Introduction
Rheumatoid arthritis (RA) is a systemic autoimmune
disease that predominantly affects multiple peripheral joints and
results in irreversible joint damage (1). Accumulating evidence suggests that the
participation of inflammation-associated cells, as well as their
production of proinflammatory mediators, including tumor necrosis
factor (TNF)-α and interleukin-1β, serve a key role in the
pathogenesis of RA (2). Although
major strides in understanding the disease have been made, the
specific mechanistic details underlying the pathogenesis of RA have
not been fully clarified (2).
In the human genome, >70% is availably
transcribed; however, only 1–2% encodes proteins (3,4). The
majority of the remainder of the genome generates non-coding RNA
(ncRNA (5,6). According to their length, ncRNA may be
divided into long ncRNA (lncRNA), which are arbitrarily defined as
transcripts longer than 200 nt, or short ncRNA, including microRNA
(miRNA) and short interfering RNA (7). It is well documented that miRNA-155,
−146 and −21 are aberrantly expressed in RA (8–10),
suggesting that miRNA serve critical roles in the regulation of RA
pathogenesis. An increasing number of studies have suggested that
lncRNA have a critical role in fundamental biological processes,
including genomic imprinting, chromosome modification, immune
response, inflammatory conditions, tumorigenesis, cellular
development and metabolism, through comprehensive mechanisms
(11,12). Emerging evidence has also indicated
that the expression of lncRNA is dysregulated in a range of
disorders, including cancer, intervertebral disc degeneration and
Alzheimer's disease (13–15). Additionally, the deregulation of
various lncRNA has been implicated in RA, including H19, Hotair,
large intergenic non-coding (linc)RNA-p21, C5T1lncRNA, LOC100652951
and LOC100506036 (16–20). Despite these notable developments,
the alteration in lncRNA and mRNA expression levels induced by RA
and the roles of these transcripts in modulating RA pathogenesis
remain unclear.
The present study determined the expression patterns
of lncRNA and mRNA in RA samples and compared them with healthy
samples. Additionally, three lncRNA and three mRNA were confirmed
using reverse transcription-quantitative polymerase chain reaction
(RT-qPCR). lncRNA classification, subgroup analysis and genomic
location analysis were performed to further analyze these
differentially expressed lncRNA. Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genome (KEGG) analysis were used to
predict the function and signaling pathways affected by the
differentially expressed mRNA, which were target genes of
aberrantly expressed lncRNA. The present results suggested that
lncRNA expression patterns may provide pivotal insights into the
pathogenesis of RA.
Materials and methods
Patients and control individuals
A total of 34 patients with RA (29 females and 5
males; mean age, 53.7 years) admitted to the Department of
Rheumatology and Immunology, The First Affiliated Hospital of
Nanchang University (Nanchang, China) between October 2014 and
April 2016, were enrolled in the present study. The diagnosis of RA
was established by the American College of Rheumatology criteria
(21). The baseline characteristics
of 10 of these patients that were used in microarray analysis are
summarized in Table I. For RA
patients, Disease activity was calculated using the disease
activity score 28 system (22),
anti-cyclic citrullinated peptide (CCP) antibody was detected by
Anti-CCP Enzyme Immunoassay Test kit (cat. no. EC170502; Shanghai
Kexin Biotech Co., Ltd., Shanghai, China) and values >25 RU/ml
were considered positive. Erythrocyte sedimentation rate was
examined by the Westergren method (23), and values ≤15 mm/h for males and ≤20
mm/h for females were considered normal. Serum C-reactive protein
(CRP) and rheumatoid factor (RF) were measured by nephelometry.
Values >8 mg/l for CRP and >20 IU/ml were considered positive
for RF. A total of 34 healthy controls (29 females and 5 males;
mean age, 50.2 years) were selected based on no history of
autoimmune disease. Among the patients and controls, 10 patients
with RA (Table I) and 10 age-matched
and sex-matched normal controls were recruited for isolating
peripheral blood mononuclear cells (PBMCs) for microarray analysis.
The remaining 24 healthy controls and 24 patients with RA were used
to validate the reliability of microarray results by RT-qPCR
analysis.
 | Table I.Demographic, clinical and laboratory
information of patients with RA. |
Table I.
Demographic, clinical and laboratory
information of patients with RA.
| Subject | Sex | Age, years | DAS28 | Erythrocyte
sedimentation rate, mm/h | C-reactive protein,
mg/l | Rheumatoid factor,
IU/ml | Anti-cyclic
citrullinated peptide, RU/ml |
|---|
| RA1 | Female | 55 | 5.03 | 37 | 8.30 | 142.00 | 670.70 |
| RA2 | Female | 54 | 4.81 | 14 | 2.15 | 124.00 | 106.30 |
| RA3 | Female | 43 | 5.50 | 12 | 2.04 | 46.50 | 155.60 |
| RA4 | Male | 68 | 6.01 | 19 | 21.60 | <20.00 | 5.66 |
| RA5 | Male | 37 | 6.39 | 33 | 16.10 | 59.60 | 20.61 |
| RA6 | Female | 68 | 4.46 | 24 | 27.90 | <20.00 | 39.90 |
| RA7 | Female | 68 | 4.12 | 10 | 5.02 | 115.00 | 175.10 |
| RA8 | Female | 49 | 7.33 | 126 | 81.50 | 1,190.00 | 493.60 |
| RA9 | Female | 43 | 7.38 | 84 | 6.95 | 32.30 | 382.00 |
| RA10 | Female | 52 | 5.25 | 40 | 4.06 | 80.90 | 1,600.00 |
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Nanchang University
(approval no. 019) and was conducted in compliance with the
Declaration of Helsinki. Informed consent was obtained from all
participants, including patients and normal controls, prior to
initiation of the study.
PBMC collection and RNA
extraction
Blood samples were collected in vacutainer tubes
containing EDTA, and PBMCs were isolated by centrifugation against
a density. Gradient (1,000 × g, 20 min, 22°C). Total RNA was
isolated from freshly obtained PBMCs using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's protocol. The integrity of the RNA
was assessed by electrophoresis on a 2% denaturing agarose gel and
visualized using ethidium bromide. The concentration and quality of
the RNA were assessed by absorbance spectrometry measuring
absorbance ratios of A260/A280 and A260/A230 using a NanoDrop
ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.).
Microarray and computational
analysis
Arraystar Human LncRNA Microarray V3.0 (8×60 K;
Arraystar, Inc., Rockville, MD, USA) is designed for the global
profiling of human lncRNA and mRNA, and is updated from the
previous Microarray V2.0. The sample preparation and microarray
hybridization were performed according to the manufacturer's
protocol. Briefly, ribosomal RNA was removed from total RNA and
subsequently mRNA was obtained using an mRNA-ONLY™ Eukaryotic mRNA
Isolation kit (Epicentre; Illumina, Inc., San Diego, CA, USA). The
random priming method (24) was
utilized to amplify each sample according to the manufacturer's
protocol (Quick Amp Labeling kit; 5190-0442 Agilent Technologies,
Inc., Santa Clara, CA, USA; amplification assay was set at 40°C for
2 h, followed by 65°C for 15 min and 0°C for 5 min), and mRNA was
transcribed into fluorescent cRNA without 3′ bias. Labeled cRNA
were hybridized to the Human LncRNA Microarray, designed for 30,586
lncRNA and 26,109 mRNA. After washing with Gene Expression Wash
Buffer 1 (Agilent Technologies, Inc.), and Gene Expression Wash
Buffer 2 (Agilent Technologies, Inc.), the Agilent Scanner G2505C
and Agilent feature extraction software (version 11.0.1.1; Agilent
Technologies, Inc.) were used to scan and analyze the arrays.
Quantile normalization and subsequent data processing were executed
using the GeneSpring GX v12.0 software package (Agilent
Technologies, Inc.). The microarray work was performed by Shanghai
Kangchen Bio-tech, Inc., (Shanghai, China).
SYBR Green I RT-qPCR
The expression levels of differentially expressed
lncRNA and mRNA were confirmed by RT-qPCR. Briefly, 5 µg of total
RNA from each sample was used for the synthesis of first strand
cDNA using a PrimeScript™ RT Reagent Kit (Takara Biotechnology Co.,
Ltd., Dalian, China) according to the manufacturer's protocol. RT
reaction was carried out in a 10 µl reaction volume containing 5X
PrimeScript™ Buffer, 1 µl RT specific primer, 0.5 µl PrimeScript™
RT Enzyme Mix, 5 µg of total RNA. RT assay was set at an initial
denaturation step at 37°C for 15 min, followed by 85°C for 5 sec.
Following first strand cDNA synthesis, PCR reaction was performed
in a 10 µl reaction volume containing 1X SYBR Green PCR Master mix
(Takara Biotechnology Co., Ltd.), 0.4 µM of each specific forward
and reverse primer, 0.5 µl of cDNA template. PCR assay was set at
an initial denaturation step at 95°C for 5 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 1 min with an ABI 7500
Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The primers were as shown in Table II. The gene expression levels were
normalized to that of GAPDH in cDNA samples, and all samples were
measured in triplicate. The relative expression levels of the genes
were calculated using the ∆Ct method where ∆Ct=Ct median gene-Ct
median GAPDH (25,26).
 | Table II.LncRNA and mRNA gene primers for
polymerase chain reaction. |
Table II.
LncRNA and mRNA gene primers for
polymerase chain reaction.
| Gene | Primer |
|---|
|
ENST00000445339 | S:
GATGGAACGAGGAATGGTAGCG A |
|
| A:
CAGTTCAGTGCATCGAGGTTTGC |
|
ENST00000569920 | S:
TAACATTCTGCATCCCTCACCCA |
|
| A:
CGCACATTACCTACCCCTTCTCA |
|
ENST00000506982 | S:
CAGTAAAAGAGCACTTGGTGGAA |
|
| A:
CCAGTGTTAGGGTCTGAGTCCAG |
| PGLYRP1 | S:
CACATGAAGACACTGGGCTGGT |
|
| A:
CATGAAGCTGATGCCAATGGAC |
| LRRN3 | S:
GGACAGCCTTTGTCAAGACTGAA |
|
| A:
CAGGGTGCAAACCTTTGGTG |
| CX3CR1 | S:
AACTCCAGTAGCTTGGGACAAATCA |
|
| A:
CCACAACTTGGGCACAGCA |
| GAPDH | S:
CCGGGAAACTGTGGCGTGATGG |
|
| A:
AGGTGGAGGAGTGGGTGTCGCTGTT |
Analysis of relationship between
lncRNA and adjacent protein-coding genes
Target genes were predicted for differentially
expressed lncRNAs. LncRNAs originate in complex transcriptional
loci and regulate gene expression through epigenetic regulation of
chromatin modification, and transcriptional and
post-transcriptional processing (27). In order to determine the underlying
roles of lncRNA in RA and shed light on the potential mechanisms of
RA, the present study mainly focused on unveiling the relationship
between lncRNA and mRNA. Abnormal lncRNA (fold change, ≥3.0) were
subjected to bioinformatics analysis for target gene prediction. To
increase the accuracy of target prediction, abnormal mRNA (fold
change, ≥3.0) were integrated with the predicted lncRNA
targets.
GO and KEGG pathway analysis based on
differentially expressed mRNA
GO (www.geneontology.org) and KEGG (www.genome.ad.jp/kegg) databases were utilized to
analyze biological functions and signaling pathways affected by
abnormally expressed mRNA (28,29).
P<0.05 was considered to indicate a statistically significant
difference.
Statistical analysis
Statistical analysis and graphic presentation were
performed using GraphPad Prism version 5.0 (GraphPad Software,
Inc., La Jolla, CA, USA). A Student's t-test was used where the
normality test passed; otherwise, the nonparametric Mann-Whitney U
test was used to analyze the data. P<0.05 was considered to
indicate a statistically significant difference.
Results
lncRNA and mRNA expression profiles in
PBMCs of the different groups
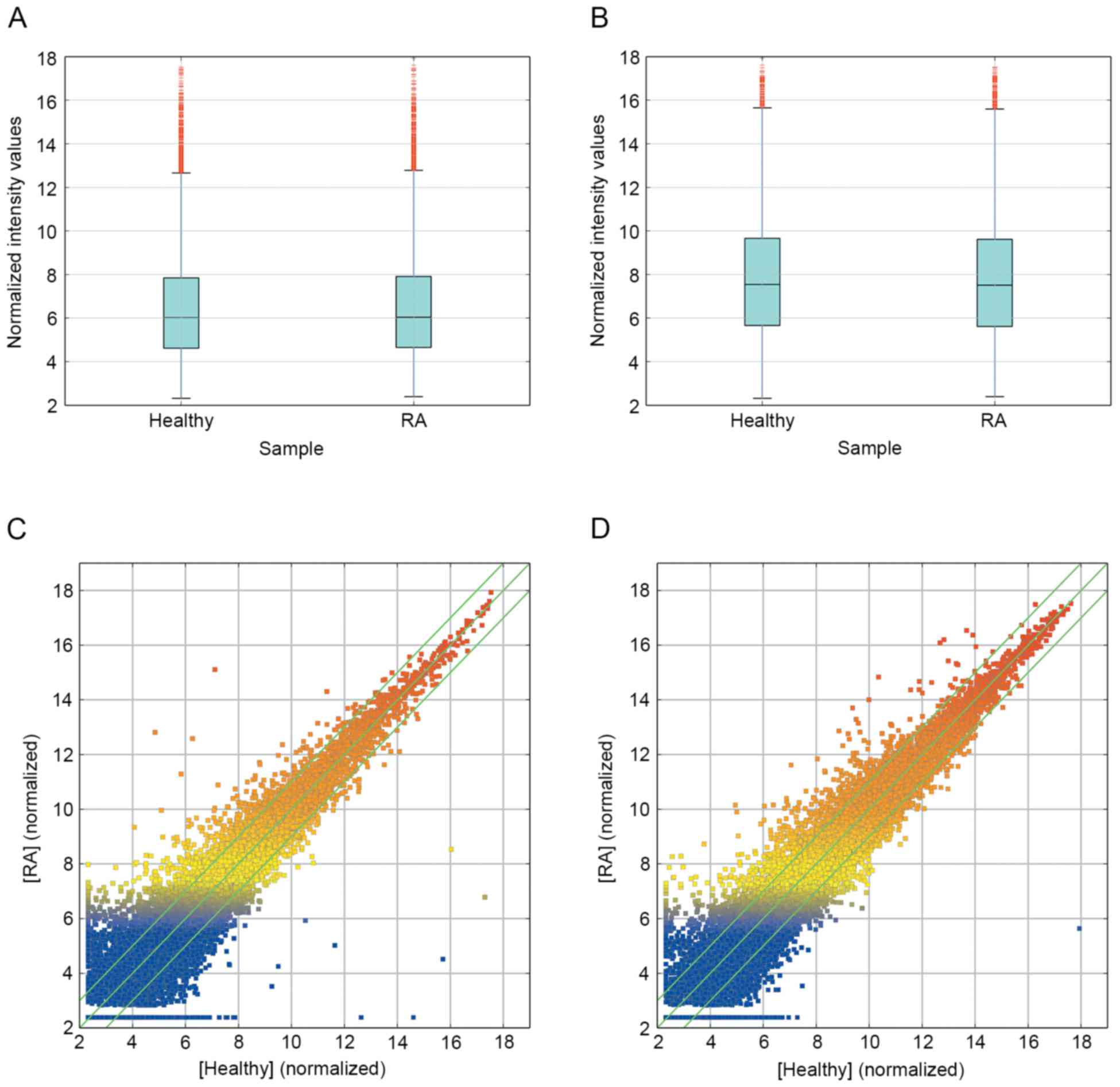
To profile differentially expressed lncRNA in RA, a
microarray analysis of lncRNA and mRNA expression in the RA group
and healthy group was performed (Figs.
1 and 2). The results revealed
that 139 lncRNA (88 up and 51 down) and 92 mRNA (67 up and 25 down)
were significantly differentially expressed between the RA group
and healthy group (fold change ≥10.0; P<0.05). Additionally,
5,045 lncRNA (2,410 up and 2,635 down) and 3,289 mRNA (1,403 up and
1,886 down) were differentially expressed with a fold change >2
(P<0.05). The most significantly deregulated lncRNA were
ENST00000583574 (up fold change, 257.7358) and ENST00000559539
(down fold change, 4726.8003). The most significantly deregulated
mRNA were ALAS2 (up fold change, 52.16745) and KCNMB3 (down fold
change, 4999.96853) (data not shown).
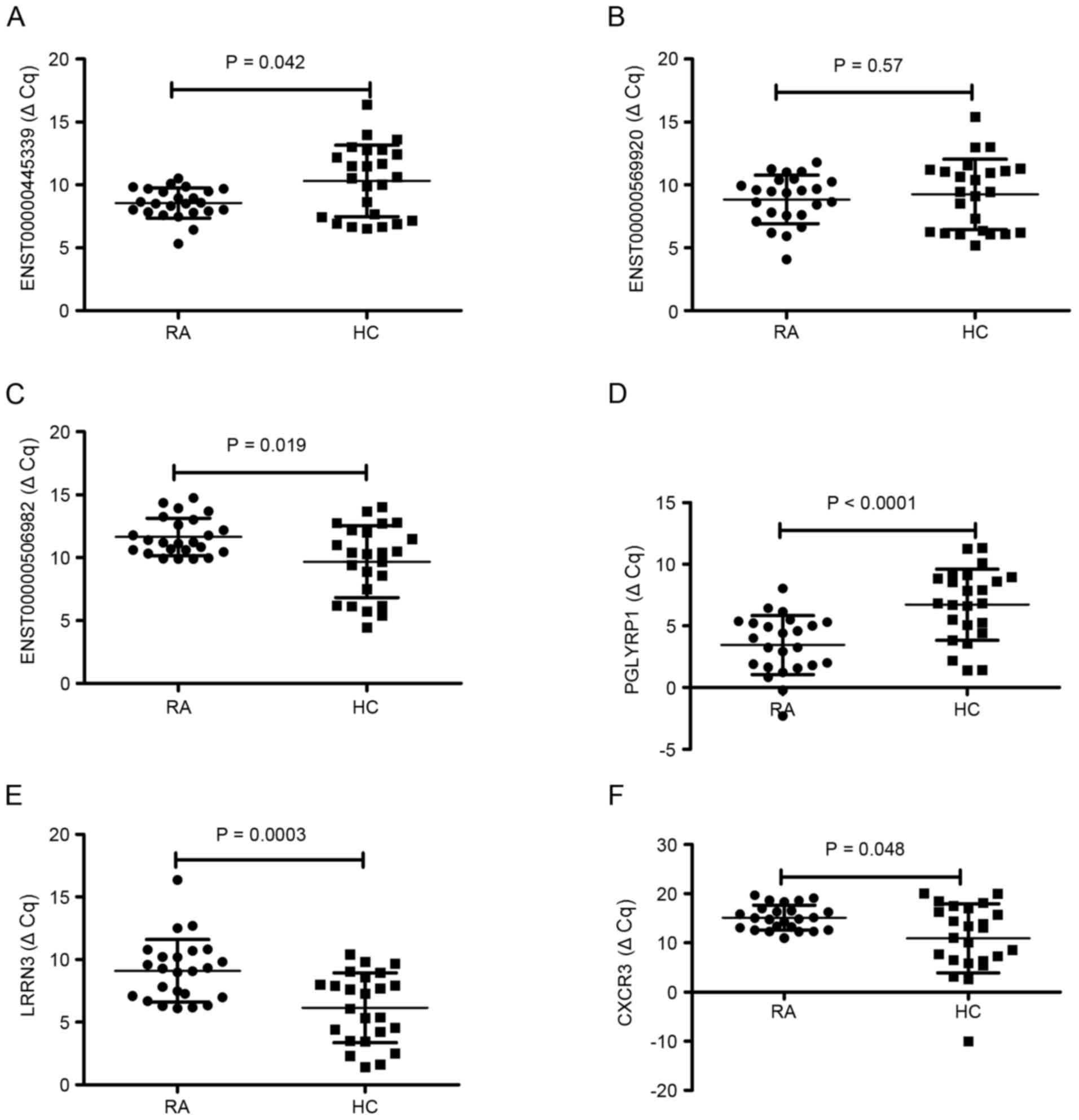
RT-qPCR validation
To validate the reliability of the microarray
results, three differentially expressed lncRNA and three
differentially expressed mRNA were randomly selected and analyzed
using RT-qPCR. For lncRNA, as demonstrated in Fig. 3, the results demonstrated that
ENST00000445339 was significantly downregulated (P=0.042) and
ENST00000506982 was significantly upregulated (P=0.019) in the 24
patients with RA compared to the levels in the healthy group
(Fig. 3A and C, respectively). No
significant difference was observed in the expression of
ENST00000569920 between RA patients and healthy controls (P=0.57;
Fig. 3B). For mRNA, the expression
of PGLYRP1 was significantly downregulated (P<0.0001), LRRN3 was
significantly upregulated (P=0.0003) and CX3CR1 was significantly
upregulated (P=0.048) in patients with RA compared with the levels
in healthy controls (Fig. 3D-F).
This demonstrated that RT-qPCR results were consistent with those
of the microarray analysis.
lncRNA classification and subgroup
analysis
Evidence from previous reports has indicated that
lncRNA may be classified into lncRNA with enhancer-like functions,
antisense lncRNA and lincRNA (26,30). The
results of the present study indicated that 610 intergenic lncRNA
(304 upregulated and 306 downregulated), 251 enhancer-like lncRNA
(138 upregulated and 113 downregulated) and 134 antisense lncRNA
(57 upregulated and 77 downregulated) were differentially expressed
(fold change ≥2.0; P<0.05) between the RA group and healthy
group. Some nearby coding genes that may be regulated by these
differentially expressed lncRNA were also identified (data not
shown).
Integration of differentially
expressed mRNA into the predicted targets of differentially
expressed lncRNA
The present results revealed that the majority of
differentially expressed lncRNA were from intergenic regions (42%),
natural antisense to protein-coding loci (19%), or intronic
antisense to protein-coding loci (15%), with the others
representing intronic sense-overlapping, exon sense-overlapping,
and bidirectional regions. In addition, 2,859 of 5,045
differentially expressed lncRNA were oriented in or around a known
protein-coding region (i.e. not intergenic). Therefore,
differentially expressed lncRNA (fold change, ≥3.0) were integrated
with predicted mRNA (fold change, ≥3.0). The result demonstrated
that 109 differentially expressed protein-coding genes were
markedly associated with each of the 85 differentially expressed
lncRNA to generate 135 lncRNA-mRNA target pairs. Among them, 81
pairs were differentially expressed unidirectionally (upregulated
or downregulated), while 54 pairs were differentially expressed
bidirectionally (data not shown).
GO and KEGG pathway analysis of
deregulated mRNA
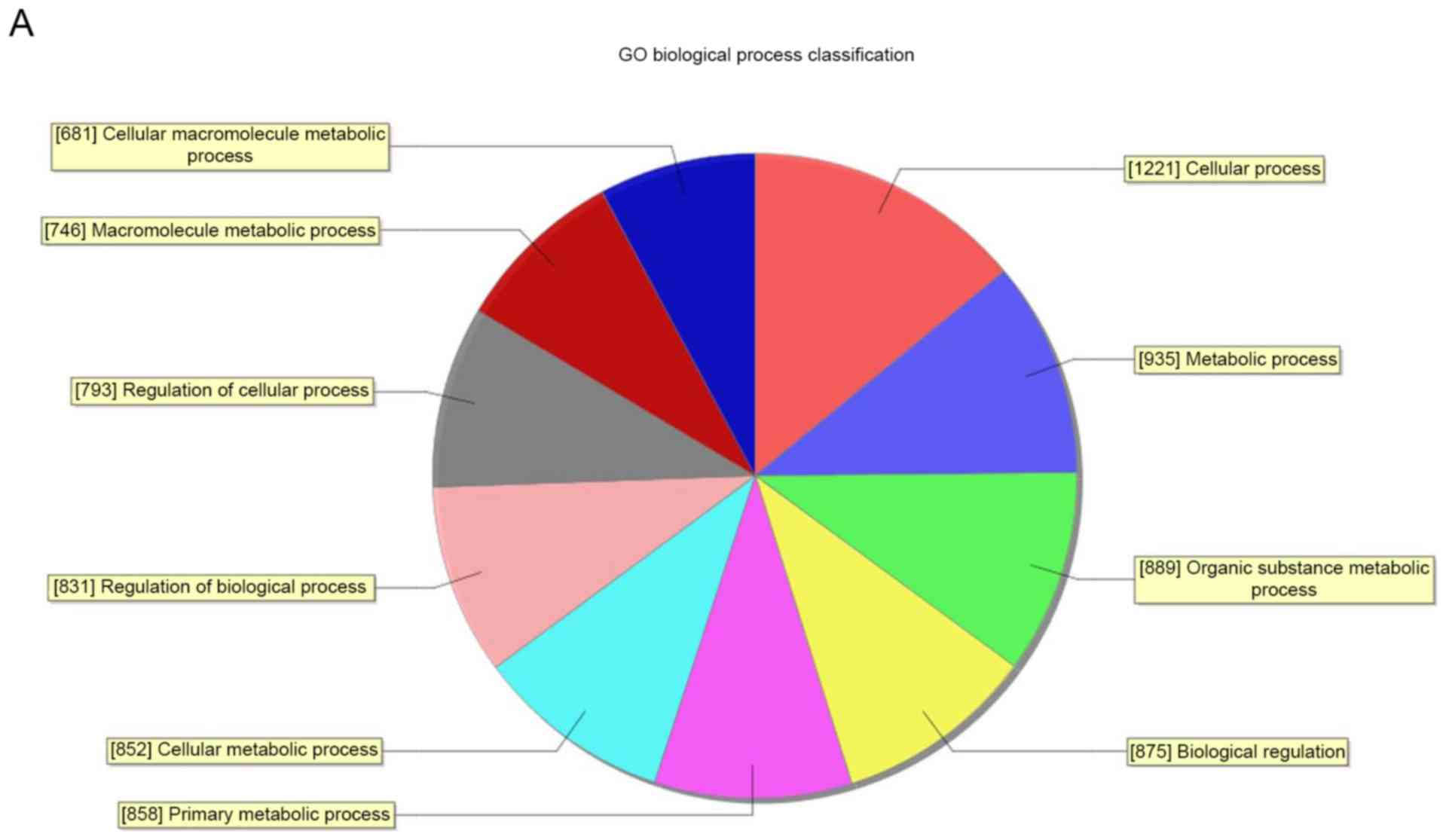
Through GO analysis, it was demonstrated that these
abnormal transcripts of lncRNA were associated with biological
process, cellular components and molecular function (Fig. 4). The analysis indicated that the
differentially expressed mRNA between the RA group and healthy
group were significantly enriched (P<0.05) in response to
wounding, DNA packaging complex, chemokine activity, RNA metabolic
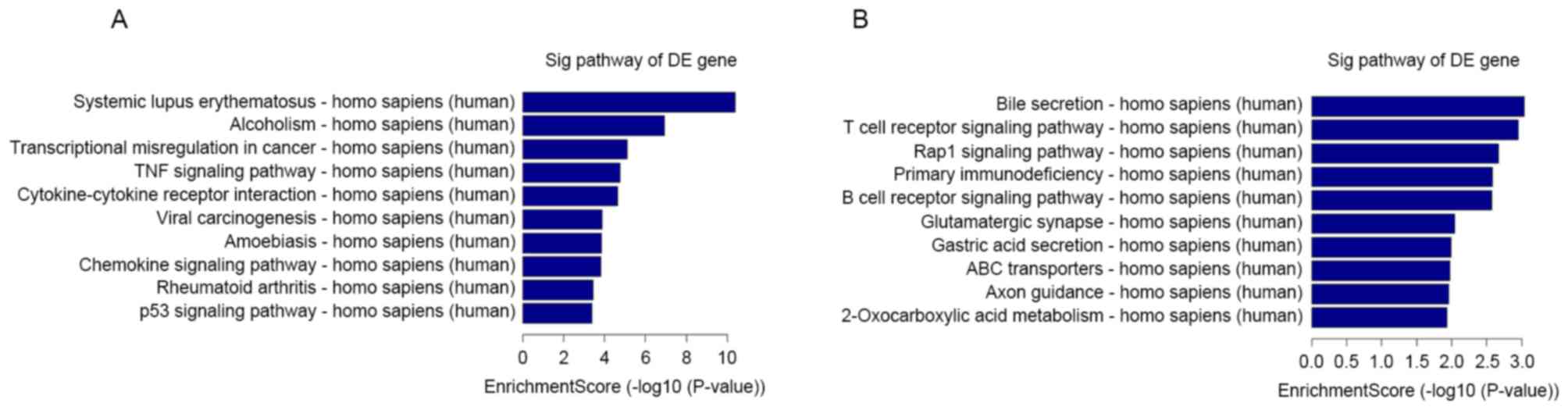
process, nuclear lumen and DNA binding. Pathway analysis indicated
that 23 upregulated pathways were identified, including systemic
lupus erythematosus and alcoholism pathways. A total of 23
downregulated pathways were identified, including bile secretion
and T cell receptor signaling pathway (Fig. 5).
Discussion
Over the past decades, expression profiles of genes
involved in the pathogenesis of RA have been extensively explored
(31,32). However, the molecular regulatory
mechanisms underlying RA have not been fully elucidated. Increasing
evidence indicates that lncRNA may serve crucial roles in various
biological processes and may regulate gene expression in human
disease (33,34). Few studies have reported that lncRNA
have an important role in RA (16–20);
however, fewer studies have investigated the expression profile of
lncRNA in RA (16–20). Therefore, the present study aimed to
better determine the potential role of lncRNA in contributing to
RA.
PBMCs are crucial sentinels of host defense
response, being used to identify novel disease mediators, disease
variants and treatment responses (35). Microarray technology has been used to
distinguish differences in gene expression profiles in PBMCs of RA
(8,32). In addition, PBMCs may simulate the
conditions of synovial tissue in RA, thus making it possible to
bypass the requirement for synovial tissue specimen, allowing the
research of larger patient populations (9). The present study analyzed the gene
expression patterns of PBMCs in patients with RA using microarrays
to uncover the possible role of lncRNA in the pathogenesis of this
disease. The results demonstrated that, compared with the healthy
group, there were notable changes in the expression of lncRNA
(2,410 upregulated and 2,635 downregulated) and mRNA (1,403
upregulated and 1,886 downregulated) in PBMCs. Furthermore, 135
matched lncRNA-mRNA pairs were identified for 85 differentially
expressed lncRNA and 109 differentially expressed mRNA. The present
study then focused on six altered lncRNA and mRNA and assessed
their expression level and the microarray results via RT-qPCR. To
the best of our knowledge, only one report exists regarding the
lncRNA pattern of RA using lncRNA array (17). The previous study analyzed the
expression profiles of 83 lncRNA and revealed that 18 lncRNA were
abnormal (17). However, 83 lncRNA
are only a fraction of the lncRNA family, and other lncRNA have yet
to be identified. Additionally, the present study comprehensively
analyzed the expression profiles of lncRNA and mRNA, using
Arraystar Human LncRNA Microarray V3.0, for 30,586 lncRNA and
26,109 mRNA.
As we know, there are five kinds of lncRNA,
including sense, antisense, bidirectional, intronic and intergenic.
Various studies have revealed a variety of mechanisms by which
lncRNA function; however, a common and key function of lncRNA is to
alter the expression of nearby encoding genes by impacting the
process of transcription (36) or
directly serving a role as enhancer-like elements (30,37). The
present study identified notably different expression patterns of
enhancer-like lncRNA (138 upregulated and 113 downregulated),
intergenic lncRNA (304 upregulated and 306 downregulated) and
antisense lncRNA (57 upregulated and 77 downregulated). In
addition, the present study identified some nearby coding genes
that may be regulated by lincRNA and enhancer-like lncRNA, and some
coding transcripts may be regulated by antisense lncRNA. These
results indicated that lncRNA could function as lincRNA,
enhancer-like lncRNA and antisense lncRNA in RA.
Furthermore, the present study revealed that not all
lncRNAs that were differentially expressed also exhibited
differential expression in their associated protein-coding genes.
This result suggested that these abnormal RA-associated lncRNA and
mRNA may be independently regulated. Future studies are required to
uncover their potential functional roles in RA disease by RNA
interference or overexpression methods in an appropriate model
system. However, the majority of the differentially expressed
lncRNA associated protein-coding genes were not deregulated,
suggesting that more complex mechanisms of regulation existed for
these lncRNA.
To understand the underlying roles of lncRNA, GO
category and KEGG pathway analyses were utilized to determine the
target gene pool. These results predicted that downregulated and
upregulated transcripts of lncRNA were associated with biological
process, cellular component and molecular function, which were
associated with 46 gene pathways that corresponded to transcripts,
including ‘rheumatoid arthritis’ and ‘TNF signaling pathway’. These
pathways may have essential roles in the occurrence and development
of RA.
The present study only detected one RA group and one
control group by microassay of gene chip. This is indeed a
limitation of the study. In order to minimize the variation between
groups, the chip assay was performed in the mode of a pooling test,
according to previous reports (26,38) in
the same research field. In future studies, we intend to
investigate the microassay of gene chip in a larger sample size in
triplicate.
In conclusion, the present study described the
global expression profiling of lncRNA and mRNA in RA by microarray
technology for the first time, which opened up a new and
interesting pathway to provide novel insights into the relationship
between lncRNA in PBMCs and RA. Additionally, the roles for the
abnormal lncRNA in the regulation of the TNF signaling pathway and
RA signaling pathways were researched.
Acknowledgements
The present study was financially supported by the
National Natural Science Foundation of China (grant no. 81360459)
and Jiangxi Provincial Natural Science Foundation of China (grant
no. 20151BAB215031). We are grateful to Dr Rui Wu (Department of
Rheumatology, The First Affiliated Hospital of Nanchang University,
Nanchang, China) for providing help.
References
|
1
|
Smolen JS, Aletaha D, Koeller M, Weisman
MH and Emery P: New therapies for treatment of rheumatoid
arthritis. Lancet. 370:1861–1874. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McInnes IB and Schett G: The pathogenesis
of rheumatoid arthritis. N Engl J Med. 365:2205–2219. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mehler MF and Mattick JS: Noncoding RNAs
and RNA editing in brain development, functional diversification,
and neurological disease. Physiol Rev. 87:799–823. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Novikova IV, Hennelly SP and Sanbonmatsu
KY: Sizing up long non-coding RNAs: Do lncRNAs have secondary and
tertiary structure? Bioarchitecture. 2:189–199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kowalczyk MS, Higgs DR and Gingeras TR:
Molecular biology: RNA discrimination. Nature. 482:310–311. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Long L, Yu P, Liu Y, Wang S, Li R, Shi J,
Zhang X, Li Y, Sun X, Zhou B, et al: Upregulated microRNA-155
expression in peripheral blood mononuclear cells and
fibroblast-like synoviocytes in rheumatoid arthritis. Clin Dev
Immunol. 2013:2961392013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pauley KM, Satoh M, Chan AL, Bubb MR,
Reeves WH and Chan EK: Upregulated miR-146a expression in
peripheral blood mononuclear cells from rheumatoid arthritis
patients. Arthritis Res Ther. 10:R1012008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dong L, Wang X, Tan J, Li H, Qian W, Chen
J, Chen Q, Wang J, Xu W, Tao C and Wang S: Decreased expression of
microRNA-21 correlates with the imbalance of Th17 and Treg cells in
patients with rheumatoid arthritis. J Cell Mol Med. 18:2213–2224.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J,
Jin Y, Zheng X, Wu Y and Fu H: The long noncoding RNA expression
profile of hepatocellular carcinoma identified by microarray
analysis. PLoS One. 9:e1017072014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wan ZY, Song F, Sun Z, Chen YF, Zhang WL,
Samartzis D, Ma CJ, Che L, Liu X, Ali MA, et al: Aberrantly
expressed long noncoding RNAs in human intervertebral disc
degeneration: A microarray related study. Arthritis Res Ther.
16:4652014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang B, Xia ZA, Zhong B, Xiong X, Sheng C,
Wang Y, Gong W, Cao Y, Wang Z and Peng W: Distinct hippocampal
expression profiles of long non-coding RNAs in an alzheimer's
disease model. Mol Neurobiol. 54:4833–4846. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stuhlmüller B, Kunisch E, Franz J,
Martinez-Gamboa L, Hernandez MM, Pruss A, Ulbrich N, Erdmann VA,
Burmester GR and Kinne RW: Detection of oncofetal h19 RNA in
rheumatoid arthritis synovial tissue. Am J Pathol. 163:901–911.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song J, Kim D, Han J, Kim Y, Lee M and Jin
EJ: PBMC and exosome-derived Hotair is a critical regulator and
potent marker for rheumatoid arthritis. Clin Exp Med. 15:121–126.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Spurlock CF III, Tossberg JT, Matlock BK,
Olsen NJ and Aune TM: Methotrexate inhibits NF-κB activity via long
intergenic (noncoding) RNA-p21 induction. Arthritis Rheumatol.
66:2947–2957. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Messemaker TC, Frank-Bertoncelj M, Marques
RB, Adriaans A, Bakker AM, Daha N, Gay S, Huizinga TW, Toes RE,
Mikkers HM and Kurreeman F: A novel long non-coding RNA in the
rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels.
Genes Immun. 17:85–92. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu MC, Yu HC, Yu CL, Huang HB, Koo M, Tung
CH and Lai NS: Increased expression of long noncoding RNAs
LOC100652951 and LOC100506036 in T cells from patients with
rheumatoid arthritis facilitates the inflammatory responses.
Immunol Res. 64:576–583. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Arnett FC, Edworthy SM, Bloch DA, McShane
DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH and Luthra
HS: The American Rheumatism Association 1987 revised criteria for
the classification of rheumatoid arthritis. Arthritis Rheum.
31:315–324. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Prevoo ML, van't Hof MA, Kuper HH, van
Leeuwen MA, van de Putte LB and van Riel PL: Modified disease
activity scores that include twenty-eight-joint counts. Development
and validation in a prospective longitudinal study of patients with
rheumatoid arthritis. Arthritis Rheum. 38:44–48. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hashemi R, Majidi A, Motamed H, Amini A,
Najari F and Tabatabaey A: Erythrocyte sedimentation rate
measurement using as a rapid alternative to the westergren method.
Emerg (Tehran). 3:50–53. 2015.PubMed/NCBI
|
|
24
|
Zhang J, Cui X, Shen Y, Pang L, Zhang A,
Fu Z, Chen J, Guo X, Gan W and Ji C: Distinct expression profiles
of LncRNAs between brown adipose tissue and skeletal muscle.
Biochem Biophys Res Commun. 443:1028–1034. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tian M, Chen R, Li T and Xiao B: Reduced
expression of circRNA hsa_circ_0003159 in gastric cancer and its
clinical significance. J Clin Lab Anal. Jun 15–2017.(Epub ahead of
print). doi: 10.1002/jcla.22281. View Article : Google Scholar
|
|
26
|
Xu G, Chen J, Pan Q, Huang K, Pan J, Zhang
W, Chen J, Yu F, Zhou T and Wang Y: Long noncoding RNA expression
profiles of lung adenocarcinoma ascertained by microarray analysis.
PLoS One. 9:e1040442014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanehisa M, Goto S, Furumichi M, Tanabe M
and Hirakawa M: KEGG for representation and analysis of molecular
networks involving diseases and drugs. Nucleic Acids Res.
38:D355–D360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang T, Jiang M, Chen L, Niu B and Cai Y:
Prediction of gene phenotypes based on GO and KEGG pathway
enrichment scores. Biomed Res Int. 2013:8707952013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Teixeira VH, Olaso R, Martin-Magniette ML,
Lasbleiz S, Jacq L, Oliveira CR, Hilliquin P, Gut I, Cornelis F and
Petit-Teixeira E: Transcriptome analysis describing new immunity
and defense genes in peripheral blood mononuclear cells of
rheumatoid arthritis patients. PLoS One. 4:e68032009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
van der Pouw Kraan TC, Wijbrandts CA, van
Baarsen LG, Voskuyl AE, Rustenburg F, Baggen JM, Ibrahim SM, Fero
M, Dijkmans BA, Tak PP and Verweij CL: Rheumatoid arthritis
subtypes identified by genomic profiling of peripheral blood cells:
Assignment of a type I interferon signature in a subpopulation of
patients. Ann Rheum Dis. 66:1008–1014. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
noncoding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Toonen EJ, Barrera P, Radstake TR, van
Riel PL, Scheffer H, Franke B and Coenen MJ: Gene expression
profiling in rheumatoid arthritis: Current concepts and future
directions. Ann Rheum Dis. 67:1663–1669. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mattick JS and Gagen MJ: The evolution of
controlled multitasked gene networks: The role of introns and other
noncoding RNAs in the development of complex organisms. Mol Biol
Evol. 18:1611–1630. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mattick JS: Linc-ing Long noncoding RNAs
and enhancer function. Dev Cell. 19:485–486. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yi Z, Li J, Gao K and Fu Y: Identifcation
of differentially expressed long non-coding RNAs in CD4+ T cells
response to latent tuberculosis infection. J Infect. 69:558–568.
2014. View Article : Google Scholar : PubMed/NCBI
|