Introduction
Cervical cancer is the second most frequent
gynecological cancer type worldwide and accounts for almost 12% of
all malignancies in women (1,2).
Cervical cancer is the result of a multistep process involving the
transformation of normal cervical epithelium to a cervical
intraepithelial pre-neoplasm, which is subsequently transformed
into cervical cancer (3). Infection
with high-risk human papillomaviruses (HPVs) has an important role
in the initiation of cervical cancer (4,5).
However, increasing evidence has indicated that HPV infection alone
is insufficient for the development and progression of cervical
cancer (5). Therefore, there is a
compelling requirement for understanding of the other molecular
mechanisms underlying the process of the genesis and progression of
the malignant phenotype.
MicroRNAs (miRNAs/miRs) are a class of endogenous,
small non-coding RNAs that are comprised of 18–25 nucleotides and
control gene expression via binding with complementary sites in the
3′-untranslated regions (UTRs) of mRNAs (6,7).
Accumulating evidence demonstrated that certain miRNAs are
deregulated in a number of human cancer types and are involved in
the regulation of numerous biological processes, including cell
growth, differentiation, invasion, migration and apoptosis
(8,9). miRNAs act as tumor suppressors or
oncogenes in cancer depending on the nature of their target genes
(10,11). Certain miRNAs have been demonstrated
to be involved in the genesis and progression of cervical cancer
(9). For instance, miR-328
suppresses cervical cancer cell proliferation and tumorigenesis
in vitro and in vivo by targeting transcription
factor 7 like 2 expression (12).
Furthermore, miR-486-3p targeting extracellular matrix protein 1
inhibits cell proliferation and metastasis in cervical cancer
(13). miR-519d contributes to the
progression and metastasis of cervical cancer by directly
regulating Smad7 (14). miR-200b was
demonstrated to suppress cell invasion and metastasis by
suppressing epithelial-mesenchymal transition in cervical carcinoma
(15).
miR-144 was originally identified to be an
erythroid-specific miRNA, which is essential for the survival and
maturation of the erythroid lineage (16,17).
Ample studies have also reported that miR-144 acts as a tumor
suppressor in different types of human cancer, including
hepatocellular carcinoma (18),
rectal cancer (19) and B-cell
lymphoma (20). A recent miRNA
microarray analysis demonstrated that miR-144 expression was
significantly downregulated in patients with cervical cancer and
lymph node metastasis as compared with those without lymph node
metastasis (21). However, the
functional role of miR-144 in cervical cancer has remained to be
elucidated.
The present study investigated the expression of
miR-144 in cervical cancer tissues and explored its potential
effects on the growth, migration and invasion of cervical cancer
cells. In addition, vascular endothelial growth factor A (VEGFA)
and VEGFC were identified as targets of miR-144 in cervical cancer
cells. These findings indicated a novel molecular mechanism
underlying in the regulatory roles of miR-144 in cervical cancer.
Thus, miR-144 may be a promising therapeutic tool for cervical
cancer in the future.
Material and methods
Cell lines and culture
The HeLa and C33A cervical cancer cell lines were
obtained from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China). Cells were cultured in RPMI1640 (Gibco; Thermo
Fisher Scientific, Inc., Waltham MA, USA) supplemented with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.),
100 g/ml streptomycin sulfate and 100 U/ml penicillin sodium in a
humidified incubator with 5% CO2 at 37°C. Cells in the
logarithmic growth phase were used for further study.
Oligonucleotide transfection
miR-144 mimics and scrambled oligonucleotide as the
negative control (NC) were chemically synthesized and purchased
from Shanghai GenePharma (Shanghai, China). miR-144 mimics or NC
(100 nM) in 2 ml RPMI1640 medium were transfected into HeLa and
C33A cells with Lipofectamine 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) after the cells reached 60–80% confluence,
according to the manufacturer's protocol. After 6 h of
transfection, the medium was replaced with RPMI1640 containing 10%
FBS. The cells were collected 48 h later, and the expression of
miR-144 in the cervical cancer cell lines was detected by
Reverse-transcription quantitative polymerase chain reaction
analysis (RT-qPCR).
Clinical samples of cervical
cancer
A total of 35 cervical cancer tissues and matched
normal tissues were collected from the Department of Gynecologic
Tumor at Fudan University Shanghai Cancer Center (Shanghai, China)
from January 2010 to December 2014. The tissues that were obtained
from surgery were immediately frozen in liquid nitrogen and
subsequently stored at −80°C for further study. Pathological
classification and staging were performed according to the
International Federation of Gynecology and Obstetrics (FIGO)
criteria (22). Based on lymph node
metastasis, 35 patients with cervical cancer were divided into two
groups, including patients with lymph node metastasis (n=9) and
those with no lymph node metastasis (n=26). All patients provided
their written informed consent to be enrolled in the study. The
present study was approved by the Ethics Committee of Fudan
University Shanghai Cancer Center (Shanghai, China) and experiments
were in accordance with the Declaration of Helsinki. The
characteristics of the cervical cancer patients are displayed in
Table I.
 | Table I.Association between miR-144
expression and clinicopathological characteristics. |
Table I.
Association between miR-144
expression and clinicopathological characteristics.
|
|
| miR-144
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | Cases (n) | Low | High | P-value |
|---|
| Age (years) |
|
|
| 0.259 |
|
<45 | 22 | 14 | 8 |
|
|
≥45 | 13 | 11 | 2 |
|
| Tumor size |
|
|
| 0.723 |
| ≤4
cm | 19 | 13 | 6 |
|
| >4
cm | 16 | 12 | 4 |
|
| Histological
type |
|
|
| 0.661 |
|
Squamous cell carcinoma | 27 | 20 | 7 |
|
|
Adenocarcinoma | 8 | 5 | 3 |
|
| Histological
differentiation |
|
|
| 0.266 |
|
Well/moderate | 15 | 9 | 6 |
|
|
Poor | 20 | 16 | 4 |
|
| FIGO stage |
|
|
| 0.004a |
| IB | 24 | 21 | 3 |
|
|
>IB | 11 | 4 | 7 |
|
| Lymph node
metastasis |
|
|
| 0.007a |
| No | 26 | 22 | 4 |
|
|
Yes | 9 | 3 | 6 |
|
RT-qPCR
Total RNA was extracted from cervical cancer tissues
and cells by using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. For
detection of miR-144 expression, RT was performed by using a
Super-Script III RT kit (cat. no. 18080085; Invitrogen; Thermo
Fisher Scientific, Inc.). To determine VEGFA and VEGFC expression,
mRNA was converted into complementary (c)DNA by using a PrimeScript
RT reagent kit (cat. no. RR047A; Takara, Tokyo, Japan). qPCR was
performed by using a SYBR® Premix Ex Taq™ II kit (cat.
no. RR036A; Takara) with analysis in an ABI 7500 Real-Time PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
qPCR conditions were 95°C for 5 min followed by 40 cycles of
denaturation at 95°C for 10 sec and annealing/elongation at 60°C
for 30 sec. U6 small nuclear RNA and GAPDH were assessed as
endogenous controls. Primer sequences were as follows: miR-144,
forward, 5′-TCATGTAGTAGATATGACAT-3′ and reverse miscript universal
primer (Qiagen GmbH, Hilden, Germany); U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′;
VEGFA forward, 5′-TTTCTGCTGTCTTGGGTGCATTGG-3′ and reverse,
5′-ACCACTTCGTGATGATTCTGCCCT-3′; VEGFC forward,
5′-GAGGAGCAGTTACGGTCTGTG-3′ and reverse,
5′-TCCTTTCCTTAGCTGACACTTGT-3′; GAPDH forward,
5′-CTGGGCTACACTGAGCACC-3′ and reverse, 5′-AAGTGGTCGTTGAGGGCAATG-3′.
The RT-qPCR was performed using the 2−ΔΔCq method
(23).
MTT assay
The viability of the HeLa and C33A cells was
evaluated by using the MTT method. The cells (5×103
cells/well) were seeded and cultured in 96-well plates, and MTT
solution (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added
to a final concentration of 0.5 mg/ml at 48 h after transfection.
The cells were incubated for another 4 h at 37°C with 5%
CO2. Subsequently, the medium was removed and 150 µl of
dimethylsulfoxide (Sigma-Aldrich; Merck KGaA) was added to each
well to dissolve the formazan crystals. Finally, the optical
density at 480 nm was measured by using an ELX-800 Microplate
Reader (BioTeke, Winooski, VT, USA).
Migration and invasion assay
A wound healing assay was performed to assess the
migration ability of cervical cancer cells. In brief, cells were
seeded in 6-well plates at a density of 106 cells/well
and cultured to confluence. Cell monolayers were scraped with a
sterile 10-µl pipette tips (EMD Millipore, Billerica, MA, USA) to
generate scratch wounds and washed three times with PBS
(Sigma-Aldrich; Merck KGaA) to remove cell debris. The cells were
incubated at 37°C with 5% CO2 for another 24 h. To
determine the migration distances, wounds were observed under a
CX41 inverted microscope (Olympus, Tokyo, Japan) at ×100
magnification and images were captured.
A cell invasion assay was performed by using
Transwell® chambers (BD Biosciences, Franklin Lakes, NJ,
USA). In brief, 300 µl cell suspension was added to the upper
chamber pre-coated with Matrigel™ (BD Biosciences) and 500 µl
RPMI1640 with 10% FBS as the chemoattractant was added to the lower
chambers. The cells were incubated at 37°C with 5% CO2
for 24 h. Following careful removal of cells remaining on the upper
surface of the membrane, those on the lower surface of the membrane
were fixed in pure methanol (Sigma-Aldrich; Merck KGaA) for 20 min
and stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA)
for 15 min. Stained cells were visualized and counted in five
randomly selected fields under a CX41 inverted microscope at ×200
magnification.
Dual luciferase reporter assay
The full-length 3′-UTRs of VEGFA and VEGFA [wild
type (WT)] were amplified by PCR from cDNA and cloned into the
PGL3-Basic Vector (Promega, Madison, WI, USA). The mutant (MUT)
construct of VEGFA and VEGFA 3′-UTRs was generated by using a
QuickChange Site-Directed Mutagenesis kit (cat. no. R407; Takara).
Co-transfection of WT or MUT VEGFA or VEGFA reporter vector with
miR-144 mimic or NC was performed by using Lipofectamine 2000.
After 48 h of transfection, dual luciferase activity was measured
by using a dual luciferase reporter assay system (Promega)
according to the manufacturer's protocols.
Protein extraction and western blot
analysis
Total protein was extracted from the cultured cells
by using radioimmunoprecipitation assay buffer (Invitrogen; Thermo
Fisher Scientific, Inc.) at 4°C. The total protein concentration
was determined by an Enhanced BCA Protein Assay kit (Beyotime
Institute of Biotechnology, Haimen, China). Equal amounts of
proteins (40 µg) were separated by 8–12% SDS-PAGE and transferred
onto a polyvinylidene difluoride membrane (EMD Millipore). The
membrane was incubated with Tris-buffered saline with Tween-20
containing 5% non-fat milk for 2 h at 37°C. The membrane was
incubated with mouse anti-human monoclonal anti-VEGFA (dilution,
1:500; cat. no. ab1316; Abcam, Cambridge, MA, USA), anti-VEGFC
(dilution, 1:1,000; cat. no. ab191274; Abcam) and anti-GAPDH
(dilution, 1:2,000; cat. no. ab9482; Abcam) antibodies at 4°C
overnight, followed by incubation with horseradish
peroxidase-conjugated secondary antibodies (1:2,000; cat. no.
A21010; WuHan AmyJet Scientific Inc., Wuhan, China) at room
temperature for 1 h. Blots were visualized by an enhanced
chemiluminescence kit (Pierce; Thermo Fisher Scientific, Inc.,
Rockford, IL, USA).
Statistical analysis
Statistical analysis was performed with SPSS
statistical software, version 17.0 (SPSS, Inc., Chicago, IL, USA).
Values are expressed as the mean ± standard deviation of at least
three independent experiments. Differences between two groups were
analyzed by a two-tailed Student's t-test or one-way analysis of
variance with a Bonferroni correction. Data plotting was performed
with GraphPad Prism, version 5 (GraphPad Software, Inc., La Jolla,
CA, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
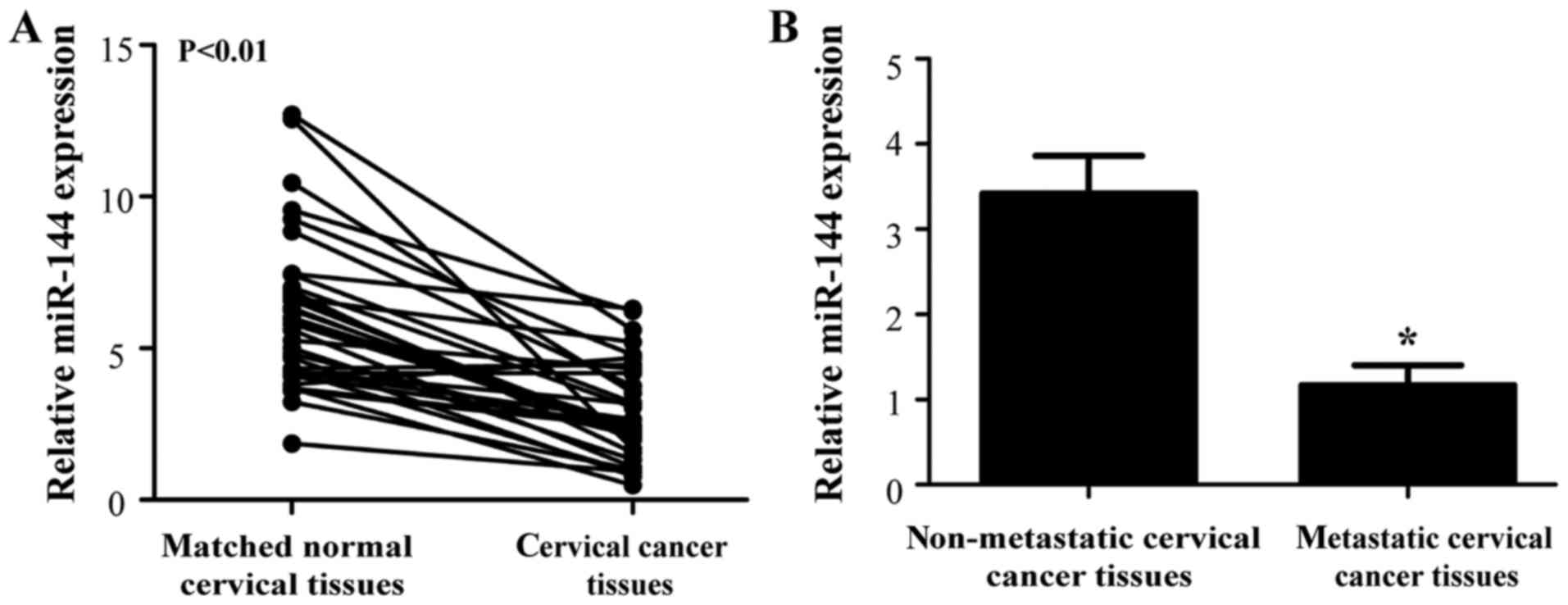
miR-144 expression is downregulated in
cervical cancer tissues
To assess the function of miR-144 in cervical
cancer, the present study first examined its expression in the
cervical tissues from patients with cervical cancer. The RT-qPCR
results demonstrated that miR-144 expression was significantly
reduced in the cervical cancer tissues compared with matched normal
cervical tissues (Fig. 1A;
P<0.01).
A previous miRNA microarray analysis reported that
miR-144 expression was downregulated in patients with lymph node
metastasis (21). Therefore, the
present study also detected the expression levels of miR-144 in
patients with lymph node metastasis. As presented in Fig. 1B, miR-144 expression in cervical
cancer tissues of patients with lymph node metastasis was also
lower than that in patients without lymph node metastasis
(P<0.05), implying that decreased expression of miR-144 may have
an important role in the progression of cervical cancer.
Association of miR-144 expression with
clinicopathological features of cervical cancer patients
In order to determine the clinical significance of
miR-144 in cervical cancer, the 35 patients were divided into two
groups based on miR-144 expression levels (low vs. high) with the
median expression level as a cut-off point. The association of
miR-144 expression with the clinicopathological features is
presented in Table I. The results
indicated that downregulation of miR-144 was significantly
associated with the FIGO stage (P=0.004) and lymph node metastasis
(P=0.007), while no significant association with any other
clinicopathological variables was observed.
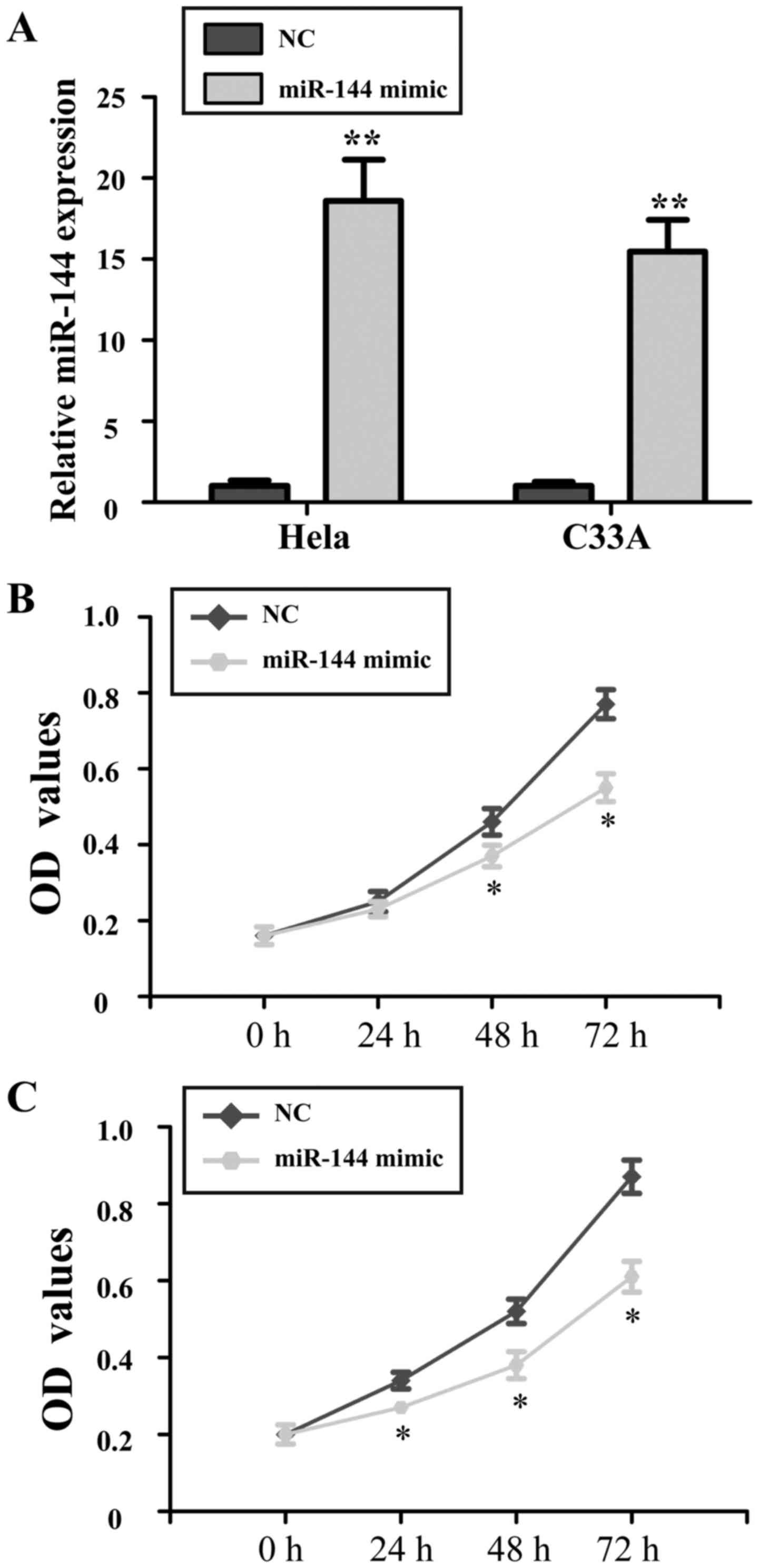
Ectopic expression of miR-144 inhibits
the proliferation of cervical cancer cells
To identify the role of miR-144 in cervical cancer
cells, miR-144 mimics or NC was introduced into HeLa and C33A
cells. The proliferation of miR-144-overexpressing cells was
evaluated by an MTT assay. Transfection with miR-144 mimics
resulted in a significantly increased expression of miR-144 in HeLa
and C33A (P<0.01; Fig. 2A). The
results of the MTT assay demonstrated that at 48 and 72 h after
transfection with miR-144 overexpression vector, the growth of HeLa
and C33A cells was significantly inhibited compared with that in
the NC group (P<0.05; Fig. 2B and
C).
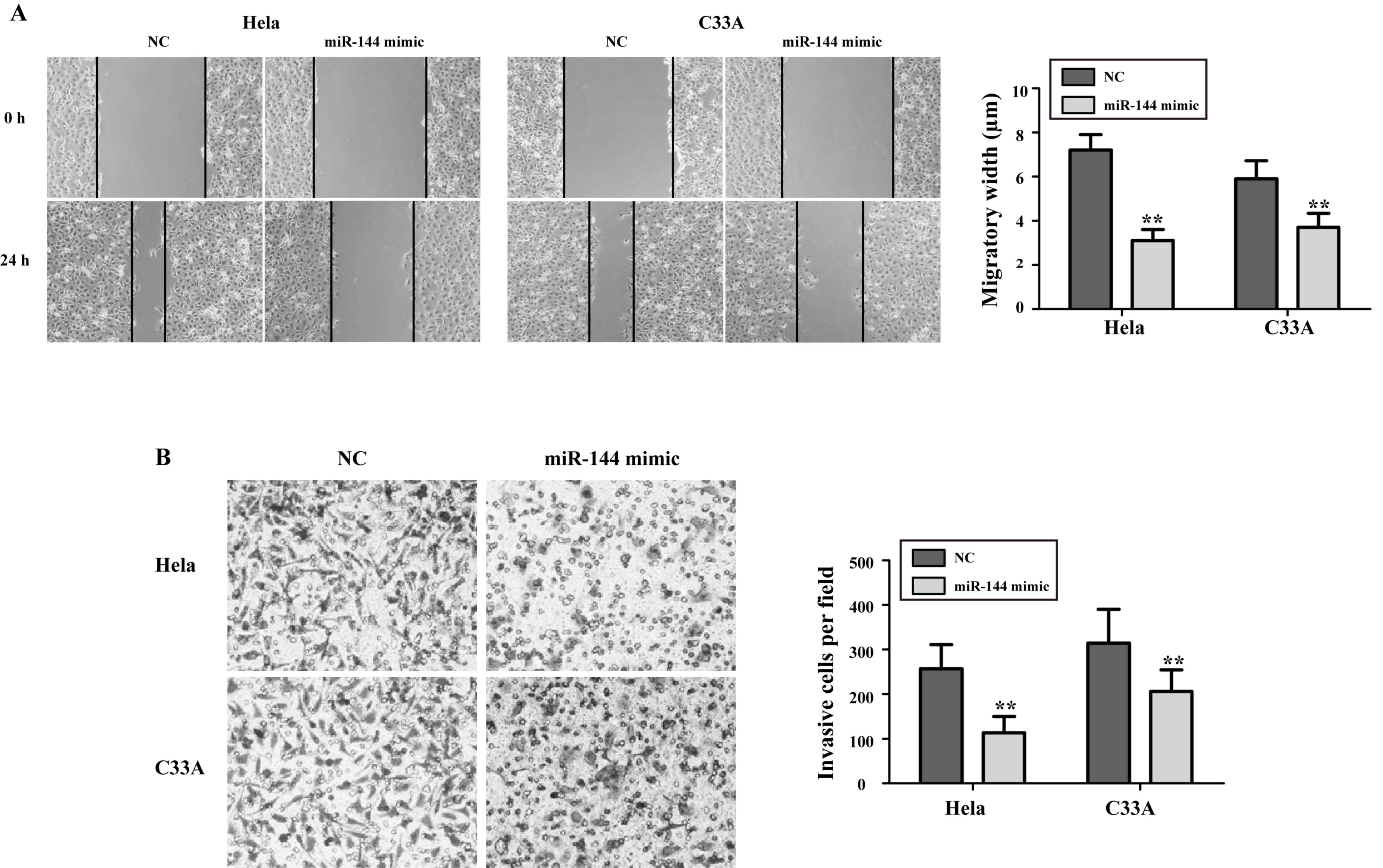
Overexpression of miR-144 decreases
the migration and invasion abilities of cervical cancer cells
Next, the present study sought to explore the effect
of miR-144 on the migration and invasion abilities of cervical
cancer cells. As presented in Fig.
3A, the migration abilities of HeLa and C33A cells treated with
miR-144 mimics were significantly decreased compared with those in
NC-transfected cells (P<0.05). As presented in Fig. 3B, the number of invasive cells in the
miR-144 mimics-transfected groups was reduced compared with that in
the NC-transfected groups (HeLa, (75.4±17.6 vs. 267.6±44.2,
P<0.01; C33A, 86.9±21.3 vs. 315.2±69.8, P<0.01).
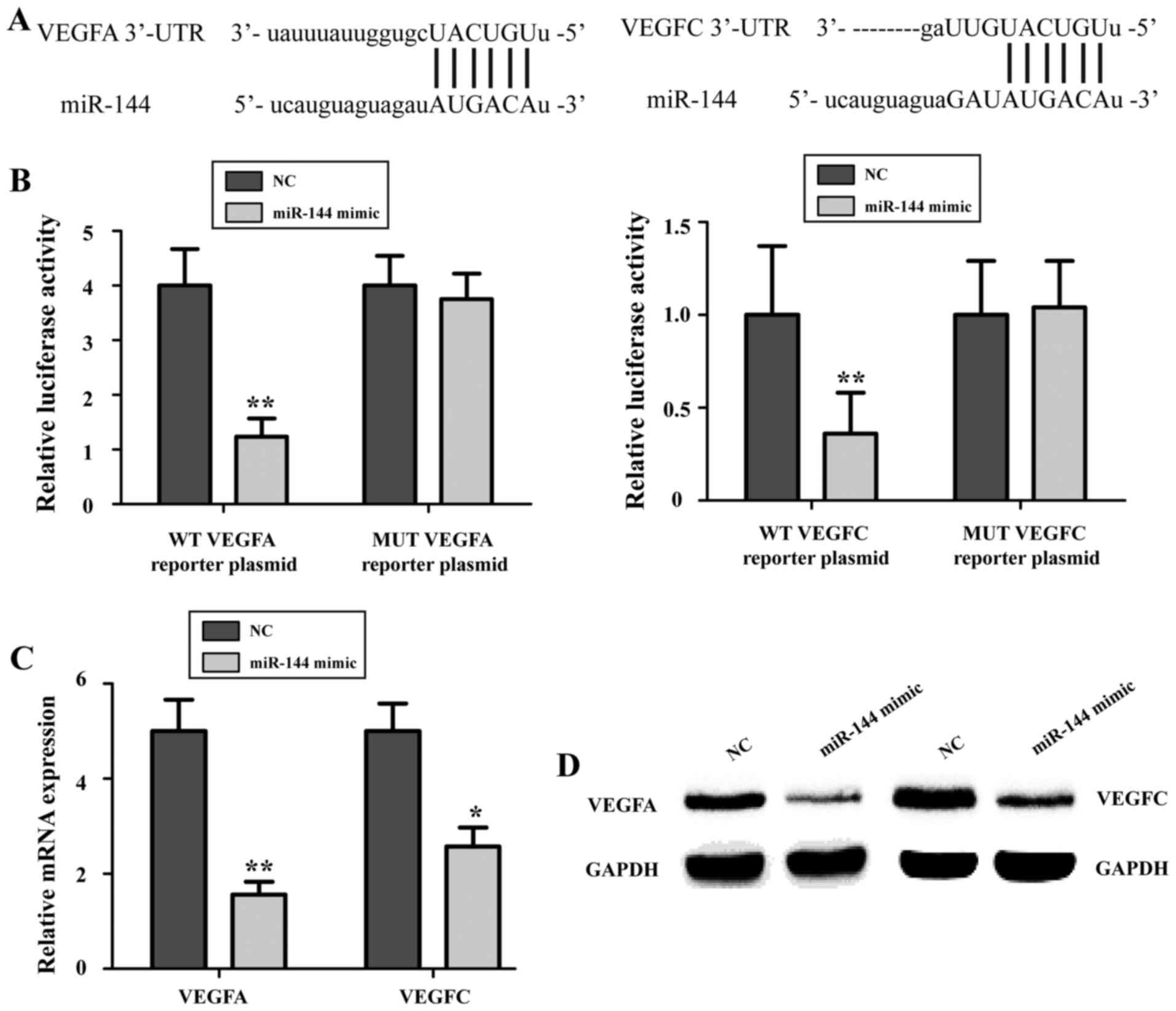
miR-144 targets the 3′-UTRs of VEGFA
and VEGFC mRNA to inhibit their expression
To further illustrate the molecular mechanisms
underlying the tumor suppressor role of miR-144 in cervical cancer
cells, a bioinformatics analysis was performed to identify targets
of miR-144. A TargetScan (http://www.targetscan.org) search revealed the
presence of a miR-144 binding site in the 3′-UTR of VEGFA mRNA as
well as that of VEGFC mRNA (Fig.
4A). To verify this, miR-144 mimics or NC and the WT or MUT
VEGFA or VEGFC luciferase reporter vectors were co-transfected into
HeLa cells. The results indicated that the relative luciferase
activity in miR-144 mimics-treated HeLa cells was significantly
lower than that in NC-transfected cells (P<0.05; Fig. 4B), whereas this inhibitory effect was
abrogated when the oligonucleotides in the binding sites of the
3′-UTRs were mutated. To further confirm that VEGFA and VEGFC are
the targets of miR-144, RT-qPCR and western blot analyses were
performed. The expression of VEGFA and VEGFC in HeLa cells was
markedly reduced following miR-144 overexpression (P<0.05;
Fig. 4C and D). These results
suggested that VEGFA and VEGFC are target genes of miR-144 in
cervical cancer cells.
Discussion
Previous studies have indicated that 99.7% of cases
with cervical cancer may be attributed to HPV infection (24). Emerging evidence has demonstrated
that certain aberrantly expressed miRNAs are associated with
tumorigenesis by upregulating the expression of oncogenes and serve
as biomarkers for the diagnosis and prognosis of various human
cancer types, including cervical cancer (25–27).
Hence, identification of miRNAs and their target genes involved in
tumorigenesis will provide key clues for the development of novel
diagnostic tools and therapies for patients with cervical
cancer.
miR-144 is located on chromosome 17q11.2 of the
human genome, which has been reported to be downregulated in
esophageal squamous cell cancer (28), laryngeal squamous cell carcinoma
(29), lung cancer (30) and bladder cancer (31). The present study assessed the
expression levels of miR-144 in cervical cancer tissues and their
matched normal cervical tissues to determine the effects of miR-144
on the malignant progression of cervical cancer. The results
indicated that miR-144 was markedly downregulated in cervical
cancer tissues, which was consistent with those of previous
studies. A miRNA microarray analysis by Ding et al (21) identified that the expression of
miR-144 was significantly downregulated in cervical cancer patients
with lymph node metastasis as compared with that in patients
without. In line with this, the present study also indicated that
the expression of miR-144 was lower in cervical cancer tissues from
patients with lymph node metastasis compared with patients without
lymph node metastasis. The downregulation of miR-144 was
significantly associated with the FIGO stage and lymph node
metastasis. These results strongly suggested that miR-144 is
involved in the progression of cervical cancer.
Epigenetic changes represent an important mechanism
for the silencing of tumor suppressor genes or activation of
oncogenes. In cervical cancer, DNA methylation-mediated silencing
of a rapidly growing number of coding and non-coding genes has been
identified and their functional relevance has been demonstrated
(32). Increased methylation has
been demonstrated for a number of miRNAs, including miR-34b,
miR-95, miR-124 and miR-375 (32).
The present study found that miR-144 expression was also repressed,
indicating that DNA methylation-mediated silencing of miR-144 may
exist in cervical cancer.
miR-144 has been indicated to have a tumor
suppressive role in the progression of cancer. For instance, Wang
et al (33) identified that
miR-144 suppressed osteosarcoma cell proliferation and metastasis
by targeting the expression of Rho-associated protein kinases 1 and
2. Matsushita et al (34)
suggested that tumor-suppressive miR-144 directly targeted the
transcripts of CCNE1/2 as potential prognostic biomarkers in
bladder cancer. Zhang et al (30) reported that miR-144-3p, a tumor
suppressive miRNA, targeted ETS-1 in laryngeal squamous cell
carcinoma. The present study expanded the current knowledge
regarding the role of miR-144 in cervical cancer and suggested a
tumor suppressive role via inhibition of cellular growth, migration
and invasion.
Since miRNAs usually exert their biological effects
via inhibiting the expression of target mRNAs, a bioinformatics
analysis was performed to predict gene targets of miR-144. VEGFA
and VEGFC were predicted as targets, which are two
cancer-associated genes containing highly evolutionarily conserved
miR-144 binding sites on the 3′-UTR of their mRNAs. A previous
study reported that VEGF contributed to endothelial cell
proliferation, division and migration (35). Knockdown of hypoxia-inducible
factor-1α was previously demonstrated to decrease the radiation
resistance of cervical cancer by reducing the expression of VEGF
(36). Anti-VEGF treatment augments
the tumor radiation response under normoxic or hypoxic conditions,
which is a novel therapeutic strategy in cervical cancer (37). VEGFA and VEGFC, two members of the
VEGF family, were recently identified as the predominant tumor
metastasis-driving factors in various human cancers types,
including renal cell carcinoma (38), hepatocellular carcinoma (39) and cervical cancer (40–42). The
present study identified VEGFA and VEGFC as targets of miR-144 via
directly binding to the 3′-UTRs of VEGFA and VEGFC mRNA in a dual
luciferase reporter assay. Furthermore, RT-qPCR and western blot
analyses confirmed that ectopic expression of miR-144 significantly
downregulated the expression of VEGFA and VEGFC. These findings
suggested that downregulation of miR-144 may contribute to aberrant
growth and metastasis of cancer cells through loss of inhibition of
VEGFA and VEGFC expression.
In conclusion, the present study demonstrated that
miR-144 exerts a suppressive effect on the growth, migration and
invasion of cervical cancer cells by directly targeting VEGFA and
VEGFC, indicating that miR-144 may be a promising therapeutic
target for cervical cancer treatment.
Acknowledgements
The present study was supported by the Leader
Training Plan of the Health Department, Shanghai Pudong New
District (grant no. PWRd2013-09), the Key Specialty Construction
Project of Pudong Health and Family Planning Commission of Shanghai
(grant no. PWZz2013-14) and the Science and Technology Development
Project of Pudong New District (grant no. PW2013A-6).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hildesheim A and Wang SS: Host and viral
genetics and risk of cervical cancer: A review. Virus Res.
89:229–240. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Castellsagué X: Natural history and
epidemiology of HPV infection and cervical cancer. Gynecol Oncol.
110(3 Suppl 2): S4–S7. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Du CX and Wang Y: Expression of P-Akt,
NFkappaB and their correlation with human papillomavirus infection
in cervical carcinoma. Eur J Gynaecol Oncol. 33:274–277.
2012.PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moreno-Moya JM, Vilella F and Simón C:
MicroRNA: Key gene expression regulators. Fertil Steril.
101:1516–1523. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartels CL and Tsongalis GJ: MicroRNAs:
Novel biomarkers for human cancer. Clin Chem. 55:623–631. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and
tumor-suppressive microRNAs in cervical cancer is required for
cancer cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang X and Xia Y: microRNA-328 inhibits
cervical cancer cell proliferation and tumorigenesis by targeting
TCF7L2. Biochem Biophys Res Commun. 475:169–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ye H, Yu X, Xia J, Tang X, Tang L and Chen
F: MiR-486-3p targeting ECM1 represses cell proliferation and
metastasis in cervical cancer. Biomed Pharmacother. 80:109–114.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou JY, Zheng SR, Liu J, Shi R, Yu HL and
Wei M: MiR-519d facilitates the progression and metastasis of
cervical cancer through direct targeting Smad7. Cancer Cell Int.
16:212016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheng YX, Zhang QF, Hong L, Pan F, Huang
JL, Li BS and Hu M: MicroRNA-200b suppresses cell invasion and
metastasis by inhibiting the epithelial-mesenchymal transition in
cervical carcinoma. Mol Med Rep. 13:3155–3160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dore LC, Amigo JD, Dos Santos CO, Zhang Z,
Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, et al: A
GATA-1-regulated microRNA locus essential for erythropoiesis. Proc
Natl Acad Sci USA. 105:pp. 3333–3338. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lawrie CH: microRNA expression in
erythropoiesis and erythroid disorders. Br J Haematol. 150:144–151.
2010.PubMed/NCBI
|
|
18
|
Cao T, Li H, Hu Y, Ma D and Cai X: miR-144
suppresses the proliferation and metastasis of hepatocellular
carcinoma by targeting E2F3. Tumour Biol. 35:10759–10764. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cai SD, Chen JS, Xi ZW, Zhang LJ, Niu ML
and Gao ZY: MicroRNA144 inhibits migration and proliferation in
rectal cancer by downregulating ROCK-1. Mol Med Rep. 12:7396–7402.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang H, Wang A, Hu Z, Xu X, Liu Z and Wang
Z: A Critical role of miR-144 in diffuse large B-cell lymphoma
proliferation and invasion. Cancer Immunol Res. 4:337–344. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ding H, Wu YL, Wang YX and Zhu FF:
Characterization of the microRNA expression profile of cervical
squamous cell carcinoma metastases. Asian Pac J Cancer Prev.
15:1675–1679. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Du L, Lei Y, Li D, Qiu X and Liang B:
Value of magnetic resonance imaging in preoperative staging of
endometrial carcinoma according to International Federation of
Gynecology and Obstetrics (2009) staging criteria. Nan Fang Yi Ke
Da Xue Xue Bao. 32:1048–1051. 2012.(In Chinese). PubMed/NCBI
|
|
23
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang LQ, Zhang Y, Yan H, Liu KJ and Zhang
S: MicroRNA-373 functions as an oncogene and targets YOD1 gene in
cervical cancer. Biochem Biophys Res Commun. 459:515–520. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schickel R, Boyerinas B, Park SM and Peter
ME: MicroRNAs: Key players in the immune system, differentiation,
tumorigenesis and cell death. Oncogene. 27:5959–5974. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen B, Hou Z, Li C and Tong Y: MiRNA-494
inhibits metastasis of cervical cancer through Pttg1. Tumour Biol.
36:7143–7149. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang N, Wei H, Yin D, Lu Y, Zhang Y, Zhang
Q, Ma X and Zhang S: MicroRNA-195 inhibits proliferation of
cervical cancer cells by targeting cyclin D1a. Tumour Biol.
37:4711–4720. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shao Y, Li P, Zhu ST, Yue JP, Ji XJ, Ma D,
Wang L, Wang YJ, Zong Y, Wu YD and Zhang ST: MiR-26a and miR-144
inhibit proliferation and metastasis of esophageal squamous cell
cancer by inhibiting cyclooxygenase-2. Oncotarget. 7:15173–15186.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu X, Cui CL, Chen WL, Fu ZY, Cui XY and
Gong X: miR-144 suppresses the growth and metastasis of laryngeal
squamous cell carcinoma by targeting IRS1. Am J Transl Res. 8:1–11.
2016.PubMed/NCBI
|
|
30
|
Zhang SY, Lu ZM, Lin YF, Chen LS, Luo XN,
Song XH, Chen SH and Wu YL: miR-144-3p, a tumor suppressive
microRNA targeting ETS-1 in laryngeal squamous cell carcinoma.
Oncotarget. 7:11637–11650. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guo Y, Ying L, Tian Y, Yang P, Zhu Y, Wang
Z, Qiu F and Lin J: miR-144 downregulation increases bladder cancer
cell proliferation by targeting EZH2 and regulating Wnt signaling.
FEBS J. 280:4531–4538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wilting SM, Miok V, Jaspers A, Boon D,
Sørgård H, Lando M, Snoek BC, van Wieringen WN, Meijer CJ, Lyng H,
et al: Aberrant methylation-mediated silencing of microRNAs
contributes to HPV-induced anchorage independence. Oncotarget.
7:43805–43819. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang W, Zhou X and Wei M: MicroRNA-144
suppresses osteosarcoma growth and metastasis by targeting ROCK1
and ROCK2. Oncotarget. 6:10297–10308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Matsushita R, Seki N, Chiyomaru T,
Inoguchi S, Ishihara T, Goto Y, Nishikawa R, Mataki H, Tatarano S,
Itesako T, et al: Tumour-suppressive microRNA-144-5p directly
targets CCNE1/2 as potential prognostic markers in bladder cancer.
Br J Cancer. 113:282–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Goel HL and Mercurio AM: VEGF targets the
tumour cell. Nat Rev Cancer. 13:871–882. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fu Z, Chen D, Cheng H and Wang F:
Hypoxia-inducible factor-1α protects cervical carcinoma cells from
apoptosis induced by radiation via modulation of vascular
endothelial growth factor and p53 under hypoxia. Med Sci Monit.
21:318–325. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tsuda N, Watari H and Ushijima K:
Chemotherapy and molecular targeting therapy for recurrent cervical
cancer. Chin J Cancer Res. 28:241–253. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cai Y, Li H and Zhang Y: Downregulation of
microRNA-206 suppresses clear cell renal carcinoma proliferation
and invasion by targeting vascular endothelial growth factor A.
Oncol Rep. 35:1778–1786. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu Z, Wang J, Mao Y, Zou B and Fan X:
MicroRNA-101 suppresses migration and invasion via targeting
vascular endothelial growth factor-C in hepatocellular carcinoma
cells. Oncol Lett. 11:433–438. 2016.PubMed/NCBI
|
|
40
|
Chen L, Wu YY, Liu P, Wang J, Wang G, Qin
J, Zhou J and Zhu J: Down-regulation of HPV18 E6, E7, or VEGF
expression attenuates malignant biological behavior of human
cervical cancer cells. Med Oncol. 28 Suppl 1:S528–S539. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu J, Cheng Y, He M and Yao S: Vascular
endothelial growth factor C enhances cervical cancer cell
invasiveness via upregulation of galectin-3 protein. Gynecol
Endocrinol. 30:461–465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen H, Suo K, Cheng Y, Zheng B and Xu L:
Vascular endothelial growth factor C enhances cervical cancer
migration and invasion via activation of focal adhesion kinase.
Gynecol Endocrinol. 29:20–24. 2013. View Article : Google Scholar : PubMed/NCBI
|