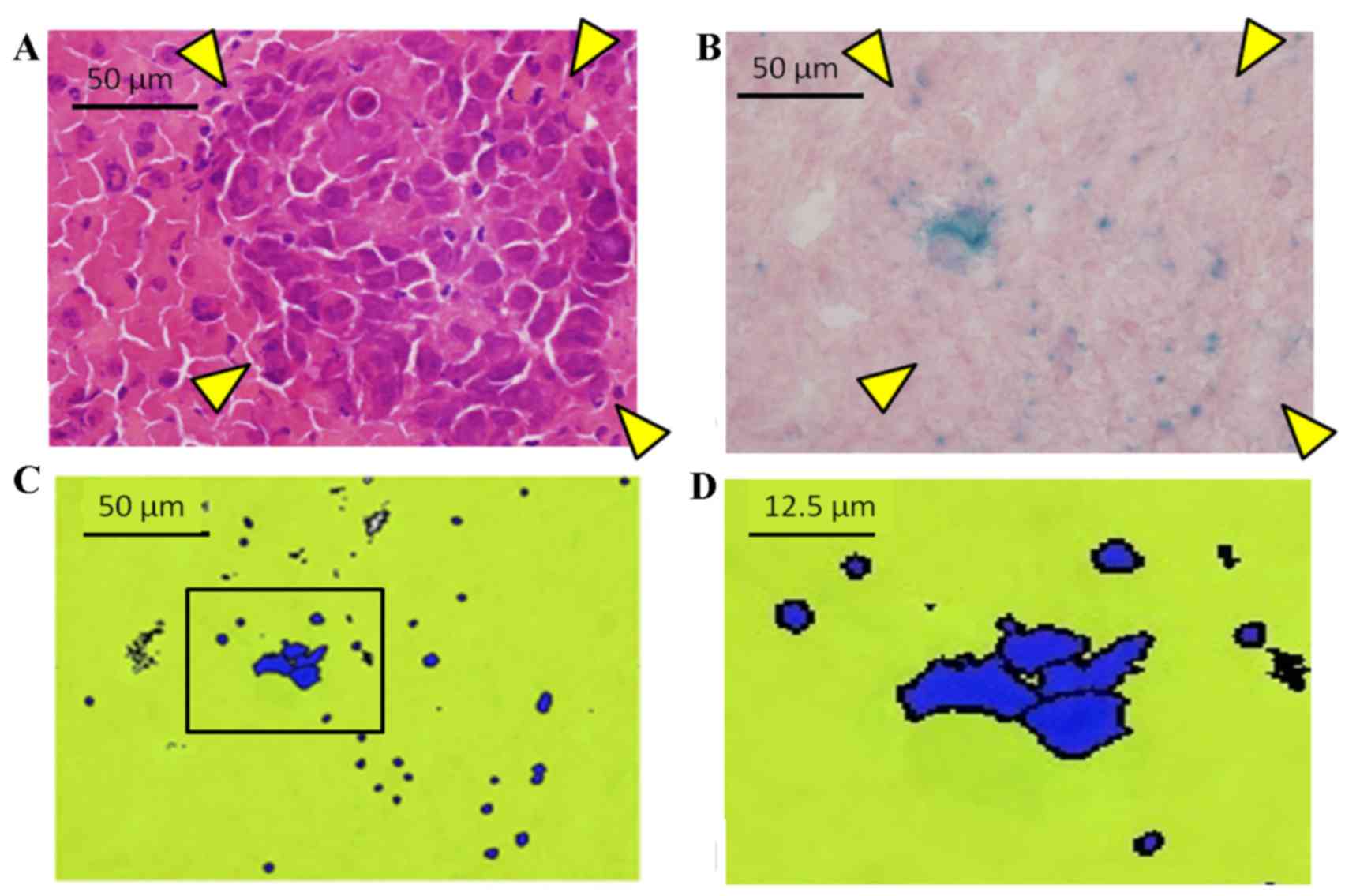
Grade-2 metastasis was not detected by microscopy.
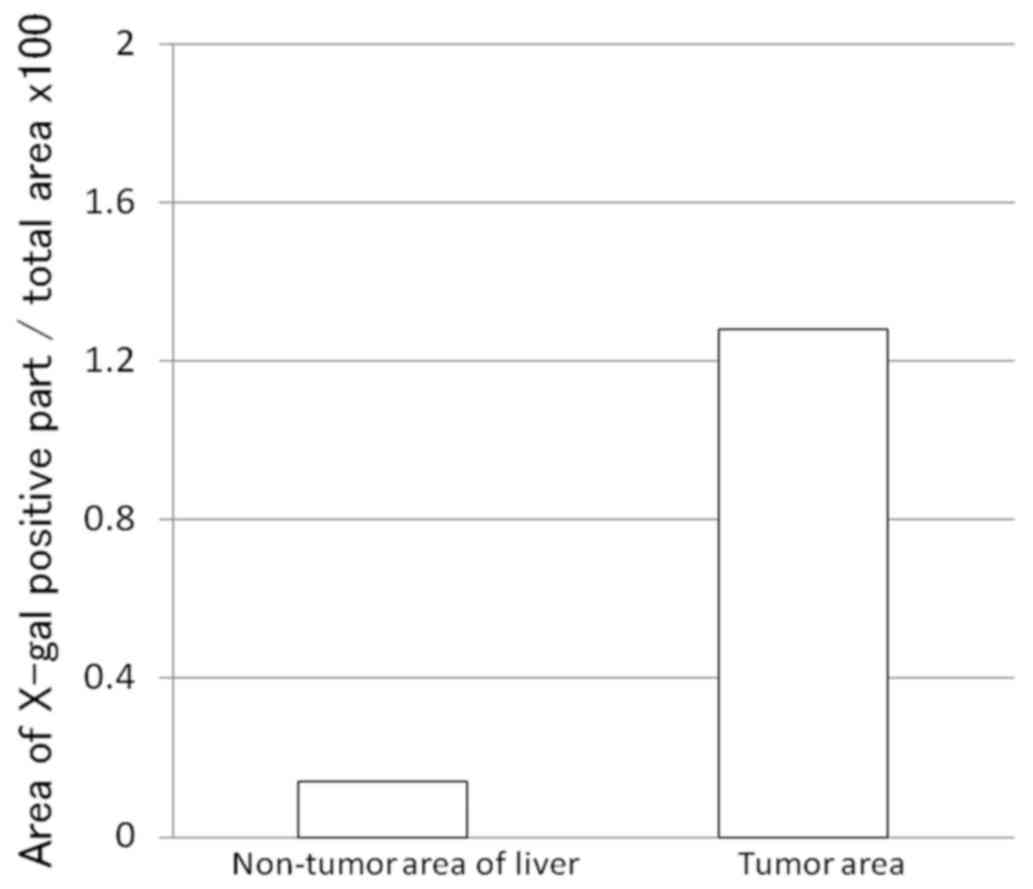
Therefore, Grade-2 metastasis was confirmed by X-gal staining
(Fig. 3) and the total amount was
calculated by the ratio of X-gal-positive cell area/liver area
×100. As a result, the LacZ-positive area ratio was 1.28% in the
tumor and 0.14% in the non-tumor section (Fig. 4).
Grade-3 metastasis detection rates were 75, 100 and
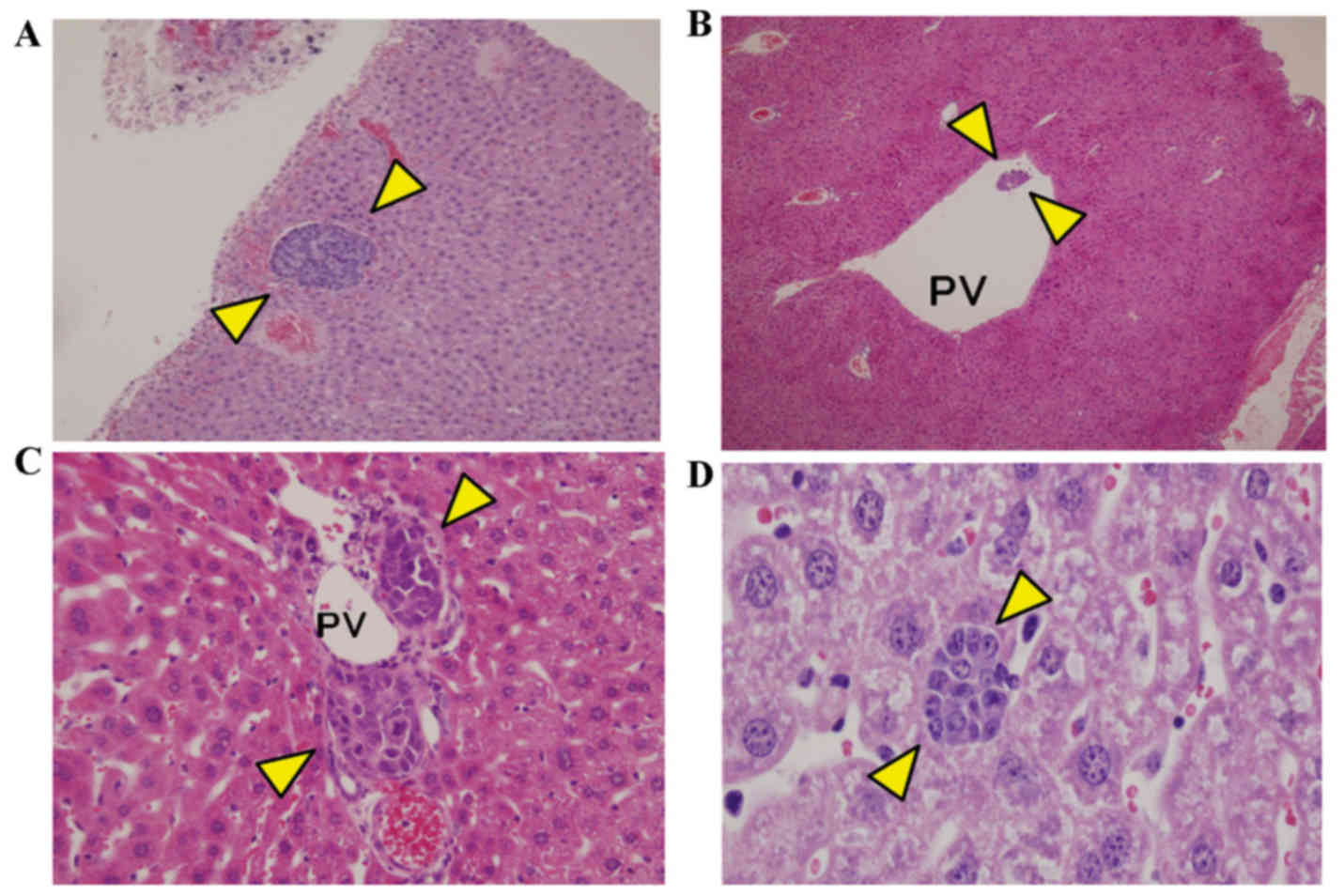
100% for 1, 2 and 3 weeks, respectively. Some lesions exhibited no
surface exposure (Fig. 5A)
Micrometastasis was observed in the portal vein lumen and wall
(Fig. 5B and C). An inflammatory
cell cluster (15–20 cells) that was similar in appearance to the
cancer cell cluster was also observed in the liver (Fig. 5D) The inflammatory cells were smaller
than hepatocytes and exhibited weak cellular adhesion and thin
nuclear chromatin, indicating the presence of plasma cells.
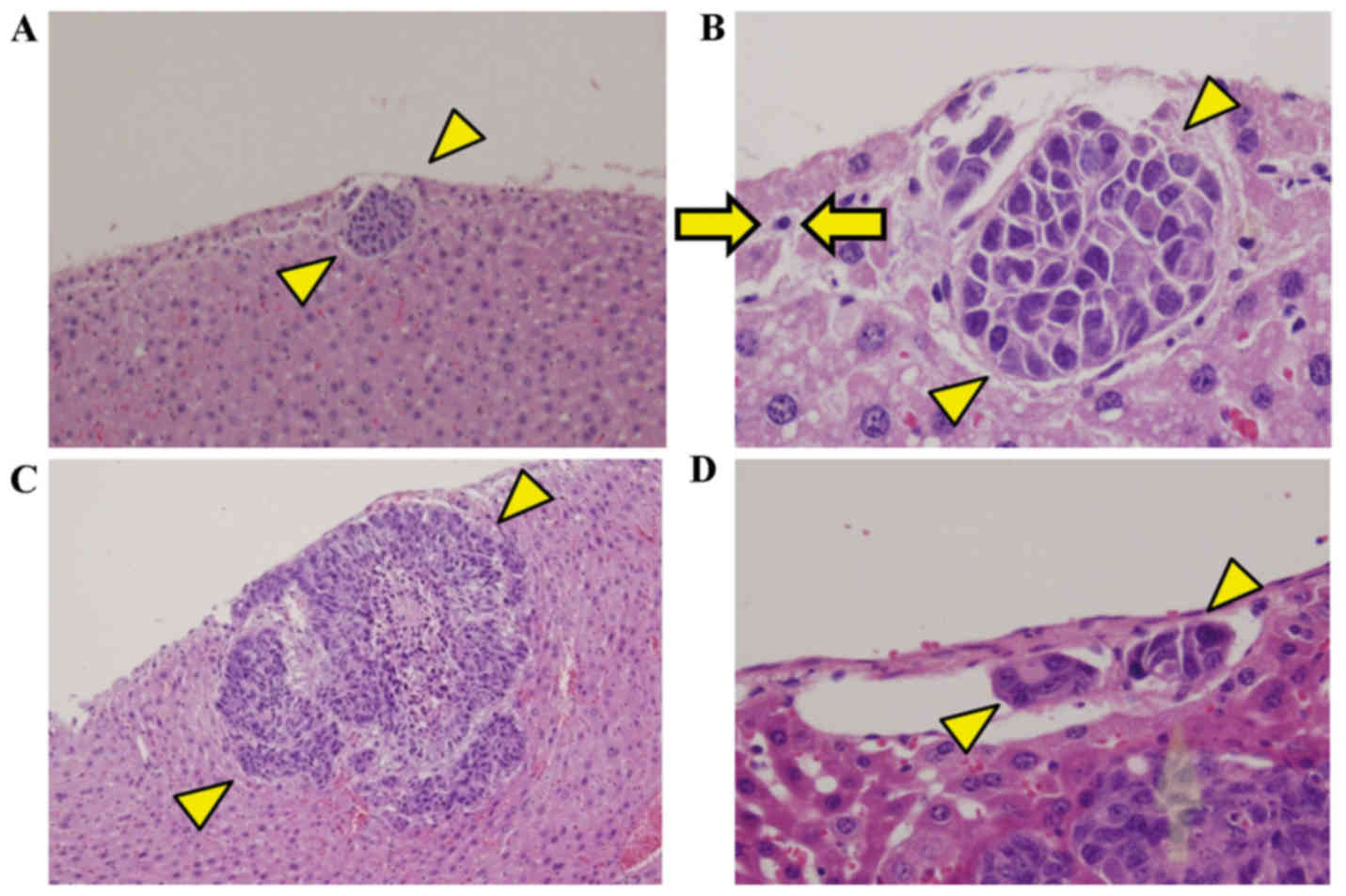
Grade-4 metastasis detection rates at 1, 2 and 3
weeks were 50, 100 and 100%, respectively, and microscopic
metastatic nodules were observed on the liver surface (Fig. 6A-C). Cancer cells were also present
in vessels surrounding the main tumor (Fig. 6D). Detection rates for each
metastatic grade are presented in Table
I.
 | Table I.Detection rate of metastases. |
Table I.
Detection rate of metastases.
|
| Week |
|---|
|
|
|
|---|
| Grade | 1 (%) | 2 (%) | 3 (%) |
|---|
| 1 | 50 | 100 | 100 |
| 2 | 0 | 0 | 0 |
| 3 | 75 | 100 | 100 |
| 4 | 50 | 100 | 100 |
Discussion
Control of liver metastasis is crucial in the
treatment of colon and rectal cancers. A number of animal models
for liver metastasis have been assessed and two methods have been
used: Tumor cell injection under the splenic capsule (splenic
injection method) or transplantation of tumor cells under the cecal
serosa (cecal method) (12–15). The splenic injection method is
advantageous as the tumor cell injection technique is relatively
easy, the formed tumor nodule does not affect survival of the
animal and it is a mouse model. The disadvantage is that the
location of the cancer cell-transplanted region is not the colon,
so the model is not orthotopic in the strict sense. By contrast, in
the cecal method the tumor is transplanted under the cecal serosa,
which is an exquisite orthotopic transplantation model. A
disadvantage of the cecal method is that tumor cell engraftment in
the cecum takes 5 weeks, it causes liver metastasis, lung and
disseminated peritoneal metastases and the handling of a rat model
is difficult (12). To investigate a
reliable ‘model with liver metastasis alone’, the present study
adopted the splenic injection method. In addition, cancer cells
were transplanted into the spleen, and the spleen was excised
following 1 week. Takiguchi et al (15) also used a KM12SM transplantation
model, employing the splenic injection method, but the spleen was
not excised and the splenic tumor grew to a very large size.
The aim of the current study was to detect ‘small
metastasis’ or ‘micrometastasis’. Although the criteria for
micrometastasis remain unclear, when a small number of cancer cells
are undetectable in conventional H&E-stained preparations but
are detected in a metastasis-target organ, it is generally
considered micrometastasis. There are two primary micrometastasis
detection methods: Detection of protein specifically expressed on
cancer or epithelial cells by immunostaining and detection of DNA
and mRNA using molecular-biological methods. The representative
method of the former is cytokeratin staining (7). Cancer cells are epithelial cells and
contain cytokeratin. By contrast, cells in blood and primary cells
of lymph nodes (lymphocytes) when the metastasis-target is a lymph
node, do not contain cytokeratin. When low levels of
cytokeratin-positive cancer cells are present in these
cytokeratin-negative environments, even a small number of cancer
cells may be clearly distinguished using cytokeratin staining. This
method is used in clinical practice to detect micro lymph node
metastasis using CAM5.2 and circulating tumor cells (CTC) (16). The representative method of the
latter is the detection of cancer-specific DNA using PCR. Nomoto
et al (17) detected
micrometastasis of pancreatic cancer using a pancreatic
cancer-specific point mutation in codon 12 of the K-ras gene in
liver tissue and regarded it as latent metastasis. However, this
method should be employed carefully because when PCR is performed
using a DNA template, fragmented or dead cancer cells and active
cancer cells are equally judged to be positive. To prevent false
positivity, a fluorescent dye strongly binding to the DNA of viable
cells, such as DAPI may be used to detect CTC (16). PCR using an mRNA template may be used
to solely judge the activity of viable cells (18), but mRNA is readily degraded and not
easy to handle. To detect cancer cells, which reached the liver
including dead cells and DNA fragments, the target to human
cell-specific chromosome was used in the present study and cancer
cells in the liver were detected at a high rate in the model. In
addition, to visualize micrometastasis of viable cells, the lacZ
gene was introduced to label cells in order to detect them in
situ. Arlt et al (19)
detected single cell-foci in the blood, withdrawn from the lung
prior to sacrifice using osteosarcoma cells stably expressing LacZ,
Dunn-LacZ and LM8-LacZ. The labeling ability of KM12SM-LacZ was
high ex vivo, but the detection of LacZ-expressing cell
aggregates at an accuracy level higher than that in conventional
pathological preparations was not possible in the liver metastasis
model in the present study. For this, two reasons were considered:
Staining of the background liver by X-gal staining; this makes
detection of the metastasis of single cells difficult and the
inability of single cells to engraft in the liver. This is due to
difficulties for single cancer cells to survive in the liver due to
rapid blood flow and the presence of many phagocytes in the portal
vein. Arlt et al (19)
attempted to detect liver metastasis of single cells, but success
or failure of the detection was not described in the study. In
H&E-stained preparations, adhered and non-adhered tumor cell
aggregates consisting of ~ten cells were observed in Grade-3
metastasis by light microscopy (Fig. 4C
and D). This finding was important to demonstrate that cancer
cell aggregates consisting of a specific number of cells moved in
the portal vein and engrafted in the liver.
The present study created a liver metastasis model
using a LacZ-labeled highly metastatic cell line, KM12SM-LacZ, and
investigated metastatic lesions formed by single cancer cells.
Arrival of DNA fragments to the liver (Grade-1 metastasis),
metastasis in pathological preparations (Grade-3) and macroscopic
metastasis on the liver surface were frequently observed, but no
intrahepatic metastasis of single cells of KM12SM-LacZ (Grade-2
metastasis) was confirmed in the X-gal-stained sections. By
contrast, micrometastasis lesions were observed in H&E-stained
preparations following light microscopy, suggesting that a specific
number of cancer cell aggregates is necessary to establish
hematogenous metastasis. However, the current model was established
within a limited environment, by injection of cancer cells into the
spleen, and thus the behavior of cancer cell aggregates in
situ remains unknown. Therefore, future studies into
unicellular metastasis in situ are warranted.
Acknowledgements
The authors of the present study thank Ms. Yukiko
Iwahara and Aimi Uchida, university students from the Department of
Clinical Pharmacy, Tokyo University of Pharmacy and Life Sciences
for their valuable technical assistance.
References
|
1
|
Nordlinger B, Guiguet M, Vaillant JC,
Balladur P, Boudjema K, Bachellier P and Jaeck D: Surgical
resection of colorectal carcinoma metastases to the liver. A
prognostic scoring system to improve case selection, based on 1568
patients. Association Française de Chirurgie. Cancer. 77:1254–1262.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Takahashi M, Hasegawa K, Oba M, Aoki T,
Sakamoto Y, Sugawara Y and Kokudo N: Repeat resection leads to
long-term survival: Analysis of 10-year follow-up of patients with
colorectal liver metastases. Am J Surg. 210:904–910. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mise Y, Imamura H, Hashimoto T, Seyama Y,
Aoki T, Hasegawa K, Beck Y, Sugawara Y, Makuuchi M, Nakajima J and
Kokudo N: Cohort study of the survival benefit of resection for
recurrent hepatic and/or pulmonary metastases after primary
hepatectomy for colorectal metastases. Ann Surg. 251:902–909. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Löwenthal D, Zeile M, Lim WY, Wybranski C,
Fischbach F, Wieners G, Pech M, Kropf S, Ricke J and Dudeck O:
Detection and characterisation of focal liver lesions in colorectal
carcinoma patients: Comparison of diffusion-weighted and
Gd-EOB-DTPA enhanced MR imaging. Eur Radiol. 21:832–840. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ueno H, Mochizuki H, Shirouzu K, Kusumi T,
Yamada K, Ikegami M, Kawachi H, Kameoka S, Ohkura Y, Masaki T, et
al: Actual status of distribution and prognostic impact of
extramural discontinuous cancer spread in colorectal cancer. J Clin
Oncol. 29:2550–2556. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fidler IJ: The pathogenesis of cancer
metastasis: The ‘seed and soil’ hypothesis revisited. Nat Rev
Cancer. 3:453–458. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Isaka N, Nozue M, Doy M and Fukao K:
Prognostic significance of perirectal lymph node micrometastases in
Dukes' B rectal carcinoma: An immunohistochemical study by CAM5.2.
Clin Cancer Res. 5:2065–2068. 1999.PubMed/NCBI
|
|
8
|
Okada K, Wada T, Ito K, Takagi Y, Aoki T
and Koyanagi Y: Investigation of the anti-tumor activity of hepatic
Kupffer cells for control of colon cancer micro-metastasis to the
liver via the Fas/Fas ligand system. J Tokyo Med Univ. 61:329–335.
2003.
|
|
9
|
Morikawa K, Walker SM, Nakajima M, Pathak
S, Jessup JM and Fidler IJ: Influence of organ environment on the
growth, selection, and metastasis of human colon carcinoma cells in
nude mice. Cancer Res. 48:6863–6871. 1988.PubMed/NCBI
|
|
10
|
Warburton PE, Greig GM, Haaf T and Willard
HF: PCR amplification of chromosome-specific alpha satellite DNA:
Definition of centromeric STS markers and polymorphic analysis.
Genomics. 11:324–333. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Becker M, Nitsche A, Neumann C, Aumann J,
Junghahn I and Fichtner I: Sensitive PCR method for the detection
and real-time quantification of human cells in xenotransplantation
systems. Br J Cancer. 87:1328–1335. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Oda H, Ogata Y and Shirouzu K: The effect
of angiogenesis inhibitor TNP-470 against postoperative lung
metastasis following removal of orthotopic transplanted human colon
cancer: An experimental study. Kurume Med J. 48:285–293. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Giavazzi R, Jessup JM, Campbell DE, Walker
SM and Fidler IJ: Experimental nude mouse model of human colorectal
cancer liver metastases. J Natl Cancer Inst. 77:1303–1308.
1986.PubMed/NCBI
|
|
14
|
Fidler IJ: Orthotopic implantation of
human colon carcinomas into nude mice provides a valuable model for
the biology and therapy of metastasis. Cancer Metastasis Rev.
10:229–243. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takiguchi S, Shimazoe T and Kono A:
Antitumor effect of camptothecin analog on liver metastatic model
of human colon cancer in nude mice. Gan To Kagaku Ryoho.
21:705–708. 1994.(In Japanese). PubMed/NCBI
|
|
16
|
Chen Y, Zou TN, Wu ZP, Zhou YC, Gu YL, Liu
X, Jin CG and Wang XC: Detection of cytokeratin 19, human
mammaglobin and carcinoembryonic antigen-positive circulating tumor
cells by three-marker reverse transcription-PCR assay and its
relation to clinical outcome in early breast cancer. Int J Biol
Markers. 25:59–68. 2010.PubMed/NCBI
|
|
17
|
Nomoto S, Nakao A, Ando N, Takeda S, Kasai
Y, Inoue S, Kaneko T and Takagi H: Clinical application of K-ras
oncogene mutations in pancreatic carcinoma: Detection of
micrometastases. Semin Surg Oncol. 15:40–46. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kutun S, Celik A, Cem Kockar M, Erkorkmaz
U, Eroğlu A, Cetin A, Erkosar B and Yakicier C: Expression of CK-19
and CEA mRNA in peripheral blood of gastric cancer patients. Exp
Oncol. 32:263–268. 2010.PubMed/NCBI
|
|
19
|
Arlt MJ, Banke IJ, Walters DK, Puskas GJ,
Steinmann P, Muff R, Born W and Fuchs B: LacZ transgene expression
in the subcutaneous Dunn/LM8 osteosarcoma mouse model allows for
the identification of micrometastasis. J Orthop Res. 29:938–946.
2011. View Article : Google Scholar : PubMed/NCBI
|