Introduction
Alzheimer's disease (AD) is the leading cause of
dementia worldwide and is characterized by the accumulation of
amyloid-β plaques (Aβ), neurofibrillary tangles, synaptic and
neuronal losses, and cognitive decline (1). Accumulation of Aβ and activation of
neuro-inflammatory cytokines are directly associated with memory
disturbances in the early stages of disease (2,3).
Although the molecular mechanisms underlying these changes remain
to be elucidated, alterations in transcription factors have been
implicated in neuronal dysfunction, excitotoxic cascades and the
production of toxic proteins in AD (4–6).
A previous RNA-sequencing analysis in acutely
isolated neurons revealed that transcription factor Myc-interacting
zinc-finger protein 1 (Miz1) was differentially expressed in
amyloid precursor protein/presenelin-1 (APP/PS1) mice compared with
age-matched control littermates (7).
Miz1, also called protein inhibitor of activated signal transducer
and activation of transcription 2 (PIAS2), was initially identified
as an interacting protein of transcription factor c-Myc (8–10). It
contains 13 zinc fingers at the C-terminus and a poxvirus and
zinc-finger domain at the N-terminus, which is required for
transcriptional activation (11,12).
Miz1 serves a critical role in proliferation, differentiation,
cell-cycle progression, apoptosis, and autophagy via
transcriptional activation and repression of its target genes
(13–19). However, few studies have investigated
the association between Miz1 and AD.
In the present study, the expression patterns of
Miz1 were examined in APP/PS1 mice and an increase in Miz1
expression was observed in the cortex but not in the hippocampus.
Miz1 was also expressed in the cell bodies and dendrites of neurons
and astrocytes, which indicated that alterations in Miz1 may be
associated with the pathophysiology of AD.
Materials and methods
Ethics statement
All animal experiments in the present study were
reviewed and approved by the Ethics Committee of Chongqing Medical
University (Chongqing, China; approval no. 0002648).
Mice
A total of 30 male APP/PS1 transgenic mice with a
C57BL/6J genetic background (n=15) and their wild-type littermates
(n=15) were purchased from the Model Animal Research Center of
Nanjing University (Nanjing, China). All mice (20–25 g; 7 months of
age) were housed in standard cages (48×26 cm2) with 5
animals per cage. Mice were kept in a room at a controlled
temperature (22±1°C) under a 12-h light/dark cycle and fed a pellet
rodent diet and water ad libitum. All mice protocols were
approved by the Chongqing Medical University Animal Welfare
Committee. Specific primers for the APP and PS1 genes were designed
by Sangon Biotech Co., Ltd. (Shanghai, China) as follows: APP
forward, 5′-GACTGACCACTCGACCAGGTTCTG-3′ and reverse,
5′-CTTGTAAGTTGGATTCTCATATCCG-3′; PS1 forward,
5′-AATAGAGAACGGCAGGAGCA-3′ and reverse,
5′-GTGGATAACCCCTCCCCCAGCCTAGACC-3′, which have been accepted for
the identification of APP/PS1 mice (19,20).
Mice were kept until they were 7 months old, as at this age mice
display both pathological and behavioral abnormalities, including
Aβ deposition and cognitive impairment (21,22). For
immunofluorescence analysis, brain tissues were fixed in 4%
paraformaldehyde, 20% sucrose in PBS, and 30% sucrose in PBS, and
subsequently sectioned into 10-mm-thick frozen slices. For western
blot analysis, the neocortex and hippocampus were dissected quickly
using RNase-free instruments and stored in liquid nitrogen for
further use.
Reagents
Antibodies against PIAS2 were from Abcam (Cambridge,
MA, USA; 1:1,000) and antibodies against Miz1 were purchased from
BIOSS (Beijing, China; bs-11234R; 1:40). Antibodies against glial
fibrillary acidic protein (GFAP) were purchased from Boster
Biological Technology (BM4393, Pleasanton, CA, USA; 1:100) and
antibodies against microtubule-associated protein 2 (MAP2) were
purchased from Boster Biological Technology (Pleasanton, CA, USA;
BM1243; 1:100). Alexa Fluor 555-labeled donkey anti-mouse IgG (H+L)
(1:200; A0460) and Alexa Fluor 488-labeled goat anti-rabbit IgG
(H+L) (1:200; A0423) were from Beyotime Institute of Biotechnology
(Haimen, China), and horseradish peroxidase (HRP)-conjugated
anti-rabbit secondary antibody (151341AP; 1:5,000) and
HRP-conjugated anti-mouse secondary antibody (HRP66008; 1:5,000)
were from ProteinTech Group, Inc. (Chicago, IL, USA). Secondary
goat anti-rabbit antibody was purchased from OriGene Technologies,
Inc. (Rockville, MD, USA; TA130021; 1:100).
Immunohistochemistry
Paraffin-embedded sections of brain tissue were
deparaffinized in xylene and rehydrated in a graded series of
ethanol prior to staining. After dewaxing and incubation in 3%
H2O2 for 30 min at 37°C, sections were washed
with PBS and boiled in 10 mmol/l sodium citrate buffer (pH 6.0;
Boster Biological Technology) for 15 min at 92–98°C for antigen
retrieval. The sections were then blocked in goat serum (Boster
Biological Technology; 10%) for 60 min at 37°C. The sections were
incubated with a 1:40 dilution of primary anti-Miz1 antibody
overnight at 4°C. Sections were then washed three times for 15 min
in PBS, incubated with secondary goat anti-rabbit antibody
(TA130021; 1:100; OriGene Technologies, Inc.) for 30 min at 37°C,
washed again, incubated with HRP-conjugated streptavidin (OriGene
Technologies, Inc.) for 30 min at 37°C and washed again. Sections
were then incubated with 3,3′-diaminobenzidine (DAB; OriGene
Technologies, Inc.) for 2 min at room temperature, a process which
was then terminated using water. Hematoxylin was used to
counterstain the nuclei at room temperature for 1 min.
Subsequently, samples were dehydrated in lithium carbonate for 1
min, incubated with dimethylbenzene for 5 min at room temperature,
mounted, and dried overnight. Finally, images were captured using
light microscopy (Nikon Corporation, Tokyo, Japan).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RTq-PCR was performed as described previously
(23). Total RNA was isolated from
brain tissues using TRIzol reagent (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and reverse transcribed using HiScript II Reverse
Transcriptase (R201-01/02; Vazyme Biotech Co., Ltd., Nanjing,
China) and random primers in a PCR reaction procedure (25°C for 5
min, 42°C for 15 min and 85°C for 5 min). qPCR was performed on the
Mastercycler ep realplex qPCR System (Eppendorf, Hamburg, Germany)
with the SYBR-Green qPCR Master Mix (Vazyme Biotech Co., Ltd.,
Jiangsu, China) with specific primers for Miz1 and β-actin as an
internal control. An Eppendorf Real Time PCR system was used with
the following parameters: One cycle at 95°C for 30 sec, followed by
40 cycles at 95°C 5 sec, 60°C for 34 sec, and one cycle at 95°C for
15 sec, 60°C for 1 min and 95°C for 15 sec. The median value of the
replicates for each sample was calculated according to the
2−ΔΔCq method (24). Data
presented were the mean from 3 independent experiments. Primers
used were as follows: Miz1 forward, 5′-AGGCACACTGTCTGAGAAGAGA-3′
and reverse, 5′-TGGTTCAGCTGCTCCAAGA-3′; and β-actin forward,
5′-ACGGTCAGGTCATCACTATCG-3′ and reverse,
5′-GGCATAGAGGTCTTTACGGATG-3′.
Western blot analysis
Proteins were extracted from mice cortex and
hippocampus using radioimmunoprecipitation assay buffer (P0013E;
Beyotime Institute of Biotechnology). Protein concentrations were
measured using a bicinchoninic acid protein assay kit (Beyotime
Institute of Biotechnology). Equal amounts of protein (50 µg) were
separated by 8% SDS-PAGE and electroblotted onto polyvinylidene
difluoride membranes (EMD Millipore, Billerica, MA, USA) using an
electrophoretic transfer system. The membranes were blocked with 5%
non-fat dry milk in Tris buffered saline and Tween-20 (TBST) for 1
h at room temperature, and incubated with primary antibodies
against Miz1 and GAPDH overnight at 4°C. The blots were
subsequently incubated with HRP-conjugated anti-rabbit and
anti-mouse secondary antibodies (both 1:5,000; ProteinTech Group,
Inc.) for 1 h at room temperature and washed in TBST 3 times for 5
min each time. The bands were visualized using an enhanced
chemiluminescence reagent (Thermo Fisher Scientific, Inc.) and a
Fusion FX5 image analysis system (Vilber Lourmat, Marne-la-Vallée,
France). Quantity One software 4.6.2 (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) was used to measure the resultant optical
density values.
Immunofluorescence
The sections of brain tissue were incubated at room
temperature in acetone for 30 min, washed with PBS, permeabilized
with 0.4% Triton-X for 10 min, and washed again with PBS. Following
antigen retrieval with 10 mM/l sodium citrate buffer by heating at
9–98°C for 20 min, sections were washed with PBS again and blocked
against non-specific antigen by incubation at room temperature with
goat serum working liquid (Boster Biological Technology; 10%) for 1
h, and incubated in primary anti-Miz1/MAP2/GFAP antibodies
overnight at 4°C. Proteins were subsequently detected by incubation
with Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 555
donkey anti-mouse IgG secondary antibodies (1:200) in the dark for
1 h at 37°C. Sections were subsequently washed and counterstained
with 4,6-diamidino-2-phenylindole (DAPI) for 15 min at room
temperature and washed with PBS. Finally, the sections were
visualized by confocal laser scanning microscopy (Nikon
Corporation).
Statistical analysis
All data are presented as the mean ± standard error
of the mean. Results were analyzed using Student's t-test. All
statistical calculations were performed using SPSS 20.0 software
(IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate
a statistically significant difference.
Results
Miz1 expression in the brain detected
by immunohistochemistry
It is known that Miz1 serves a regulatory role in
inflammation, which may provide a critical transcriptional
checkpoint to prevent excessive inflammatory responses and tissue
damage in the host. To determine the role of Miz1 in Alzheimer's
disease, the expression of Miz1 was analyzed in the brains of
wild-type (WT) mice and APP/PS1 (AD) mice at the age of 7 months
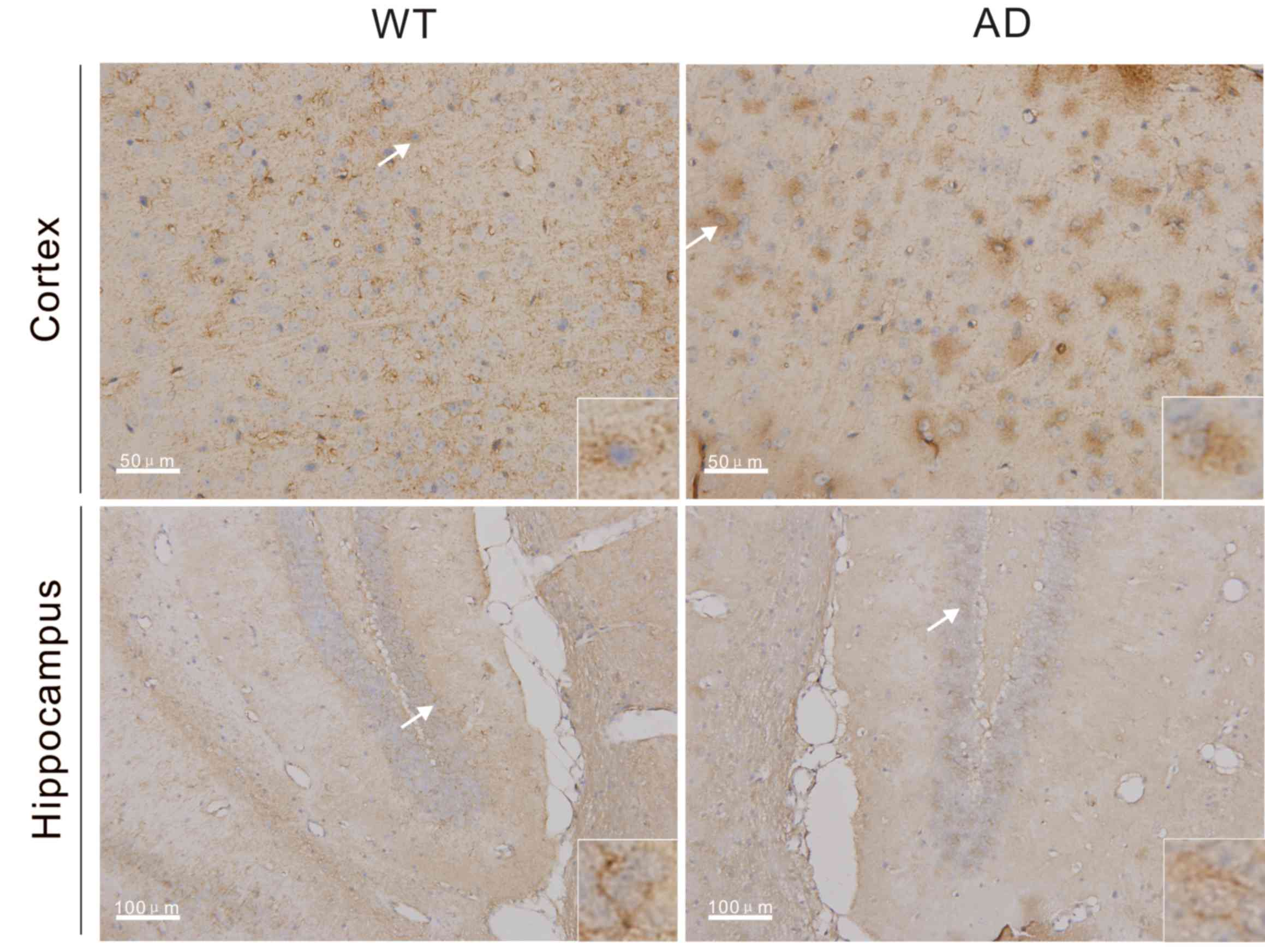
using immunohistochemistry, and brown staining indicated that cells
were positive for Miz1 expression. Miz1 expression was detected in
brain tissues from the WT and AD mice (Fig. 1). Compared with WT mice, expression
of Miz1 was markedly increased in the cortex of AD mice at 7 months
of age. However, no marked differences were observed in Miz1
expression in the hippocampus of WT and AD mice. In cortex and
hippocampus tissues from both groups, Miz1 was primarily located in
the cytoplasm (Fig. 1).
Elevated Miz1 expression in the cortex
of AD mice
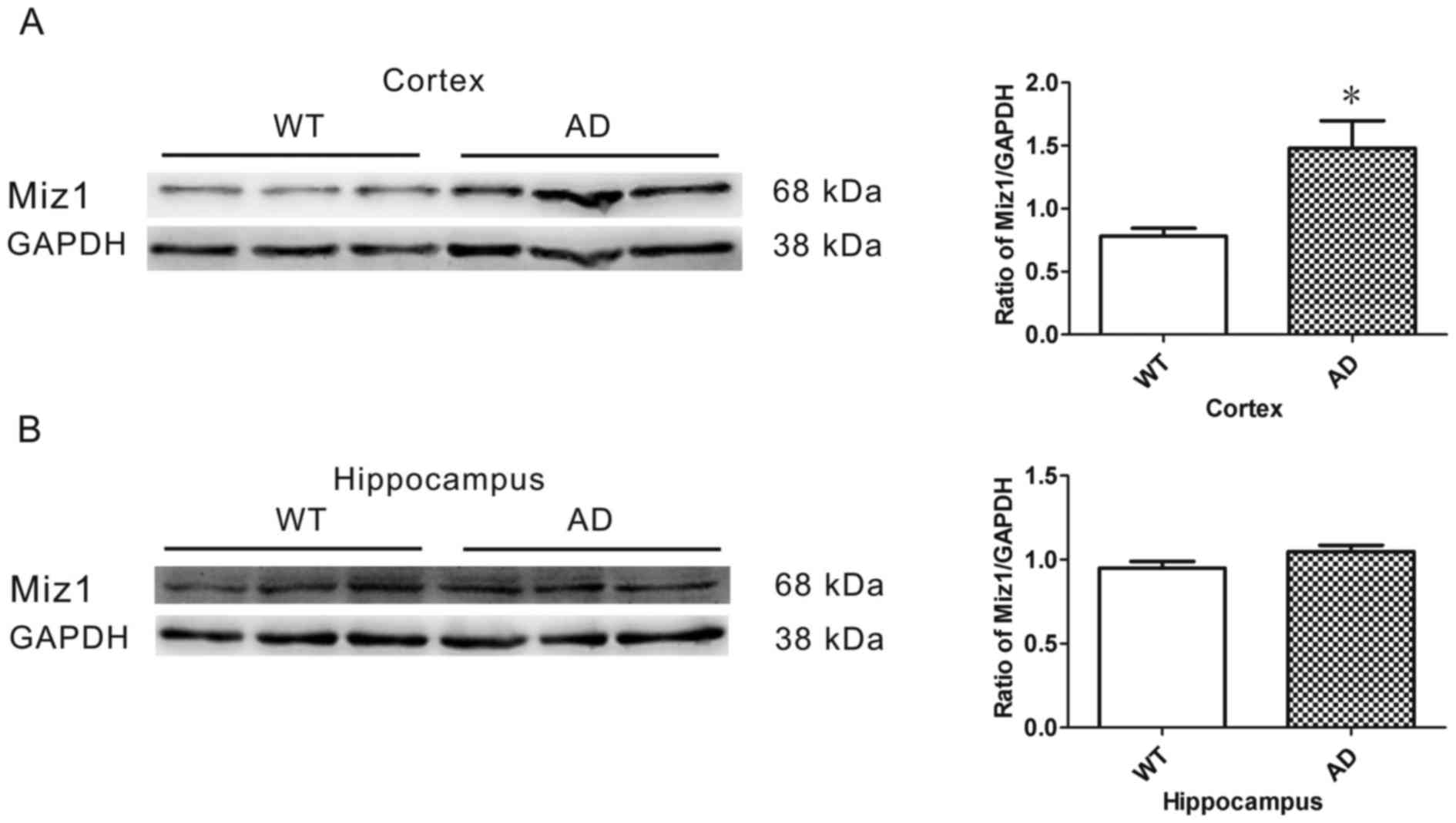
Western blot analysis was conducted to determine the
expression of Miz1 protein in the brains of WT (n=6) and AD (n=6)
mice. GAPDH was used as an internal loading control (Fig. 2A). The results revealed that the
expression of Miz1 was significantly higher in the cortex of AD
mice compared with WT mice (P<0.05; Fig. 2A). No significant difference was
observed in Miz1 expression in the hippocampus between the groups
(Fig. 2B).
Elevated Miz1 mRNA expression in the
cortex of AD mice
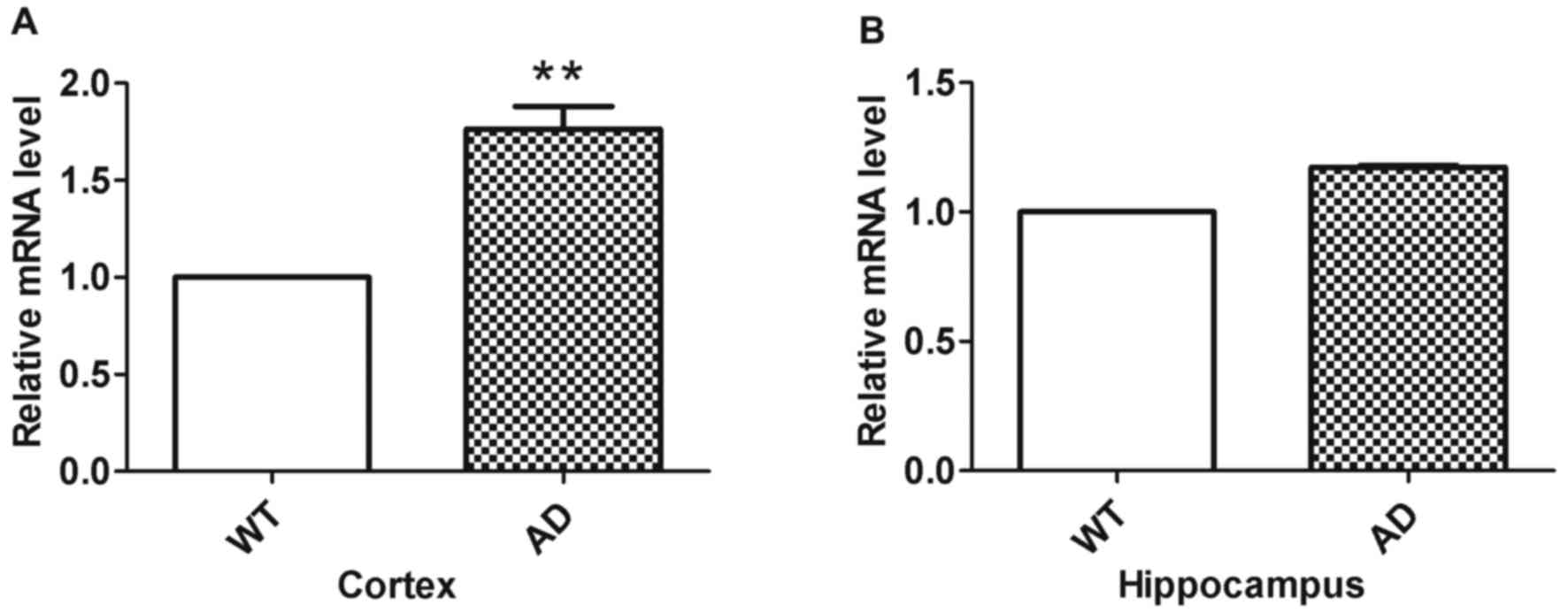
RT-qPCR was used to analyze Miz1 mRNA fold changes
in the groups (n=3; Fig. 3). The
results indicated that Miz1 expression was significantly higher in
the cortex of AD mice compared with WT mice (Fig. 3A; P<0.01). However, no significant
difference was observed in the hippocampus between WT and AD mice
(Fig. 3B).
Miz1 expression in mouse brain tissue
detected by immunofluorescence
The role of inflammation in the pathophysiology of
AD is well-established (21,22). Astrocytes are regarded as the most
important inflammation cells (25).
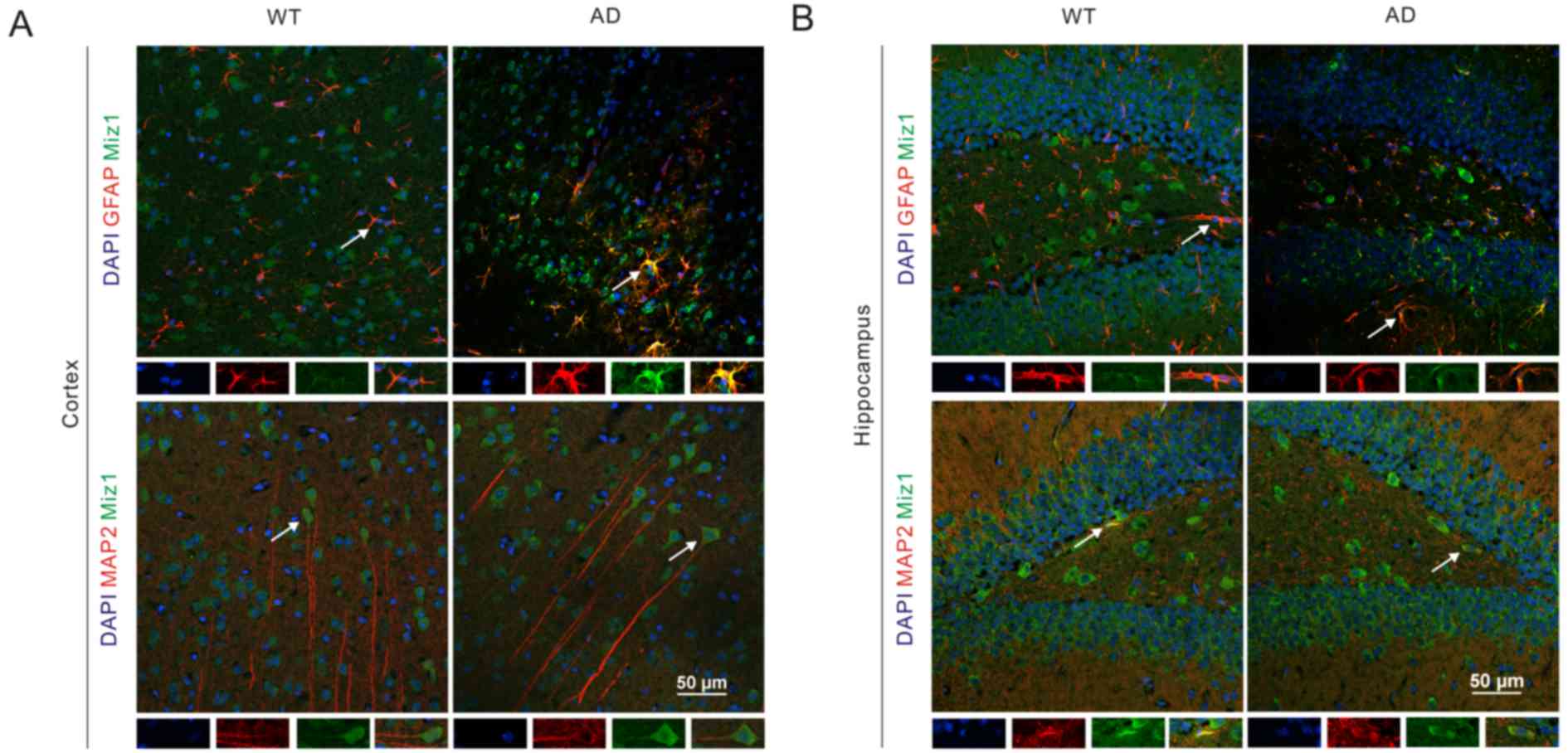
The location of Miz1 in the WT and AD mouse brain tissues was
assessed via immunofluorescence assay. The results indicated that
Miz1 (green) was co-expressed with GFAP (red) in both the cortex
and hippocampus of WT and AD mice (Fig.
4). It was also observed that Miz1 was co-expressed with MAP2
in the brains of WT and AD mice. Furthermore, Miz1 was
predominantly found in the cytoplasm of neurons. These results
indicate that the expression of Miz1 in astrocytes may be
associated with inflammation in the pathology of AD (26–29).
Discussion
AD is an age-related neurodegenerative disorder
characterized by accumulation of Aβ plaques, neurofibrillary
tangles (NFTs), synaptic and neuronal loss, and cognitive decline
(30). However, the underlying
physiological mechanisms remain to be elucidated. It has been
postulated that Aβ deposition and neurofibrillary tangles are
critical factors in the pathogenesis of AD (31). Unfortunately, animal studies and
clinical trials over the past 20 years have failed to manipulate
levels of Aβ and NFTs as targets for AD treatment (32–34).
Therefore, there is a need to identify novel treatment targets and
mediators associated with AD.
Miz1 serves a critical role in regulating
proliferation, differentiation, cell cycle progression, and
apoptosis (35–37). It also mediates DNA damage responses,
lymphoid development and inflammation via transcriptional
activation and repression of target genes (29). However, the cell cycle-independent
functions of Miz1 are poorly understood (10). Previous observations have indicated
that Miz1 has a broad expression profile, and it has also been
detected in neural precursor cells and in different parts of the
adult brain, indicating that the functions of Miz1 extend beyond
regulation of cell proliferation (38,39). In
the present study, it was demonstrated that Miz1 was broadly
expressed in the cortex and hippocampus. Furthermore, Miz1 was
detected in both neurons and astrocytes, suggesting that it may
serve an important regulatory role in a wide variety of processes,
including neuronal plasticity, neurotransmission and
neuroinflammation. In the present study, it was observed that Miz1
levels were significantly increased in the cortex of AD mice.
Consistent with this finding, qPCR analysis revealed that the mRNA
levels of Miz1 were also increased in the cortex of AD mice. It is
interesting to note that no significant differences were observed
in Miz1 expression in the hippocampus. Such region-specific
alterations in Miz1 in AD mice may reflect region-specific
signaling events associated, at least in part, with basal
differences in distinct brain structures (40,41).
The expression changes of Miz1 in AD mice suggest
that Miz1 may participate in the progression of AD. To the best of
our knowledge, the present study is the first to indicate increased
expression of Miz1 in the cerebrum. However, the specific roles of
Miz1 in AD remain unknown. Double-label immunofluorescence analysis
revealed that Miz1 was co-expressed with Map-2 in neurons,
indicating that Miz1 may be associated with neural plasticity. As a
transcription factor, Miz1 activates a variety of target genes in
the brain. Multiple proteins encoded by target genes of Miz1 are
associated with synaptic surface receptor trafficking, synaptosome
transport and endocytosis (10).
Based on this, the increased Miz1 expression in AD mice observed in
the present study may be associated with synaptic dysfunction and
thus impairments of cognition. However, Miz1 was also observed in
astrocytes, and it is known that astrocytes are the main effectors
in the inflammatory process of the central nervous system (42). In lung tissue, Miz1 in the cytoplasm
suppresses lipopolysaccharide- and tumor necrosis factor-induced
inflammatory responses by specifically interfering with c-Jun
N-terminal kinase activation independently of its transcriptional
activity, indicating that Miz1 provides a critical transcriptional
checkpoint preventing the host from excessive inflammatory response
(26). However, whether Miz1 located
in astrocytes in the central nervous system is associated with the
anti-inflammatory process in AD remains unclear. Notably, the
disruption of normal posttranslational modification-based signaling
at the synapse is a pathological mechanism that likely contributes
to cognitive dysfunction in diseases such as AD (43). A previous study reported that Miz1
may participate in small ubiquitin-like modifier (SUMO) conjugation
and SUMO ligase activity (44).
Therefore, Miz1 may also participate in AD progression through
multiple mechanisms.
In summary, the results of the present study
demonstrate that the expression of Miz1, which is a protein
associated with a variety of functions including autophagy and
inflammation, was altered in APP/PS1 mice. Subcellular localization
suggests that Miz1 in neurons and astrocytes may serve a role in
the pathology of AD. Since Miz1 promotes autophagy and inhibits
inflammation and apoptosis, this suggests that the increased
expression of Miz1 in APP/PS1 mice may be a compensatory response.
However, the exact function of Miz1 in AD requires further study to
be fully understood.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81171197 and
81220108010).
References
|
1
|
Portelius E, Zetterberg H, Skillbäck T,
Törnqvist U, Andreasson U, Trojanowski JQ, Weiner MW, Shaw LM,
Mattsson N and Blennow K: Alzheimer's Disease Neuroimaging
Initiative: Cerebrospinal fluid neurogranin: Relation to cognition
and neurodegeneration in Alzheimer's disease. Brain. 138:3373–3385.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ossenkoppele R, Pijnenburg YA, Perry DC,
Cohn-Sheehy BI, Scheltens NM, Vogel JW, Kramer JH, van der Vlies
AE, La Joie R, Rosen HJ, et al: The behavioural/dysexecutive
variant of Alzheimer's disease: Clinical, neuroimaging and
pathological features. Brain. 138:2732–2749. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Haas LT, Salazar SV, Kostylev MA, Um JW,
Kaufman AC and Strittmatter SM: Metabotropic glutamate receptor 5
couples cellular prion protein to intracellular signalling in
Alzheimer's disease. Brain. 139:526–546. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hendrickx A, Pierrot N, Tasiaux B,
Schakman O, Kienlen-Campard P, De Smet C and Octave JN: Epigenetic
regulations of immediate early genes expression involved in memory
formation by the amyloid precursor protein of Alzheimer disease.
PLoS One. 9:e994672014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Parr C, Mirzaei N, Christian M and Sastre
M: Activation of the Wnt/β-catenin pathway represses the
transcription of the β-amyloid precursor protein cleaving enzyme
(BACE1) via binding of T-cell factor-4 to BACE1 promoter. FASEB J.
29:623–635. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lai YJ, Liu L, Hu XT, He L and Chen GJ:
Estrogen Modulates ubc9 expression and synaptic redistribution in
the brain of APP/PS1 mice and cortical neurons. J Mol Neurosci.
61:436–448. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang J, Liu AY, Tang B, Luo D, Lai YJ, Zhu
BL, Wang XF, Yan Z and Chen GJ: Chronic nicotine differentially
affects murine transcriptome profiling in isolated cortical
interneurons and pyramidal neurons. BMC Genomics. 18:1942017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu J, Zhao Y, Eilers M and Lin A: Miz1 is
a signal- and pathway-specific modulator or regulator (SMOR) that
suppresses TNF-alpha-induced JNK1 activation. Proc Natl Acad Sci
USA. 106:pp. 18279–18284. 2009, View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang Y, Do H, Tian X, Zhang C, Liu X, Dada
LA, Sznajder JI and Liu J: E3 ubiquitin ligase Mule ubiquitinates
Miz1 and is required for TNFalpha-induced JNK activation. Proc Natl
Acad Sci USA. 107:pp. 13444–13449. 2010, View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wolf E, Gebhardt A, Kawauchi D, Walz S,
von Eyss B, Wagner N, Renninger C, Krohne G, Asan E, Roussel MF and
Eilers M: Miz1 is required to maintain autophagic flux. Nat Commun.
4:25352013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Peukert K, Staller P, Schneider A,
Carmichael G, Hänel F and Eilers M: An alternative pathway for gene
regulation by Myc. EMBO J. 16:5672–5686. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu J, Chen M, Ren XR, Wang J, Lyerly HK,
Barak L and Chen W: Regulation of hedgehog signaling by
Myc-interacting zinc finger protein 1, Miz1. PLoS One.
8:e633532013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu L, Wu H, Ma L, Sangiorgi F, Wu N, Bell
JR, Lyons GE and Maxson R: Miz1, a novel zinc finger transcription
factor that interacts with Msx2 and enhances its affinity for DNA.
Mech Dev. 65:3–17. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Adhikary S, Peukert K, Karsunky H, Beuger
V, Lutz W, Elsässer HP, Möröy T and Eilers M: Miz1 is required for
early embryonic development during gastrulation. Mol Cell Biol.
23:7648–7657. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Herold S, Hock A, Herkert B, Berns K,
Mullenders J, Beijersbergen R, Bernards R and Eilers M: Miz1 and
HectH9 regulate the stability of the checkpoint protein, TopBP1.
EMBO J. 27:2851–2861. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saito M, Novak U, Piovan E, Basso K,
Sumazin P, Schneider C, Crespo M, Shen Q, Bhagat G, Califano A, et
al: BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse
large B cell lymphoma. Proc Natl Acad Sci USA. 106:pp. 11294–11299.
2009, View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Campaner S, Doni M, Verrecchia A, Fagà G,
Bianchi L and Amati B: Myc, Cdk2 and cellular senescence: Old
players, new game. Cell Cycle. 9:3655–3661. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Herkert B, Dwertmann A, Herold S, Abed M,
Naud JF, Finkernagel F, Harms GS, Orian A, Wanzel M and Eilers M:
The Arf tumor suppressor protein inhibits Miz1 to suppress cell
adhesion and induce apoptosis. J Cell Biol. 188:905–918. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kosan C, Saba I, Godmann M, Herold S,
Herkert B, Eilers M and Möröy T: Transcription factor miz-1 is
required to regulate interleukin-7 receptor signaling at early
commitment stages of B cell differentiation. Immunity. 33:917–928.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He XL, Yan N, Chen XS, Qi YW, Yan Y and
Cai Z: Hydrogen sulfide down-regulates BACE1 and PS1 via activating
PI3K/Akt pathway in the brain of APP/PS1 transgenic mouse.
Pharmacol Rep. 68:975–982. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Liu P, Zhu H, Xu Y, Ma C, Dai X,
Huang L, Liu Y, Zhang L and Qin C: miR-34a, a microRNA up-regulated
in a double transgenic mouse model of Alzheimer's disease, inhibits
bcl2 translation. Brain Res Bull. 80:268–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Li N, Ma J, Gu Z, Yu L, Fu X, Liu
X and Wang J: Effects of an amyloid-beta 1–42 oligomers antibody
screened from a phage display library in APP/PS1 transgenic mice.
Brain Res. 1635:169–179. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Murdock DG, Bradford Y, Schnetz-Boutaud N,
Mayo P, Allen MJ, D'Aoust LN, Liang X, Mitchell SL, Zuchner S,
Small GW, et al: KIAA1462, a coronary artery disease associated
gene, is a candidate gene for late onset Alzheimer disease in APOE
carriers. PLoS One. 8:e821942013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nam Y, Kim JH, Seo M, Kim JH, Jin M, Jeon
S, Seo JW, Lee WH, Bing SJ, Jee Y, et al: Lipocalin-2 protein
deficiency ameliorates experimental autoimmune encephalomyelitis:
The pathogenic role of lipocalin-2 in the central nervous system
and peripheral lymphoid tissues. J Biol Chem. 289:16773–16789.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Do-Umehara HC, Chen C, Urich D, Zhou L,
Qiu J, Jang S, Zander A, Baker MA, Eilers M, Sporn PH, et al:
Suppression of inflammation and acute lung injury by Miz1 via
repression of C/EBP-δ. Nat Immunol. 14:461–469. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Heneka MT, Carson MJ, El Khoury J,
Landreth GE, Brosseron F, Feinstein DL, Jacobs AH, Wyss-Coray T,
Vitorica J, Ransohoff RM, et al: Neuroinflammation in Alzheimer's
disease. Lancet Neurol. 14:388–405. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Heppner FL, Ransohoff RM and Becher B:
Immune attack: The role of inflammation in Alzheimer disease. Nat
Rev Neurosci. 16:358–372. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen P, Wang W, Zhang Y, Yuan Y and Wu Y:
Decreased MIZ1 expression in severe experimental acute
pancreatitis: A rat study. Dig Dis Sci. 61:758–766. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gjoneska E, Pfenning AR, Mathys H, Quon G,
Kundaje A, Tsai LH and Kellis M: Conserved epigenomic signals in
mice and humans reveal immune basis of Alzheimer's disease. Nature.
518:365–369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Reddy PH, Manczak M, Yin X, Grady MC,
Mitchell A, Kandimalla R and Kuruva CS: Protective effects of a
natural product, curcumin, against amyloid β induced mitochondrial
and synaptic toxicities in Alzheimer's disease. J Investig Med.
64:1220–1234. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Giacobini E and Gold G: Alzheimer disease
therapy-moving from amyloid-β to tau. Nat Rev Neurol. 9:677–686.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Herrup K, Carrillo MC, Schenk D, Cacace A,
Desanti S, Fremeau R, Bhat R, Glicksman M, May P, Swerdlow R, et
al: Beyond amyloid: Getting real about nonamyloid targets in
Alzheimer's disease. Alzheimers Dement. 9:452–458.e1. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vandenberghe R, Rinne JO, Boada M,
Katayama S, Scheltens P, Vellas B, Tuchman M, Gass A, Fiebach JB,
Hill D, et al: Bapineuzumab for mild to moderate Alzheimer's
disease in two global, randomized, phase 3 trials. Alzheimers Res
Ther. 8:182016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Si J, Yu X, Zhang Y and DeWille JW: Myc
interacts with Max and Miz1 to repress C/EBPdelta promoter activity
and gene expression. Mol Cancer. 9:922010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kerosuo L and Bronner ME: Biphasic
influence of Miz1 on neural crest development by regulating cell
survival and apical adhesion complex formation in the developing
neural tube. Mol Biol Cell. 25:347–355. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Haikala HM, Klefstrom J, Eilers M and
Wiese KE: MYC-induced apoptosis in mammary epithelial cells is
associated with repression of lineage-specific gene signatures.
Cell Cycle. 15:316–323. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lein ES, Hawrylycz MJ, Ao N, Ayres M,
Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ,
et al: Genome-wide atlas of gene expression in the adult mouse
brain. Nature. 445:168–176. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sanz-Moreno A, Fuhrmann D, Zankel A,
Reingruber H, Kern L, Meijer D, Niemann A and Elsässer HP: Late
onset neuropathy with spontaneous clinical remission in mice
lacking the POZ domain of the transcription factor Myc-interacting
zinc finger protein 1 (Miz1) in Schwann cells. J Biol Chem.
290:727–743. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Schneider JS, Mettil W and Anderson DW:
Differential effect of postnatal lead exposure on gene expression
in the hippocampus and frontal cortex. J Mol Neurosci. 47:76–88.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Abdul-Rahman O, Sasvari-Szekely M, Ver A,
Rosta K, Szasz BK, Kereszturi E and Keszler G: Altered gene
expression profiles in the hippocampus and prefrontal cortex of
type 2 diabetic rats. BMC Genomics. 13:812012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xu L, He D and Bai Y: Microglia-mediated
inflammation and neurodegenerative disease. Mol Neurobiol.
53:6709–6715. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lee L, Dale E, Staniszewski A, Zhang H,
Saeed F, Sakurai M, Fa' M, Orozco I, Michelassi F, Akpan N, et al:
Corrigendum: Regulation of synaptic plasticity and cognition by
SUMO in normal physiology and Alzheimer's disease. Sci Rep.
5:117822015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cheong MS, Park HC, Hong MJ, Lee J, Choi
W, Jin JB, Bohnert HJ, Lee SY, Bressan RA and Yun DJ: Specific
domain structures control abscisic acid-, salicylic acid-, and
stress-mediated SIZ1 phenotypes. Plant physiol. 151:1930–1942.
2009. View Article : Google Scholar : PubMed/NCBI
|