Introduction
Malignant tumors are a life-threatening disease
globally and in China. In 2015, 25% of total mortalities were
caused by cancer (1). The morbidity
and mortality of cancer has been increasing for a number of years.
In 2009, colorectal cancer was the cause of 8% of total mortalities
caused by cancer (2). Furthermore,
colorectal cancer is one of the most common tumor types. According
to data published by the National Cancer Institute in 2016,
colorectal cancer was the third most common tumor type (1). In developing countries, the rate of
colorectal cancer is also growing rapidly. From 2010–2012
developing countries contributed to 52% of the total number of
mortalities caused by colorectal cancer, and limited medical
resources meant patients had a poor prognosis and survival rate
(3). Treatment for colorectal cancer
remains limited to traditional methods, such as surgical
operations, radiotherapies and chemotherapies.
The emerging targeted molecular therapies were
gradually accepted by doctors and demonstrated particular
advantages in clinical treatment for colorectal cancer (4). The combination of emerging and
traditional therapies improves the level of disease-free survival,
survival rate and prognosis in patients with colorectal cancer.
However, disease recurrence following surgery or chemotherapy, drug
resistance and deterioration remains inevitable (5,6).
The occurrence and development of colorectal cancer
is a process controlled by multiple genes and variable factors. For
example, the migration inhibitory factor/cluster of differentiation
74 signalling axis has recently been identified as a novel
therapeutic target for colon cancer (7). Furthermore, loss of periplakin has been
demonstrated to be associated with tumorigenesis of colon cancer
(8). The lymph node is the primary
defence against the metastasis of colorectal tumors, and is also
where deterioration of health begins in patients with colorectal
cancer.
Lymph node metastasis may worsen the prognosis,
reduce the survival rate and even make patients more susceptible to
the recurrence of colorectal cancer (3). Furthermore, lymph node metastasis may
worsen the postoperative curative effect and enhance
drug-resistance to chemotherapies for colorectal cancer. Previous
research based on clinical practical experience demonstrated that
the 5-year survival rate may reach 60–80% in patients without lymph
node metastasis; however, the 5-year survival rate of patients with
lymph node metastasis may only reach 30% (9). Surgery is unable to improve the
prognosis of patients with colorectal cancer.
With the development of molecular biology, the
understanding of colorectal cancer has progressed beyond the
cellular level and further to elucidate the role of genetic
biomarkers. There are two types of genetic alterations in
colorectal cancer, chromosomal instability (CIN) and microsatellite
instability (MSI). Aneuploidy and polyploidy are common phenotypes
in CIN and contribute to 80–85% of the morbidity of colorectal
cancer (10). MSI is primarily
caused by errors in the DNA repairing process, and contributes to
15–20% of the total morbidity (10).
There are multiple chromosome sites with copy number variation
(CNV) in CIN-type colorectal cancer (11). If CNV occurs inside or around the
tumor-associated gene sequences, oncogenes may be activated and
anti-oncogenes may be inactivated, which eventually induces
tumorigenesis (12). A previous
study indicated that increased CNVs may be associated with the
progression of colitis gravis to colorectal cancer (13). Another study demonstrated that CNVs
were able to determine the lymph node metastasis in colorectal
cancer (14). Evidence now indicates
that the occurrence of CNVs in chromosome 4 may seriously induce
lymph node metastasis in colorectal cancer (14–16).
Single nucleotide polymorphisms (SNPs) are a primary
cause for variation in the human race. However, the association
between SNPs and colorectal cancer remains unclear. In the present
study, 1,053 associated genes on chromosome 4 were screened for
lymph node metastasis-associated SNPs and CNVs and the mRNA level
of lymph node metastasis on these genes was further investigated.
The current study aimed to provide a molecular basis for clinical
tests and treatment.
Materials and methods
Subjects
A total of 78 tissue samples (39 colorectal tumor
and 39 normal tissues) from 39 patients were collected between
January 2013 and September 2014 following tumor reduction surgery
in Shenzhen Second People's Hospital (Shenzhen, China). The
collection of tissues was approved by the Ethics Committee of
Shenzhen Second People's Hospital. Written informed consent was
provided by all patients. All experiments using human blood samples
were conducted in accordance with the Clinical Sample Collection
and Treatment Guidelines outlined by the Ethics Committee of
Shenzen Second People's Hospital. Among the 39 patients, 19 were
female and 20 were male. The age of the subjects ranged from 29–84,
with a mean age of 61.4 years. There were 19 cases with tumor
diameters ≤5 cm and 20 cases with tumor diameters >5 cm. In the
39 patients, 29 exhibited tumors in the colon, whereas 10 patients
exhibited tumors in the rectum. There were 6 cases with
highly-differentiated tumors, 25 with medium-differentiated tumors
and 8 with low-differentiated tumors, according to a
differentiation scale determined by the Pathology Department of
Shenzhen Second People's Hospital. There were 17 cases presenting
lymph node metastasis, 8 cases were identified with distant
metastasis and 14 cases exhibited no tumor metastasis. Pathological
staging demonstrated that 19 cases were at phase I–II and 20 cases
were at phase III–IV. Pathological staging was assessed according
to the TNM colorectal cancer staging system of the American Joint
Committee on Cancer (AJCC; AJCC staging system 7th edition 2011;
cancerstaging.org) and the Union for
International Cancer Control (uicc.org)
standards. All patients included in the study were tested to
exclude other diseases, such as gastroenteritis.
DNA and RNA extraction
During surgery, 0.2 g tissue was harvested from each
patient, pre-treated with liquid nitrogen and processed into a
powder. The DNA extraction procedure was performed using a DNeasy
Blood & Tissue kit (Qiagen GmbH, Hilden, Germany), according to
the manufacturer's instructions. RNA extraction was performed using
TRIzol reagent (Ambion; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), following the manufacturer's instructions. The nucleotide
concentration was determined using a Qubit 3.0 fluorometer and the
integrity of DNA molecules were examined by electrophoresis, using
1% agarose gel stained with 0.01% acridine orange.
Exosome sequencing and bioinformatics
analysis
The exosome sequencing was conducted using the Hiesq
2000 System (Illumina, Inc., San Diego, CA, USA) with a NimbleGen
4.6 microarray chip (Roche Diagnostics, Basel, Switzerland). The
raw data were acquired and filtered according to signal intensity,
gene annotation and sequence clustering. The sequences were then
compared and statistically analyzed. Comparative genome
hybridization (CGH) data were analyzed using R 3.3.2 rCGH software
obtained from Bioconductor (bioconductor.org). Log ratios of CNVs were calculated
by comparing normalized data from the sequencing of tumor and
normal tissues.
Gene expression analysis via reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted from the tumor and normal tissues,
as described above. RNA samples were reverse transcribed using a
PrimeScript Reverse Transcription kit (Takara Biotechnology Co.,
Ltd., Dalian, China), according to the manufacturer's instructions.
A SYBR Premix Ex Taq kit (Takara Biotechnology Co., Ltd.) was used
for qPCR, according to the manufacturer's instructions. The RT-qPCR
primers for vascular endothelial growth factor C (VEGFC), cyclin-A2
(CCNA2), interleukin-2 (IL2), ATP-binding cassette sub-family G
member 2 (ABCG2), epidermal growth factor (EGF), nuclear factor
kappa B subunit 1 (NFKB1) and glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) are presented in Table I. Blank controls using only primers
or templates were included in the RT-qPCR experiment. An Applied
Biosystems 7900HT Fast Real-Time PCR machine (Applied Biosystems;
Thermo Fisher Scientific, Inc.) was used to run the following
reaction conditions: 95°C for 15 min, then 40 cycles of 95°C for 10
sec and 60°C for 30 sec. Relative gene expression levels were
normalized to GAPDH, according to the 2−ΔΔCq method
(10,17). Experiments were performed in
triplicate.
 | Table I.Primer sequences for polymerase chain
reaction. |
Table I.
Primer sequences for polymerase chain
reaction.
| Gene | Sequence, 5′→3′ | Product length,
bp |
|---|
| GAPDH | F:
GGGTGTGAACCATGAGAAGT | 149 |
|
| R:
CAGTGATGGCATGGACTGTG |
|
| VEGFC | F:
TGGGGAAGGAGTTTGGAGTC | 181 |
|
| R:
GTTACTGGTTTGGGGCCTTG |
|
| CCNA2 | F:
TGCTGACCCATACCTCAAGT | 167 |
|
| R:
GGTAGGTCTGGTGAAGGTCC |
|
| IL2 | F:
AACTCACCAGGATGCTCACA | 159 |
|
| R:
TGCTGATTAAGTCCCTGGGT |
|
| ABCG2 | F:
ACGCATCCTGAGATCCTGAG | 155 |
|
| R:
CAGGTCATTGGAAGCTGTCG |
|
| EGF | F:
CAGGGAAGATGACCACCACT | 168 |
|
| R:
TCTCGGTACTGACATCGCTC |
|
| NFKB1 | F:
TGTCCAGCTTCGGAGGAAAT | 182 |
|
| R:
CACTACCAAACATGCCTCCG |
|
Statistical analysis
SPSS version 16.0 (SPSS, Inc., Chicago, IL, USA) was
used to run data normalization and statistical tests. CGH data were
analyzed using R 3.3.2 rCGH software obtained from Bioconductor
(bioconductor.org). Fisher's exact test was
applied to evaluate the association among SNPs, CNVs and lymph node
metastasis in colorectal cancer. Student's t-test was applied to
test the statistical significance of gene expression alteration.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Quality control of sequencing
data
Raw data were filtered and adapters were removed.
Sequencing results demonstrated that the target sequence was
4.5×1012 base pairs, with coverage of >99% and the
sequencing depth of ×250. A quality check was automatically
performed on the raw data by the R 3.3.2 rCGH package, and the
qualified data were used for subsequent analysis.
Association between SNPs and lymph
node metastasis colorectal cancer
All 20,000 SNPs in the 1,053 cancer related genes
were analyzed, 10,000 nonsense mutations were filtered out and the
remaining SNPs were analyzed by statistical tests. Results
indicated that 21 SNPs in 16 genes were significantly associated
with colorectal cancer (P<0.05). Patients with lymph node
metastasis exhibited higher mutation rates of solute carrier family
28 member 3 (SLC28A3; rs10868138, rs56350726), breast cancer 1
(BRCA1; rs16941, rs16942, rs799917, rs1799966), ribonucleotide
reductase regulators subunit M2 (RRM2; rs1130609), PMS1 homolog 2
(PMS2; rs1805323), cytidine deaminase (CDA; rs2072671), epoxide
hydrolase 1 (EPHX1; rs2234922), heterogeneous ribonucleoprotein
particle-associated with lethal yellow (RALY; rs2281209), Siglec-3
(CD33; rs2455069), B cell lymphoma 10 (BCL10; rs3768235) and ETS
variant 1 (ETV1; rs9639168) than patients without lymph node
metastasis (P<0.05). Patients with lymph node metastasis had a
lower mutation frequency of macrophage stimulating 1 receptor 1
(MST1R; rs1062633), lysine methyltransferase 2B (KMT2B;
rs16970649), B cell lymphoma 2 (BCL2; rs1800477), U6 small nuclear
RNA-associated Sm-like protein 3 (rs1870134), thyroid transcription
factor 1 (TTF1; rs3739914, rs8999) and mitogen-activated protein 3
kinase 1 (MAP3K1; rs702689) than patients without lymph node
metastasis (P<0.05). These results are presented in Table II.
 | Table II.Association between SNPs and lymph
node metastasis colorectal cancer. |
Table II.
Association between SNPs and lymph
node metastasis colorectal cancer.
|
|
|
| Lymph node
metastasis, n (%) |
|
|---|
|
|
|
|
|
|
|---|
| Gene | SNP, rs ID | Gene type | With lymph node
metastasis | Without lymph node
metastasis | P-value |
|---|
| MST1R | rs1062633 | TC/CC C | 3 (27.3) | 10 (71.4) | 0.047 |
|
|
| TT | 8 (72.7) | 4 (28.6) |
|
| SLC28A3 | rs10868138 | TC | 6 (54.5) | 0 (0) | 0.003a |
|
|
| TT | 5 (45.5) | 14 (100) |
|
| SLC28A3 | rs56350726 | TA/AA | 7 (63.6) | 0 (0.0) | 0.001a |
|
|
| TT | 4 (36.4) | 14 (100.0) |
|
| RRM2 | rs1130609 | TG/GG | 8 (72.7) | 4 (28.6) | 0.047 |
|
|
| TT | 3 (27.3) | 10 (71.4) |
|
| BRCA1 | rs16941 | TC/CC | 10 (90.9) | 7 (50.0) | 0.042 |
|
|
| TT | 1 (9.1) | 7 (50.0) |
|
| BRCA1 | rs16942 | TC/CC | 10 (90.9) | 7 (50.0) | 0.042 |
|
|
| TT | 1 (9.1) | 7 (50.0) |
|
| BRCA1 | rs799917 | GA/AA | 10 (90.9) | 7 (50.0) | 0.042 |
|
|
| GG | 1 (9.1) | 7 (50.0) |
|
| BRCA1 | rs1799966 | TC/CC | 10 (90.9) | 7 (50.0) | 0.042 |
|
|
| TT | 1 (9.1) | 7 (50.0) |
|
| KMT2B | rs16970649 | CT | 0 (0) | 5 (35.7) | 0.046 |
|
|
| CC | 11 (100) | 9 (64.3) |
|
| BCL2 | rs1800477 | CT/TT | 0 (0.0) | 5 (35.7) | 0.046 |
|
|
| CC | 11 (100) | 9 (64.3) |
|
| PMS2 | rs1805323 | GT/TT | 10 (90.9) | 7 (50.0) | 0.042 |
|
|
| GG | 1 (9.1) | 7 (50.0) |
|
| LSM3 | rs1870134 | GC/CC | 1 (9.1) | 8 (57.1) | 0.033 |
|
|
| GG | 10 (90.9) | 6 (42.9) |
|
| CDA | rs2072671 | AC/CC | 7 (63.6) | 3 (21.4) | 0.049 |
|
|
| AA | 4 (36.4) | 11 (78.6) |
|
| EPHX1 | rs2234922 | AG/GG | 4 (36.4) | 0 (0.0) | 0.026 |
|
|
| AA | 7 (63.6) | 14 (100.0) |
|
| RALY | rs2281209 | GA | 4 (36.4) | 0 (0.0) | 0.026 |
|
|
| GG | 7 (63.6) | 14 (100.0) |
|
| CD33 | rs2455069 | AG | 4 (36.4) | 0 (0.0) | 0.026 |
|
|
| AA | 7 (63.6) | 14 (100.0) |
|
| TTF1 | rs3739914 | AG/GG | 2 (18.2) | 9 (64.3) | 0.042 |
|
|
| AA | 9 (81.8) | 5 (35.7) |
|
| TTF1 | rs8999 | CA/AA | 2 (18.2) | 10 (71.4) | 0.015 |
|
|
| CC | 9 (81.8) | 4 (28.6) |
|
| BCL10 | rs3768235 | CT | 6 (54.5) | 1 (7.1) | 0.021 |
|
|
| CC | 5 (45.5) | 13 (92.9) |
|
| MAP3K1 | rs702689 | GA/AA | 1 (9.1) | 9 (64.3) | 0.012 |
|
|
| GG | 10 (90.9) | 5 (35.7) |
|
| ETV1 | rs9639168 | TC/CC | 9 (81.8) | 5 (35.7) | 0.042 |
|
|
| TT | 2 (18.2%) | 9 (64.3%) |
|
Clustering analysis of lymph node
metastasis-associated SNPs in colorectal cancer
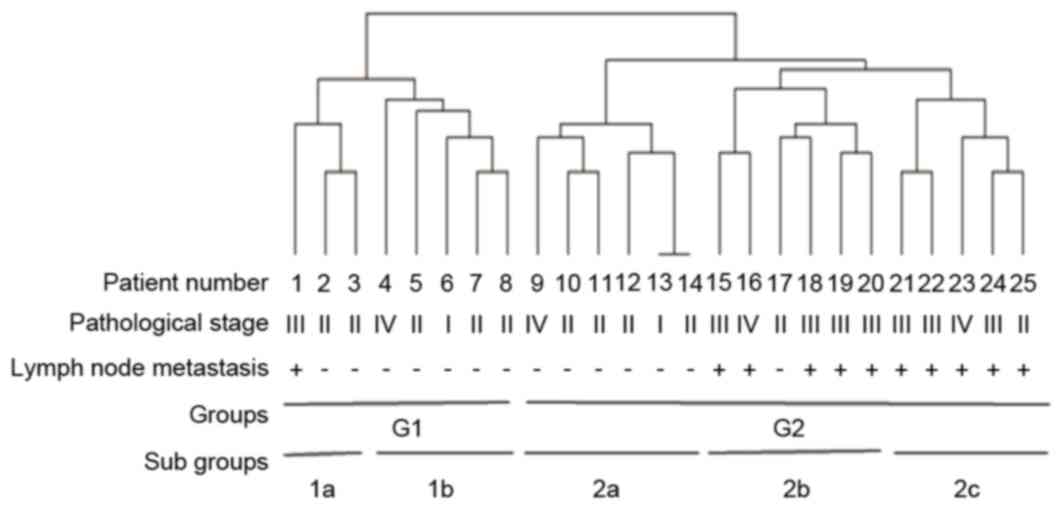
The mutations present in patients were analyzed via
hierarchical clustering. The results divided patients into two
groups (G1 and G2) and five sub-groups (1a, 1b, 2a, 2b and 2c).
Results demonstrated an association between colorectal cancer stage
and mutation frequency, with a higher rate of lymph node metastasis
being present in the later stages of colorectal cancer (Fig. 1). Clustering of 21 lymph node
metastasis-associated SNPs in colorectal cancer indicated that the
mutation frequencies of SLC28A3, BRCA1, RRM2, PMS2, CDA and ETV1
were associated with the G2 group and the mutation frequencies of
EPHX1, RALY, CD33 and BCL10 were associated with groups 2b and 2c
(Fig. 2). Multiple mutations in
BRCA1 were closely clustered, indicating that BRCA1 may be a
potential marker for lymph node metastasis in colon cancer.
However, a larger sample size is required to verify genetic
hallmarks for lymph node metastasis.
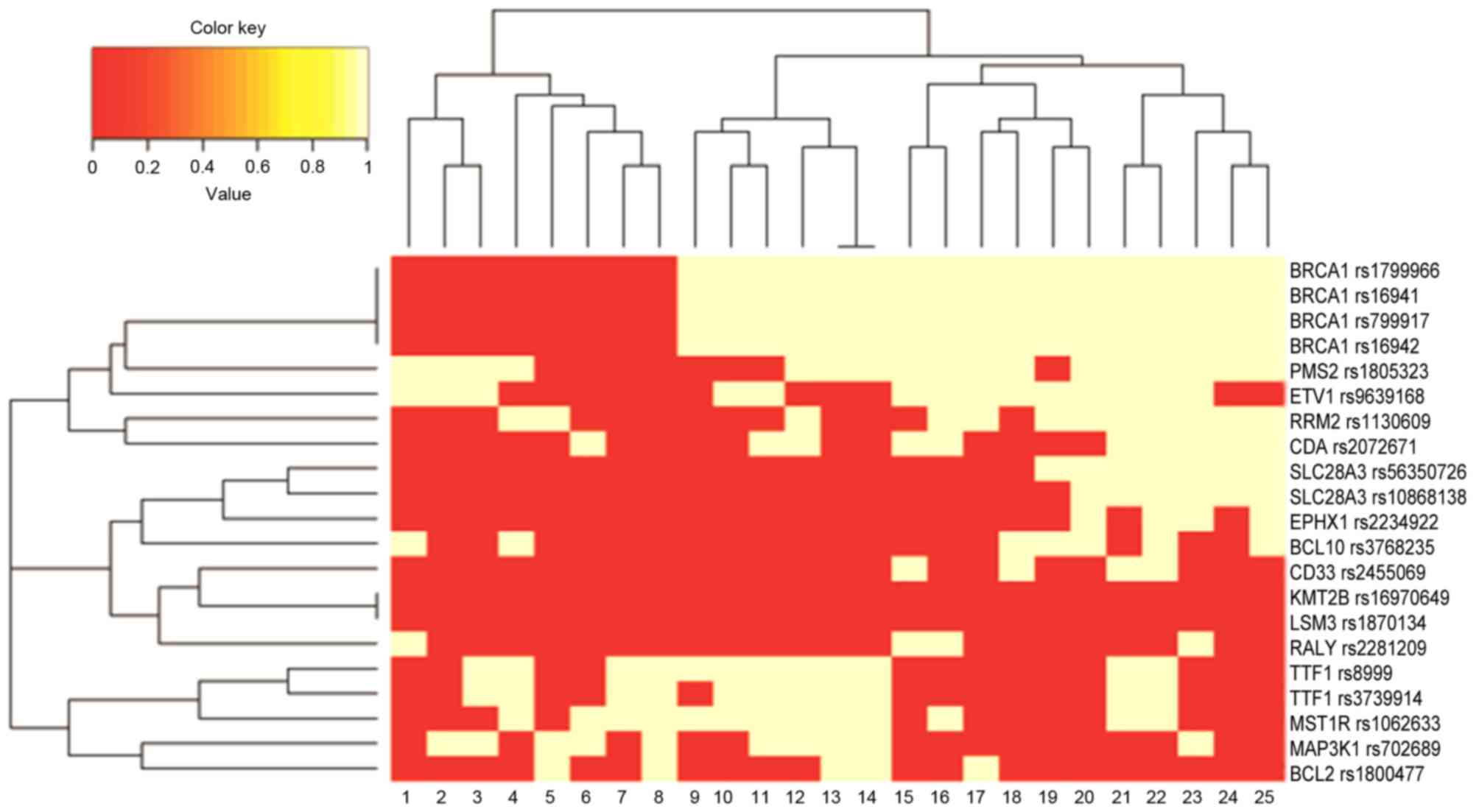 | Figure 2.Clustering of correlations between
single nucleotide polymorphisms and groups of patients with
colorectal cancer. The color key indicates the correlation
coefficient, with red being the most-associated. BRCA1, breast
cancer 1; PMS2, PMS1 homolog 2; ETV1, ets variant 1; RRM2,
ribonucleotide reductase regulators subunit M2; CDA, cytidine
deaminase; SLC28A3, solute carrier family 28 member 3; EPHX1,
epoxide hydrolase 1; BCL10, B cell lymphoma 10; CD33, Siglec-3;
KMT2B, lysine methyltransferase 2B; LSM3, U6 small nuclear
RNA-associated Sm-like protein 3; RALY, ribonucleoprotein
particle-associated with lethal yellow; TTF1, thyroid transcription
factor 1; MST1R, macrophage stimulating 1 receptor 1; MAP3K1,
mitogen-activated protein 3 kinase 1; BCL2, B cell lymphoma 2. |
Association between CNVs and lymph
node metastasis in colorectal cancer
Of all cases, 15 presented with copy number
alterations, which accounted for 60% of all subjects. In the 1,503
candidate genes, 80 were identified to have CNVs. However, only one
of the CNVs in the 80 genes (DDR1) was associated with lymph node
metastasis in colorectal cancer, although this was not
statistically significant (P=0.072).
Gene expression alteration in lymph
node metastasis of colorectal cancer
The relative mRNA expression level of EGF in tumor
tissues (1.00±0.28) was significantly lower than in normal tissues
(4.89±1.56; P<0.05). The relative mRNA expression level of NFKB1
in tumor tissues (3.23±0.80) was significantly higher than in
normal tissues (1.25±0.25; P<0.05).
Discussion
In the present study, 21 SNPs in 16 genes associated
with lymph node metastasis of colorectal cancer were screened. Only
1 CNV in the DDR1 gene was identified to be associated with lymph
node metastasis in colorectal cancer, although this difference was
not statistically significant. EGF and NFKB1 were abnormally
expressed in colorectal tumor tissues.
EGF is a multi-functional growth factor; it is able
to bind specific receptors on the cell surface and further induce
signal transduction. In tumor cells, EGF may mediate proliferation
by activating the EGF receptor pathway which leads to tumor cells
survival and metastasis (18).
Previous studies demonstrated that EGF is able to induce tumor
metastasis through matrix metalloproteinases (19), tyrosine kinase PK2 (20), Podoplanin (21), Rictor binding protein (22), epithelial mesenchymal transition
(23) and improving blood vessel
(24) and lymph gland growth
(25,26). In the present study, the mRNA level
of EGF was significantly upregulated in patients with lymph node
metastasis, which indicated that EGF may be associated with lymph
node metastasis in colorectal cancer. A previous study demonstrated
that EGF is able to induce lymph gland metastasis by promoting
lymph gland progression (19).
NFKB was initially identified in B lymphocytes, it
is associated with a number of transcriptional processes by binding
to promoter sites. NFKB is rarely mentioned as being associated
with lymph node metastasis. The current study indicates that a
significant upregulation of NFKB occurs during lymph node
metastasis in colorectal cancer, indicating that NFKB may be
associated with lymph node metastasis. The present study may
provide a basis for further validation and identification of
genetic markers.
In conclusion, the results of the present study
indicate a number of potential genetic biomarkers associated with
lymph node metastasis which may provide insight into early
prognosis of colorectal cancer. However, a limitation of the
present study was that the sample size was small. Therefore, a
larger sample size is required for further validation of genetic
biomarkers for colorectal cancer.
Acknowledgements
The present study was supported by the Project of
Basic Research Plan Program of Shenzhen (grant no.
JCYJ20160328161613864), the Innovation Program of Shenzhen (grant
no. JCYJ20150330102720122), the Project of Shenzhen Basic
Development Program (grant nos. JCYJ20120613171430264 and
JCYJ20130329110928684), the Project of Shenzhen International
Cooperation Foundation (grant nos. GJHZ20130412153906740 and
GJHZ20160301163138685), the Nature Science Foundation of Guangdong
(grant no. 2016A030313029) and the Project of Guangdong Scientific
Plan Foundation (grant nos. 2013B021800097 and 2014A020212038).
References
|
1
|
American Cancer Society, . Cancer facts
and figures 2016. American Cancer Society; Atlanta, GA: 2016
|
|
2
|
Ilic M and Ilic I: Colorectal cancer
mortality trends in Serbia during 1991–2010: An age-period-cohort
analysis and a joinpoint regression analysis. Chin J Cancer.
35:552016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bishehsari F, Mahdavinia M, Vacca M,
Malekzadeh R and Mariani-Costantini R: Epidemiological transition
of colorectal cancer in developing countries: Environmental
factors, molecular pathways, and opportunities for prevention.
World J Gastroenterol. 20:6055–6072. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Abramson RG: Overview of targeted
therapies for cancer. My Cancer Genome. https://www.mycancergenome.org/content/molecular-medicine/overview-of-targeted-therapies-for-cancer/July
29–2016
|
|
5
|
Gupta A, Kaur CD and Saraf S and Saraf S:
Targeting of herbal bioactives through folate receptors: A novel
concept to enhance intracellular drug delivery in cancer therapy. J
Recept Signal Transduct Res. 1–10. 2017.
|
|
6
|
Matsumoto K, Umitsu M, De Silva DM, Roy A
and Bottaro DP: HGF-MET in cancer progression and biomarker
discovery. Cancer Sci. Jan 8–2017.(Epub ahead of print). View Article : Google Scholar
|
|
7
|
Bozzi F, Mogavero A, Varinelli L, Belfiore
A, Manenti G, Caccia C, Volpi CC, Beznoussenko GV, Milione M, Leoni
V, et al: MIF/CD74 axis is a target for novel therapies in colon
carcinomatosis. J Exp Clin Cancer Res. 36:162017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li X, Zhang G, Wang Y, Elgehama A, Sun Y,
Li L, Gu Y, Guo W and Xu Q: Loss of periplakin expression is
associated with the tumorigenesis of colorectal carcinoma. Biomed
Pharmacother. 87:366–374. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen W, Chen X and Lou R: Clinical study
on lymph node metastasis of colorectal cancer. Chin J Cancer.
19:479–480. 2000.(in Chinese).
|
|
10
|
Grady WM and Carethers JM: Genomic and
epigenetic instability in colorectal cancer pathogenesis.
Gastroenterology. 135:1079–1099. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sen S: Aneuploidy and cancer. Curr Opin
Oncol. 12:82–88. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pérez-Torras S, Vidal-Pla A, Cano-Soldado
P, Huber-Ruano I, Mazo A and Pastor-Anglada M: Concentrative
nucleoside transporter 1 (hCNT1) promotes phenotypic changes
relevant to tumor biology in a translocation-independent manner.
Cell Death Dis. 4:e6482013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shivakumar BM, Rotti H, Vasudevan TG,
Balakrishnan A, Chakrabarty S, Bhat G, Rao L, Pai CG and
Satyamoorthy K: Copy number variations are progressively associated
with the pathogenesis of colorectal cancer in ulcerative colitis.
World J Gastroenterol. 21:616–622. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen Z, Liu Z, Deng X, Warden C, Li W and
Garcia-Aguilar J: Chromosomal copy number alterations are
associated with persistent lymph node metastasis after
chemoradiation in locally advanced rectal cancer. Dis Colon Rectum.
55:677–685. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang JW, Shi ZZ, Zhang TT, Hao JJ, Wang
Z, Wang XM, Yang H, Wang MR, Zhou ZX and Zhang Y: Analysis of
genomic aberrations associated with the clinicopathological
parameters of rectal cancer by array-based comparative genomic
hybridization. Oncol Rep. 29:1827–1834. 2013.PubMed/NCBI
|
|
16
|
Sawada T, Yamamoto E, Suzuki H, Nojima M,
Maruyama R, Shioi Y, Akasaka R, Kamimae S, Harada T, Ashida M, et
al: Association between genomic alterations and metastatic behavior
of colorectal cancer identified by array-based comparative genomic
hybridization. Genes Chromosomes Cancer. 52:140–149. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-tie quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maurer G, Tarkowski B and Baccarini M: Raf
kinases in cancer-roles and therapeutic opportunities. Oncogene.
30:3477–3488. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kajanne R, Miettinen P, Mehlem A, Leivonen
SK, Birrer M, Foschi M, Kähäri VM and Leppä S: EGF-R regulates MMP
function in fibroblasts through MAPK and AP-1 pathways. J Cell
Physiol. 212:489–497. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Verma N, Keinan O, Selitrennik M, Karn T,
Filipits M and Lev S: PYK2 sustains endosomal-derived receptor
signalling and enhances epithelial-to-mesenchymal transition. Nat
Commun. 6:60642015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Inoue H, Miyazaki Y, Kikuchi K, Yoshida N,
Ide F, Ohmori Y, Tomomura A, Sakashita H and Kusama K: Podoplanin
expression during dysplasia-carcinoma sequence in the oral cavity.
Tumour Biol. 33:183–194. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang F, Zhang X, Li M, Chen P, Zhang B,
Guo H, Cao W, Wei X, Cao X, Hao X and Zhang N: mTOR complex
component Rictor interacts with PKCzeta and regulates cancer cell
metastasis. Cancer Res. 70:9360–9370. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hardy KM, Booth BW, Hendrix MJ, Salomon DS
and Strizzi L: ErbB/EGF signaling and EMT in mammary development
and breast cancer. J Mammary Gland Biol Neoplasia. 15:191–199.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim HD, Meyer AS, Wagner JP, Alford SK,
Wells A, Gertler FB and Lauffenburger DA: Signaling network state
predicts twist-mediated effects on breast cell migration across
diverse growth factor contexts. Mol Cell Proteomics.
10:M111.008433. 2011. View Article : Google Scholar
|
|
25
|
Bracher A, Cardona AS, Tauber S, Fink AM,
Steiner A, Pehamberger H, Niederleithner H, Petzelbauer P, Gröger M
and Loewe R: Epidermal growth factor facilitates melanoma lymph
node metastasis by influencing tumor lymphangiogenesis. J Invest
Dermatol. 133:230–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rovenská E: Importance of
lymphangiogenesis and ultrastructure of lymphatic capillaries in
metastasis of malignant melanoma. Vnitr Lek. 60:582–585.
2014.PubMed/NCBI
|